Abstract
The vasotocin receptor family is homologous to the mammalian vasopressin G-protein coupled receptor (GPCR) family. The vasotocin receptor 2 (VT2R) and 4 (VT4R) have recently been shown to play important role(s) in the neuroendocrine regulation of stress in birds. A homology-based structural model of VT4R of the domestic chicken, Gallus gallus, was built using the sophisticated SYBYL-X suite. The structure of VT4R built with and without extra- and intracellular unstructured loops showed a seven-helix transmembrane domain, which is a characteristic feature of GPCRs. Several agonists and antagonists were screened by molecular docking to map their potential binding sites on the structure of VT4R. Interestingly, the presence of the N-terminal, intracellular and extracellular loops and C-terminal amino acid sequences emerging from the transmembrane domains during molecular docking appeared to influence the binding interface of the peptide agonists and peptide/non-peptide antagonists on the VT4R. The presence of unstructured loops, however, did not affect the relative binding affinity ranking of the peptide antagonists to VT4R. In general, the natural ligand, arginine vasotocin and the peptide/non-peptide antagonists were observed to be more deeply buried in the receptor. Results of in vitro inhibition experiments, using cultured anterior pituitary cells, showed excellent agreement with the binding affinity of the antagonists predicted by molecular docking. The results of this study provide valuable clues for the rational design of novel pharmaceutical compounds capable of blocking or attenuating the stress response.
Keywords: vasotocin receptor 4, 3D structure, homology modelling, stress, antagonist
1. Introduction
The study of G protein-coupled receptors (GPCR) is of immense interest in medicinal chemistry and comparative physiology. Focusing upon the docking of the natural ligand to its receptor as well as a series of analogues provide a rational approach to drug design and possible clues regarding the evolution of this family of receptors. Availability of the complete sequence of the genomes of several species has provided an avenue to compare the functional evolution of GPCRs. The arginine vasotocin/mesotocin (AVT/MT) group of GPCR belongs to the rhodopsin-like receptor sub-family, which includes the mammalian vasopressin/oxytocin family. Recently, the vasotocin sub-type four receptor (VT4R) isolated from Gallus gallus has been studied (Kuenzel, Kang, & Jurkevich, 2013; Selvam et al., 2013). The chicken is a useful biomodel to study a variety of factors/conditions influencing the stress response. DNA and amino acid sequence comparisons show that the avian VT4R is homologous to the mammalian V1aR (Baeyens & Cornett, 2006). Functionally, the avian VT4R appears to be involved in the stress response and has been shown to be localized in corticotropes, which in birds occur specifically in the cephalic region of the anterior pituitary (AP) (Selvam et al., 2013). Chickens, when subjected to immobilization stress, showed significant changes (p < .05) in pituitary pro-opiomelanocortin (POMC) heteronuclear (hn) RNA. The gene product, POMC hnRNA, is subsequently spliced producing adrenocorticotropic hormone (ACTH), which activates adrenal production of the stress hormone corticosterone (CORT) (Kang, Jayanthi, Kumar, & Kuenzel, 2012; Kuenzel, Kang, & Jurkevich, 2012; Selvam et al., 2013).
Very useful information exists on the structure–activity relationship of the mammalian V1aR (Breton et al., 2001; Cotte et al., 2000; Mouillac et al., 1995), frog VTR (Acharjee et al., 2004; Cho et al., 2007; Kohno, Kamishima, & Iguchi, 2003; Mahlmann et al., 1994) and fish VTR (Hausmann et al., 1996; Mahlmann et al., 1994) using 3D modelling techniques, chimeric receptor approaches and/site-directed mutagenesis. The structural interactions that are important for ligand binding and subsequent activation of these receptors, however, are still not clearly understood. Determination of the 3D structures of the individual subtypes of the vasotocin receptors and characterization of the agonist and antagonist binding interfaces on these receptors is critical to understand the significance of differential tissue localization, expression and function(s) of these receptors. Given the current experimental challenges involved in the determination of 3D structures of transmembrane receptors, use of reliable molecular-modelling methods presents a rational approach toward characterization of the structure – activity relationship of transmembrane receptors such as, the VT4R. In this context, we have developed a 3D structural model of VT4R from the chicken (G. gallus) and have mapped the agonist and antagonist binding sites on the modelled VT4R structure. Results of the in vitro inhibition assays completely corroborate the conclusions drawn based on molecular docking. Availability of the 3D structural model is expected to trigger more intensive studies aimed towards characterizing structure–activity relationships of the VT4R using a variety of experimental approaches.
2. Materials and methods
2.1. Materials
The primary antibody against CORT was purchased from Fitzferald Inc. (Concord, MA, USA). The secondary antibody and [125I] CORT tracer were purchased from MP Biomedicals Inc. (Orangeburg, NY, USA). The selected VT4R antagonists (SR-49059, OPC-21268, H-5350) were purchased from Sigma-Aldrich (St. Louis, MO, USA), and H-6722 was purchased from Bachem Americas Inc. (Torrance, CA, USA).
2.2. Homology modelling of VT4R
The sequence of the chicken VT4R was obtained from the Uni-Prot database (ID: A8CWP8). The first step in homology modelling was to identify the appropriate template for building the 3D structure of the target sequence. BLAST (Basic Local Alignment Search Tool) (Altschul et al., 1997) and the VT4R amino acid sequence as a query, a search against the Protein Data Bank (PDB) (Berman et al., 2000) yielded several positive hits. Of the list of the PDB structures, bovine rhodopsin (PDB code: 1U19) was chosen as the best template to build the 3D model, since it not only satisfied the criteria of having a high sequence identity match with the target sequence and also showed a greater length of sequence coverage. Apart from rhodopsin, β-Adrenergic receptor (PDB ID: 2Y00) and NFQ Opioid receptor (PDB ID: 4EA3) structures were also selected as potential templates to build the homology model for VT4R. Sequence alignment of the template to target was carried out using the Clustal_W algorithm (Thompson, Gibson, Plewniak, Jeanmougin, & Higgins, 1997) which showed a sequence identity of 21% and a maximum coverage of 77% between the template and the target. A 3D model of VT4R was built using the program Composer in the Biopolymer module of the SYBYL-X suite (SYBYL-X, Tripos International). Loop conformations (N-terminus, C-terminus and the regions between the seven transmembrane helices) were automatically generated by the program. A sequence to structure alignment of target and template amino acid sequence resulted in the identification of the structurally conserved regions (SCRs) and structurally variable regions from the amino acid sequence of the VT4R. A four-step minimization procedure was performed to generate energy minimal conformations of VT4R. Hydrogen atoms were initially added to the VT4R molecule and their position(s) were refined by a 1000-step minimization keeping the rest of the VT4R structure fixed. Subsequently, a second cycle of 1000-step minimization was carried out by keeping the SCRs fixed. The final two steps of energy minimization (each of 1000-steps) were performed with the entire polypeptide backbone. However, subsequent total geometric optimization of the polypeptide was achieved without confining any portion of the protein. All minimization procedures were carried out using the Tripos force field with steepest descent for the first two steps and the Powell algorithm with a conjugate gradient for the last two steps. Molecular dynamics calculations were performed under periodic boundary conditions for a total time of 1 ns with a time step of 20 ps. RMSD plots were generated using the frames collected at every interval. Similar modelling procedure was employed to generate the 3D structure model of VT4R wherein the unstructured N- and C-terminal segments and the loop connecting the transmembrane helices were selectively cropped. This 3D structure is hereafter referred to as TM-VT4R.
2.3. Insertion of VT4R structure in lipid bilayer
Embedding of VT4R into the lipid bilayer and minimization was performed as per the method reported by Jo, Lim, Klauda, and Im (2009) using the Membrane Builder in CHARMM-GUI (Jo, Kim, & Im, 2007; Woolf & Roux, 1996) site. Minimized VT4R structure was inserted into a membrane bilayer to generate the membrane/protein complex. The modelled VT4R structure was embedded into a rectangular homogenous lipid system (consisting of DMPC (1,2-dimyristoyl-snglycero-3-phosphocholine), DPPC (1,2-dipalmitoyl-snglycero-3-phosphocholine), DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), POPC (1-palmitoyl,2-oleoyl-snglycero-3-phosphocholine), DLPE (1,2-dilauroyl-sn-glycero-3-phosphoethanolamine) and POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine). The protein receptor was oriented perpendicular with respect to the plane of the lipid bilayer. Explicit water molecules were included, and the system was neutralized by the addition of sodium and potassium ions (at 150 mM concentration). All the components were assembled, and the entire system was subjected to equilibration for 375 ps using CHARMM force field.
2.4. Validation of the 3D model of VT4R and the VT4R-ligand binary complex
The energy minimized VT4R model was validated and evaluated by PROCHECK (Laskowski, MacArthur, Moss, & Thornton, 1993). PROCHECK is a structure verification program that determines the stereochemical quality of the homology-based structural model by assessing various parameters such as lengths, angles and planarity of the peptide bonds, geometry of the hydrogen bonds and side chain conformations of the amino acids as a function of atomic resolution, which relies on the Ramachandran plot statistics (Ramachandran & Sasisekharan, 1968). Further, the 3D homology model of VT4R was superimposed on the template 1U19 to calculate for the root mean square deviation (RMSD) between the template and target structures. Electrostatic potential of the 3D model of VT4R was analysed using APBS (Adaptive Poisson–Boltzmann solver) based on the adaptive multilevel finite element method for solving the Poisson–Boltzmann equation (Baker, Sept, Joseph, Holst, & McCammon, 2001). All the images and the electrostatic potential of the surface were visualized using PyMOL (DeLano, 2004). The final optimized structural model of the VT4R was used for docking the potential agonists and antagonists.
2.5. Molecular docking
Both the non-peptide and peptide-based agonists and antagonists were both generated using a small molecule sketching tool and peptide builder modules available on SYBYL-X suite. All the ligands generated were subjected to the multi-staged energy minimization process similar to the one used for the receptor molecule. Docking of the agonists and antagonists was performed using AutoDock tools and Autodock Vina programs (Trott & Olson, 2010). Ligands were built on SYBYL-X software suite using the small molecule and peptide-building modules and were further subjected to optimization using the Molecular Mechanics method by the Tripos force field and Powell FF algorithm. The termination gradient was set to .05 kcal/(mol* Å). AVT (agonist) and 60 potential antagonists were screened for binding to VT4R and TM-VT4R. Only the antagonists that showed significant binding affinity to the VT4R/TM-VT4R are discussed in this study. The antagonists are classified as small benzylamine derivatives (2-Fluoro-3-(trifluoromethyl) benzylamine & 3-(Trifluoromethyl)benzylamine), non-peptide (SR-49059 ((2S)-1-[(2R,3S)-5-chloro-3-(2-chlorophenyl)-1-3,4-dimethoxyphenyl)sulfonyl-3-hydroxy-2H-indole-2-carbonyl]pyrrolidine-2-carboxamide), YM-087 – 4′-(2-methyl-1,4,5,6-tetrahydroimidazo[4,5-d][1]benzoazepine-6-carbonyl)-2-phenylbenzanilide monohydrochloride and OPC-21268 – N-[3-[4-[4-(2-oxo-3,4-dihydroquinolin-1-yl) piperidine-1-carbonyl]phenoxy]propyl]acetamide) and peptide (H-6722(Deamino-Cys1,D-Tyr(Et)2,Thr4,Orn8)-Oxytocin) and H-5350 – ((d(CH2)51, Tyr(Me)2,Arg8)-Vasopressin)) antagonist. Using AutoDock tools, polar hydrogen atoms were added to the protein and ligand molecules, and the non-polar hydrogens were merged. All the bonds in the ligand were set as rotatable, using the Lamarckian Genetic Algorithm method. Docking calculations were performed in a protein-fixed and ligand-flexible mode. A grid box with a dimension of 30*42*26(X*Y*Z) points was used to cover the entire receptor molecule that could facilitate the ligands to interact freely. Two important criteria were used for the validation of the docking procedure which included the binding energy of the VT4R/ligand (agonist/antagonist) complex and the location or position of the docked ligand inside the receptor binding pocket. These two factors determined the quality of the fitting, which further related to the biological activity of the docked agonist or antagonist. The best docking conformation with the lowest docking score (ΔGbinding) was selected for ranking. Protein/ligand conformations, including bond lengths and hydrogen bonds were analysed and represented using PyMOL (DeLano, 2004).
2.6. Radioimmunoassay
Thirty 7–8-week-old male birds were randomly allocated to each of 5 groups (Control, 5 min stress, 15 min stress, 1 h stress, 1 h stress repeated for 10 consecutive days; n = 6 birds/treatment). Birds were housed in individual cages under a 16-h light, 8-h dark lighting cycle (LD16:8) with light on at 07:00 and given food and water ad libitum. The immobilization stress was performed by wrapping the birds in a cloth harness secured with velcro to restrain their wings from flapping. Birds were positioned on their ventral surface and their legs (tarsometatarsal bone area) were secured with a velcro strap. All birds had access to water during their period of restraint. Upon completion of stress treatment, blood sample was collected from the brachial vein using heparinized syringe. Blood was centrifuged, plasma collected and stored at −20 °C until further use. Birds were immediately decapitated after blood sampling, AP was removed and snap-frozen for total RNA extraction. Plasma CORT was extracted by adding 2 mL ethyl ether to 200 μL plasma samples. CORT levels were determined by radioimmunoassay (RIA) modified from the procedure of Proudman and Opel (Madison, Jurkevich, & Kuenzel, 2008; Proudman & Opel, 1989). The intra- and interassay coefficients of variation were 9 and 14%, respectively. All experimental animal procedures followed protocols approved by the University of Arkansas Institutional Animal Care and Use Committee.
2.7. Primary anterior pituitary cell culture
Primary AP cells from male birds were obtained using a modified trypsin/neuraminidase procedure, as described previously (Fehrer, Silsby, Behnke, & el Halawani, 1985; Hopkins & Farkuhar, 1973; Kang, Gazzillo, You, Wong, & El Halawani, 2004; Kang, Youngren, & El Halawani, 2002). Cultures were maintained at 39 °C in a humidified 5% CO2/95% air incubator for 4 days. Cell viability (85–95%) was determined by trypan blue dye exclusion and quantified using a haemocytometer. Dispersed AP cells (.5 × 105) were treated with different combinations of corticotropin-releasing hormone (CRH: 0, .1, 1, 2 and 10 nM; Bachem Americas Inc. Torrance, CA, USA) and arginine vasotocin (AVT: 0, .1, 2, 10 and 50 nM; Bachem Americas Inc. Torrance, CA, USA) for 15 min, 1 h and 6 h. At the conclusion of incubation, cells were collected by centrifugation, washed with 2 mL of phosphate-buffered saline (PBS) and dissolved in 1 mL Trizol® reagent (Life Technologies, Palo Alto, CA, USA) and frozen at −80 °C.
2.8. Real-time RT-PCR for POMC RNAs
Total RNA was extracted from the AP gland or cultured primary cells using Trizol® reagent (Life Technologies, Palo Alto, CA, USA) as recommended by the manufacturer with minor modifications. To eliminate genomic DNA contamination, total RNA was treated with RNase-free DNase I (Invitrogen, Carlsbad, CA, USA, 1 U/μg RNA) for 30 min at 37 °C. After DNA digestion, total RNA was purified using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and total RNA concentration was determined using NanoDrop 1000 (Thermo Scientific, Wilmington, DE, USA). Single-stranded cDNA was synthesized from 2 μg total RNA using oligo (dT)16 primer and superscript II (Invitrogen, Carlsbad, CA, USA), as previously described (Kang, Thayananuphat, Bakken, & El Halawani, 2007; Kang et al., 2010). The best primer pairs for hnRNA and mRNA of POMC were initially selected from several pairs based on PCR product quality and lengths after electrophoresis on a 3% agarose gel. Real-time RT-PCR was performed using the Power SYBR green and Applied Biosystems 7300 Real-Time PCR system under the following conditions: 40 cycles of denaturation (at 95 °C) for 60 s, annealing (at 60 °C) for 30 s, extending (at 72 °C) for 60 s, and finally extending at 72 °C for 10 min. A non-template control and an endogenous loading control (chicken GAPDH and β-actin) were used for relative quantification. The fold change values for the stressed groups compared with controls were determined by the ΔΔCT method.
2.9. Statistical analysis
A two-way fixed analysis of variance using MS-excel 2010 was used to evaluate CORT plasma levels and relative changes of POMC RNAs. p < .05 was considered statistically significant.
3. Results and discussion
3.1. Homology modelling of VT4R
Vasotocin receptor 4 (VT4R) is one of the four vasotocin receptor subtypes found in the chicken (G. gallus) (Baeyens & Cornett, 2006). A BLAST search using the VT4R amino acid sequence as query against the PDB resulted in only a few hits and most of them showed a sequence identity less than 20%. A reliable homology model can be generated even though the sequence identity between template and target sequence is as low as 20% (Lushington et al., 2008). Length of the sequence coverage between the template and the target sequence is another important criterion that needs to be satisfied to obtain a reliable homology-based structural model. In total, seven positive hits were recorded of which the crystal structure of bovine rhodopsin (PDB code – 1U19) which showed a sequence identity of 21% and a reasonable sequence coverage of 77% was chosen as the suitable template for building the 3D homology model of the VT4R. Interestingly, despite the low sequence identity, the secondary structural regions between the template and target (VT4R) were well-aligned with a backbone RMSD of .301 Å (Supplementary Figure S1). The final energy of the minimized structure(s) calculated using the TRIPOS force field using the Powell algorithm was about −1490 kcal mol−1 (Figure 1(A)).
Figure 1.
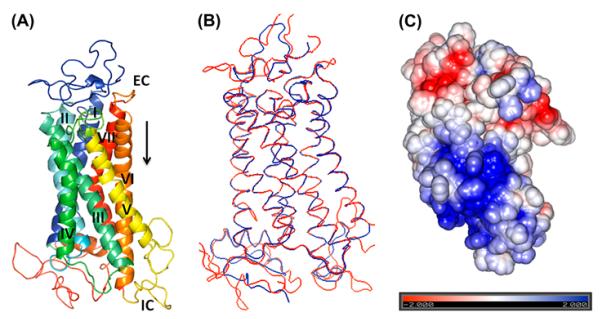
A 3D homology structure of the VT4R (A) Homology Model built using the template 1JFP/1U19 (bovine rhodopsin). Seven transmembrane helices (TM-I-VII), each shown with a different spectral colour are labelled with Roman numbers. EC – Extracellular side and IC intracellular side of the receptor. (B) Superimposition of the template (1U19) shown in red and target (VT4R) shown in blue structures are represented by ribbon diagram. (C) Electrostatic potential map of VT4R positively and negative charged residues are represented in blue and red, respectively.
3.2. Validation of the 3D structural model of VT4R and the VT4R–ligand complex
The energy-minimized model was evaluated using the PROCHECK algorithm that relies on Ramachandran plot statistics. PROCHECK analysis of the minimized model showed that 93.0% of the Φ and ψ angles of the nonglycine and non-proline residues were located in the core regions of the Ramachandran plot, 6.1% were in the additionally allowed zones, .8% of the residues were in the generously allowed region and none of the residues found in the disallowed region (Supplementary Figure S2). Superimposition of the template and target structures showed that weighted RMSD of Cα trace between VT4R and 1U19 was .301 Å and most of the observed differences were in the loop regions as these structures are known to be highly variable. Results suggest that the model built was highly reliable with little or no bad long- or short-distance structural contacts (Figure 1(B)). The energy-minimized structure of the VT4R was embedded in the coarse-grain of lipid molecules to examine structural changes in the receptor. Comparison of the RMSD values of the backbone Cα atoms suggests that explicit inclusion of the non-polar lipid bilayer did not significantly alter the orientation of the transmembrane helices (TM). It should be worthwhile to mention that 1 ns molecular dynamics simulation of the VT4R structure, with the explicit inclusion of water, showed maximum RMSD of 1.4 Å (Supplementary Figure S3).
3.3. Description of the 3D structure of VT4R
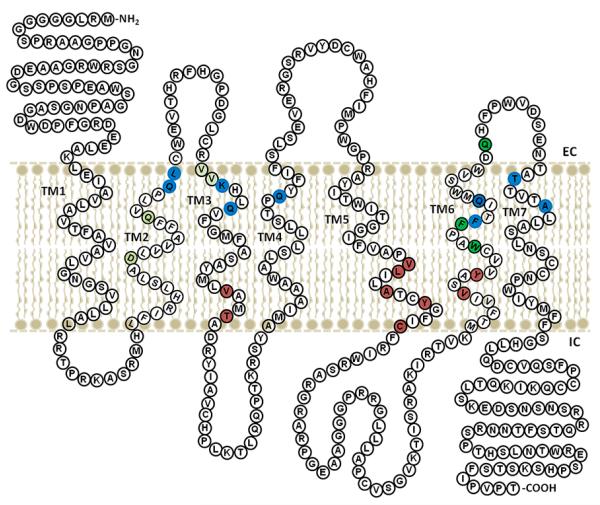
The structure of VT4R consists of seven TMs, a feature characteristic of the structure of GPCRs (Figure 1(A)). Both the extra- and intracellular loops (non-transmembrane loops) of VT4R are predominantly unstructured. The length and the orientation of the TM are not affected by the presence of extra- and intracellular loops, and this aspect is obvious from the comparison of VT4R and TM-VT4R structures (Figure 1(A) and (B)). With the exception of TMIII and TMV, the TMs are about the same length and are oriented perpendicularly to the plane of the lipid bilayer in both VT4R and TM-VT4R structures (Figure 5). TMIII is tilted away from TMII at an angle of ~120° to the plane of the membrane in the structure of TM-VT4R (Figure 1(A)). In addition, the most prominent difference observed between the VT4R and TM-VT4R structures is the more prominent tilt of TMV towards TMVI in VT4R. The helical tilt along with the prominent kink introduced due to the presence of Pro235 creates a cavity conducive for the binding of agonists and antagonists (to be discussed later). Electrostatic potential surface of the VT4R shows a dense positively charged cluster consisting of arginines (R249, R252, R255, R257, R 259, R286 and R290) and lysines (K288 and K293) located on the intracellular loop III (Figure 1(C)). This cationic cluster can be a potential target sites for specifically designed negative charge on antagonists. Similarly, negative charges contributed by residues D319, D326, and E 328 located on extracellular loop III, form a rim at the apical portion of the receptor that is proximal to the extracellular surface of the membrane (Figure 1(C)). The presence of such closely placed negative charges is significant and can plausibly serve as specific initial recognition site(s) for the conserved positively charged arginine residue present in the ligand, AVT. Interestingly, D104 located on TMII, D155, R156 (present in TMIII and TMVI, respectively) and Y157 (located in the intracellular loop) are well conserved from fish to the mammals. Although the exact interplay of structural events involved in the activation of VT receptors is not clear, these residues have been shown to be important for the activation of the VT receptors (Bockaert & Pin, 1999) or perhaps stabilize both the inactive and the activated conformation of receptors in this sub-family of the rhodopsin-like GPCRs (Rovati, Capra, & Neubig, 2007).
Figure 5.
Two-dimensional cartoon of VT4R representation of the amino acid residues involved in interaction with agonist and antagonist. Antagonist binding sites in the modelled VT4R structure with and without the non-transmembrane loops are shown in brown and blue coloration respectively. The well-conserved residues in the VT receptor family are shown in green.
Docking of ligand into the VT4R structure revealed that the ligands docked into the receptor in different conformations and distinct poses were obtained after clustering of RMSD structural deviation (2.0 Å cutoff) each time from their initial position. The most reliable conformation (s) of the VT4R–ligand binary complex were selected based on the most favourable interaction energy between the pose of ligand and the receptor. A low-docking score (ΔGbinding) for the ligand–receptor binding signifies that the binding interaction is strong (Table 1).
Table 1.
Representation of the ligands that show the significant binding to VT4R.
| S.No. | Ligand | ΔGbinding (kcal/mol) |
|---|---|---|
| 1 | YM-087-4′-(2-methyl-1,4,5,6-tetrahydroimidazo[4,5-d][1]benzoazepine-6-carbonyl)-2- phenylbenzanilide monohydrochloride |
−8.9 |
| 2 | SR-49059 ((2S)-1-[(2R,3S)-5-chloro-3-(2-chlorophenyl)-1–3,4- dimethoxyphenyl)sulfonyl-3-hydroxy-2H-indole-2-carbonyl]pyrrolidine-2-carboxamide |
−8.3 |
| 3 | 2-Fluoro-3-(trifluoromethyl)benzylamine | −8.1 |
| 4 | OPC-21268 - N-[3-[4-[4-(2-oxo-3,4-dihydroquinolin-1-yl) piperidine-1-carbonyl] phenoxy]propyl]acetamide |
−8 |
| 5 | 3-(Trifluoromethyl)benzylamine | −7.9 |
| 6 | H-6722(Deamino-Cys1,D-Tyr(Et)2,Thr4,Orn8)-Oxytocin | −7.2 |
| 7 | H-5350 - ((d(CH2)51,Tyr(Me)2,Arg8)-Vasopressin | −6.4 |
3.4. Agonist and antagonistic binding sites on VT4R
Molecular docking results were evaluated based on the ligand–protein interactions including properties like the steric, electrostatic and the intramolecular energy. The GPCR family of protein receptors contains critical residues that are known to play an important role in the recognition and binding of agonist(s) and antagonist(s).
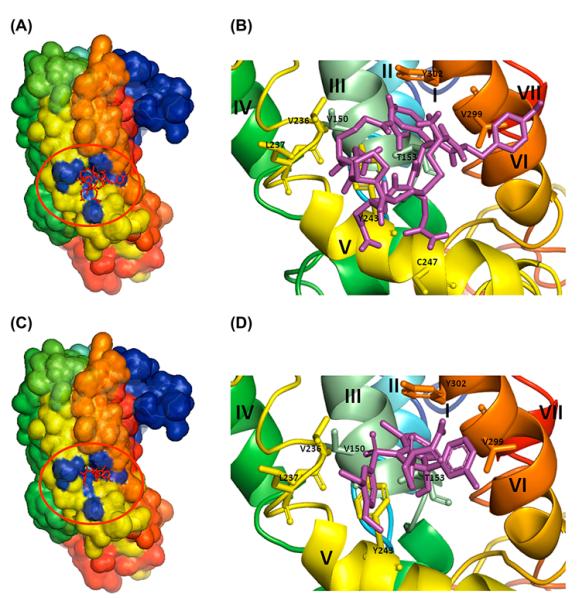
Various peptide- and non-peptide-based compounds are known to interact with the mammalian V1aR and its avian homologue, VT4R (Thibonnier, Coles, Conarty, Plesnicher, & Shoham, 2000). The natural agonist for VT4R is AVT. Based on the available literature, a selective number of diverse ligands were screened for binding to the modelled VT4R structure (containing the non-transmembrane loops). Some of the ligands that were screened showed strong binding affinity to the VT4R as evidenced by the high negative ΔGbinding values (Table 1). A closer look at the binding sites of the ligands clearly shows that the ligands bind to the same generalized region, as well as similar amino acid residues on the receptor (Figures 2(A–D) and 3(A–D)). The binding sites for the peptide and non-peptide ligands are located in a deep pocket contributed by residues in TMIII, TMV, TMVI and few amino acids located proximal to the intracellular loops (Figures 2 (A–D), 4(A) and (B)). It is worthwhile to note that both the agonist and the peptide/non-peptide antagonists bind in the same binding pocket. A common binding site for the agonists, peptide and non-peptide antagonist SR-49059 suggests that the antagonists can be expected to show inhibition kinetics similar to those exhibited by competitive inhibitors. In comparison, small molecule antagonists such as benzylamine derivatives, exhibit relatively weaker binding affinity to VT4R (Table 1). It is important to mention that unlike the VT4R-peptide/non-peptide ligand interaction sites, which lie deeply buried in the receptor, the peptide/non-peptide antagonist binding sites in the other related receptors are reported to be located proximally to the extracellular surface of the membrane.
Figure 2.
Docking of ligands onto the modelled VT4R structure with loops. (A) Agonist (vasotocin) (C) antagonist (SR-49059) shown as red sticks (in the red circle) were docked onto the modelled VT4R with loops and shown in surface view. Panels (B) and (D) are close-up views of the binding site of the agonist and antagonist shown as sticks (magenta) and the amino acid residues involved in the interaction are represented in single letter codes. The seven TM helices are labelled in Roman numerals starting from I to VII.
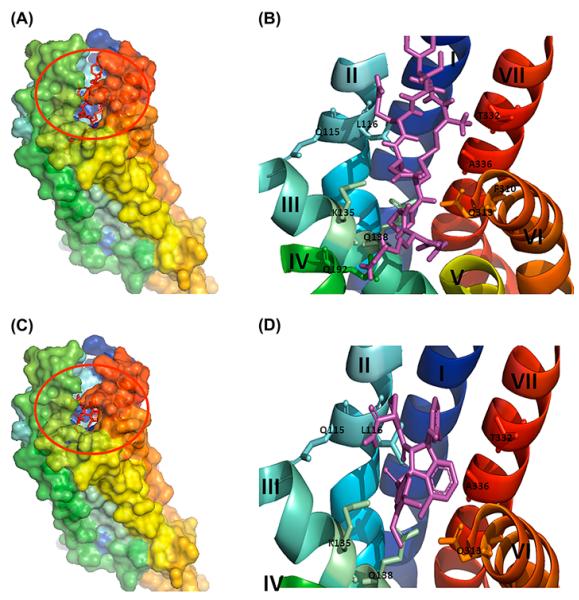
Figure 3.
Docking of ligands onto the modelled VT4 without loops (TM-VT4R) (A) Agonist (vasotocin) (C) antagonist (SR-49059) shown in red sticks (in the red circle) were docked onto the modelled VT4 receptor without loops and shown in surface view. (B) and (D) are close-up view of the binding site of the agonist and antagonist shown in sticks (magenta) amino acid residues involved in the interaction are represented in single letter codes. 7TM helices are labelled in Roman numerals starting from I to VII.
Figure 4.
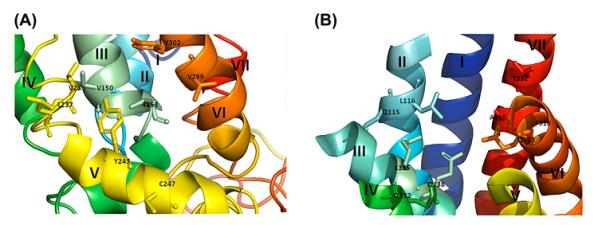
Binding interactions of agonist and antagonist with the VT4R models. (A) and (C) Agonist, (B) and (D) Antagonist of the model with and without loops (TM-VT4R). List of interactions and the bond distances between the atoms of the binding pocket of receptor and ligands (vasotocin/SR-49059) are highlighted. Ligands are shown in magenta and the bond distances are shown in yellow dotted lines. The amino acid residues involved in the interaction are represented in single letter codes. 7TM helices are labelled in roman letters starting from I to VII.
In contrast, small molecule antagonists such as, 2-Fluoro-3-(trifluoromethyl) benzylamine and 3-(trifluoromethyl) benzylamine, bind at a site close to the extracellular side of the receptor. This class of antagonists screened is consistently found to bind to a predominantly hydrophobic pocket contributed by non-polar residues M142, F143 W306, F309, Q313 and M314 located on helices TMIII and TMVI. This observation is consistent with previous studies wherein benzylamine derivatives have been shown to dock at a site totally different from the binding site of the peptide agonists and antagonists (Gieldon et al., 2001).
It is known that extracellular loops play an important role in the activation of the receptor. Tyrosine 115 is located in the extracellular loop region of vasopressin receptors and is well conserved in mammals and frogs (Acharjee et al., 2004). Site-directed mutagenesis studies revealed that the Y115 is critical for conferring selectivity to the agonist (Chini et al., 1995). Interestingly, Tyr115 is missing in VT4R and replaced by a basic amino acid, His115 (actual number is H125), in the ECL-1 region of VT4R. With respect to Tyr residues, one of the most intensely studied conservative motifs in GPCRs is the E/DRY sequence in intracellular loop #2 (Acharjee et al., 2004; Rovati et al., 2007; Audet & Bouvier, 2012). The DRY sequence exists at the end of TMIII facing the intracellular region. Of relevance is that of the three amino acids, the least conserved one is tyrosine and is generally considered not important for receptor function (Flanagan, 2005). Similarly, in the chicken, the tyrosine residue (actual number, if present would be Y125 in ECL-1) is not critical since His125 is substituted for tyrosine at that site. In addition, modelling studies suggest that ECL-1 of the VT4R appears less important for ligand docking since the loop is located remotely from the agonist/antagonist binding pocket.
3.5. Non-transmembrane loops influence the binding of the ligands to the receptor
Extracellular and intracellular loops in transmembrane proteins are largely variable in structure and therefore most of the in silico receptor-ligand studies are performed with modelled transmembrane receptors without their N- and C-terminal segments and the TM loops. Therefore, ligand-docking studies were also performed with the VT4R comprising solely of its transmembrane domains (TM-VT4R). The ΔGbinding values obtained for the peptide agonist (AVT) (ΔGbinding = −7.0 kcal/mol) and non-peptide antagonist(SR-49059) (ΔGbinding = −8.7 kcal/mol) interactions are in the same order of magnitude as observed for the peptide–ligand interactions with its complete receptor sequence containing all the loops and N- and C-terminal amino acid segments. Importantly, the TM-VT4R/peptide ligand binding interface was significantly different from that observed in the peptide ligand – complete VT4R (containing all loops) binary complex (Figure 3(A–D)). Nonetheless, the peptide ligand-TM-VT4R binary complex is largely consistent with the ligand-binding sites reported for other VT receptors. Residues that are in close proximity to both the peptide agonist (AVT) and non-peptide antagonist (SR-49059) binding site are Q115, K135, Q138, Q192 and Q313 (Figure 4(C) and (D)). These residues are well conserved in the VT family and are located close to the extracellular face of the membrane bilayer. A 2D representation of the structure of VT4R with residues highlighted in Figure 5 provides a picture of the differences observed in the peptide agonist and non-peptide antagonist interaction sites on the VT4R receptor with and without loops. Minor differences between the ligand-binding sites in TM-VT4R and other mammalian vasopressin receptors are observed. The amino acids W306, F310 and F309 in the mammalian vasopressin (V1a) receptor were shown to provide interaction sites for the binding of antagonist, SR-49059 (Figure 5). Note that only F310 was observed to contribute to the binding of SR-49059 in the modelled structure of the TM-VT4R /SR-49059 binary complex (Figure 4(B)). The residue A337 (located in TMVII) in the human vascular vasopressin receptor (V1aR) has been shown to be critical for the binding of the non-peptide antagonist OPC-21268 (Thibonnier et al., 2000). Interestingly, the modelled structure of the TM-VT4R/SR-49059 binary complex shows that residues, T332 and A336 (in TMVII), provide binding sites for the non-peptide antagonist. Importantly, the observed discrepancies are limited to peptide and non-peptide ligands (Figure 5). Small ligands such as the benzylamine derivatives were found to be bound to the same ligand-binding pocket regardless of whether docking studies were executed with receptor containing the loop regions or not. These results clearly suggest that extra – and intracellular non-transmembrane loops interfere with peptide/non-peptide ligand binding. It may not be unreasonable to suggest that in silico peptide ligand docking studies should be conducted both in the presence and absence of transmembrane loops to obtain an unbiased holistic picture of ligand–receptor interactions.
3.6. Immobilization stress enhances plasma CORT and transiently activates POMC hnRNA in the anterior pituitary gland
In order to confirm the results from 3D modelling/docking studies for screening VT4R antagonist, we used male chickens as a stress model system as previously (Madison et al., 2008; Selvam et al., 2013). Plasma CORT was measured by RIA to confirm activation of the HPA axis in male birds exposed for different time intervals to immobilization stress (Figure 6(A)). Immediately after immobilization, stress CORT levels increased by more than twofold (1369 pg/mL, ±67) when compared to unstressed male birds (611 pg/mL, ±78) and increased time-dependently up to 1 h stress (2442 pg/mL, ±990) and 1 h/10 days (2731 pg/mL, ±1107). This result suggests that male birds have the rapid (within 5 min) and time-dependent immobilization stress-induced activation of the HPA axis and persistent activation of the HPA axis in chronic stress (1 h, 10 days). To develop a stress-induced marker gene in the AP gland, hnRNA and mRNA of the POMC gene known to be regulated by glucocorticoids (Drouin, Sun, & Nemer, 1989) were measured in the AP gland from time-course stressed birds by real-time RT-PCR (Figure 6(B)). Five-minute immobilization stress induced a rapid increase in POMC hnRNA (100%) but not mRNA. This rapid induction of hnRNA of POMC gene by immobilization stress was observed in AP gland of mammals too (Ginsberg et al., 2006). Both hnRNA and mRNA expression were blunted at 15 min stress (24, 65% compared with controls, respectively) and recovered to normal levels at 1 h following immobilization stress. Chronic immobilization stress (1 h 10 consecutive days) caused significant decreases in both hnRNA and mRNA expression (69% and 61% compared with controls, respectively). Results suggest that stress-induced POMC activation in the AP gland was transient and the in vivo feedback inhibition of the POMC gene at 15 min was induced through a possible negative glucocorticoid response element response in the POMC gene promoter by increased plasma CORT (1994 pg/ml, ±671). Therefore, we used POMC hnRNA as a stress-induced marker RNA in the in vitro AP cell culture system, which lacked the feedback inhibition by stress-induced that occurs in vivo by stress-induced CORT.
Figure 6.
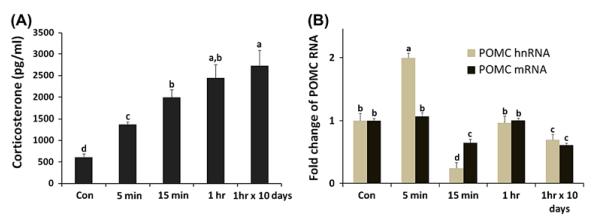
Plasma CORT and expression changes of pro-opiomelanocortin (POMC) RNAs by immobilization stress. (A) Plasma CORT concentrations (pg/ml) were measured by RIA after immobilization stress. (B) Total RNAs from the AP gland were used for real-time RT-PCR of POMC heteronuclear (hn) and mRNA. Data (mean ± SEM) are presented as the fold changes of relative expression levels compared with the unstressed control group (Con) set at a value of 1.0. Significant differences (p < .05) are identified by different letters. n = 8 birds/group.
3.7. Antagonist effects of selected compounds on the POMC hnRNA expression stimulated by CRH/AVT treatment in anterior pituitary primary cells
In general, in the primary AP cells, 15-min and 1-h treatment of AVT and/or CRH with various dosages did not significantly induce hn RNA of POMC (Figure 7(A)). Moreover, high doses of CRH (>1.0 nM) and AVT (>2.0 nM) caused reduction in hnRNA. A combination of .1, 1.0 and 2.0 nM of AVT, and 0 and .1 nM of CRH treatment for 6 h induced a significant stimulation of hnRNA of POMC with the maximum stimulation (173% increase) of POMC hnRNA at .1 nM CRH and 1 nM AVT following 6-h incubation (Figure 7(A)). Therefore, we used this in vitro primary AP cell stress condition protocol to investigate the antagonistic effect of selected VT4R antagonists utilized in the 3D model/docking study. Pre-treatment of AP cells before CRH/AVT stimulation (.1/1.0 nM) with each antagonist for 30 min significantly reduced POMC hnRNA expression 55% (SR-49059), 39% (OPC-21268), 44% (H-6722) and 35% (H-5350) compared with CRH/AVT induced hnRNAexpression dose-dependently (p < .05) (Figure 7(B)).
Figure 7.
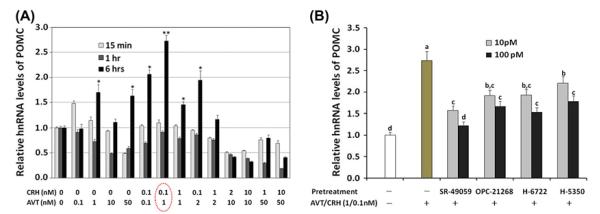
CRH/AVT treatments affecting POMC hnRNA stimulation in primary AP cells and antagonistic effects of selected VT4R antagonists. (A) Dose and time dependent effect of CRH/AVT altering POMC hnRNA expression. Total RNAs from AP were used for real-time RT-PCR of POMC hnRNA. Star markers (* and **) show group means significantly different (p < .05). (B) The selected antagonists were applied 30 min before CRH (0.1nM)/AVT (1.0nM) treatment using 10 and 100pM concentrations to the AP cells. Data (mean ± SEM) are expressed as fold differences from controls (no treatment of CRH/AVT) set at 1.0. Significant differences (p < .05) are identified by different letters.
4. Conclusion
Vasotocin receptor 4 (VT4R), a member of the GPCR family, appears to play an important role in the neuroendocrine hypothalamo/pituitary/adrenal axis regulating the avian stress response. The lack of detailed structural information for the receptor stimulated the development of a model for the 3D structure of the VT4R. The ligand (agonist/antagonist) binding sites have been successfully mapped for the first time. Significant differences in the ligand-binding sites were observed when molecular docking was performed with the VT4R structure constructed with transmembrane domains only compared with the complete receptor sequence including the N-terminal, loops and C-terminal amino acid sequence. Results suggest that the complete receptor sequence should be included in receptor models as the intracellular and extracellular loops influence the binding interactions with ligands within the transmembrane domains. The non-peptide antagonist, SR-49059 showed the strongest binding affinity to the VT4R which was in excellent agreement with the results of in vitro inhibition assays performed using pituitary cells and POMC hnRNA associated with the stress response. Detailed mutation studies in combination with structural and functional analyses can be expected to pave the way for a clearer understanding of the molecular mechanism(s) underlying the stress response.
Supplementary Material
Acknowledgments
This study is supported in part by NSF Grant #IOS-0842937, the Arkansas Biosciences Institute (ABI) Grant, the Division of Agriculture, University of Arkansas, NIH (P30 GM 103450), and DOE (DE-02-01ER15161) grants.
List of abbreviations
- GPCR
G Protein coupled receptor
- VT4R
Vasotocin 4 receptor
- AVT/MT
Arginine vasotocin/mesotocin
- POMC
Pro-opiomelanocortin
- ACTH
Adrenocorticotropic hormone
- AVT
Arginine vasotocin
- RIA
Radioimmunoassay
- BLAST
Basic local alignment search tool
- APBS
Adaptive Poisson-Boltzmann Solver
- CORT
Corticosterone
- EC
Extracellular side
- IC
Intracellular side
- TM-VT4R
VT4R without the non-transmembrane loops, C and N terminal amino acid segments
- CRH
corticotropin releasing hormone
Footnotes
Supplementary material The supplementary material for this paper is available online at http://dx.doi.org/10.1080/07391102.2013.787025.
References
- Acharjee S, Do-Rego JL, Oh DY, Ahn RS, Choe H, Vaudry H, Kwon HB. Identification of amino acid residues that direct differential ligand selectivity of mammalian and nonmammalian V1a type receptors for arginine vasopressin and vasotocin: Insights into molecular coevolution of V1a type receptors and their ligands. Journal of Biological Chemistry. 2004;279:54445–54453. doi: 10.1074/jbc.M408909200. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audet M, Bouvier M. Restructuring G-protein-coupled receptor activation. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Baeyens DA, Cornett LE. The cloned avian neurohypophysial hormone receptors. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2006;143:12–19. doi: 10.1016/j.cbpb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Bourne PE. The protein data bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO Journal. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C, Chellil H, Kabbaj-Benmansour M, Carnazzi E, Seyer R, Phalipou S, Mouillac B. Direct identification of human oxytocin receptor-binding domains using a photoactivatable cyclic peptide antagonist: Comparison with the human V1a vasopressin receptor. Journal of Biological Chemistry. 2001;276:26931–26941. doi: 10.1074/jbc.M102073200. [DOI] [PubMed] [Google Scholar]
- Chini B, Mouillac B, Ala Y, Balestre MN, Trumpp-Kall-meyer S, Hoflack J, Jard S. Tyr115 is the key residue for determining agonist selectivity in the V1a vasopressin receptor. EMBO Journal. 1995;14:2176–2182. doi: 10.1002/j.1460-2075.1995.tb07211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Acharjee S, Moon MJ, Oh DY, Vaudry H, Kwon HB, Seong JY. Molecular evolution of neuropeptide receptors with regard to maintaining high affinity to their authentic ligands. General and Comparative Endocrinology. 2007;153:98–107. doi: 10.1016/j.ygcen.2006.12.013. [DOI] [PubMed] [Google Scholar]
- Cotte N, Balestre MN, Aumelas A, Mahe E, Phalipou S, Morin D, Mouillac B. Conserved aromatic residues in the transmembrane region VI of the V1a vasopressin receptor differentiate agonist vs. antagonist ligand binding. European Journal of Biochemistry. 2000;267:4253–4263. doi: 10.1046/j.1432-1033.2000.01472.x. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2004. Retrieved from http://pymol.sourceforge.net. [Google Scholar]
- Drouin J, Sun YL, Nemer M. Glucocorticoid repression of pro-opiomelanocortin gene transcription. Journal of Steroid Biochemistry. 1989;34:63–69. doi: 10.1016/0022-4731(89)90066-6. [DOI] [PubMed] [Google Scholar]
- Fehrer SC, Silsby JL, Behnke EJ, el Halawani ME. Hypothalamic and serum factors influence on prolactin and luteinizing hormone release by the pituitary gland of the young turkey (Meleagris gallopavo) General and Comparative Endocrinology. 1985;59:73–81. doi: 10.1016/0016-6480(85)90420-4. [DOI] [PubMed] [Google Scholar]
- Flanagan CA. A GPCR that is not “DRY”. Molecular Pharmacology. 2005;68(1):1–3. doi: 10.1124/mol.105.014183. [DOI] [PubMed] [Google Scholar]
- Gieldon A, Kazmierkiewicz R, Slusarz R, Ciarkowski J. Molecular modeling of interactions of the non-peptide antagonist YM087 with the human vasopressin V1a, V2 receptors and with oxytocin receptors. Journal of Computer-Aided Molecular Design. 2001;15:1085–1104. doi: 10.1023/a:1015905822671. [DOI] [PubMed] [Google Scholar]
- Ginsberg AB, Frank MG, Francis AB, Rubin BA, O’Conner KA, Spencer RL. Specific and time-dependent effects of glucocorticoid receptor agonist RU28362 on stress-induced pro-opiomelanocortin hnRNA, c-fos mRNA and zif268 mRNA in the pituitary. Journal of Neuroendocrinology. 2006;18:129–138. doi: 10.1111/j.1365-2826.2005.01396.x. [DOI] [PubMed] [Google Scholar]
- Hausmann H, Richters A, Kreienkamp HJ, Meyerhof W, Mattes H, Lederis K, Richter D. Mutational analysis and molecular modeling of the nonpeptide hormone binding domains of the [Arg8]vasotocin receptor. Proceedings of the National academy of Sciences of the United States of America. 1996;93:6907–6912. doi: 10.1073/pnas.93.14.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins CR, Farquhar MG. Hormone secretion by cells dissociated from rat anterior pituitaries. Journal of Cell Biology. 1973;59:276–303. doi: 10.1083/jcb.59.2.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE. 2007;2:e880-1–e880-9. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophysical Journal. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SW, Jayanthi S, Kumar TKS, Kuenzel WJ. Identification of vasotocin 4 receptor (VT4R) antagonists by homology modeling/docking analysis and primary anterior pituitary cell culture. 10th International Symposium on Avian Endocrinology; Gifu, Japan. 2012. P08:52. [Google Scholar]
- Kang SW, Gazzillo LC, You S, Wong EA, El Halawani ME. Turkey prolactin gene regulation by VIP through 35-bp cis-acting element in the proximal promoter. General and Comparative Endocrinology. 2004;138(2):157–165. doi: 10.1016/j.ygcen.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Kang SW, Leclerc B, Kosonsiriluk S, Mauro LJ, Iwasawa A, El Halawani ME. Melanopsin expression in dopamine–melatonin neurons of the premammillary nucleus of the hypothalamus and seasonal reproduction in birds. Neuroscience. 2010;170:200–213. doi: 10.1016/j.neuroscience.2010.06.082. [DOI] [PubMed] [Google Scholar]
- Kang SW, Thayananuphat A, Bakken T, El Halawani ME. Dopamine–melatonin neurons in the avian hypothalamus controlling seasonal reproduction. Neuroscience. 2007;150:223–233. doi: 10.1016/j.neuroscience.2007.08.031. [DOI] [PubMed] [Google Scholar]
- Kang SW, Youngren OM, El Halawani ME. Influence of VIP on prolactinemia in turkey anterior pituitary cells: Role of cAMP second messenger in VIP-induced prolactin gene expression. Regulatory Peptides. 2002;109:39–44. doi: 10.1016/s0167-0115(02)00168-4. [DOI] [PubMed] [Google Scholar]
- Kohno S, Kamishima Y, Iguchi T. Molecular cloning of an anuran V(2) type [Arg(8)] vasotocin receptor and mesotocin receptor: Functional characterization and tissue expression in the Japanese tree frog (Hyla japonica) General and Comparative Endocrinology. 2003;132:485–498. doi: 10.1016/s0016-6480(03)00140-0. [DOI] [PubMed] [Google Scholar]
- Kuenzel WJ, Kang SW, Jurkevich A. Neuroendocrine regulation of stress in birds with an emphasis on vasotocin receptors (VTRs) General Comparative Endocrinology. 2013 doi: 10.1016/j.ygcen.2013.02.029. doi: 10.1016/j.ygcen.2013.02.029 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Lushington GH. Comparative modeling of proteins. Methods in Molecular Biology. 2008;443:199–212. doi: 10.1007/978-1-59745-177-2_11. [DOI] [PubMed] [Google Scholar]
- Madison FN, Jurkevich A, Kuenzel WJ. Sex differences in plasma corticosterone release in undisturbed chickens (Gallus gallus) in response to arginine vasotocin and corticotropin releasing hormone. General and Comparative Endocrinology. 2008;155:566–573. doi: 10.1016/j.ygcen.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Mahlmann S, Meyerhof W, Hausmann H, Heierhorst J, Schonrock C, Zwiers H, Richter D. Structure, function, and phylogeny of [Arg8]vasotocin receptors from teleost fish and toad. Proceedings of the National academy of Sciences of the United States of America. 1994;91:1342–1345. doi: 10.1073/pnas.91.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillac B, Chini B, Balestre MN, Elands J, Trumpp-Kallmeyer S, Hoflack J, Barberis C. The binding site of neuropeptide vasopressin V1a receptor. Evidence for a major localization within transmembrane regions. Journal of Biological Chemistry. 1995;270:25771–25777. doi: 10.1074/jbc.270.43.25771. [DOI] [PubMed] [Google Scholar]
- Proudman JA, Opel H. Daily changes in plasma prolactin, corticosterone, and luteinizing hormone in the unrestrained, ovariectomized turkey hen. Poultry Science. 1989;68:177–184. doi: 10.3382/ps.0680177. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Advances in Protein Chemistry. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: Beyond the ground state. Molecular Pharmacology. 2007;71:959–964. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- Selvam R, Jurkevich A, Kang SW, Mikhailova MV, Cornett LE, Kuenzel WJ. Distribution of the vasotocin subtype four receptor (VT4R) in the anterior pituitary gland of the chicken, Gallus gallus and its possible role in the avian stress response. Journal of Neuroendocrinology. 2013;25:56–66. doi: 10.1111/j.1365-2826.2012.02370.x. [DOI] [PubMed] [Google Scholar]
- Thibonnier M, Coles P, Conarty DM, Plesnicher CL, Shoham M. A molecular model of agonist and nonpeptide antagonist binding to the human V(1) vascular vasopressin receptor. Journal of Pharmacology and Experimental Therapeutics. 2000;294:195–203. [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf TB, Roux B. Structure, energetics, and dynamics of lipid–protein interactions: A molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996;24:92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.