Abstract
Purpose
Rapid diagnosis of urinary tract infection would have a significant beneficial impact on clinical management, particularly in patients with structural or functional urinary tract abnormalities who are highly susceptible to recurrent polymicrobial infections. We examined the analytical validity of an electrochemical biosensor array for rapid molecular diagnosis of urinary tract infection in a prospective clinical study in patients with neurogenic bladder.
Materials and Methods
The electrochemical biosensor array was functionalized with DNA probes against 16S rRNA of the most common uropathogens. Spinal cord injured patients at a Veterans Affairs hospital were recruited into the study. Urine samples were generally tested on the biosensor within 1 to 2 hours of collection. Biosensor results were compared with those obtained using standard clinical microbiology laboratory methods.
Results
We successfully developed a 1-hour biosensor assay for multiplex identification of pathogens. From July 2007 to December 2008 we recruited 116 patients, yielding a total of 109 urine samples suitable for analysis and comparison between biosensor assay and standard urine culture. Of the samples 74% were positive, of which 42% were polymicrobial. We identified 20 organisms, of which Escherichia coli, Pseudomonas aeruginosa and Enterococcus species were the most common. Biosensor assay specificity and positive predictive value were 100%. Pathogen detection sensitivity was 89%, yielding a 76% negative predictive value.
Conclusions
To our knowledge we report the first prospective clinical study to successfully identify pathogens within a point of care time frame using an electrochemical biosensor platform. Additional efforts to improve the limit of detection and probe design are needed to further enhance assay sensitivity.
Keywords: urinary bladder, neurogenic, spinal cord injuries, urinary tract infections, biosensing techniques, RNA, ribosomal, 16S
Urinary tract infection, which is among the most common bacterial infections, carries a significant health care burden.1,2 Patients with SCI and neurogenic bladder are at high risk for UTI due to impaired bladder emptying and the frequent need for instrumentation or indwelling catheters.3–5 Judicious management for UTI in patients with SCI is critical given the risk of complications, including urolithiasis and urosepsis. Diagnosing UTI in patients with SCI is challenging because signs and symptoms are frequently nonspecific, infections are polymicrobial and it is difficult to differentiate bacterial colonization from infection. Standard urine culture typically requires 2 to 3 days from sample collection to delivery of the microbiology report. Rapid identification and quantitation of multiple pathogens in urine samples is of great benefit to understand the pathogenesis of polymicrobial UTI and enable evidence-based clinical decision making.
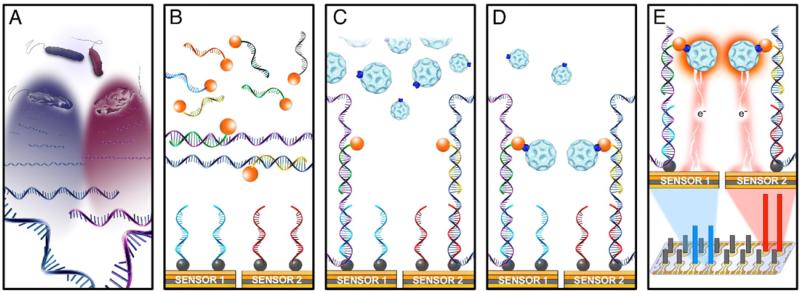
We previously reported the development of a 1-hour electrochemical biosensor assay for direct pathogen identification from urine samples. The detection strategy relies on translating species specific hybridization of bacterial 16S rRNA with capture and detector probe pairs into a measurable electrical current (fig. 1).6,7 For bacterial identification 16S rRNA sequences are well characterized molecular targets. A sandwich assay with capture and detector probes obviates the need to label the target. Signal amplification is achieved using an enzyme tag with recyclable catalytic properties, enabling ultrasensitive detection without nucleic acid amplification.
Figure 1.
Multiplex detection of pathogens using electrochemical biosensor array. A, simultaneous lysis of different bacteria releasing 16S rRNA target. B, hybridization of 16S rRNA targets with cocktail of detector probes labeled with fluorescein (orange circles). C, deposition of target detector probe mix on sensor surface for sandwich hybridization with capture probes. Each sensor was functionalized with biotin labeled (gray circles) capture probes of different specificity. D, anti-fluorescein horseradish peroxidase enzyme tag binding to sandwich hybrid. E, horseradish peroxidase substrate oxidation under fixed voltage generated amperometric signal measured in nA from each sensor. Magnitude of signal output corresponded to starting concentration of each pathogen. Each experimental condition was performed in duplicate.
We report a prospective clinical study of the bio-sensor assay in patients with SCI, who are susceptible to complicated UTI. With the frequent need to access health care resources patients with SCI are exposed to more diverse and multidrug resistant uropathogens than typical community acquired uropathogens.8,9 A reliable diagnostic test that enables pathogen identification within a point of care time frame would provide a valuable tool to manage UTI in these challenging cases.
MATERIALS AND METHODS
Study Design
Protocol approval was obtained from the Stanford University institutional review board, and the VAPAHCS research and development committee. Informed consent was obtained from all research subjects. From July 2007 to December 2008 patients were prospectively recruited from the SCI service at VAPAHCS. The decision to collect urine was made by the treating clinicians as part of routine clinical evaluation or because UTI was suspected. From each patient 2 urine samples were collected by voiding, straight catheterization or directly from the indwelling catheter with assistance from the SCI nursing staff. One sample was sent for standard clinical laboratory culture and susceptibility analysis at the VAPAHCS clinical microbiology laboratory (67%) or elsewhere (33%). The second sample was sent directly to our laboratory for biosensor assay. Quantitative plating was done at our laboratory to determine bacterial concentrations. Urine samples received after July 2008 were evaluated at our laboratory by plating 5 [H9262]l samples on BBL™ CHROMagar™ Orientation and TSA with 5% sheep blood and incubation overnight at 35C.10,11 For biosensor testing 1.5 ml of each urine sample were pelleted by centrifugation and the supernatant was discarded. Biosensor signals were comparable in fresh and frozen pellets (data not shown). Therefore, when biosensor assay was not immediately done, sample pellets were stored at –70C for later testing.
Electrochemical Biosensor Assay
Unmodified 16-sensor arrays were functionalized with capture probes at our laboratory, as previously described.6,7 Urine pel-lets were lysed by 5-minute room temperature incubation in 0.1% Triton X-100, 20 mM tris-HCl (pH 8.0), 2 mM ethylenediaminetetraacetic acid (pH 8.0) and 5 mg/ml lysozyme with the addition of 1M NaOH. Hybridization of the detector probe and electrochemical detection were done as previously described.6,7 All samples were analyzed on duplicate sensors.
Probe Sequences
Oligonucleotide probes were synthesized elsewhere, including capture probes with a 5’-biotin modification and detector probes with 3’-fluorescein modifications. Probe pairs were designed to hybridize with species specific regions of the 16S rRNA of EC, EF, PM, PA and AB as well as the KE group, EB family and UNI probes. Sequences of the capture probes UNI782C, EB1172C, EC449C, KE434C, PM187C, PA102C and EF207C, and the detector probes UNI751D, EB1137D, EC408D, KE399D, PM147D, PA74D and EF171D were previously reported.6,7 AB probes were capture AB456C (AGTAACGTCCACTATCTCTAGGTATTAACTAAAGT) and detector AB421D (AGGCTCCTCCTCGCTTAAAGRAGTGCTTTACAACCATA).
Data Analysis
Sensor-to-sensor variation of log10 transformed biosensor measurements was estimated by the pooled SD between duplicate measurements in all samples. The SD was approximately constant at 0.104. Positive signals were defined as the average signals of the duplicates greater than 3 SD (log10 units) over NC. This threshold yielded the highest concordance between clinical laboratory and bio-sensor data, as indicated on an empirical ROC curve with AUC calculated using the trapezoidal rule. Signals derived from sensors containing the AB probe functioned as the NC because this organism was not found in any of our clinical urine samples.
Clinical microbiology laboratory results served as the gold standard to compare our biosensor results. Clinical microbiology reported samples with 1 or 2 organisms of 1,000 to 10,000 cfu/ml as gram-positive or gram-negative and organisms at 10,000 cfu/ml or greater were identified by species. Polymicrobial samples with more than 2 organisms at 100,000 cfu/ml or greater were identified as “mixed, no predominant organism” and were not further identified unless requested by the physician. In polymicrobial samples with 1 or 2 organisms and mixed flora the predominant organisms were speciated and reported as “culture is mixed, predominant organisms worked up.” Clinical laboratory results and UNI probe signals were compared to estimate biosensor sensitivity, specificity, and positive and negative predictive values.
RESULTS
Study Population and Urine Characteristics
From July 2007 to December 2008 we recruited 116 patients from the VAPAHCS SCI unit as study participants. A total of 126 urine samples were collected. Of the patients 108 provided 1, 6 provided 2 and 2 provided 3 samples. Multiple samples were collect at least 1 month apart. Table 1 lists patient demographics. Of the study population 97% were male and most required assistance for bladder emptying through intermittent catheterization or an in-dwelling catheter.
Table 1.
Study patient characteristics
| No. Pts (%) | |
|---|---|
| Male | 112 (97) |
| Female | 4 (3) |
| Total | 116 |
| Inpt | 63 (53)* |
| Outpt | 55 (47) |
| Urine collection method: | |
| Voided | 31 (27)† |
| Straight catheterization | 26 (22) |
| Indwelling urethral catheter | 46 (40) |
| Indwelling suprapubic catheter | 12 (10) |
| Ileal conduit | 1 (1) |
In 2 patients from whom we collected 2 samples each 1 sample was collected at an outpatient clinic visit and 1 was collected during an inpatient stay.
Including 9 patients with a chronic condom catheter.
To quantitate the bacterial concentration at sample collection we determined the plate count of serially diluted samples and found excellent agreement with clinical microbiology laboratory results. Discrepancies were found in only 3 of the 126 samples. In each case our plating indicated a lower bacterial concentration than reported by the clinical labora tory. All samples with discrepancies in bacterial concentration were plated at our laboratory within 1 hour of collection while clinical laboratory analysis was done elsewhere, raising the possibility of bacterial overgrowth during sample transport. The results of our internal BBL CHROMagar Orientation medium plating for pathogen identification were consistent with the clinical microbiology laboratory reports in all samples.
Of the 126 urine samples collected 17 were excluded from analysis due to the lack of comparable data from the clinical laboratory, including 4 samples that were not analyzed, 7 that were “mixed, no predominant organism,” 3 with “mixed skin/genital flora” and 3 containing yeast. Table 2 lists clinical laboratory results in the remaining 109 samples included in analysis. The rate of positive urine cultures (74%) and polymicrobial samples (42% of positive samples) were significantly greater than in our previous study, which was drawn from a general population.6
Table 2.
Clinical microbiology laboratory sample characterization
| No. Samples (%) | |
|---|---|
| Number of urine samples | 109 |
| No growth* | 28 (26) |
| 1 Organising:† | 47 (43) |
| E. coli | 16 |
| Enterococcus spp. | 7 |
| P. aeruginosa | 6 |
| K. pneumoniae | 5 |
| E. cloacae | 2 |
| P. mirabilis | 1 |
| K. oxytoca | 1 |
| Methicillin resistant S. aureus | 1 |
| S. saprophyticus | 1 |
| C. brakii | 1 |
| C. freundii | 1 |
| S. agalactiae | 1 |
| Nonspeciated gram-pos | 3 |
| Nonspeciated gram-neg | 1 |
| 2 Organisms: | 19 (17) |
| E. coli, P. aeruginosa | 2 |
| E. coli, P. mirabilis | 1 |
| E. coli, C. koseri | 1 |
| E. coli, Streptococcus viridans | 1 |
| E. coli, α-hemolytic Streptococcus | 1 |
| K. pneumoniae, P. aeruginosa | 1 |
| K. pneumoniae, P. mirabilis | 1 |
| K. pneumoniae, S. marcescens | 1 |
| K. pneumoniae, Enterococcus spp. | 1 |
| K. pneumoniae, lactose pos gram-neg rod | 1 |
| M. morganii, P. aeruginosa | 1 |
| M. morganii, gram-pos | 1 |
| P. mirabilis, gram-pos | 1 |
| P. stuartii, gram-pos | 1 |
| S. marcescens, Enterococcus spp. | 1 |
| P. aeruginosa, S. aureus | 1 |
| C. youngae, gram-pos | 1 |
| Enterococcus spp., S. agalactiae | 1 |
| Mixed culture predominant organism(s):* | 15 (14) |
| E. coli | 2 |
| P. aeruginosa | 1 |
| P. stuartii | 1 |
| P. aeruginosa, Enterococcus spp. | 4 |
| K. pneumoniae, Enterococcus spp. | 1 |
| E. coli, P. rettgeri | 1 |
| E. coli, P. aeruginosa, Enterococcus spp. | 2 |
| E. coli, P. mirabilis, Enterococcus spp. | 2 |
| K. pneumoniae, P. aeruginosa, Enterococcus spp. | 1 |
No growth considered after 18-hour to 2-day plate incubation.
Positive samples with 1 or more organism or mixed culture with 1 or more predominant organisms.
Biosensor Clinical Validation
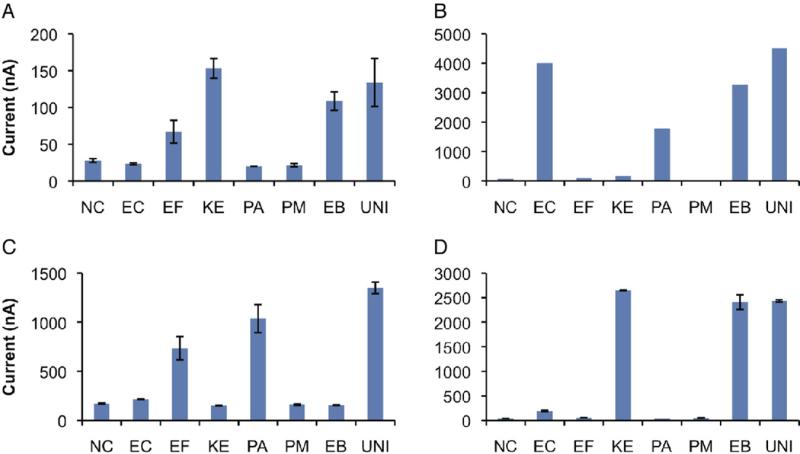
Figure 2 shows representative assay results in 3 polymicrobial urine samples and a single pathogen sample. Different organism combinations were detected with species specific as well as broad-spectrum probes for EB and the UNI probe for all species.
Figure 2.
Pathogen detection in clinical urine samples using electrochemical biosensors. Each 16-sensor array was modified with 8 capture probes in duplicate and tested against urine samples from patients with SCI. Electrochemical sensor results completely agreed with clinical microbiology laboratory reports. Sensors modified with species specific probes (x axis) were appropriately positive with magnitude of signal output (y axis) corresponding to pathogen concentration. UNI probe was appropriately positive in all 3 samples. AB probe served as NC since no clinical samples contained AB. A, sample containing 60,000 cfu/ml K. pneumoniae and 20,000 cfu/ml EF with appropriately positive EB probe. B, sample containing 1 × 107 cfu/ml EC and 8 × 106 cfu/ml PA with appropriately positive EB probe. C, sample containing 1 × 106 cfu/ml PA and 5 × 105 cfu/ml EF species. D, urine sample containing 1 × 107 cfu/ml K. pneumoniae with appropriately positive EB probe. Error bars represent average of duplicate sensors.
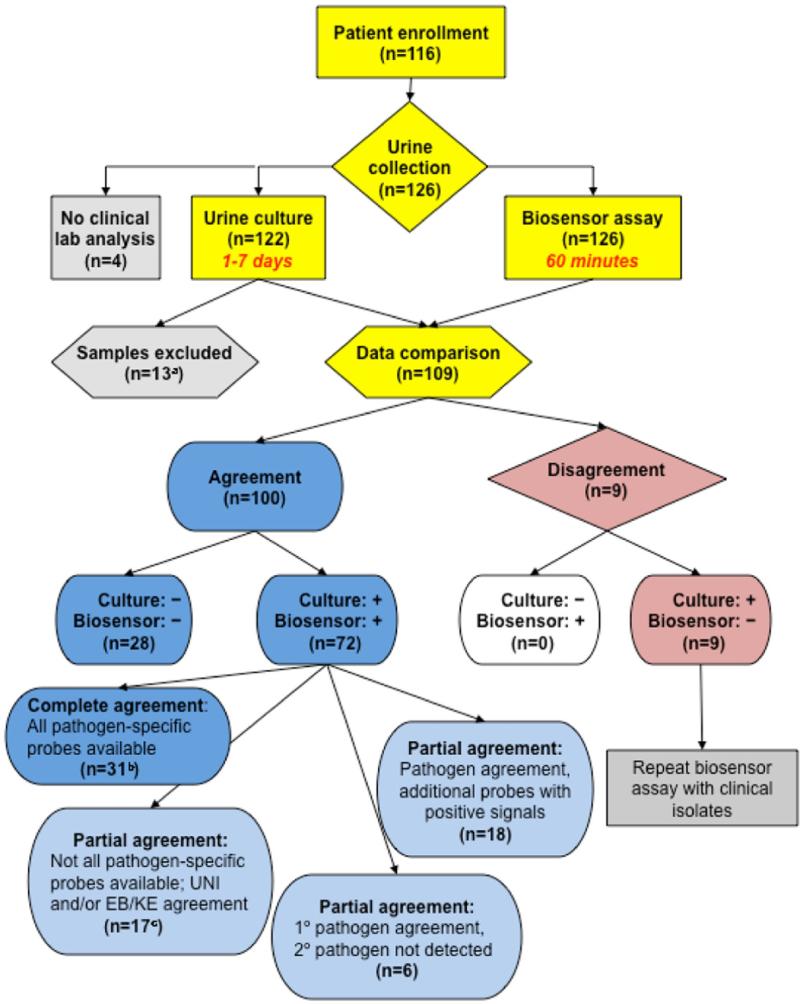
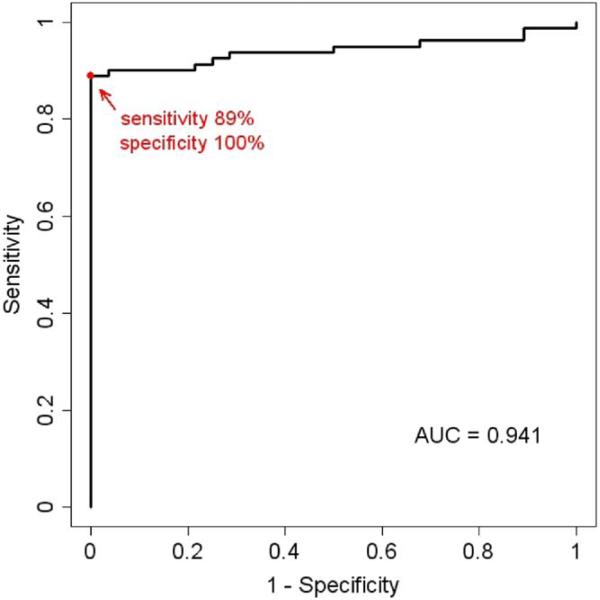
Figure 3 shows biosensor and clinical laboratory results. In the 109 samples with data available we determined that the highest concordance between clinical laboratory data and biosensor data was at a cutoff for a positive biosensor signal of 3 SD above NC using ROC analysis (fig. 4). Biosensor results agreed with clinical laboratory findings in all 28 samples reported as having no growth, yielding 100% specificity and 100% positive predictive value. Since the biosensor was not configured with probes for all possible pathogens, the UNI was used to determine sensitivity. Of the 81 samples reported to be culture positive by clinical laboratory findings the biosensor was positive in 72, yielding 89% overall sensitivity and 76% negative predictive value. In the 9 samples missed by the biosensor the pathogen concentration was 4 × 104 cfu/ml or less. By re-inoculating the clinical isolates from these samples and growing them to a concentration of 105 cfu/ml we successfully detected the pathogens using the biosensor (data not shown). The overall limit of detection in the range of 104 cfu/ml in clinical samples was consistent with that in our previous reports.6,7
Figure 3.
Clinical study. Asterisk indicates 13 samples with no specific pathogen identified at clinical laboratory were excluded from biosensor comparison analysis, including 7 mixed cultures with no predominant organism, 3 with mixed skin flora and 3 with yeast. b, samples were considered in complete agreement when specific probes for all organisms in sample were present and appropriately positive on biosensor. c, samples with 1 or more organisms for which specific probe was not present on biosensor were considered in agreement to best probe diversity when UNI and other appropriate probes were positive.
Figure 4.
ROC curve analysis of pathogen detection using electrochemical biosensor. Maximum concordance was 0.917 at cutoff of 3 SD above background.
In 31 concordant positive samples the biosensor was in complete agreement with clinical laboratory results in regard to species specificity and the number of pathogens. In 17 concordant positive samples not all species specific probes were available on the biosensor but signals from the positive control probes (EB and/or UNI) were in agreement with clinical laboratory findings. For example, the clinical laboratory reported that sample 98 contained Staphylococcus aureus and the biosensor appropriately showed that only the UNI probe was positive.
In all polymicrobial samples biosensor and clinical laboratory results agreed on the primary pathogen with some discrepancies on secondary pathogens. In 6 samples 2 or more pathogens were identified at the clinical laboratory but the biosensor only identified the primary pathogen. For example, the clinical laboratory reported that sample 33 had greater than 100,000 cfu/ml Klebsiella pneumoniae and 30,000 cfu/ml Pseudomonas aeruginosa but the biosensor identified only K. pneumoniae. In 18 samples the biosensor was in agreement with clinical laboratory findings for the primary and secondary pathogens but the biosensor had additional low level signals that were statistically significant. Plating at our laboratory confirmed the additional organisms in 5 cases and the remaining 13 were likely due to probe cross reactivity. Figure 2, D shows a representative example for which the clinical laboratory reported K. pneumoniae. The biosensor confirmed K. pneumoniae but the signal from the EC probe was also statistically significant. Furthermore, for some pathogens, particularly Klebsiella and Enterobacter species, there is significant intraspecies variation in 16S rRNA sequences (D. A. Haake, unpublished data). Consequently the KE probe contained numerous degenerate bases to target Klebsiella and Enter-obacter species.6,7 At high bacterial concentrations the KE probe cross-reacted with additional pathogens, including Serratia, Morganella, Citrobacter, and occasionally Escherichia coli and Proteus mira-bilis. This lack of specificity for the KE probe was not observed during our initial probe development using clinical isolates. These findings clearly suggest the need to improve probe design for Klebsiella and Enterobacter species, and illustrate the importance of validating biosensor technology with clinical samples, rather than simply with clinical isolates.
DISCUSSION
We report a prospective clinical study of a rapid molecular assay for uropathogen identification in patients with SCI, a challenging patient cohort susceptible to recurrent polymicrobial UTIs. This 1-hour assay is based on the detection of 16S rRNA with an electrochemical biosensor. We obtained a snapshot of the bacterial constituency in urine at collection, an approach that is arguably more clinically relevant than current methods, which are limited by the time required for specimen storage and transport. Furthermore, polymicrobial UTI is generally poorly understood and difficult to study using culture based approaches. Our multiplex biosensor assay provides an attractive platform for studying the pathogenesis of polymicrobial infections.
Our results compared favorably with conventional clinical microbiology laboratory analysis with 89% overall sensitivity, 100% specificity, 76% negative predictive value and 100% positive predictive value. With the current protocol and probe design pathogens at concentrations as low as 2 × 104 cfu/ml were detected. All pathogens that the biosensor missed were at 4 × 104 cfu/ml or less. In 18 samples the biosensor had significant positive signals for additional pathogens not identified at the clinical laboratory. In all of these samples the strongest biosensor signal corresponded to the primary pathogen reported by the clinical laboratory.
This study improves on our initial pilot study.6 1) Patient characteristics and polymicrobial urine samples were previously not considered. 2) Urine samples were previously obtained from the clinical microbiology laboratory, frequently more than 24 hours after collection. In the current study we processed and assayed freshly collected urine samples.
There are limitations to our study. We noted pathogens for which we did not have species specific probes. In these situations the UNI and EB probes were capable of identifying samples as positive. In addition, our KE probe recognized not only Klebsiella and Enter-obacter species, but also Citrobacter, Serratia and Morganella species. Efforts are under way to design more specific Klebsiella and Enterobacter probes, and species specific probes for the other less common pathogens.
Traditional clinical microbiology and commercially available automated systems require initial isolation of pathogens before phenotypic identification. The need to propagate bacteria on laboratory medium for identification is the primary reason for the time-consuming nature of clinical bacteriology. Also, the need for culture may result in failure to identify pathogens that do not grow well in the laboratory setting.12 Important laboratory tests are increasingly outsourced, which can lead to additional errors related to sample handling and prolong overall sample processing time.13
In contrast, our genotypic approach relies on molecular probes and is compatible with direct detection from clinical samples without bacterial isolation. Furthermore, the electrochemical biosensor is based on computer chip technology, which is amenable to precise, cost-effective production.
Patients with SCI are susceptible to complicated UTIs caused by a wide spectrum of uropathogens. This population as well as urological patients with anatomically and neurologically impaired bladder emptying, and pediatric patients in whom history and physical examination may be unreliable, are among those most likely to benefit from this technology. Also, polymicrobial UTI is poorly understood and the relationship between the primary and secondary pathogens is unclear (synergistic, neutral or antagonistic). Potential applications of our assay could include dynamic monitoring of the pathogen concentration in patients at high risk or assessing the efficacy of antibiotic treatment.
While our assay provides quantitative information on bacterial species and concentrations, it does not differentiate between infection and colonization. We are currently investigating concomitant measurement of urinary inflammatory biomarkers to facilitate this differentiation. In addition, we are developing a biosensor assay for antibiotic susceptibility determination. Other efforts are under way for integration with microfluidic technology and automation of sample processing for point of care application.14 A point of care device capable of delivering accurate, expeditious identification of pathogens directly from body fluids would be a significant advance in diagnostic microbiology.
CONCLUSIONS
This simple, rapid assay is ideally suited to UTI diagnosis in patients at risk for polymicrobial infection. In conjunction with a complementary antibiotic susceptibility assay and microfluid based automation under development our novel approach toward point of care UTI diagnosis may lead to more judicious and effective management for UTI.
ACKNOWLEDGMENTS
Victoria Wolfe, Christina Hirsch and Mey Yip, SCI Unit, Veterans Affairs Palo Alto Health Care System, assisted with urine collection; Simon Kimm assisted with figure 1; Vincent Gau provided technical support; 33% of urine samples were analyzed at Specialty Laboratories, Valencia, California; and oligonucleotide probes were synthesized at IDT, Coralville, Iowa.
Study received approval from the Stanford University institutional review board, and VAPAHCS research and development committee.
Supported by Department of Veterans Affairs Merit Review B4872R (JCL) and National Institute of Allergy and Infectious Diseases Cooperative Agreement Award AI075565 (DAH).
Abbreviations and Acronyms
- AB
Acinetobacter baumannii
- EB
Enterobacteriaceae family
- EC
Escherichia coli
- EF
Enterococcus species
- KE
Klebsiella-Enterobacter
- NC
negative control
- PA
Pseudomonas aeruginosa
- PM
Proteus mirabilis
- SCI
spinal cord injury
- UNI
universal eubacterial
- UTI
urinary tract infection
- VAPAHCS
Veterans Affairs Palo Alto Health Care System
REFERENCES
- 1.Griebling TL. Urologic Diseases in America project: trends in resource use for urinary tract infections in men. J Urol. 2005;173:1288. doi: 10.1097/01.ju.0000155595.98120.8e. [DOI] [PubMed] [Google Scholar]
- 2.Griebling TL. Urologic Diseases in America project: trends in resource use for urinary tract infections in women. J Urol. 2005;173:1281. doi: 10.1097/01.ju.0000155596.98780.82. [DOI] [PubMed] [Google Scholar]
- 3.Biering-Sorensen F. Bagi P and Hoiby N: Urinary tract infections in patients with spinal cord lesions: treatment and prevention. Drugs. 2001;61:1275. doi: 10.2165/00003495-200161090-00004. [DOI] [PubMed] [Google Scholar]
- 4.Cardenas DD, Hoffman JM, Kirshblum S, et al. Etiology and incidence of rehospitalization after traumatic spinal cord injury: a multicenter analysis. Arch Phys Med Rehabil. 2004;85:1757. doi: 10.1016/j.apmr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Esclarin De Ruz A. Garcia Leoni E and Herruzo Cabrera R: Epidemiology and risk factors for uri-nary tract infection in patients with spinal cord injury. J Urol. 2000;164:1285. [PubMed] [Google Scholar]
- 6.Liao JC, Mastali M, Gau V, et al. Use of electro-chemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. J Clin Microbiol. 2006;44:561. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao JC, Mastali M, Li Y, et al. Development of an advanced electrochemical DNA biosensor for bacterial pathogen detection. J Mol Diagn. 2007;9:158. doi: 10.2353/jmoldx.2007.060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girard R, Mazoyer MA, Plauchu MM, et al. High prevalence of nosocomial infections in rehabilitation units accounted for by urinary tract infections in patients with spinal cord injury. J Hosp Infect. 2006;62:473. doi: 10.1016/j.jhin.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Hinkel A, Finke W, Botel U, et al. Increasing resistance against antibiotics in bacteria isolated from the lower urinary tract of an outpatient population of spinal cord injury patients. Urol Int. 2004;73:143. doi: 10.1159/000079695. [DOI] [PubMed] [Google Scholar]
- 10.D'Souza HA, Campbell M, Baron EJ. Practical bench comparison of BBL CHROMagar Orienta tion and standard two-plate media for urine cultures. J Clin Microbiol. 2004;42:60. doi: 10.1128/JCM.42.1.60-64.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hengstler KA, Hammann R, Fahr AM. Evaluation of BBL CHROMagar orientation medium for detection and presumptive identification of uri-nary tract pathogens. J Clin Microbiol. 1997;35:2773. doi: 10.1128/jcm.35.11.2773-2777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton W, Veeh R, Shirtliff M, et al. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. 2003;112:1466. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Procop GW, Winn W. Outsourcing microbiology and offsite laboratories. Implications on patient care, cost savings, and graduate medical education. Arch Pathol Lab Med. 2003;127:623. doi: 10.5858/2003-127-0623-OMAOL. [DOI] [PubMed] [Google Scholar]
- 14.Mai JDH, Gaster RS, Wu A, et al. A microfluidic system for rapid bacterial pathogen detection. Presented at IEEE International Conference on Nanotechnology, Hong Kong Special Administrative Region of People's Republic of China. 2007 Aug 2–5;:1330. [Google Scholar]