Abstract
The muscle regulatory factor MRF4 is expressed in both embryonic and adult vertebrate skeletal muscle cells. In mammals the MRF4 gene has a complex cis-regulatory structure, with many kilobases (kb) of upstream sequence required for embryonic expression in transgenic mice. Here, initial functional comparison between Xenopus and mammalian MRF4 genes revealed that 610 base pairs (bp) of the XMRF4a proximal promoter drove substantial transgenic expression in X. laevis myogenic cells, from somites of neurula embryos through adult myofibers, and as little as 180 bp gave detectable expression. Over 300 bp of XMRF4a proximal promoter sequence is highly conserved among three X. laevis and X. tropicalis MRF4 genes, but only about 150 bp shows significant identity to mammalian MRF4 genes. This most-conserved XMRF4a region contains a putative MEF2 binding site essential for expression both in transgenic embryos and in transfected mouse muscle cells. A rat MRF4 minimal promoter including the conserved region also was active in transgenic X. laevis embryos, demonstrating a striking difference between the mouse and Xenopus transgenic systems. The longest XMRF4a promoter construct tested, with 9.5 kb of 5′-flanking sequence, produced significantly greater expression in transfected mouse cells than did promoters 4.3-kb or shorter, suggesting that the intervening region contains an enhancer, although no increased expression was evident when this region was included in transgenic X. laevis embryos. Further identification and analysis of Xenopus MRF4 transcriptional control elements will offer insights into the evolution of this gene and of the myogenic gene regulatory network.
Keywords: Xenopus, transgenic, muscle, MRF4, promoter
Introduction
Within the genetic and epigenetic regulatory network that governs skeletal myogenesis in mammals, one transcription factor family, comprising MRF4, MyoD, myogenin and Myf5, occupies a central position (Pownall et al., 2002; Berkes and Tapscott, 2005; Bryson-Richardson and Currie, 2008). These four closely related myogenic regulatory factors (MRFs) control gene expression during myoblast specification and myofiber differentiation, maintenance, hypertrophy, repair, and regeneration, but the MRFs’ individual roles in these cellular events remain incompletely understood. Though MRF4 was for some years regarded primarily as a myofiber differentiation factor and unimportant for myoblast determination, which was thought to depend instead on MyoD and Myf5, more precise gene targeting showed that MRF4 can function as a determination gene when MyoD and Myf5 are absent (Kassar-Duchossoy et al. 2004). The revised model, in which either Myf5 or MRF4 acts upstream of MyoD to direct embryonic multipotent mesodermal cells into the myogenic lineage, is more consistent with the observations that MRF4 transcripts appear before those of MyoD and precede or are contemporaneous with those of Myf5 in somites of both mouse (Summerbell et al., 2002) and chick (Lin-Jones and Hauschka, 1996).
In vitro and transgenic approaches have both contributed to our current understanding of mammalian MRF4 regulation. Transfection of cells in vitro showed that E-box motifs (CANNTG) in rat and mouse MRF4 proximal promoters are required for their activation by MyoD, myogenin, or Myf5. MEF2 binding, at a site encompassing the TATA box, is also required for maximal muscle-specific expression (Black et al., 1995; Naidu et al., 1995). Although 130 base pairs (bp) of rat MRF4 proximal promoter suffices for expression in transfected muscle cells, greater expression is seen with 5 kilobases (kb) of upstream sequence (Hinterberger et al., 1992), which includes an enhancer region comprising several E-boxes and a consensus MEF2 site (Kerkvliet and Hinterberger, 1997). The proximal promoter by itself has no activity in myotomes and very little in myofibers of transgenic mice, but 8.5 kb of 5′-flanking sequence activates expression in thoracic somitic myocytes and in fetal myofibers (Pin et al., 1997). When added to the proximal promoter, the enhancer at −5 kb drives increases expression chiefly in fast muscle fibers at fetal stages but fails to activate expression in the embryonic somites (Pin and Konieczny, 2002). Similarly for the mouse MRF4 gene, a 7.5-kb promoter fragment drives partial expression in somites (Fomin et al. 2004), but fuller expression depends on an enhancer located between −88 kb and −140 kb from the MRF4 coding region (Carvajal et al., 2001). Recent work (Carvajal et al., 2008) shows that numerous enhancers 5′ of the MRF4 coding region interact in mutually exclusive ways with the MRF4 promoter and the closely linked Myf5 promoter in mouse. One particular enhancer located at −8 kb from the MRF4 coding region was previously shown to direct temporally and spatially distinct activity from the two promoters (Chang et al., 2004, 2007).
The MRF family is conserved throughout the vertebrate classes, yet the genes’ individual expression patterns differ among representatives of the classes that have been examined. Several differences between rodents and Xenopus exemplify this. In mice, myogenin is expressed in the myotomes, while in Xenopus, Xmyogenin mRNA reportedly appears only in the secondary myogenesis of limbs and dorsal body muscles during metamorphosis (Nicolas et al., 1998). In rats (Hinterberger et al., 1991) and in mice (Miner and Wold, 1990), MRF4 mRNA is more abundant than MyoD mRNA in adult muscle, whereas in Xenopus the reverse holds true (Jennings, 1992). Muscle denervation in rats leads to increased MRF4 transcript levels (Adams et al., 1995), but denervation of Xenopus muscle leads to decreased levels of XMRF4 mRNA (Jennings, 1992; Nicholas et al., 2000). These observations indicate that the MRF gene regulatory network must function somewhat differently in Xenopus than it does in mammals.
Here, I show that Xenopus myogenic cells require only a short MRF4 promoter for transgenic activity. Approximately 180 bp 5′ to the start codon of an X. laevis MRF4 gene sufficed for transgenic expression in embryonic myotomes. A rat MRF4 minimal promoter, containing a core sequence that is conserved in all vertebrate MRF4 genes, also drove expression in transgenic Xenopus embryos, clearly demonstrating a major functional difference between the mouse and frog transgenic assay systems. Postmetamorphic transgenic animals bearing either the rat or the Xenopus minimal promoter displayed reporter expression in trunk, limb and cranial muscles. Including additional sequence up to 610 bp 5′ of the start codon resulted in greater embryonic expression. Although transgenesis assays gave no evidence of any cis-regulatory activity located 5′ of the 610-bp promoter, sequence comparison among Xenopus MRF4 genes showed strong conservation of several upstream regions, and transient transfection of mouse C2C12 myoblasts pointed to an enhancer between 4.3 kb and 9.5 kb 5′ of the coding region.
Results
Gene cloning and sequence analysis
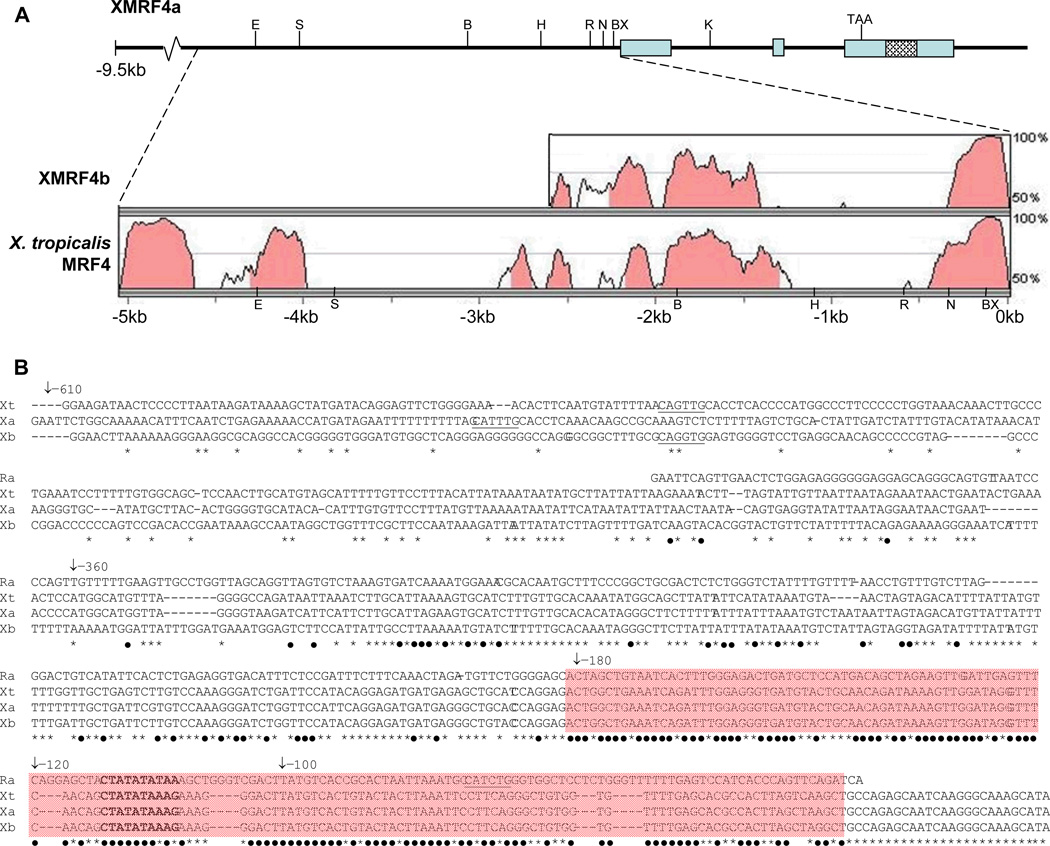
Two distinct XMRF4 sequences, apparently corresponding to the two MRF4 loci in the duplicated genome of X. laevis, were represented in multiple clones from a lambda genomic library. One clone of XMRF4a (gene designation consistent with Della Gaspera et al., 2006) contained the full coding sequence in three exons, approximately 9.5 kb of 5′-flanking sequence, and approximately 1 kb of 3′-flanking sequence (Fig. 1A). An XMRF4b (Della Gaspera et al., 2006) clone with all three exons was partially sequenced but the lengths of its flanking regions were not determined. The two XMRF4 genes displayed greater than 93% identity for approximately 330 bp 5′ to the start codon. This region included a TATA box that is conserved in all available vertebrate MRF4 gene sequences. The mammalian, lizard and chicken MRF4 genes also contain a MEF2 binding site (CTATATATAA) that overlaps the TATA box; in X. laevis and X. tropicalis MRF4 genes, the corresponding site (CTATATAAAG) deviated at one nucleotide from a MEF2 consensus site [YTA(A/T4)TAR (Black and Olson, 1998)]. A 150-bp region flanking the TATA box and putative MEF2 site in the Xenopus MRF4 genes displayed 71% identity with the corresponding region of the rat MRF4 promoter (Fig. 1B). No E boxes were present within the conserved proximal 330-bp region of any of the three Xenopus MRF4 genes. In XMRF4a, three E boxes lie at −450 bp, −475 bp and −555 bp from the first codon, and one is found in a similar location in XMRF4b and in X. tropcalis MRF4.
Figure 1. Structure of the XMRF4a genomic clone and comparison to other MRF4 gene sequences.
(A) Upper portion of the figure shows the 5′-flanking region, exons, introns, and 3′-flanking sequence of the 13-kb clone. Restriction enzyme sites mentioned in the text (B, BamH I; BX, BstX I; E, EcoR V; H, Hind III; K, Kpn I; N, Nco I; R, EcoR I; S, Sac I) and the stop codon are indicated. Cross-hatching identifies the P2X7 gene homology region (see text). Lower portion shows results of VISTA alignments of 5066 bp of XMRF4a 5′-flanking sequence versus XMRF4b and X. tropicalis MRF4 loci. Shading indicates conservation of 70% or greater. For XMRF4b, only 2325 bp of sequence was available for comparison. (B) Alignment of nucleotide sequences from rat MRF4 (Ra), X. tropicalis MRF4 (Xt), XMRF4a (Xa), and XMRF4b (Xb) promoters. Numbering refers to positions 5′ from the XlMRF4a start codon. −610 indicates the EcoR I site, −360 the Nco I site, −180 the BstX I site, and −120 and −100 indicate the 5′ ends of PCR-generated deletions. The MEF2 site is in bold type and E boxes (potential MRF binding sites) are underlined. Black dots beneath nucleotides indicate conservation in all four species, while asterisks indicate conservation only among the three Xenopus genes. Shading covers the 150-bp highly conserved region described in the text.
Another region of conservation between XMRF4a and -b began in XMRF4a approximately 1.1 kb upstream of the end of the 330-bp proximal region and extended at least 1.2 kb upstream from there. It was conserved between XMRF4a and X. tropicalis MRF4 as well. VISTA alignment with the X. tropicalis sequence indicated the existence of an even more strongly conserved region beginning about 4 kb upstream from the XMRF4a start codon and continuing to the end of the available 9673-bp XMRF4a sequence (Fig. 1A). No identity with the Xenopus MRF4 conserved upstream regions was indicated in any other available vertebrate genome sequence by VISTA or BLAST analysis.
A BLASTN search of GenBank using the XMRF4a sequence as a query showed that the 3′ untranslated region of XMRF4a contains a segment nearly identical to a portion of the coding sequence of an unrelated gene in X. laevis (Fig. 1A). A 289-bp sequence region, flanked by TA repeats, displayed 96% identity to a fragment of the cDNA for the P2X7 ATP-gated ion channel (Paukert et al., 2002).
Endogenous MRF4 expression in X. laevis
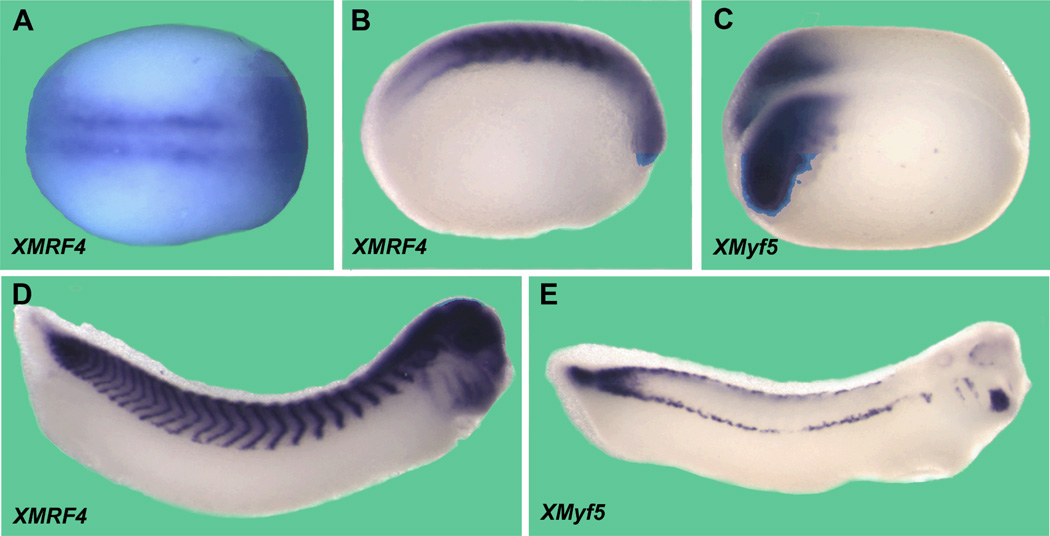
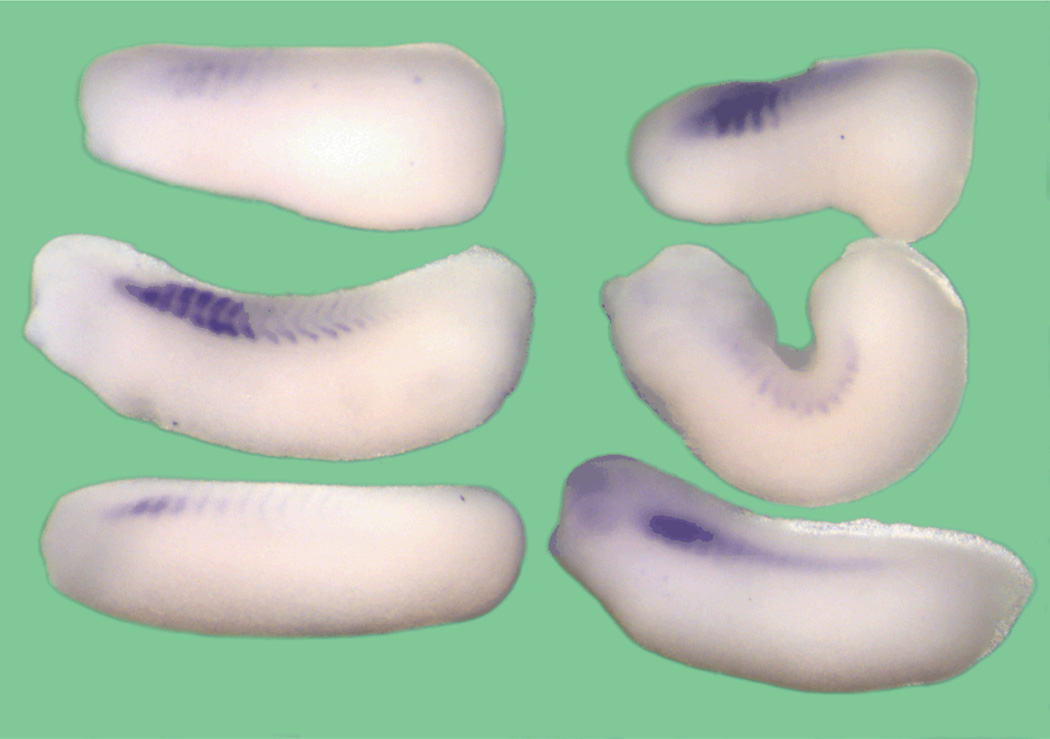
In situ hybridizations showed XMRF4 transcripts in the presomitic mesoderm beginning by at least stage 15 (Fig. 2A), consistent with our previous RT-PCR results (Ataian et al., 2003). Expression continued in somites and myotomes at all stages examined (Fig. 2B, D) with transcripts confined to the perinuclear region of myocytes. Unexpectedly, staining was also seen in the anterior of embryos, apparently in the eyes, brain, and other structures (Fig. 2B, D); this expression will be described more fully elsewhere.
Figure 2. Whole mount in situ hybridization of X. laevis embryos with the full-length XMRF4 probe or XMyf5 probe.
All are oriented anterior to the right. (A) Stage 14–15 early neurula, dorsal view. XMRF4 expression is seen in presomitic mesoderm prior to segmentation, as well as in the anterior. (B) Stage 20 neurula, lateral view. XMRF4 staining is most intense in the somites but also evident in the eye primordia. (C) Stage 16 neurula, dorsolateral view. XMyf5 expression is confined to the posterior mesoderm. (D) Stage 31–32 tailbud embryo, lateral view. In addition to the myotomal staining, XMRF4 expression is evident in the eyes, brain, branchial arches, otic vesicles, and head mesoderm. (E) Stage 31–32 tailbud embryo, lateral view. XMyf5 expression is seen in tailbud mesoderm, dorsal and ventral myotomal cells, and in primordia of some cranial muscles.
Because MRF4 and Myf5 genes are closely linked in the genomes of mammals and share overlapping regulatory regions (Carvajal et al., 2001, 2008), it was of interest to examine the XMyf5 expression pattern in conjunction with that of XMRF4. As demonstrated by other investigators (e.g., (Martin and Harland, 2001; Polli and Amaya, 2002), XMyf5 transcripts were detected in the posterior mesoderm at neurula (Fig. 2C) and tailbud (Fig. 2E) stages, becoming restricted to dorsal and ventral lips of the myotomes. XMyf5 expression was also seen in branchial arches and in primordia of cranial and extraocular muscles of late tailbud embryos; however, no XMyf5 expression was detected in neural structures, and at every stage its pattern was clearly distinct from that of XMRF4.
Cell transfections
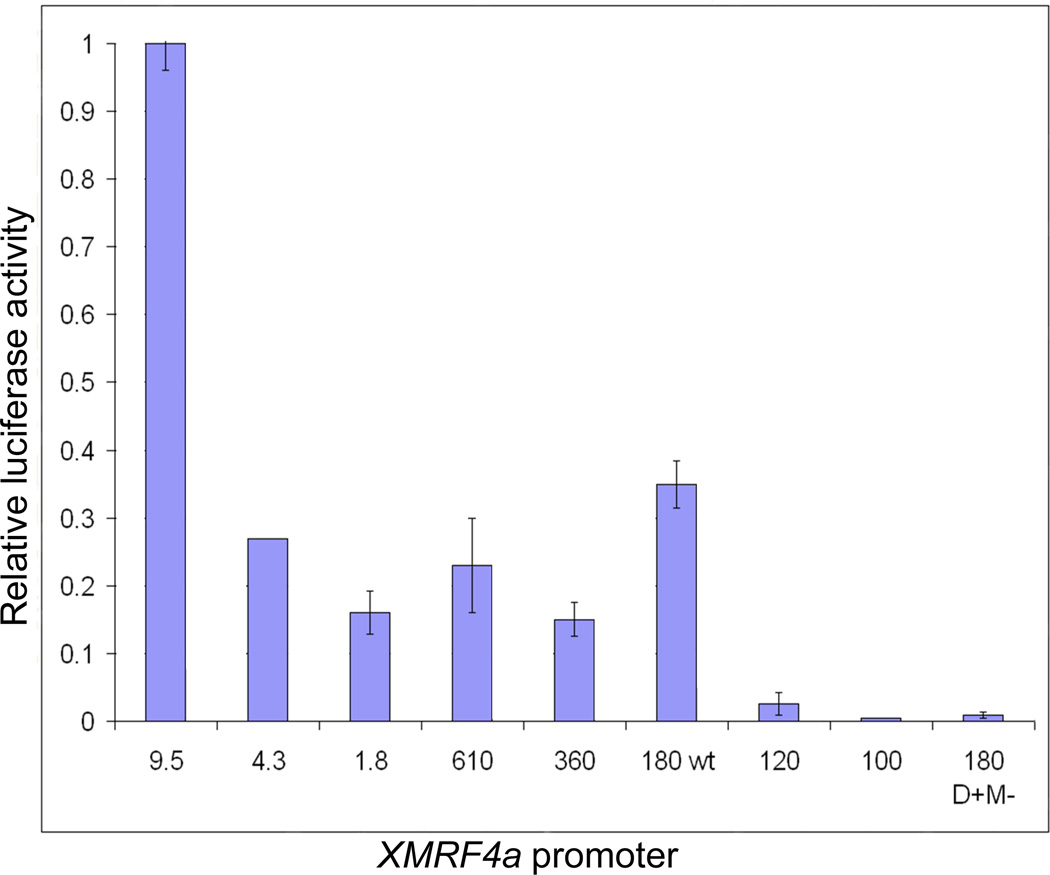
Transient transfection of C2C12 mouse myoblasts with XMRF4a 5′-flanking sequence fragments (hereafter called “promoters” even if several kb in length) linked to a luciferase reporter demonstrated that the TATA/MEF2 site plus approximately 60 bp of additional 5′ sequence (180wt) constitutes a minimal promoter in myotubes that differentiated from these cells (Fig. 3). Constructs that included only 6 bp of XMRF4a sequence 5′ to the TATA/MEF2 site (120) had greatly reduced expression, while constructs in which the promoter was truncated 10 bp downstream of the site (100) showed no activity. When the TATA/MEF2 site was mutated to abolish MEF2 binding (180 D+M−), promoter activity also was lost. Longer constructs that included genomic sequence as far as 4.3 kb 5′ from the first XMRF4a codon resulted in slightly less expression than did the minimal promoter. However, when the entire available 9.5 kb of XMRF4a 5′-flanking sequence was included, expression occurred at levels approximately threefold greater than that from the minimal promoter, implying the presence of an enhancer located between −4.3 kb and −9.5 kb that is active in mammalian myotubes.
Figure 3. Relative activity of XMRF4a promoters in mouse myotubes.
The graph shows averages of values ± s.e.m. from two to four independent transient transfection experiments for each XMRF4a construct, except for 4.3-kb which is a single datum. Each experimental value is the average of two separate C2C12 cell transfections, expressed as the ratio of test construct to RL-SV40 control, normalized to the value obtained for a positive control construct.
Transgenic animals
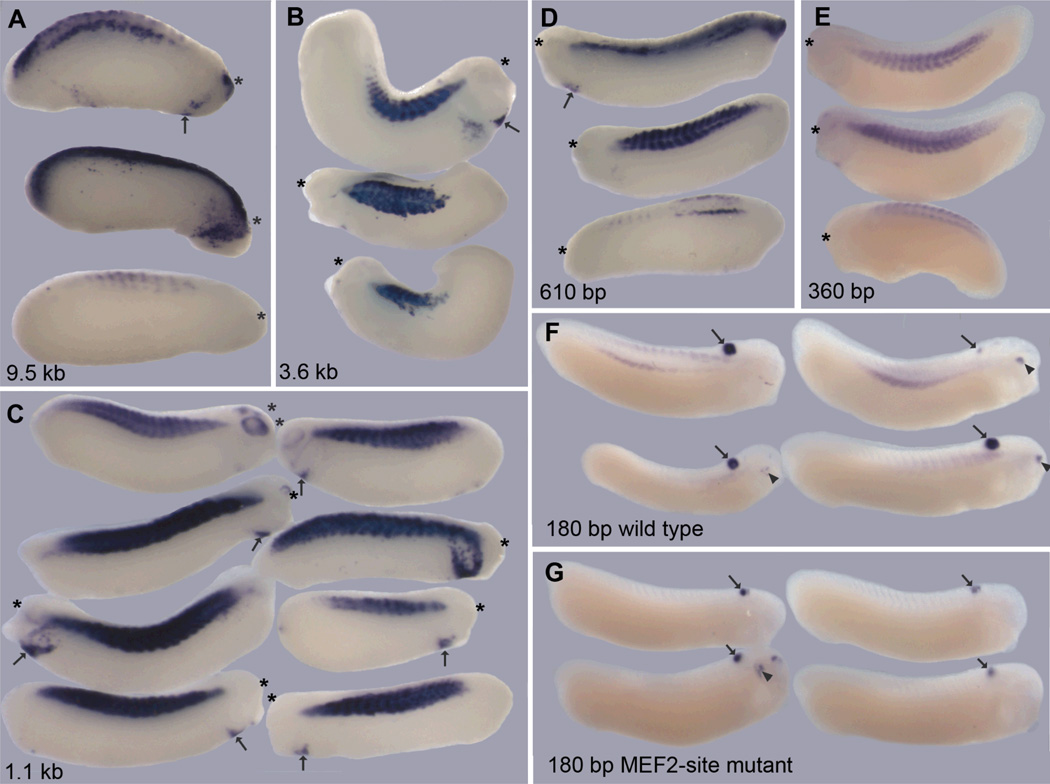
To study XMRF4a transcriptional regulation in X. laevis embryonic cells in vivo, test promoters were linked to a green fluorescent protein (GFP) cDNA and inserted as transgenes. Tailbud embryos were examined rather than earlier stages in order to allow for accumulation of detectable transcript levels from low-activity insertion sites. Constructs containing 9.5 kb, 3.6 kb, 1.1 kb, 610 bp, 360 bp, or 180 bp of wild-type XMRF4a promoter sequence all were expressed in myotomal muscle (Fig. 4A–F), at frequencies ranging from 8% to 31% (Table 1). In repeated experiments with the same construct, I obtained large variations in the percentage of positive embryos, preventing statistically valid quantitative ranking among these promoters based on their expression frequencies alone. When the gamma-crystallin-GFP construct was included as a control, successful transgene insertion could be inferred from GFP expression in the hindbrain and (at late tailbud stages) in the lenses of the eyes. Among these reporter-positive embryos, the 3.6-kb and 1.1-kb XMRF4a promoters gave myotomal expression in large majorities of cases, whereas the 180-bp promoter gave expression in less than a third of cases (Table 2). The intensity of in situ hybridization staining in the myotomes varied greatly within all groups of embryos carrying the same XMRF4a-GFP construct. However, the intensity of staining seen with the 360-bp or 180-bp promoters never reached the maximal level seen with the longer promoters. There was also a tendency for expression from the two shorter promoters to be confined to the dorsal and ventral ends of myotomes (Fig. 4E, F). (Unlike the endogenous XMRF4 staining, GFP reporter transcripts were not localized to the perinuclear region of myocytes, possibly because they lack a 3′ untranslated region.) One other qualitative difference was that the 610-bp and longer promoters occasionally produced expression in the branchial arches, in the cardiogenic region and the vicinity of eyes (Fig. 4A–D), a result not seen with the two shorter promoters. Transgenic embryos in which GFP was under the control of a 2.3-kb XMRF4b promoter showed myotomal expression identical to that seen with similar-sized XMRF4a constructs (data not shown).
Figure 4. Relative activity of XMRF4a promoters in transgenic X. laevis embryos.
Lengths of the promoters (described in the text) are indicated. Each panel shows lateral views of representative positive individuals from a single experiment. All specimens are oriented with dorsal sides towards the top of the figure, but because expression often was asymmetric, either the left or right side may be shown, whichever had more intense staining, and the anterior end is marked with an asterisk. In panels A–D, arrows indicate ectopic expression in cardiogenic regions. Embryos in F and G are co-transgenic for both the indicated XMRF4a-GFP construct and the gamma-crystallin-GFP construct; all are oriented with the anterior to the right side of the figure. Staining of the hindbrain (arrows) and, in some cases, the lens of the eye (arrowheads), results from gamma-crystallin-GFP expression.
Table 1.
Frequency of transgenic myotomal expression by MRF4 promoters
| Promoter | Experiment no. | GFP+ embryos | |
|---|---|---|---|
| XMRF4a promoters | |||
| 9.5 kb | 1 | 24% (11/46) | |
| 2 | 22% (15/69) | ||
| total | 23% (26/115) | ||
| 3.6 kb | 1 | 27% (25/94) | |
| 2 | 15% (8/53) | ||
| 3 | 59% (10/17) | ||
| total | 26% (43/164) | ||
| 1.1 kb | 1 | 36% 87/239 | |
| 2 | 8% (10/126) | ||
| 3 | 55% (40/73) | ||
| total | 31% (137/438) | ||
| 610 bp | 1 | 67% (8/12) | |
| 2 | 7% (3/44) | ||
| 3 | 57% (12/21) | ||
| total | 30% (23/77) | ||
| 360 bp | 1 | 20% (26/129) | |
| 2 | 25% (1/4) | ||
| total | 20% (27/133) | ||
| 180 bp (wt) | 1 | 8% (13/158) | |
| 2 | 6% (9/147) | ||
| 3 | 20% (5/25) | ||
| total | 8% (27/330) | ||
| 180 bp (D+M−) | 1 | 0% (0/120) | |
| 2 | 0% (1*/228) | ||
| total | 0% (1*/348) | ||
| *faint, questionable staining | |||
| rat MRF4 promoter | |||
| 410 bp | 1 | 7% (6/87) | |
| 2 | 12% (15/123) | ||
| total | 10% (21/210) | ||
Table 2.
Frequency of co-expression of γ-crystallin and XMRF4a promoters
| XMRF4a promoter length | % crys-GFP+ that were also XMRF4a-GFP+ |
|---|---|
| 3.6 kb | 88% (14/16) |
| 1.1 kb | 93% (67/72) |
| 180 bp (wild-type) | 30% (13/43) |
| 180 bp (D+M−) | 2% (1*/60) |
faint, questionable staining
Just as with mouse myoblasts transfected in vitro, mutation of the MEF2 site in the 180-bp XMRF4a promoter essentially abolished reporter gene expression in myotomal muscle of transgenic embryos. In one such experiment, inclusion of the gamma-crystallin-GFP control resulted in expression in brain or lens in 26% of the specimens, confirming that transgene insertion had taken place in those embryos despite the absence of myotomal expression (Fig. 4G).
To test the hypothesis that a minimal mammalian MRF4 promoter, due to its strong identity with the Xenopus proximal promoters, would be active in X. laevis embryos, 0.4 kb of rat MRF4 promoter sequence (Hinterberger et al., 1992; Naidu et al., 1995) was linked to the GFP cDNA. The rat promoter produced expression similar to that seen with the shorter XMRF4a promoters both in frequency (Table 1) and in pattern and intensity (Fig. 5).
Figure 5. Activity of the rat MRF4 minimal promoter in transgenic X. laevis embryos.
In situ hybridization shows GFP expression in representative positive individuals, oriented with dorsal side towards the top of the figure, anterior towards the left.
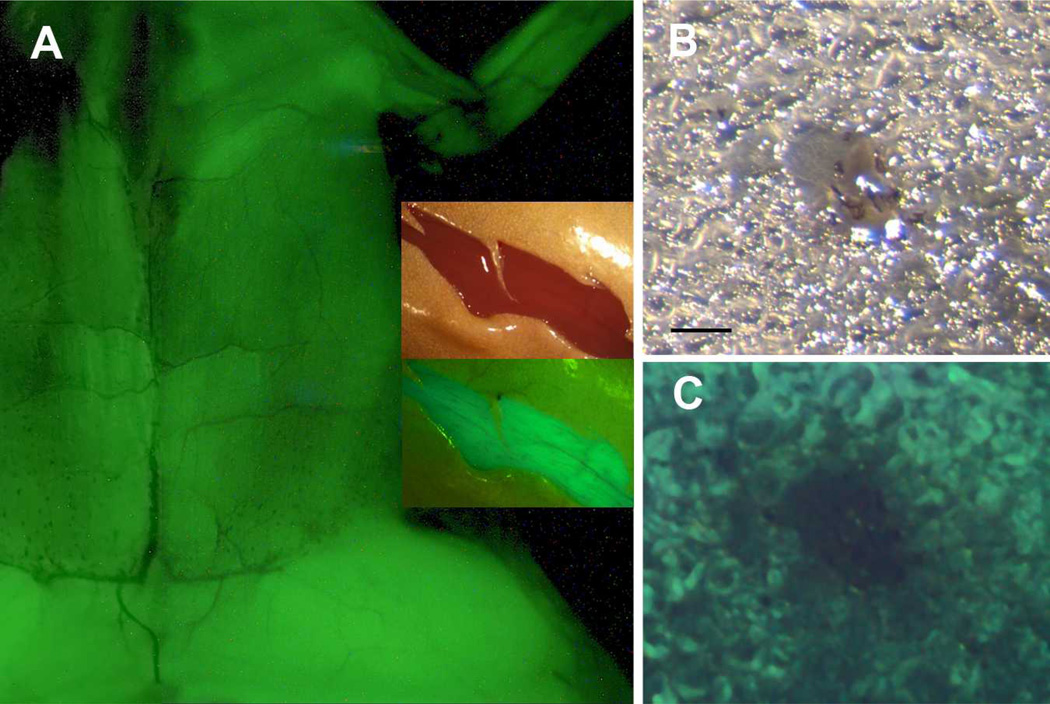
Transgenic tadpoles carrying the XMRF4a-GFP constructs were raised through metamorphosis in order to determine whether their transgenes would be expressed in limb muscles. The 3.6-kb, 1.1-kb, 610-bp (Fig. 6A) and 180-bp promoters all drove GFP expression in post-metamorphic skeletal muscles in the trunk, limbs, and head (animals bearing the 360-bp promoter have not yet been fully examined). In sections of transgenic hindlimb muscle, GFP expression was apparently restricted to myofibers (Fig. 6B, C).
Figure 6. Postmetamorphic expression of XMRF4a transgenes.
(A) GFP fluorescence driven by the 610-bp promoter in skeletal muscle, ventral view, anterior at top, skin removed. This is a composite of two separate exposures of the same animal. Most of the trunk and the left forelimb and proximal hindlimb are shown, but the head is out of the frame. Inset shows reflected-light (upper) and fluorescence (lower) images of a ventral skin incision in the hindlimb of a frog carrying the 1.1-kb promoter. GFP fluorescence in muscle is clearly distinguishable by color from autofluorescence in the skin. (B) Reflected-light image of a 50-µm frozen section of hindlimb muscle from an XMRF4a-1.1-GFP transgenic frog. Note the neurovascular bundle in the center. (C) GFP fluorescence in the same section as in B. Scale bar = 100µm.
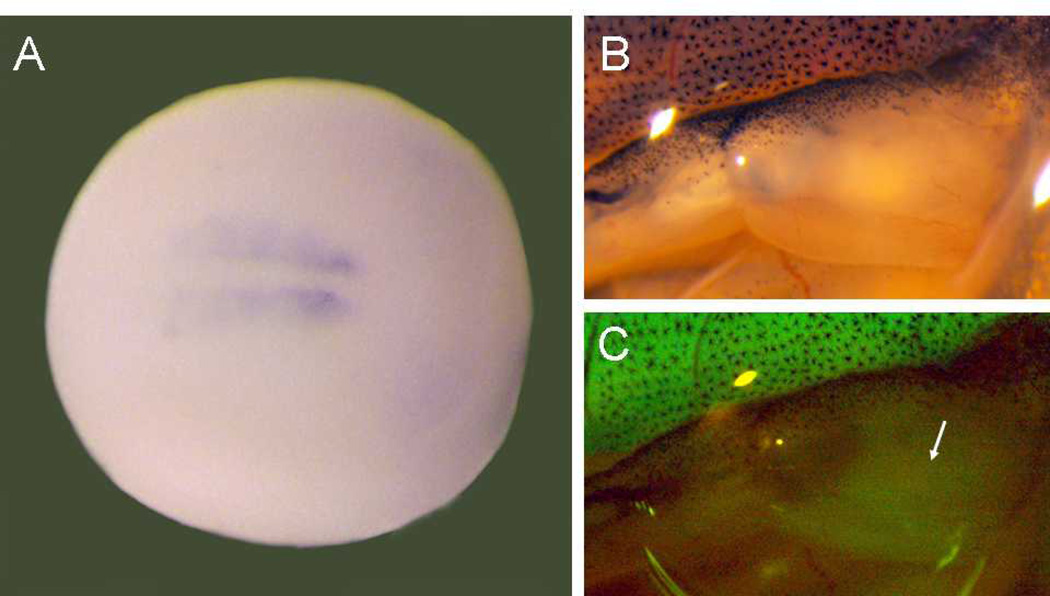
In order to compare the onset of XMRF4a-GFP transgene expression to that of the endogenous XMRF4 genes, strongly-expressing transgenic adults were mated with wild types to provide embryos for analysis of early stages. F1 offspring carrying the XMRF4a 610-bp promoter construct had detectable GFP expression in somites by at least stage 20 (Fig. 7A). F1 embryos carrying the 3.8-kb and the 1.1-kb transgenes showed similar results (data not shown). During metamorphosis of these animals, GFP fluorescence became detectable in hindlimb muscle at approximately stage 56 (Fig. 7B, C).
Figure 7. Onset of XMRF4a transgene expression.
(A) In situ hybridization showing GFP expression under control of the 610-bp promoter in the early somites of an F1 individual at stage 18–20, dorsal view, anterior to the right. (B) Reflected-light image of the right hindlimb of a stage-56 F1 animal carrying the 1.1-kb promoter construct. (C) GFP fluorescence from the same limb as in B. Note that the developing muscle (arrow) in the limb shows much lower intensity of fluorescence than do the tail myotomes at the top of the figure.
Discussion
With this study, Xenopus laevis MRF4 becomes the second member of the X. laevis MRF gene family, after XMyf5 (Yang et al., 2002), subjected to transgenic analysis of its cis-regulatory regions. Embryonic myotomes and, later, limb muscles expressed an XMRF4a proximal promoter as short as −180 bp from the first codon, while a 610-bp promoter produced somewhat stronger embryonic expression.
Promoter expression in any transgenic founder reflects a balance between the inherent cell-type-specific transcriptional activity of the test sequence and the repressive properties of the chromosomal site into which it randomly inserts. In principle, a stronger promoter will show detectable activity even in relatively repressive genomic sites, and it will appear more active than weaker promoters when it lands in permissive sites. The longer promoters tested here, ranging from 610 bp up to 9.5 kb, all gave detectable expression in myotomes at similar frequencies, while the 360-bp and 180-bp wild-type promoters displayed lower expression frequencies. Large batch-to-batch variances reduced the statistical significance of these differences when test constructs alone were inserted, but inclusion of the gamma-crystallin-GFP construct as a marker of transgene co-insertion served to filter the data so that much larger differences were seen between the 1.1-kb and 180-bp constructs’ expression frequencies, consistent with their relative intensities of in situ hybridization staining. Taken together, the expression frequencies and the staining intensities suggest that the 610-bp promoter contains the all regulatory elements required for XMRF4a expression in myotomes of tailbud embryos, although its initial expression appeared somewhat delayed and more restricted than the endogenous gene (compare Figures 2A and B with Figure 7A).
These results stand in striking contrast to those obtained with mouse or rat MRF4 promoters in transgenic mice, where the regulatory activity required for detectable somitic expression is found at least 5.6 kb upstream from the coding region (Fomin et al., 2004). Yet as shown here, a rat MRF4 0.4-kb proximal promoter is at least minimally effective in transgenic Xenopus, demonstrating an important functional difference between the two transgenic systems. This conclusion differs from previous studies of mammalian promoters in transgenic X. laevis (e.g., Beck and Slack, 1999; Lim et al., 2004) which emphasized the similarities of transgene expression in the two systems. In those studies, however, proximal promoters that lacked activity in mice were not tested in for activity Xenopus.
XMRF4a promoter fragments were examined for their efficacy driving reporter expression in mouse muscle cells in vitro as well as in transgenic X. laevis. Here too, the two types of data are not congruent. Luciferase assays of transfected mouse myotubes showed that the 9.5-kb promoter was very effective, while the 4.3-kb to 180-bp promoters were less but about equally effective. This result suggests the existence of an enhancer located between −4.3 kb and −9.5 kb from the coding region. Paradoxically, however, this X. laevis sequence region failed to display enhancer activity in transgenic embryos; promoters from 9.5 kb down to 610 bp in length performed indistinguishably. In addition, the in vitro results point more strongly to a central role for the −180 promoter than do the transgenics, where this promoter displayed minimal activity.
Within the 180-bp XMRF4a promoter, only the 150 bp flanking the putative MEF2 site has significant sequence identity to mammals, and truncation of the rat 0.4-kb promoter has shown that this conserved region accounts for all of its in vitro activity as well (Naidu et al., 1995). Deletion of the 5′ end of this region from the XMRF4a −180 promoter to make the −120 promoter essentially eliminated its in vitro activity, suggesting that these approximately 60 bp have a key role both in mammals and in Xenopus. As it is in the mammalian MRF4 (Black et al., 1995, Naidu et al., 1995) and Xenopus MyoDa (Leibham et al., 1994) genes, the putative MEF2 binding site in the XMRF4a minimal promoter was obligatory for expression. Loss of MEF2 binding to the XMRF4a −180D+M− promoter was not tested biochemically in this study; however, electrophoretic mobility shift assays of an identical mutant MEF2 site in Xenopus MyoDa showed that in vitro synthesized XMEF2A protein, as well as nuclear extracts from developing embryos or from C2 myotubes, bound to the wild-type but not to the mutant site (Leibham et al., 1994; Wong et al., 1994).
In rat and mouse MRF4 promoters as well as in many other muscle genes, MEF2 acts synergistically with MRF proteins, which bind at E-boxes in the proximal promoter. The three E-boxes in the 610-bp XMRF4a promoter have not been tested functionally, and it is unknown whether they are binding sites for Xenopus MRF proteins, but the lower transgenic activity observed for the 360-bp promoter, which lacks these sites, is consistent with a possible role of the E-boxes in activating transcription.
X. laevis XMRF4a and XMRF4b and X. tropicalis MRF4 genes show conserved and non-conserved regions within their first few kb of 5′-flanking sequence, yet expression differences between XMRF4a constructs did not coincide with the boundaries of conserved regions. The 1.1-kb promoter lacks the conserved regions found between −1 kb and −3 kb in the 3.6-kb promoter and upstream of −4 kb in the 9.5-kb promoter, yet all three constructs gave very similar patterns and intensities of reporter gene expression. While evolutionary sequence constraint is associated with an observed genome function in many cases (Hardison, 2000), other constrained regions likely have critical functions not detected by experimental assays employed thus far (de la Calle-Mustienes et al., 2005; Margulies et al., 2007). If the Xenopus MRF4 and Myf5 genes have overlapping regulatory regions as they do in mammals (Carvajal et al., 2001, 2008), some of the conserved sequence 5′ to their coding region will likely contain regulatory elements functionally associated with the Myf5 promoter rather than with MRF4. Whatever their functions, the presence of strikingly similar patterns of sequence conservation between the X. laevis MRF4a and –b loci and between X. laevis and X. tropicalis MRF4, with no similarity to mammals, strongly suggests that they exert control over aspects of skeletal muscle development unique to frogs. Conversely, non-conserved as well as conserved sequence elements must contribute to the regulation of XMRF4a and –b because their transcript levels differ during the course of muscle development and during regeneration (Della Gaspera et al., 2006).
The 610-bp and longer XMRF4a promoters showed non-myotomal transgenic expression in some cases, albeit in a pattern that differed from endogenous XMRF4, which showed extrasomitic expression domains in brain, eye, branchial arches, and other anterior structures. Because it followed a consistent pattern in multiple founder embryos, transgene expression in the cardiogenic and branchial regions at tailbud stage may not represent simply random ectopic expression resulting from the transgenesis procedure (i.e., misregulated transcription of randomly inserted or extrachromosomal DNA, or aberrant cell movements due to embryo manipulation). Non-myotomal XMRF4a transgene expression may instead represent an abnormal manifestation, in the experimental context, of normal XMRF4a activity in anterior structures. This context includes the absence of potential intronic and 3′ regulatory regions and the absence of a linked Myf5 promoter, which in transgenic mice results in abnormal MRF4 promoter activity (Carvajal et al., 2008). Furthermore, sequence upstream of −9.5 kb may be required for activating a normal XMRF4a expression pattern in the brain, eyes, and other anterior structures.
This initial study of the cis-regulatory structure of the XMRF4a gene in comparison to mammalian MRF4 has identified a highly-conserved core promoter that sufficed for transgenic myotomal and postmetamorphic expression in X. laevis; demonstrated that additional elements within −610 bp of the XMRF4a coding region enhanced myotomal expression; pointed to an upstream enhancer within the −9.5-kb region; and shown that use of different transgenic and in vitro assays can lead to very different results. Identification and analysis of the putative upstream enhancer, as well as finer-scale deletion and point-mutation analysis of the XMRF4a 610-bp promoter, to fully understand their activities both in mammalian cells and in transgenic Xenopus embryos, should provide insight into the evolution of transcriptional control of this gene. Establishment of additional breeding lines carrying these transgenic constructs will facilitate discovery of the regulatory elements involved in XMRF4a transcriptional responses to physiological events such as denervation and muscle injury.
Materials and Methods
Isolation and sequence analysis of XMRF4 promoters
A 230-bp NotI-PvuII fragment at the 5′ end of a Xenopus laevis MRF4 cDNA (Jennings, 1992) was used to screen a genomic library (Lambda Fix II, Stratagene). Recovered inserts were subcloned into pBluescript (Stratagene). One insert of about 13 kb in length, designated XMRF4a, was sequenced through 9673 bp (GenBank Accession No. EU045573) including 5066 bp 5′ to the start codon. From another insert, designated XMRF4b, a 6-kb EcoR I fragment was partially sequenced including 2325 bp 5′ to the start codon (GenBank Accession No. EU045574). The XMRF4a 5′ sequence was used as a query for a BLAST search of the X. tropicalis v4.1 sequence (Joint Genome Institute, http://www.jgi.doe.gov) that identified the orthologous region of the X. tropicalis genome (scaffold 5, 1883000 to 1888563). The VISTA computational tool [http://genome.lbl.gov./vista (Brudno et al., 2007)], with the calculation window set to 100 bp, was used for comparisons between these 5′ sequence regions. An alignment of the X. tropicalis region against other vertebrate genomes was inspected at the UCSC Bioinformatics Site [http://genome.ucsc.edu (Kent et al., 2002)].
XMRF4a promoter-reporter gene constructs
After subcloning a 1.9-kb EcoR I-Kpn I fragment containing 610 bp of sequence 5′ to the start codon into pBluescript, a BamH I site was introduced just ahead of the start codon by PCR with proofreading polymerase (Vent, New England Biolabs). This 610-bp EcoR I-BamH I fragment was then used to generate shorter promoters, by restriction digestion at an Nco I site (−360 bp) or a BstX I site (−180 bp), and by PCR to introduce a Kpn I site about 6 bp upstream of the MEF2/TATA site (−120 bp) or about 6 bp downstream of the MEF2/TATA site (−100 bp). PCR-based modifications utilized primers consisting of 18–20 bp of XMRF4a sequence with an additional restriction enzyme recognition site. A mutated version of the 180-bp promoter designed to prevent binding of MEF2 to its site (D+M−), as previously demonstrated in the Xenopus XMyoDa promoter (Leibham et al., 1994), was made by site-directed mutagenesis (MORPH kit, 5-prime-to-3-prime Inc., Boulder, CO). The mutagenic oligonucleotide (CAACAGgTATATAAgcAAAGGG) differed from the wild-type sequence at the nucleotides shown in lower-case letters. Other promoter constructs were generated from the full-length XMRF4a clone by using PCR with proofreading polymerase (DyNAzyme, Finnzymes Oy., Espoo, Finland) to introduce an Xho I site just ahead of the start codon and a Kpn I site at the 5′ end of the clone. The resulting 9.5-kb product was inserted into a plasmid and subsequently digested with EcoR V, Sac I, BamH I, or with Hind III to create a 4.3-kb, a 3.6-kb, a 1.8-kb, and a 1.1-kb promoter, respectively.
Green fluorescent protein reporter constructs were made by excising the GFP5 (Siemering et al., 1996):SV40 polyadenylation cassette from XNkx2.5-GFP5 (Sparrow et al., 2000) with Kpn I and Not I and inserting it into pBluescript. XMRF4a promoter fragments were then inserted at the Kpn I site. A mammalian MRF4-GFP5 promoter-reporter construct was produced by inserting the −336 to +71 fragment of rat MRF4 (Naidu et al., 1995). Luciferase reporter constructs for cell transfection were made by ligating the XMRF4a promoter fragments into pGL2-Basic (Promega). Procedural details are available on request. All mutants and constructs were verified by sequencing.
Generation and analysis of transgenic Xenopus laevis
MRF4-GFP5 and gamma-crystallin-GFP3 (Offield et al., 2000) plasmids were digested with restriction enzymes to release the promoter-reporter cassettes, which were then separated from the vector by agarose gel electrophoresis and extracted from the gel with clean-up mini-columns (Qiagen or Promega). For a positive control in some experiments, the gamma-crystallin-GFP3 construct was included along with the XMRF4a-GFP5 construct, as embryos incorporating one construct have a high probability of incorporating another co-injected construct as well (Hartley et al., 2001). Approximately 100 ng of each DNA was used per transgenesis reaction by the method of Smith et al. (2006), which is based on the original REMI method (Kroll and Amaya, 1996) but omits egg extract and restriction enzyme from the pre-injection incubation of DNA with sperm nuclei. GFP mRNA was routinely visualized in embryos fixed at tailbud stage by in situ hybridization (Sive et al., 2000) with a digoxigenin-labeled probe transcribed from the first 380 bp of the GFP5 cDNA, with BM Purple (Roche) as the substrate for alkaline phosphatase. In some experiments, living GFP+ embryos were identified by epifluorescence microscopy (Leica MZFLIII) and raised through metamorphosis. In order to examine transgene expression in early embryos, sexually mature transgenic individuals were mated with wild-type individuals. For visualization of GFP in adult tissues, post-metamorphic animals expressing GFP were anesthetized deeply with benzocaine, skinned, and photographed with epifluorescence. The animals were then sacrificed. Animal care and euthanasia followed protocols approved by University of Alaska Anchorage IACUC.
In situ hybridization probes for endogenous XMRF4 mRNA and for XMyf5 mRNA
An antisense digoxigenin-labeled RNA probe was transcribed from the full-length XMRF4a cDNA followed by alkaline hydrolysis to produce fragments several hundred bp in length. A probe for XMyf5 was transcribed from a 470-bp EcoR I–Pst I fragment of the cDNA (Hopwood et al., 1991). In situ hybridization was performed as for the GFP probe.
Cell culture and transfection
C2C12 mouse myoblasts were grown in DMEM with 20% fetal bovine serum (FBS) and were plated at approximately 50% confluence on gelatin-coated 24-well plates prior to transfection. Equal molar amounts of the different XMRF4a-GL2 plasmids were used, together with a constant amount of renilla luciferase plasmid RL-SV40 (Promega) as a control for transfection efficiency. Following transfection using Qiagen Superfect or GTS GenePorter2 reagent, cells were maintained in 20% FBS for 6 hours and then switched to 2% horse serum to promote differentiation for 5 days before harvest and assay (Dual-Luciferase Kit, Promega).
Acknowledgements
I thank Sylvia Evans and Rob Grainger for providing me with opportunities to learn frog transgenesis in their laboratories and for plasmids. Many of my students at the University of Alaska Anchorage contributed to aspects of this project. This work was supported by grants from NIH (HD35326) and from the Alaska BRIN program.
Footnotes
References
- Adams L, Carlson BM, Henderson L, Goldman D. Adaptation of nicotinic acetylcholine receptor, myogenin, and mrf4 gene expression to long-term muscle denervation. J Cell Biol. 1995;131:1341–1349. doi: 10.1083/jcb.131.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataian Y, Owens J, Hinterberger T. Mrf4 gene expression in Xenopus embryos and aneural myofibers. Dev Dyn. 2003;226:551–554. doi: 10.1002/dvdy.10233. [DOI] [PubMed] [Google Scholar]
- Beck CW, Slack JM. Gut specific expression using mammalian promoters in transgenic xenopus laevis. Mech Dev. 1999;88:221–227. doi: 10.1016/s0925-4773(99)00217-8. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. Myod and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Black BL, Martin JF, Olson EN. The mouse mrf4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J Biol Chem. 1995;270:2889–2892. doi: 10.1074/jbc.270.7.2889. [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (mef2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Brudno M, Poliakov A, Minovitsky S, Ratnere I, Dubchak I. Multiple whole genome alignments and novel biomedical applications at the vista portal. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nat Rev Genet. 2008;9:632–646. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- Carvajal JJ, Cox D, Summerbell D, Rigby PW. A bac transgenic analysis of the mrf4/myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development. 2001;128:1857–1868. doi: 10.1242/dev.128.10.1857. [DOI] [PubMed] [Google Scholar]
- Carvajal JJ, Keith A, Rigby PW. Global transcriptional regulation of the locus encoding the skeletal muscle determination genes mrf4 and myf5. Genes Dev. 2008;22:265–276. doi: 10.1101/gad.442408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH, Primig M, Hadchouel J, Tajbakhsh S, Rocancourt D, Fernandez A, Kappler R, Scherthan H, Buckingham M. An enhancer directs differential expression of the linked mrf4 and myf5 myogenic regulatory genes in the mouse. Dev Biol. 2004;269:595–608. doi: 10.1016/j.ydbio.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Chang TH, Vincent SD, Buckingham ME, Zammit PS. The a17 enhancer directs expression of myf5 to muscle satellite cells but mrf4 to myonuclei. Dev Dyn. 2007;236:3419–3426. doi: 10.1002/dvdy.21356. [DOI] [PubMed] [Google Scholar]
- De la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Gaspera B, Sequeira I, Charbonnier F, Becker C, Shi DL, Chanoine C. Spatio-temporal expression of mrf4 transcripts and protein during xenopus laevis embryogenesis. Dev Dyn. 2006;235:524–529. doi: 10.1002/dvdy.20628. [DOI] [PubMed] [Google Scholar]
- Fomin M, Nomokonova N, Arnold HH. Identification of a critical control element directing expression of the muscle-specific transcription factor mrf4 in the mouse embryo. Dev Biol. 2004;272:498–509. doi: 10.1016/j.ydbio.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hartley KO, Hardcastle Z, Friday RV, Amaya E, Papalopulu N. Transgenic xenopus embryos reveal that anterior neural development requires continued suppression of bmp signaling after gastrulation. Dev Biol. 2001;238:168–184. doi: 10.1006/dbio.2001.0398. [DOI] [PubMed] [Google Scholar]
- Hinterberger TJ, Mays JL, Konieczny SF. Structure and myofiber-specific expression of the rat muscle regulatory gene mrf4. Gene. 1992;117:201–207. doi: 10.1016/0378-1119(92)90730-d. [DOI] [PubMed] [Google Scholar]
- Hinterberger TJ, Sassoon DA, Rhodes SJ, Konieczny SF. Expression of the muscle regulatory factor mrf4 during somite and skeletal myofiber development. Dev Biol. 1991;147:144–156. doi: 10.1016/s0012-1606(05)80014-4. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. Xenopus myf-5 marks early muscle cells and can activate muscle genes ectopically in early embryos. Development. 1991;111:551–560. doi: 10.1242/dev.111.2.551. [DOI] [PubMed] [Google Scholar]
- Jennings CG. Expression of the myogenic gene mrf4 during xenopus development. Dev Biol. 1992;151:319–332. [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at ucsc. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkvliet CM, Hinterberger TJ. Distal regulatory regions of the rat mrf4 gene. Biochem Biophys Res Commun. 1997;237:170–176. doi: 10.1006/bbrc.1997.6980. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic xenopus embryos from sperm nuclear transplantations reveal fgf signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Leibham D, Wong MW, Cheng TC, Schroeder S, Weil PA, Olson EN, Perry M. Binding of tfiid and mef2 to the tata element activates transcription of the xenopus myoda promoter. Mol Cell Biol. 1994;14:686–699. doi: 10.1128/mcb.14.1.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W, Neff ES, Furlow JD. The mouse muscle creatine kinase promoter faithfully drives reporter gene expression in transgenic xenopus laevis. Physiol Genomics. 2004;18:79–86. doi: 10.1152/physiolgenomics.00148.2003. [DOI] [PubMed] [Google Scholar]
- Lin-Jones J, Hauschka SD. Myogenic determination factor expression in the developing avian limb bud: An rt-pcr analysis. Dev Biol. 1996;174:407–422. doi: 10.1006/dbio.1996.0084. [DOI] [PubMed] [Google Scholar]
- Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, Harland RM. Hypaxial muscle migration during primary myogenesis in xenopus laevis. Dev Biol. 2001;239:270–280. doi: 10.1006/dbio.2001.0434. [DOI] [PubMed] [Google Scholar]
- Naidu PS, Ludolph DC, TO RQ, Hinterberger TJ, Konieczny SF. Myogenin and mef2 function synergistically to activate the mrf4 promoter during myogenesis. Mol Cell Biol. 1995;15:2707–2718. doi: 10.1128/mcb.15.5.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas N, Gallien CL, Chanoine C. Expression of myogenic regulatory factors during muscle development of xenopus: Myogenin mrna accumulation is limited strictly to secondary myogenesis. Dev Dyn. 1998;213:309–321. doi: 10.1002/(SICI)1097-0177(199811)213:3<309::AID-AJA7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Offield MF, Hirsch N, Grainger RM. The development of xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development. 2000;127:1789–1797. doi: 10.1242/dev.127.9.1789. [DOI] [PubMed] [Google Scholar]
- Paukert M, Hidayat S, Grunder S. The p2×(7) receptor from xenopus laevis: Formation of a large pore in xenopus oocytes. FEBS Lett. 2002;513:253–258. doi: 10.1016/s0014-5793(02)02324-4. [DOI] [PubMed] [Google Scholar]
- Pin CL, Konieczny SF. A fast fiber enhancer exists in the muscle regulatory factor 4 gene promoter. Biochem Biophys Res Commun. 2002;299:7–13. doi: 10.1016/s0006-291x(02)02571-8. [DOI] [PubMed] [Google Scholar]
- Pin CL, Ludolph DC, Cooper ST, Klocke BJ, Merlie JP, Konieczny SF. Distal regulatory elements control mrf4 gene expression in early and late myogenic cell populations. Dev Dyn. 1997;208:299–312. doi: 10.1002/(SICI)1097-0177(199703)208:3<299::AID-AJA2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Polli M, Amaya E. A study of mesoderm patterning through the analysis of the regulation of xmyf-5 expression. Development. 2002;129:2917–2927. doi: 10.1242/dev.129.12.2917. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of xenopus laevis: A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Smith SJ, Fairclough L, Latinkic BV, Sparrow DB, Mohun TJ. Xenopus laevis transgenesis by sperm nuclear injection. Nat Protoc. 2006;1:2195–2203. doi: 10.1038/nprot.2006.325. [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Cai C, Kotecha S, Latinkic B, Cooper B, Towers N, Evans SM, Mohun TJ. Regulation of the tinman homologues in xenopus embryos. Dev Biol. 2000;227:65–79. doi: 10.1006/dbio.2000.9891. [DOI] [PubMed] [Google Scholar]
- Summerbell D, Halai C, Rigby P. Expression of the myogenic regulatory factor mrf4 precedes or is contemporaneous with that of myf5 in the somitic bud. Mech Dev. 2002;117:331. doi: 10.1016/s0925-4773(02)00208-3. [DOI] [PubMed] [Google Scholar]
- Wong MW, Pisegna M, Lu MF, Leibham D, Perry M. Activation of xenopus myod transcription by members of the mef2 protein family. Dev Biol. 1994;166:683–695. doi: 10.1006/dbio.1994.1347. [DOI] [PubMed] [Google Scholar]
- Yang J, Mei W, Otto A, Xiao L, Tao Q, Geng X, RUPP RA, DING X. Repression through a distal tcf-3 binding site restricts xenopus myf-5 expression in gastrula mesoderm. Mech Dev. 2002;115:79–89. doi: 10.1016/s0925-4773(02)00121-1. [DOI] [PubMed] [Google Scholar]