Abstract
After injury to the central nervous system, a glial scar develops that physically and biochemically inhibits axon growth. In the scar, activated astrocytes secrete inhibitory extracellular matrix, of which chondroitin sulfate proteoglycans (CSPGs) are considered the major inhibitory component. An inhibitory interface of CSPGs forms around the lesion and prevents axons from traversing the injury, and decreasing CSPGs can enhance axon growth. In this report, we established an in vitro interface model of activated astrocytes and subsequently investigated gene delivery as a means to reduce CSPG levels and enhance axon growth. In the model, a continuous interface of CSPG producing astrocytes was created with neurons seeded opposite the astrocytes, and neurite crossing, stopping, and turning were evaluated as they approached the interface. We investigated the efficacy of lentiviral delivery to degrade or prevent the synthesis of CSPGs, thereby removing CSPG inhibition of neurite growth. Lentiviral delivery of RNAi targeting two key CSPG synthesis enzymes, chondroitin polymerizing factor and chondroitin synthase-1, decreased CSPGs, and reduced inhibition by the interface. Degradation of CSPGs by lentiviral delivery of chondroitinase also resulted in less inhibition and more neurites crossing the interface. These results indicate that the interface model provides a tool to investigate interventions that reduce inhibition by CSPGs, and that gene delivery can be effective in promoting neurite growth across an interface of CSPG producing astrocytes.
Keywords: CSPG inhibition, astrocyte/neuron co-culture, gene delivery, astrocyte interface
Introduction
One of the main barriers to regeneration in the central nervous system is the formation of the glial scar, which consists largely of reactive astrocytes and inhibitory extracellular matrix (ECM) molecules. Axons cannot regenerate past the scar, the ends become dystrophic, and growth is stalled (Li and Raisman, 1995; Silver and Miller, 2004; Tom et al., 2004). The formation of the glial scar also has beneficial effects, such as isolating the injury site, minimizing the area of inflammation and cellular degeneration, and helping to restore the blood brain barrier (BBB; Rolls et al., 2009; Stichel and Muller, 1998; Yiu and He, 2006).
Chondroitin sulfate proteoglycans (CSPGs) are among the ECM molecules found in the glial scar and are potent inhibitors of axonal elongation (Asher et al., 2001; Snow et al., 1990). CSPGs are a family of molecules consisting of a core protein to which sulfated glycosaminoglycan (GAG) side chains are bound. The synthesis of CSPGs begins with the core protein; to which multiple enzymes add sugar moieties, creating the chondroitin sulfate GAGs (CS-GAGs). The CS-GAG chain is polymerized by the addition of the repeating disaccharide unit, N-acetyl galactosamine and glucuronic acid, by an enzyme complex consisting of chondroitin synthases (ChSy1-3), and chondroitin polymerizing factor (ChPF; Izumikawa et al., 2007, 2008; Kitagawa et al., 2001, 2003). Chondroitin polymerization is achievable by multiple combinations of ChSy1-3 and ChPF resulting in differing lengths of CS-GAG chains (Chau et al., 2004; Izumikawa et al., 2007, 2008). CSPGs are widely distributed in the developing and adult CNS and serve an important role in the developing nervous system by guiding axons to their correct targets through the creation of inhibitory boundaries that define the path and prevent axons from growing in inappropriate directions (Bandtlow and Zimmermann, 2000; Schwartz and Domowicz, 2004). In the glial scar, CSPGs are expressed by both astrocytes and oligodendrocyte precursor cells (Asher et al., 2000, 2002; Haas et al., 1999; Jones et al., 2002, 2003; McKeon et al., 1999). Numerous in vitro and in vivo studies have demonstrated that CSPGs are inhibitory to axon growth (Davies et al., 1997, 1999; McKeon et al., 1991; Smith-Thomas et al., 1994; Snow et al., 1990) and strategies to eliminate CSPGs are being pursued to promote spinal cord regeneration.
In this report, we developed an in vitro model of the glial scar and investigated gene delivery for localized, sustained production of factors to prevent deposition or promote degradation of CSPGs. In the glial scar model, a continuous, defined interface of CSPG-producing astrocytes was created that mimics the barrier that regenerating axons encounter at the edge of a lesion in vivo. Neurons were seeded opposite this interface and neurite behavior was evaluated as they approached the interface. Gene delivery to the astrocytes initially employed chondroitinase (ch’ase), which digest the CS-GAG side chains to attenuate the inhibitory activity of the molecule in vitro (McKeon et al., 1995; Powell et al., 1997; Sango et al., 2003; Yu and Bellamkonda, 2001) and has also been reported to increase axonal regeneration and functional recovery in vivo (Bradbury et al., 2002; Houle et al., 2006). Additionally, gene delivery was investigated to prevent the synthesis of CSPGs through lentiviral delivery of RNAi against two CSPG synthesis enzymes (ChPF and ChSy-1). This interface model combined with gene delivery may identify targets that most effectively reduce the glial scar and allow axon growth.
Materials and Methods
Construction of RNAi Vectors
The miRNA lentiviruses (LVs) were constructed using the Block-iT HiPerform Lentiviral Pol II miR RNAi Expression System with EmGFP (Life Technologies, Carlsbad, CA) following the manufacturer’s instructions. Ten different miRNA target sequences were designed against both ChPF and ChSy1 using the BLOCK-iT RNAi Designer (Life Technologies) and used to produce 10 different RNAi lentiviruses for each gene. RNAi lentiviruses were constructed with co-cistronic expression of emerald green fluorescent protein (EmGFP) and the miRNA sequences under the control of the human cytomegalovirus (CMV) promoter. A negative control RNAi virus was also produced using a plasmid provided in the kit that contains a miRNA sequence that forms a hairpin, is processed into mature miRNA, but does not target any known vertebrate gene. Knockdown was evaluated using qPCR (described in a subsequent section) and the two viruses that knocked down mRNA levels of their respective targets the most were selected for further studies. The most effective ChPF sense target sequence was 5′-AGGTATGGTACACACTAGA-3′and ChSy1 was 5′-CCAGTACACTACACAGTTA-3′. A virus with co-cistronic expression of the selected ChPF and ChSy1 miRNA sequences was also produced to evaluate knockdown when both sequences were expressed in one primary transcript. The final lentiviral plasmids were sequenced to ensure the correct sequence was produced.
Production of Lentivirus
Lentivirus was prepared using established techniques in HEK293T cells grown in Dulbecco’s modified Eagle’s medium (Life Technologies) with 10% fetal bovine serum (FBS) at 37°C and 5% CO2 The lentiviral packaging vectors (pMDL-GagPol, pRSV-Rev, and pIVS-VSV-g), previously described by Dull et al. (1998), were co-transfected along with pBOB-IgK-Chase-CE, plenti6.4-EmGFP-miR-ChPF, plenti6.4-EmGFP-miR-ChSy1, plenti6.4-EmGFP-miR-ChPF/ChSy1, or plenti6.4-EmGFP-miR-neg (negative control vector with a scrambled RNAi sequence) into 293T cells using Lipofectamine 2000 (Life Technologies). After 48 h of transfection, the supernatant was collected and filtered (0.45 µm). Viruses were concentrated using PEG-it (System Biosciences, Mountain View, CA), with the precipitated lentiviruses resuspended in PBS. The titer of lentivirus was determined by HIV-1 p24 Antigen ELISA Kit (ZeptoMetrix, Buffalo, NY Co.).
RNA Extraction and Quantitative PCR
RNA was extracted using the Absolutely RNA miniprep kit following the manufacturer’s protocol (Agilent Technologies, Santa Clara, CA) including DNase I treatment to remove genomic DNA. cDNA was synthesized from 1 µg of RNA using Superscript III First-Strand Synthesis SuperMix (Life Technologies) and quantitative PCR (qPCR) was performed in triplicate for all samples. qPCR was performed on an iCycler (Bio-Rad, Hercules, CA) with Power SYBR Green PCR Master Mix (Life Technologies). The primer sequences were as follows: ChPF forward, 5′-GACCGCACATACCAGGAGAT-3′; ChPF reverse, 5′-GAAAGTGTGCTGTTCCGTGA-3′; ChSy1 forward, 5′-GC AGACCTTCAGCAAGATCC-3′; ChSy1 reverse, 5′-TTCTTTGGGCTCTTTGTGCT-3′; β-actin forward, 5′-CA CCCGCGAGTACAACCTTC-3′; β-actin reverse, 5′-CCCATACCCACCATCACACC-3′. qPCR started with a 10 min hot start at 95°C followed by 40 cycles of the following: 95°C for 15 s, 60°C for 1 min. A serial dilution of cDNA was used to determine the amplification efficiency for each gene on each qPCR plate. The Pfaffl method (Pfaffl, 2001) was used to calculated fold changes in ChPF and ChSy1 mRNA levels compared to miR-neg transduced Neu7s using β-actin as the internal control gene.
Construction of Co-Culture Plates
The co-cultures were carried out in 12-well cell culture plates that were custom altered with removable, liquid tight, glass interfaces separating the wells into two equal compartments. No. 1 coverglass were cut to fit in the wells with the manufacturer’s edge down, against the bottom of the well, and secured into place with polydimethylsiloxane (PDMS; Fisher Scientific, Pittsburgh, PA), creating the interface. A temporary lid was made with guides to hold the glass interface vertical in each well while the PDMS cured. To ensure a liquid tight seal, a magnetic system was employed. Metal clips were secured to the top of each glass interface. These clips were attracted to magnets temporarily adhered to the outside bottom of the plate in the center of each well. Uncured PDMS was applied to the intersection of the well wall and the edges of the glass interface and cured overnight at 60°C. The magnets, metal clips, and temporary lid were removed. The plates with intact interfaces were sterilized with UV light and coated with poly-l-lysine (PLL; Sigma–Aldrich, St. Louis, MO).
Co-Culture
Neu7 cells, a rat astrocyte cell line produced and characterized by Fok-Seang et al. (1995), were maintained in DMEM with 10% FBS at 37°C and 5% CO2. Neu7s were transduced with lentiviruses encoding either ch’ase, miR-ChPF, miR-ChSy1, miR-ChPF/ChSy1, or miR-neg (5 × 104 physical particles/cell) and subcultured for at least 1 week before being used for experiments. To determine transduction efficiency, flow cytometry was performed for GFP-positive Neu7s. A BD LSR II and the FACSDiva software (BD Biosciences, San Jose, CA) were used for data acquisition and analysis. Neu7s were seeded at 20,000 cells/cm2 on one side of the co-culture well. Neu7s were cultured for 24 h before seeding DRG neurons on the other half of the well.
To obtain primary neurons, dorsal root ganglia (DRG) were isolated from E9 white leghorn chicken embryos (Phil’s Fresh Eggs, Inc., Forreston, IL) and maintained in Hank’s balanced salt solution buffer supplemented with 6 g/L glucose until the isolation was complete. DRG were incubated for 30 min at 37°C in 0.25% trypsin, followed by trituration with fire-polished glass Pasteur pipettes to dissociate the ganglia. Non-neuronal and neuronal cells were separated by panning for 2 h at 37°C. Dissociated DRG neurons were seeded (5,000 cells/cm2) in media containing 2 ng/mL nerve growth factor (NGF) on the opposite half of the co-culture well than the Neu7s were seeded 24 h previously. The glass interfaces were removed 4 h after neuron seeding and cultures were fixed for staining 48 h after the interfaces were removed. For the chondroitinase (ch’ase) positive control experiments, at the time of neuron seeding, the media was aspirated from the Neu7 side of the well and replaced with fresh media. For ch’ase-positive wells, the fresh media was supplemented with 0.4 units/mL chondroitinase ABC (Seikagaku). The above procedures were also used in the control experiments with NIH/3T3 cells (ATCC).
Immunocytochemistry
The neurons were stained for neuron-specific class III β-tubulin by incubating fixed cells in TUJ1 antibody (Covance) diluted in 5% normal goat serum (Vector Labs, Burlingame, CA) in PBS followed by incubation in TRITC or FITC-conjugated goat anti-mouse secondary antibody (Jackson Immunoresearch, West Grove, PA) in PBS. Cells were counterstained with DAPI to visualize cell nuclei.
Neu7s were stained for CSPGs by incubating fixed cells in CS-56 (Sigma–Aldrich) diluted in 1% normal goat serum in PBS followed by incubation in Alexa Fluor 546 (Life Technologies) in PBS. Cells were counterstained with DAPI to visualize cell nuclei.
Quantification of Neurite Behavior
Neurite behavior at the interface was evaluated live at 100× with a Leica DMIRB microscope with a CoolSnap HQ2 camera (Photometrics, Tucson, AZ) and MetaVue imaging software (Molecular Devices) by a blinded observer. No neurites that originated from neuron somas in contact with any other cells were quantified. Primary neurites within 1mm of the Neu7 interface were quantified as either crossing or turning, defined as follows: Crossing: A neurite that crossed the Neu7 interface without connecting to another neuron on the Neu7 monolayer. Turning: Neurites that, at any point, grew towards the interface, but terminated pointing away from it Stopping: A neurite that, at any point, grew towards the interface, and terminated pointing in the direction of the interface. The percentage of neurites that crossed, turned, and stopped was calculated based on the total number of neurites within 1mm of the Neu7 cell interface.
Statistics
Statistical analysis was performed using GraphPad Prism software. Comparative analyses for co-culture data were executed using a Welch’s t-test at a 95% confidence level. Mean values with SEM are reported. For multiple comparisons, an ANOVA with post-hoc testing was performed.
Results
Decreased ChPF and ChSy1 mRNA Levels After Treatment With RNAi Lentiviruses
The ability of RNAi against ChPF and ChSy1, which are both key biosynthetic enzymes involved in assembling the CS-GAG chains onto core proteins, to reduce mRNA levels was investigated. Lentiviral delivery of miRNA targeting ChPF, ChSy1, or both was evaluated by qPCR after transduction and at least 1 week of subculture of Neu7 cells. Ten different miRNA sequences were screened for both genes and the sequence that maximally reduced target mRNA levels was used for further studies. Neu7 cells are derived from rat astrocyte cells and produce an extracellular matrix (ECM) that contains a large amount of CSPGs (Fok-Seang et al., 1995). Over 95% transduction efficiency was obtained in Neu7 cells with all RNAi lentiviruses measured by flow cytometry for co-cistronic expression of GFP. The miR-ChPF LV knocked down ChPF mRNA levels to 27% of that found in the negative control (Neu7 cells transduced with a lentivirus encoding a scrambled miRNA sequence—miR-neg; Fig. 1a). The miR-ChSy1 lentivirus reduced ChSy1 mRNA levels to 26% compared to the negative control (Fig. 1b). A dual virus was constructed with co-cistronic expression of both the ChPF and ChSy1 miRNA sequences (referred to as miR-ChPF/ChSy1) and reduced ChPF levels in Neu7 transduced cells to 6% and ChSy1 levels to 23% compared to the negative control (Fig. 1a and b).
Figure 1.
ChPF and ChSy1 mRNA knockdown in Neu7s after transduction with miR-LVs. a: ChPF and (b) ChSy1 mRNA knockdown in miR-transduced Neu7 cells. Knockdown was assessed by qPCR (n=3).
Decreased CS-56 Immunolabeling After Treatment With Lentiviruses
Monolayers of treated Neu7 cells were immunostained for CS-56, which recognizes intact chondroitin sulfate proteoglycans (Avnur and Geiger, 1984), in order to visualize and evaluate CSPG deposition (Table I). Lentiviruses encoding for RNAi targeting the biosynethetic enzymes or a lentivirus encoding for ch’ase were investigated. Untreated Neu7 cells and cells transduced with the negative control miR-neg LV had substantial CS-56 staining (Fig. 2a and b). Degradation of CS-GAG chains by treatment of Neu7 cells with ch’ase resulted in little to no CS-56þ staining (Fig. 2c), indicating low CSPG levels. Similarly, low CSPG levels were observed for Neu7 cells transduced with the ch’ase LV (Fig. 2d). Although miR-ChPF LV treated Neu7 cells had low ChPF mRNA levels, CS-56 immunostaining was comparable to both untreated Neu7s and miR-neg LV treated Neu7s (Fig. 2a, b, and e). In contrast, CS-56 staining intensity was reduced in miR-ChSy1 LV treated Neu7 cells (Fig. 2f). Treatment with the dual virus that targeted both ChPF and ChSy1 resulted in significantly lower CS-56 immunolabeling than controls (Fig. 2g). As a control for cell type, monolayers of NIH/3T3 cells, which do not secrete significant quantities of CSPGs, with and without the addition of ch’ase were also stained for CS-56 and no difference was observed (Fig. 2h and i).
Table I.
Quantification of CS-56 staining.
| Cell type, condition | Level of CS56+ staining |
|---|---|
| Neu7, no treatment | +++ |
| Neu7, miR-neg LV | +++ |
| Neu7, ch’ase | − |
| Neu7, ch’ase LV | − |
| Neu7, miR-ChPF LV | +++ |
| Neu7, miR-ChSy1 LV | ++ |
| Neu7, dual miR-ChPF/ChSy1 LV | + |
| NIH3T3, no treatment | − |
| NIH3T3, ch’ase | − |
The level of staining was categorized as: (i) − indicates no or negligible staining, (ii) + indicates low levels of staining, (iii) ++ indicates intermediate levels of staining, and (iv) +++ indicates high levels of staining.
Figure 2.
CS-56 stained Neu7 and NIH3T3 cells. Representative images of Neu7 cells stained for CS proteoglycans that were (a) untreated (b) transduced with the scrambled, negative control miR-neg LV, (c) treated with ch’ase, (d) transduced with the ch’ase LV, (e) transduced with the miR-ChPF LV, (f) transduced with the miR-ChSy1 LV, and (g) transduced with the dual miR-ChPF/ChSy1 LV. Representative images of NIH3T3 cells stained for CS proteoglycans that were (h) not treated and (i) treated with ch’ase. Scale bar is 150 µm.
Co-Culture Interface Model of the Glial Scar
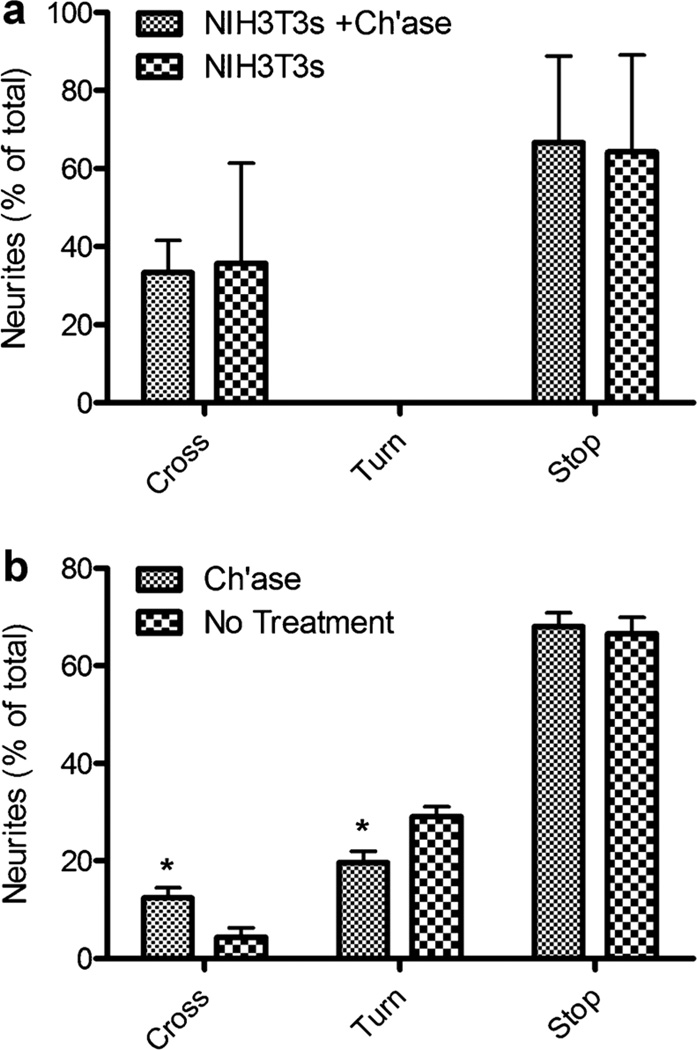
An in vitro model of the glial scar consisting of a defined interface between CSPG producing astrocytes and neurons was established to investigate axonal behavior at the inhibitory barrier created by the interface (Fig. 3). Co-cultures of neurons with Neu7 cells resulted in 4.3 ± 2.0% of neurites crossing the interface, 30 ± 2.0% turning away from the interface, and 66 ± 3.3% stopping at or before encountering the interface. Co-cultures with Neu7 cells pretreated with ch’ase were used as a positive control to validate the co-culture model developed in this report. The percentage of neurites that crossed the Neu7 cell interface when treated with ch’ase was significantly higher (12.4 ± 2.0%) than untreated Neu7 cells (Fig. 4b). Additionally, a smaller percentage of neurites turned away from the ch’ase treated Neu7 cell interface (19.6 ± 2.4%) than untreated cells (Fig. 4b). No difference was observed in the percentage of neurites that stopped at the interface with or without ch’ase treatment of Neu7 cells. Control co-cultures of neurons with NIH/3T3 cells were performed to evaluate neurite behavior at an interface of cells that do not produce a large amount of CSPGs (Fig. 2h). No difference in the percentage of neurites that crossed onto or stopped before encountering the NIH/3T3 monolayer were observed when the cells were either untreated or pretreated with ch’ase (Fig. 4a). No neurites were observed turning away from the interface in either condition.
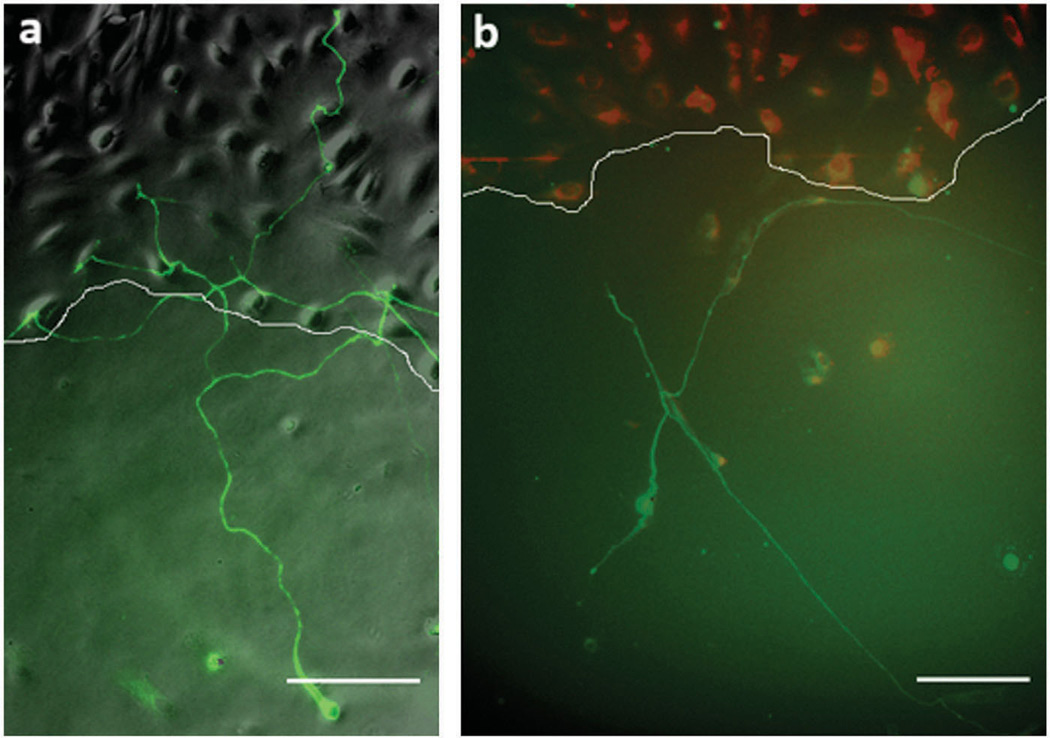
Figure 3.
Examples of neurite behavior at the interface. a: A representative image that shows a neurite crossing the interface of Neu7 cells and (b) a neurite turning as it encounters the Neu7 cell interface. The white line delineates the border of the Neu7 cell interface. Scale bar is 100 µm.
Figure 4.
The percentage of neurites that crossed and turned in control cocultures. a: Co-cultures performed with NIH3T3 cells (n=3) that were treated with and without (negative control) ch’ase. b: Co-culture performed for Neu7 cells either treated with ch’ase (n=17) or untreated (negative control, n=8) as a control. The symbol ‘‘*’’ indicates statistical significance between ch’ase treated and untreated (P<0.05).
Decreased Neurite Inhibition by Chondroitinase Lentivirus Treated Neu7 Cells
Lentiviral delivery of the ch’ase gene was subsequently investigated using the interface model. Treatment of Neu7 cells with the ch’ase LV produced similar results to the ch’ase positive control co-cultures described previously. A significantly greater percentage of neurites crossed the interface when Neu7 cells were transduced with ch’ase LV (26.8 ± 3.4%) compared to untreated Neu7s (10.1 ± 2.0%) and a significantly lower percentage turned away from the interface in the ch’ase LVþ co-cultures (15.6 ± 3.2%) compared to untreated Neu7 cells (23.4 ± 2.9%; Fig. 5a). No significant difference was observed in the percentage of neurites that stopped.
Figure 5.
Co-culture results with ch’ase LV treated Neu7 cells. a: Ch’ase LV transduction of cells (n=23) resulted in an increased percentage of neurites crossing and fewer neurites turning upon encountering the Neu7 interface compared to untreated cells (negative control, n=20). The symbol ‘‘*’’ indicates statistical significance between ch’ase treated and untreated (P<0.05). b: Co-culture results with varying ratios of ch’ase LV to negative control (GFP) LV treated Neu7 cells. The ratios of LV:GFP were 50:50 (n=23), 10:90 (n=21), and 0:100 (n=21). The symbol ‘‘*’’, indicates significant difference relative to 0:100.
Co-cultures were subsequently performed to evaluate neurite behavior at the interface with differing numbers of cells transduced with ch’ase LV. Ch’ase LV treated Neu7 cells were mixed in varying ratios with Neu7 cells transduced with a control lentivirus encoding GFP. For a 50:50 ratio of ch’ase LV to GFP LV Neu7 cells used for co-cultures, a greater percentage of neurites crossed onto the Neu7 monolayer (50:50, 20.3 ± 3.2%) relative to control (0:100, 6.4 ± 1.2%), but no significant difference in the percentage of neurites that turned away from the interface was observed (50:50, 18.5 ± 2.1%) relative to control (0:100, 23.3 ± 2.4%; Fig. 5b). A significantly greater percentage of neurites turned (30.3 ± 2.5%) in the 10:90 ratio co-culture compared to the 0:100 ratio co-culture (Fig. 5b). The number of crossing neurites in the 10:90 ratio co-culture was less than observed in the 50:50 ratio co-culture. Taken together, decreasing the amount of ch’ase LV transduced Neu7 cells led to reductions in the number of axons that crossed the interface.
Decreased Neurite Inhibition by RNAi Lentivirus Treated Neu7 Cells
LV delivery to promote axon crossing was investigated in co-culture experiments using RNAi to prevent the formation of CS-GAGs as an alternative to degradation via ch’ase. RNAi against two CS-GAG synthesis enzymes (ChPF and ChSy1) was used. No significant differences in neurite behavior were observed when miR-ChPF LV-treated Neu7 cells were used in co-culture experiments (Fig. 6a). Contrastingly, a significantly greater percentage of neurites crossed (17.4 ± 2.3%) and a significantly lower percentage turned away (14.6 ± 1.8%) from the interface of miR-ChSy1 LV treated Neu7 cells compared to Neu7 cells treated with the negative control miRNA LV (Cross—9.1 ± 1.6%; Turn—28.7 ± 2.3%) in co-culture experiments (Fig. 6b).
Figure 6.
Co-culture results with miR-LV transduced Neu7 cells. a: Co-culture data with miR-ChPF transduced Neu7 cells (n=14) and negative control (n=15). b: miR-ChSy1 LV transduced Neu7 cells (n=33) and negative control (n=32). c: Dual miR-ChPF/ChSy1 LV transduced Neu7 cells (n=36) and negative control (n=36). d: Co-culture results with a 50:50 ratio of dual miR-ChPF/ChSy1 LV to miR-neg LV transduced Neu7 cells (n=21), and a 0:100 ratio (n=22). For all cases, the negative control is scrambled miRNA LV. The symbol ‘‘*’’ indicates statistical significance relative to control P<0.05.
The efficacy of combining the two RNAi sequences to reduce inhibition to neurite extension was evaluated by delivery of the virus with co-cistronic expression of both ChPF and ChSy1 miRNA sequences. A significantly greater percentage of neurites crossed the interface of miR-ChPF/ChSy1 LV transduced Neu7 cells (31.9 ± 2.9%) compared to Neu7 cells treated with the scrambled negative control miRNA LV (7.3 ± 0.9%; Fig. 6c). Turning and stopping were also statistically different when the dual virus (Turn—11.3 ± 1.2%; Stop—56.8 ± 2.8%) was compared to negative controls (Turn—20.7 ± 1.6%; Stop—72.1 ± 1.9%; Fig. 6c). The effect of transduction efficiency for this intracellular gene target was investigated using a 50:50 ratio of dual LV transduced Neu7 cells mixed with Neu7 cells transduced with the negative control miRNA LV. The percentage of neurites crossing the interface was not significantly different between the 50% dual LV infected Neu7 and the control, but the percentage of neurites that turned from the 50% dual LV infected Neu7 cell interface (9.8 ± 3.1%) was statistically lower than control (27.0 ± 3.7%; Fig. 6d).
Discussion
In this report we developed a novel in vitro model to simulate the in vivo glial scar and evaluate viral gene delivery for reduced CSPG inhibition. Reactive astrocytes within the glial scar in vivo create an interface that prevents axons from regenerating past the barrier (Chau et al., 2004; Davies et al., 1997). The model in this report aims to recreate the astrocyte interface observed in vivo by creating a continuous interface of reactive astrocytes that produce CSPGs. This model mimics the inhibitory boundary that forms in vivo at the interface of an injury or a biomaterial implant. The astrocyte cell line used, Neu7s produces a large amount of CSPGs but are still able to support axon extension when neurons are cultured on monolayers (Muir et al., 1998; Powell et al., 1997; Smith-Thomas et al., 1994). Here, neurons were seeded opposite the astrocyte interface, not on top of the astrocyte monolayers as in other models, which lack an interface (Fidler et al., 1999; Fok-Seang et al., 1995; Muir et al., 1998; Smith-Thomas et al., 1994) or on surfaces of immobilized conditioned media from astrocytes (Laabs et al., 2007; Snow et al., 1990; Tom et al., 2004). Using live cells to create an interface allows for continuous, sustained manipulation of CSPG production that would not be possible with conditioned media; however, the use of lives cells produces a less defined system than alternative in vitro models that deposit CSPGs on the culture surface. The astrocyte boundary created in this model was likely less dense than the border that develops in vivo. Future studies that adapt this interface model to 3D cell culture have the potential to mimic the in vivo environment and astrocyte activation more closely (East et al., 2009). However, this simplified, 2D model allowed for interventions such as gene delivery to be investigated for their ability to reduce inhibition of axon growth, which could ultimately be translated to in vivo models.
Lentiviral delivery of ch’ase to the astrocytes in the model reduced axon inhibition that enhanced axonal crossing of the interface. Ch’ase degrades the CS-GAG chains of CSPGs which have been shown to be the major inhibitory competent of the molecule (Fidler et al., 1999; Fok-Seang et al., 1995) and increases axon growth in vitro (Snow et al., 1990) and in vivo, resulting in some functional improvements after injury to the CNS (Barritt et al., 2006; Bradbury et al., 2002). Delivery of ch’ase has been challenging, as a single dose of ch’ase may degrade CS-GAGs initially present, yet may be ineffectual against the eventual reestablishment of the inhibitory ECM (Garcia-Alias et al., 2008). This shortterm efficacy necessitates multiple injections of ch’ase or invasive continuous delivery systems such as intrathecal catheters that can disrupt the injury site, and these systems are prone to complications and infections (Jones and Tuszynski, 2001). Additionally, sustained delivery of active ch’ase is further complicated by the thermal instability of the molecule at body temperature (Tester et al., 2007). Viral delivery of ch’ase could overcome these limitations and provide a local, sustained supply of active enzyme after injury. We report that ch’ase LV can produce similar results to ch’ase enzyme in an in vitro model of the glial scar. Decreased axon inhibition was observed for both viral and non-viral ch’ase treated astrocyte interfaces. Increased neurite crossing of the interface was observed when 50% of the astrocytes composing the interface were transduced with ch’ase LV. After transduction with ch’ase LV, astrocytes were subcultured for several weeks and still had reduced levels of CSPGs, suggesting that active enzyme expression can be sustained for long time periods through LV delivery.
Viral delivery of RNAi to prevent the production of CSPGs reduced deposition by transduced cells and also reduced neurite inhibition in the in vitro model. RNAi approaches were evaluated as ch’ase degradation does not completely remove the inhibitory sugar side chains from CSPGs (Lemons et al., 2003); and methods to prevent the synthesis of the side chains provide the opportunity to overcome the limitations of ch’ase in reducing CSPG inhibition (Grimpe and Silver, 2004; Schwartz and Domowicz, 2004; Smith-Thomas et al., 1994). CS can have sulfation motifs that bind, sequester, or present signaling molecules that modulate cell behavior. The RNAi strategy could be employed to limit expression of patterns that are inhibitory (Properzi et al., 2005; Silver and Miller, 2004), without affecting the patterns that are permissive to axonal extension after CNS lesion (Lin et al., 2011). While the inhibitory properties of CS can be overcome with Ch’ase (Garcia-Alias et al., 2009; Wang et al., 2011), this approach likely removes both inhibitory and permissive sulfation patterns. However, RNAi methods may not be specific to CSPGs and could affect heparan sulfate proteoglycans, which are important for growth factor binding and presentation (Kinnunen et al., 1996; Moon et al., 2002). Short interfering RNA (siRNA) against the CSPG specific synthesis enzyme, ChPF, was reported to reduce axon inhibition by conditioned media from treated Neu7s (Laabs et al., 2007). Yet, siRNA delivery in vivo is transient, limiting the efficacy of this method (Brummelkamp et al., 2002). Additionally, several isoforms of the CS-GAG enzymes involved in the polymerization complex (Izumikawa et al., 2007, 2008; Kitagawa et al., 2001, 2003) may necessitate targeting multiple molecules with RNAi. In this report, lentiviral delivery of miRNA provided the opportunity for sustained expression and the targeting of more than one transcript fromthe same vector (ChPF and ChSy1). In the in vitro model of the glial scar, miRNA LV against ChPF reduced ChPF mRNA levels in transduced Neu7s but did not significantly reduce CS-56 immunoreactivity, or result in differences in axon behavior at the interface of transduced Neu7s. The miR-ChSy1 LV decreased ChSy1 mRNA, CSPGs, and axon inhibition by the interface. A LV was created with co-cistronic expression of miRNA against both ChPF and ChSy1 from the same transcript. This dual LV knocked down both ChPF and ChSy1 mRNA, reduced CSPGs further than just the miR-ChSy1 LV, and significantly decreased axon inhibition at the interface. Because of the ability for the enzyme isoforms to compensate for each other, targeting more than two could result in further reduction in CS-GAGs and axon inhibition.
One of the challenges associated with gene delivery in vivo is transduction efficiency and the resulting amount of transgene expression. We evaluated multiple ratios of treated to negative control astrocytes in the glial scar model to mimic variable transduction efficiencies. Depending on whether the target of the vector is intracellular (RNAi) or extracellular (ch’ase) the effect of transduction efficiency varied. Both the 50:50 and 10:90 (ch’ase LV:GFP LV Neu7s) ratios resulted in reduced axon inhibition, although to a lesser extent than 100:0 (ch’ase LV:untreated Neu7s). These results suggest that lower transduction efficiencies, often seen in vivo, might be adequate to degrade CS-GAGs because the transcript is secreted from transduced cells with the ability to act on neighboring cells in the environment. Because RNAi acts intracellularly, higher transduction efficiencies may be necessary for the treatment to be effective. A 50:50 ratio of dual (miR-ChPF/ChSy1) transduced Neu7s mixed with Neu7s transduced with the negative control miRNA LV was able to reduce inhibition at the interface, but to a lesser extent. Crossing was not significantly different however, a significantly lower percentage of neurites turned away from the interface. This result suggests that in order for RNAi to be an effective treatment in vivo, high transduction efficiency in CSPG producing cells may be required. Neurite behavior in cocultures performed with varying ratios of treated to negative control cells, and therefore intermediate levels of CSPGs, suggests that the method used to reduce CSPGs (degradation or synthesis prevention) influences neurite behavior in different ways. The model developed in this report could elucidate biological differences between cues that direct neurite crossing, turning, and stopping upon encountering an inhibitory interface.
In conclusion, lentiviral delivery of ch’ase and RNAi against CSPG synthesis enzymes reduced axon inhibition in a novel in vitro model of the glial scar. Targeting two of the enzymes involved in assembling CSPGs was more effective than just one. Transduction efficiency and whether the target is intra- or extracellular are important considerations when determining the potential of these methods in vivo. Lentiviral delivery of ch’ase and RNAi against CS-GAG specific synthesis could synergize to reduce CS inhibition of axon regeneration by both degrading the initially deposited CSPGs and preventing synthesis of new CSPGs. The sustained expression achieved with lentiviral delivery combined with the flexibility to deliver multiple genes (including growth factors) from a single or combination of vectors make this technique attractive for further investigation.
Acknowledgments
Financial support for this research was provided by the NIH (RO1 EB005678, RO1 EB 003806). We thank Dr. G.M. Smith for the ChAC lentiviral construct, Dr. J.W. Fawcett for the Neu7 cell line, and the Keck Biophysics Facility at Northwestern University for equipment use.
References
- Asher RA, Morgenstern DA, Fidler PS, Adcock KH, Oohira A, Braistead JE, Levine JM, Margolis RU, Rogers JH, Fawcett JW. Neurocan is upregulated in injured brain and in cytokine-treated astrocytes. J Neurosci. 2000;20(7):2427–2438. doi: 10.1523/JNEUROSCI.20-07-02427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Moon LD, Fawcett JW. Chondroitin sulphate proteoglycans: Inhibitory components of the glial scar. Prog Brain Res. 2001;132:611–619. doi: 10.1016/S0079-6123(01)32106-4. [DOI] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Shearer MC, Adcock KH, Pesheva P, Fawcett JW. Versican is upregulated in CNS injury and is a product of oligodendrocyte lineage cells. J Neurosci. 2002;22(6):2225–2236. doi: 10.1523/JNEUROSCI.22-06-02225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnur Z, Geiger B. Immunocytochemical localization of native chondroitin-sulfate in tissues and cultured cells using specific monoclonal antibody. Cell. 1984;38(3):811–822. doi: 10.1016/0092-8674(84)90276-9. [DOI] [PubMed] [Google Scholar]
- Bandtlow CE, Zimmermann DR. Proteoglycans in the developing brain: New conceptual insights for old proteins. Physiol Rev. 2000;80(4):1267–1290. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- Barritt AW, Davies M, Marchand F, Hartley R, Grist J, Yip P, McMahon SB, Bradbury EJ. Chondroitinase ABC promotes sprouting of intact and injured spinal systems after spinal cord injury. J Neurosci. 2006;26(42):10856–10867. doi: 10.1523/JNEUROSCI.2980-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416(6881):636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Chau CH, Shum DK, Li H, Pei J, Lui YY, Wirthlin L, Chan YS, Xu XM. Chondroitinase ABC enhances axonal regrowth through Schwann cellseeded guidance channels after spinal cord injury. FASEB J. 2004;18(1):194–196. doi: 10.1096/fj.03-0196fje. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Fitch MT, Memberg SP, Hall AK, Raisman G, Silver J. Regeneration of adult axons in white matter tracts of the central nervous system. Nature. 1997;390(6661):680–683. doi: 10.1038/37776. [DOI] [PubMed] [Google Scholar]
- Davies SJ, Goucher DR, Doller C, Silver J. Robust regeneration of adult sensory axons in degenerating white matter of the adult rat spinal cord. J Neurosci. 1999;19(14):5810–5822. doi: 10.1523/JNEUROSCI.19-14-05810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East E, Golding JP, Phillips JB. A versatile 3D culture model facilitates monitoring of astrocytes undergoing reactive gliosis. J Tissue Eng Regen Med. 2009;3(8):634–646. doi: 10.1002/term.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler PS, Schuette K, Asher RA, Dobbertin A, Thornton SR, Calle-Patino Y, Muir E, Levine JM, Geller HM, Rogers JH, Faissner A, Fawcett JW. Comparing astrocytic cell lines that are inhibitory or permissive for axon growth: The major axon-inhibitory proteoglycan is NG2. J Neurosci. 1999;19(20):8778–8788. doi: 10.1523/JNEUROSCI.19-20-08778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok-Seang J, Smith-Thomas LC, Meiners S, Muir E, Du JS, Housden E, Johnson AR, Faissner A, Geller HM, Keynes RJ, et al. An analysis of astrocytic cell lines with different abilities to promote axon growth. Brain Res. 1995;689(2):207–223. doi: 10.1016/0006-8993(95)00575-b. [DOI] [PubMed] [Google Scholar]
- Garcia-Alias G, Lin R, Akrimi SF, Story D, Bradbury EJ, Fawcett JW. Therapeutic time window for the application of chondroitinase ABC after spinal cord injury. Exp Neurol. 2008;210(2):331–338. doi: 10.1016/j.expneurol.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Garcia-Alias G, Barkhuysen S, Buckle M, Fawcett JW. Chondroitinase ABC treatment opens a window of opportunity for task-specific rehabilitation. Nat Neurosci. 2009;12(9):1145–1151. doi: 10.1038/nn.2377. [DOI] [PubMed] [Google Scholar]
- Grimpe B, Silver J. A novel DNA enzyme reduces glycosaminoglycan chains in the glial scar and allows microtransplanted dorsal root ganglia axons to regenerate beyond lesions in the spinal cord. J Neurosci. 2004;24(6):1393–1397. doi: 10.1523/JNEUROSCI.4986-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas CA, Rauch U, Thon N, Merten T, Deller T. Entorhinal cortex lesion in adult rats induces the expression of the neuronal chondroitin sulfate proteoglycan neurocan in reactive astrocytes. J Neurosci. 1999;19(22):9953–9963. doi: 10.1523/JNEUROSCI.19-22-09953.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle JD, Tom VJ, Mayes D, Wagoner G, Phillips N, Silver J. Combining an autologous peripheral nervous system ‘‘bridge’’ and matrix modification by chondroitinase allows robust, functional regeneration beyond a hemisection lesion of the adult rat spinal cord. J Neurosci. 2006;26(28):7405–7415. doi: 10.1523/JNEUROSCI.1166-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa T, Uyama T, Okuura Y, Sugahara K, Kitagawa H. Involvement of chondroitin sulfate synthase-3 (chondroitin synthase-2) in chondroitin polymerization through its interaction with chondroitin synthase-1 or chondroitin-polymerizing factor. Biochem J. 2007;403(3):545–552. doi: 10.1042/BJ20061876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa T, Koike T, Shiozawa S, Sugahara K, Tamura J, Kitagawa H. Identification of chondroitin sulfate glucuronyltransferase as chondroitin synthase-3 involved in chondroitin polymerization: Chondroitin polymerization is achieved by multiple enzyme complexes consisting of chondroitin synthase family members. J Biol Chem. 2008;283(17):11396–11406. doi: 10.1074/jbc.M707549200. [DOI] [PubMed] [Google Scholar]
- Jones LL, Tuszynski MH. Chronic intrathecal infusions after spinal cord injury cause scarring and compression. Microsc Res Tech. 2001;54(5):317–324. doi: 10.1002/jemt.1144. [DOI] [PubMed] [Google Scholar]
- Jones LL, Yamaguchi Y, Stallcup WB, Tuszynski MH. NG2 is a major chondroitin sulfate proteoglycan produced after spinal cord injury and is expressed by macrophages and oligodendrocyte progenitors. J Neurosci. 2002;22(7):2792–2803. doi: 10.1523/JNEUROSCI.22-07-02792.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LL, Margolis RU, Tuszynski MH. The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury. Exp Neurol. 2003;182(2):399–411. doi: 10.1016/s0014-4886(03)00087-6. [DOI] [PubMed] [Google Scholar]
- Kinnunen T, Raulo E, Nolo R, Maccarana M, Lindahl U, Rauvala H. Neurite outgrowth in brain neurons induced by heparin-binding growth-associated molecule (HB-GAM) depends on the specific interaction of HB-GAM with heparan sulfate at the cell surface. J Biol Chem. 1996;271(4):2243–2248. doi: 10.1074/jbc.271.4.2243. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Uyama T, Sugahara K. Molecular cloning and expression of a human chondroitin synthase. J Biol Chem. 2001;276(42):38721–38726. doi: 10.1074/jbc.M106871200. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Izumikawa T, Uyama T, Sugahara K. Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J Biol Chem. 2003;278(26):23666–23671. doi: 10.1074/jbc.M302493200. [DOI] [PubMed] [Google Scholar]
- Laabs TL, Wang H, Katagiri Y, McCann T, Fawcett JW, Geller HM. Inhibiting glycosaminoglycan chain polymerization decreases the inhibitory activity of astrocyte-derived chondroitin sulfate proteoglycans. J Neurosci. 2007;27(52):14494–14501. doi: 10.1523/JNEUROSCI.2807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons ML, Sandy JD, Anderson DK, Howland DR. Intact aggrecan and chondroitin sulfate-depleted aggrecan core glycoprotein inhibit axon growth in the adult rat spinal cord. Exp Neurol. 2003;184(2):981–990. doi: 10.1016/S0014-4886(03)00383-2. [DOI] [PubMed] [Google Scholar]
- Li Y, Raisman G. Sprouts from cut corticospinal axons persist in the presence of astrocytic scarring in long-term lesions of the adult rat spinal cord. Exp Neurol. 1995;134(1):102–111. doi: 10.1006/exnr.1995.1041. [DOI] [PubMed] [Google Scholar]
- Lin R, Rosahl TW, Whiting PJ, Fawcett JW, Kwok JC. 6-Sulphated chondroitins have a positive influence on axonal regeneration. PLoS ONE. 2011;6(7):e21499. doi: 10.1371/journal.pone.0021499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon RJ, Schreiber RC, Rudge JS, Silver J. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J Neurosci. 1991;11(11):3398–3411. doi: 10.1523/JNEUROSCI.11-11-03398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon RJ, Hoke A, Silver J. Injury-induced proteoglycans inhibit the potential for laminin-mediated axon growth on astrocytic scars. Exp Neurol. 1995;136(1):32–43. doi: 10.1006/exnr.1995.1081. [DOI] [PubMed] [Google Scholar]
- McKeon RJ, Jurynec MJ, Buck CR. The chondroitin sulfate proteoglycans neurocan and phosphacan are expressed by reactive astrocytes in the chronic CNS glial scar. J Neurosci. 1999;19(24):10778–10788. doi: 10.1523/JNEUROSCI.19-24-10778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon LD, Asher RA, Rhodes KE, Fawcett JW. Relationship between sprouting axons, proteoglycans and glial cells following unilateral nigrostriatal axotomy in the adult rat. Neuroscience. 2002;109(1):101–117. doi: 10.1016/s0306-4522(01)00457-2. [DOI] [PubMed] [Google Scholar]
- Muir E, Du JS, Fok-Seang J, Smith-Thomas LC, Housden ES, Rogers J, Fawcett JW. Increased axon growth through astrocyte cell lines transfected with urokinase. Glia. 1998;23(1):24–34. [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell EM, Fawcett JW, Geller HM. Proteoglycans provide neurite guidance at an astrocyte boundary. Mol Cell Neurosci. 1997;10(1–2):27–42. doi: 10.1006/mcne.1997.0629. [DOI] [PubMed] [Google Scholar]
- Properzi F, Carulli D, Asher RA, Muir E, Camargo LM, van Kuppevelt TH, ten Dam GB, Furukawa Y, Mikami T, Sugahara K, et al. Chondroitin 6-sulphate synthesis is up-regulated in injured CNS, induced by injury-related cytokines and enhanced in axon-growth inhibitory glia. Eur J Neurosci. 2005;21(2):378–390. doi: 10.1111/j.1460-9568.2005.03876.x. [DOI] [PubMed] [Google Scholar]
- Rolls A, Shechter R, Schwartz M. The bright side of the glial scar in CNS repair. Nat Rev Neurosci. 2009;10(3):235–241. doi: 10.1038/nrn2591. [DOI] [PubMed] [Google Scholar]
- Sango K, Oohira A, Ajiki K, Tokashiki A, Horie M, Kawano H. Phosphacan and neurocan are repulsive substrata for adhesion and neurite extension of adult rat dorsal root ganglion neurons in vitro. Exp Neurol. 2003;182(1):1–11. doi: 10.1016/s0014-4886(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Schwartz NB, Domowicz M. Proteoglycans in brain development. Glycoconj J. 2004;21(6):329–341. doi: 10.1023/B:GLYC.0000046278.34016.36. [DOI] [PubMed] [Google Scholar]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5(2):146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- Smith-Thomas LC, Fok-Seang J, Stevens J, Du JS, Muir E, Faissner A, Geller HM, Rogers JH, Fawcett JW. An inhibitor of neurite outgrowth produced by astrocytes. J Cell Sci. 1994;107(Pt 6):1687–1695. doi: 10.1242/jcs.107.6.1687. [DOI] [PubMed] [Google Scholar]
- Snow DM, Lemmon V, Carrino DA, Caplan AI, Silver J. Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp Neurol. 1990;109(1):111–130. doi: 10.1016/s0014-4886(05)80013-5. [DOI] [PubMed] [Google Scholar]
- Stichel CC, Muller HW. The CNS lesion scar: New vistas on an old regeneration barrier. Cell Tissue Res. 1998;294(1):1–9. doi: 10.1007/s004410051151. [DOI] [PubMed] [Google Scholar]
- Tester NJ, Plaas AH, Howland DR. Effect of body temperature on chondroitinase ABC’s ability to cleave chondroitin sulfate glycosaminoglycans. J Neurosci Res. 2007;85(5):1110–1118. doi: 10.1002/jnr.21199. [DOI] [PubMed] [Google Scholar]
- Tom VJ, Steinmetz MP, Miller JH, Doller CM, Silver J. Studies on the development and behavior of the dystrophic growth cone, the hallmark of regeneration failure, in an in vitro model of the glial scar and after spinal cord injury. J Neurosci. 2004;24(29):6531–6539. doi: 10.1523/JNEUROSCI.0994-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Ichiyama RM, Zhao R, Andrews MR, Fawcett JW. Chondroitinase combined with rehabilitation promotes recovery of forelimb function in rats with chronic spinal cord injury. J Neurosci. 2011;31(25):9332–9344. doi: 10.1523/JNEUROSCI.0983-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7(8):617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Bellamkonda RV. Dorsal root ganglia neurite extension is inhibited by mechanical and chondroitin sulfate-rich interfaces. J Neurosci Res. 2001;66(2):303–310. doi: 10.1002/jnr.1225. [DOI] [PubMed] [Google Scholar]