Summary
Nucleosome remodelers of the DDM1/Lsh family are required for DNA methylation of transposable elements, but the reason for this is unknown. How DDM1 interacts with other methylation pathways, such as small RNA-directed DNA methylation (RdDM), which is thought to mediate asymmetric methylation through DRM enzymes, is also unclear. Here, we show that most asymmetric methylation is facilitated by DDM1 and mediated by the methyltransferase CMT2 separately from RdDM. We find that heterochromatic sequences preferentially require DDM1 for DNA methylation, and that this preference depends on linker histone H1. RdDM is instead inhibited by heterochromatin and absolutely requires the nucleosome remodeler DRD1. Together, DDM1 and RdDM mediate nearly all transposon methylation, and collaborate to repress transposition and regulate the methylation and expression of genes. Our results indicate that DDM1 provides DNA methyltransferases access to H1-containing heterochromatin to allow stable silencing of transposable elements in cooperation with the RdDM pathway.
Introduction
DNA methylation in flowering plants occurs in three sequence contexts: CG, CHG and CHH, where H is any nucleotide except G. Methylation in each context is believed to be primarily catalyzed by a specific family of DNA methyltransferases: MET1 homologous to animal Dnmt1) for CG, chromomethylases (CMT) for CHG, and DRM (homologous to animal Dnmt3) for CHH (Law and Jacobsen, 2010). The majority of plant methylation is found in transposable elements (TEs), where methylation occurs in all sequence contexts and is crucial for the repression of TE expression and transposition (Law and Jacobsen, 2010). Substantial methylation is also found in the bodies of active genes, where methylation is generally restricted to the CG context (Law and Jacobsen, 2010; Zemach et al., 2010b).
The establishment of plant DNA methylation in all sequence contexts, and the maintenance of CHH methylation is mediated by a specialized branch of the RNA interference pathway referred to as RNA-directed DNA methylation (RdDM) (Law and Jacobsen, 2010). RdDM relies on two plant-specific homologs of RNA polymerase II – pol IV and pol V. The pol IV branch of RdDM is thought to synthesize the long RNA molecules that are made double-stranded by RNA-dependent RNA polymerase 2 (RDR2) and processed by Dicer-like nucleases into small interfering RNA (sRNA). RNA pol V and associated factors are believed to produce nascent transcripts from target loci that are recognized by sRNA-containing AGO4 complexes that target DNA methylation via DRM enzymes (Haag and Pikaard, 2011).
DNA methylation is also influenced by chromatin factors: CHG methylation by CMT3 requires dimethylation of histone H3 at lysine 9 (H3K9me2), to which CMT3 binds via its chromo and bromo adjacent homology domains (Du et al., 2012; Law and Jacobsen, 2010), and CHG methylation is kept out of genes by a histone demethylase, IBM1, which removes H3K9me2 from gene bodies (Saze and Kakutani, 2011).
A more enigmatic chromatin factor that is essential for normal DNA methylation is the Snf2 family nucleosome remodeler DDM1 (Jeddeloh et al., 1999; Lippman et al., 2004). Snf2 remodelers hydrolyze ATP to move along DNA, altering nucleosome composition and placement and allowing other proteins to access the DNA (Ryan and Owen-Hughes, 2011). DDM1 can shift nucleosomes in vitro (Brzeski and Jerzmanowski, 2003), and its mutation has been reported to cause a profound loss of methylation from some TEs and repeats (Jeddeloh et al., 1999), but not from genes (Lippman et al., 2004). Mutation of Lsh, the mouse homolog of DDM1, causes a similar methylation phenotype (Tao et al., 2011), indicating that DDM1 remodelers are ancient components of the DNA methylation pathway. The loss of DDM1 leads to strong transcriptional activation of TEs (Lippman et al., 2004), and inbred ddm1 mutant lines have increased rates of transposition (Tsukahara et al., 2009). sRNAs correspond to the TEs hypomethylated in ddm1 mutant plants, leading to the suggestion that DDM1 participates in RdDM (Lippman et al., 2004). However, DDM1 and RdDM synergize to silence rDNA loci (Blevins et al., 2009), indicating that RdDM can function without DDM1, and DDM1 can mediate CHH methylation independently of RdDM at some TEs (Teixeira et al., 2009). Thus, the related questions of how DDM1 interacts with the methyltransferase pathways, including RdDM, and why some sequences require DDM1 for methylation and others do not, remain largely unanswered.
Results
DDM1 and RdDM separately mediate nearly all DNA methylation in TEs
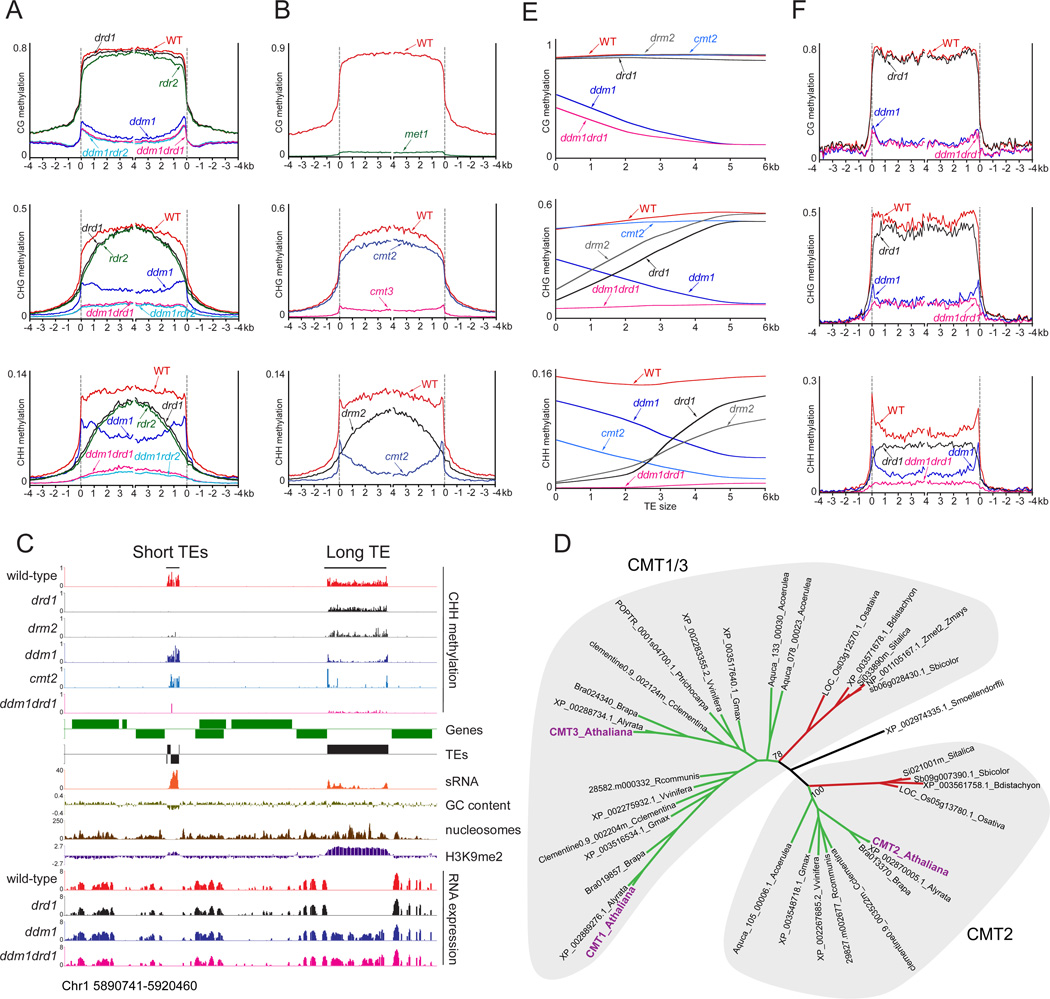
To understand how DDM1 mediates DNA methylation, we quantified genomic methylation of ddm1 mutant Arabidopsis thaliana plants (Table S1). Lack of DDM1 caused a 58%, 57% and 32% overall reduction of CG, CHG and CHH methylation, respectively (Figure S1A), reflected in much lower TE methylation in all sequence contexts (Figure 1A). The strong loss of CHH methylation might be caused by a dependence of RdDM on methylation in other contexts, as proposed to explain the loss of non-CG methylation in met1 mutant plants (Figure S1A) (Lister et al., 2008), or may indicate that a large fraction of plant CHH methylation is mediated by DDM1 independently of RdDM (Teixeira et al., 2009). To answer this question, we determined DNA methylation in plants with a mutation in RdDM pathway gene RDR2, which is required for the production of all endogenous sRNA molecules (Xie et al., 2004). We also analyzed methylation in plants lacking DRD1, a Snf2 remodeler that positively regulates RdDM at a number of loci (Kanno et al., 2004; Law et al., 2010) and forms a complex with RdDM pathway proteins that is thought to facilitate pol V activity (Law et al., 2010; Zhong et al., 2012). Lack of RDR2 and DRD1 caused a relatively modest loss of CHH methylation (Figures 1A and S1A), demonstrating that the majority of CHH methylation does not require RdDM (Wierzbicki et al., 2012). Combining the ddm1 mutation with either rdr2 or drd1 caused a nearly complete loss of CHH methylation (Figures 1A and S1A), as well as of CG and CHG methylation in TEs (Figure 1A), demonstrating that a great deal of Arabidopsis CHH methylation is mediated by DDM1 separately from RdDM, and that the two pathways together are responsible for almost all DNA methylation of TEs. The methylation phenotypes of drd1 and ddm1drd1 mutants are virtually indistinguishable from those of rdr2 and ddm1rdr2, respectively (Figure 1A), indicating that DRD1 is absolutely required for RdDM.
Figure 1. DDM1 and CMT2 mediate RdDM-independent CHH methylation of long TEs.
(A) Patterns of TE DNA methylation (CG, CHG and CHH) in wild-type and indicated mutants. Arabidopsis TEs were aligned at the 5′ end or the 3′ end, and average methylation for all cytosines within each 100-bp interval is plotted. The dashed lines represent the points of alignment. (B) Patterns of TE methylation in met1, cmt3, cmt2 and drm2 plants. (C) CHH methylation, sRNA, GC content, nucleosomes, H3K9me2 and RNA levels of a representative region. Genes and TEs oriented 5′ to 3′ and 3′ to 5′ are shown above and below the line, respectively. (D) Phylogenetic tree of angiosperm chromomethylases, with Selaginella moellendorffii (black) as an outgroup. Dicots are shown in green and monocots in red. (E) LOWESS fit of CG, CHG and CHH methylation levels in wild-type and indicated mutants calculated in 50-bp windows and plotted against TE size. (F) DNA methylation in wild-type and indicated mutants was averaged specifically in long TEs (> 4 kb) as described in (A). See also Figure S1 and Table S1.
DDM1-dependent CHH methylation is catalyzed by CMT2
The presence of extensive RdDM-independent CHH methylation raises the question of which DNA methyltransferase is responsible. Previous studies have demonstrated that MET1 and CMT3 mediate virtually all Arabidopsis CG and CHG methylation, respectively (Cokus et al., 2008; Lister et al., 2008), and our data confirm these results (Figures 1B and S1A). However, even plants lacking DRM1 (which appears to be specifically active during early seed development (Jullien et al., 2012)), DRM2, and CMT3 have substantial residual CHH methylation (Cokus et al., 2008; Lister et al., 2008), indicating that another methyltransferase must be involved. Mutation of DRM2 causes CHH methylation loss that closely resembles that in RdDM mutants (Figures 1A–C), consistent with the established link between DRM2 and RdDM (Law and Jacobsen, 2010). Unexpectedly, lack of CMT2, a homolog of CMT3 (Figure 1D), has little impact on CHG methylation but causes a major loss of CHH methylation (Figures 1B and S1A–B). The observation that DRM2 accounts for RdDM (Figures 1A–C) indicates that CMT2 is responsible for the DDM1-mediated, sRNA-independent CHH methylation (Figure 1A). In support of this conclusion, residual methylation in cmt2 plants is correlated with that in ddm1 and anticorrelated with that in RdDM mutants (Table S2), whereas residual methylation in drm2 plants (mediated by CMT2) is uncorrelated with sRNA abundance (Table S2). CMT2 appears to have evolved prior to the radiation of angiosperms (Figure 1D) to methylate a different sequence context than canonical chromomethylases (Cokus et al., 2008; Du et al., 2012; Zemach et al., 2010b).
DDM1 and RdDM mediate methylation of distinct TE sizes and domains
The ddm1 and drd1 mutant lines exhibit a similar absolute level but very different patterns of CHH methylation loss (Figures 1A and S1A). The drd1 mutation strongly reduces TE CHH methylation near the points of alignment, whereas ddm1 has a larger effect away from the points of alignment (Figure 1A). The same distinction is evident for CG and CHG methylation (Figure 1A), and for the CHH methylation phenotypes of drm2 and cmt2 (Figure 1B). Lack of DDM1 and RdDM thus affects TEs very differently.
More than 80% of annotated Arabidopsis TEs are shorter than 1000 bp (Buisine et al., 2008), and such TEs would only contribute to the patterns of TE methylation shown in Figures 1A and 1B close to the points of alignment, suggesting that the differences between ddm1 and drd1, as well as between cmt2 and drm2, may be caused by differential hypomethylation of short and long TEs. Indeed, the ddm1 and cmt2 hypomethylation effects are positively correlated with TE size, whereas drd1 and drm2 hypomethylation is negatively correlated with TE size (Figures 1C and 1E). Pericentric heterochromatin and chromosome arms are enriched for long and short TEs, respectively (Figure S1C) (Ahmed et al., 2011), and consequently DDM1 preferentially mediates DNA methylation near the centromeres, whereas RdDM predominantly functions along the chromosome arms (Figures S1D–E).
The methylation patterns in Figure 1A are also consistent with DDM1 and DRD1 mediating methylation differently at the edges and inside the bodies of TEs. To examine this issue, we averaged methylation across TEs longer than 4 kb, so that short TEs would not influence the methylation level near the points of alignment. The hypomethylation induced by drd1 is strongest at TE edges, whereas ddm1 hypomethylation is greater within TE bodies (Figures 1C and 1F). The little remaining non-CG methylation in the ddm1drd1 double mutant is evenly distributed across the entire TE sequence (Figure 1F). Similarly, cmt2 nearly eliminates CHH methylation of TEs longer than 4 kb except at the edges, where CHH methylation is mediated by DRM2 (Figures 1C and S1F). Taken together, our results indicate that DDM1 is preferentially required for DNA methylation within the bodies of long TEs, which is catalyzed by MET1 (CG), CMT3 (CHG) and CMT2 (CHH), whereas RdDM mostly targets short TEs and TE edges through DRM2 (Figures 1C, 1E–F and S1F), consistent with the observed enrichment of pol V at such sequences (Zhong et al., 2012).
Heterochromatin requires DDM1 for DNA methylation and inhibits RdDM
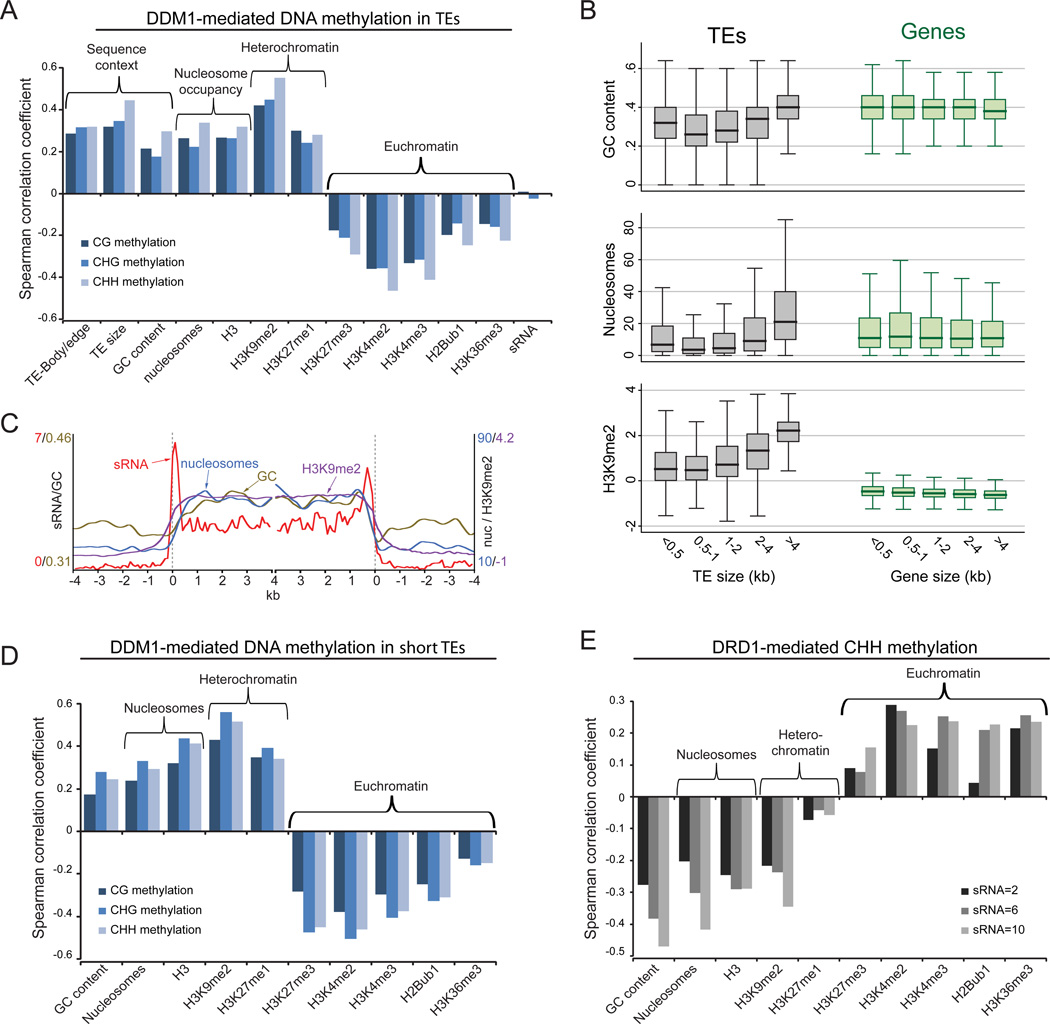
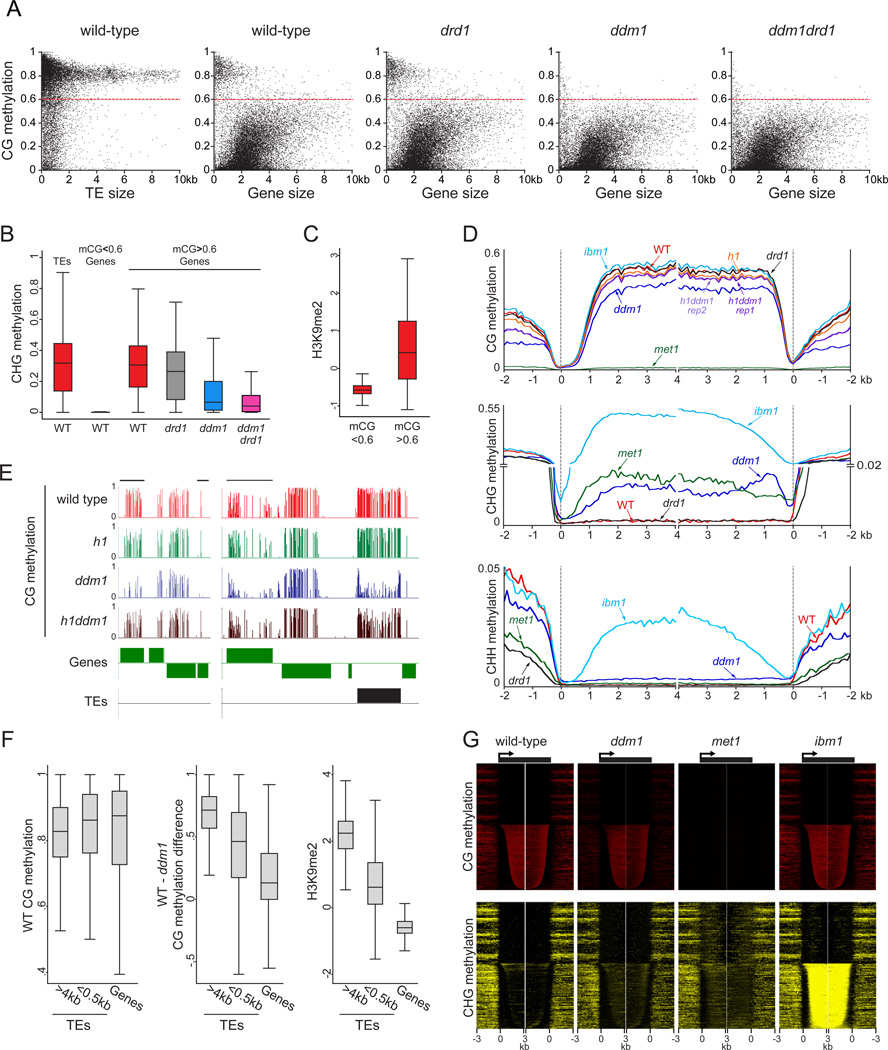
Because DDM1 can remodel nucleosomes, we asked whether chromatin features are responsible for the differential requirement of DDM1 for DNA methylation. Sequence composition is thought to be a major determinant of the nucleosome landscape (Iyer, 2012), and indeed ddm1 TE demethylation in all sequence contexts is strongly correlated with nucleosome occupancy and GC content (Figure 2A). Short TEs and TE edges are relatively AT-rich and nucleosomedepleted (Figures 2B–C and S2A), consistent with the preferential requirement of DDM1 to maintain DNA methylation in the bodies of long TEs (Figures 1E–F and 2A). However, nucleosome occupancy alone is unlikely to determine the dependence of DNA methylation on DDM1 because genic GC content is similar to that of long TEs and nucleosome occupancy in genes is higher than in most TEs (Figure 2B), presumably because genes and long TEs are composed largely of protein-coding sequences. The properties of nucleosomes in euchromatic genes do differ from those of heterochromatic TEs, as exemplified by different sets of posttranslational histone modifications (Roudier et al., 2011). Somewhat unexpectedly, we find that H3K9me2, the most well-studied histone modification associated with plant heterochromatin, is enriched in the bodies of long TEs compared to short TEs (Figures 2B–C). More generally, DDM1-mediated TE DNA methylation – the residual methylation in the drd1 mutant – is correlated with heterochromatic histone modifications, anticorrelated with euchromatic ones, and not correlated with sRNA abundance (Figures 2A and S2B). Furthermore, the dependence of short TE methylation on DDM1 also correlates with GC content, nucleosome occupancy and histone modifications (Figure 2D) just like methylation of all TEs (Figure 2A), indicating that the chromatin environment, rather than TE size, ultimately determines the extent to which DDM1 is required for maintenance of DNA methylation.
Figure 2. Heterochromatin requires DDM1 for DNA methylation and inhibits RdDM.
(A) Spearman correlation coefficients between DDM1-mediated methylation (drd1 methylation minus ddm1drd1 methylation) and DNA sequence and chromatin features of TEs in 50-bp windows. (B) Box plots showing GC content, nucleosome occupancy and H3K9me2 levels in 50-bp windows within TEs and genes of the indicated size. (C) sRNA, GC content, nucleosome occupancy and H3K9me2 levels were averaged in long TEs (> 4 kb) as described in Figure 1. (D) Spearman correlation coefficients between DDM1-mediated methylation in short TEs (<500 bp) and chromatin features. (E) Spearman correlation coefficients between DRD1-mediated CHH methylation (ddm1 methylation minus ddm1drd1 methylation) and chromatin features, calculated for 50-bp windows with three different levels of sRNA. See also Figure S2 and Table S2.
RdDM would be expected to occur at sRNA-associated loci. Indeed, DRD1-mediated methylation – the residual methylation in the ddm1 mutant – is positively correlated with sRNA abundance (Figure S2C), and TE edges, which are hypermethylated at CHH sites in comparison to internal sequences in wild-type plants (Figure 1F), are preferentially targeted by sRNA (Figure 2C) (Lee et al., 2012). However, sRNA molecules are also abundantly derived from TE bodies (Figure 2C). To understand how chromatin structure affects RdDM, we analyzed DRD1-mediated DNA methylation at sequences with similar levels of sRNA. With sRNA levels held constant, DRD1-mediated methylation is negatively correlated with GC content, nucleosome occupancy and heterochromatic histone modifications (Figures 2E, S2B and S2D). Thus, RdDM is inhibited by heterochromatin, as has been suggested by (Schoft et al., 2009).
Histone H1 mediates the dependence of heterochromatic DNA methylation on DDM1
Our results indicate that DDM1 is preferentially required for methylation of heterochromatic sequences in all contexts, most likely by allowing methyltransferases access to the DNA. To determine which component of heterochromatin blocks enzyme access, we examined histone H1, which binds to the nucleosome core and the linker DNA that separates nucleosomes (Thomas, 1999), condenses chromatin and inhibits nucleosome mobility and transcription in vitro (Pennings et al., 1994; Robinson and Rhodes, 2006), and is associated with more compact, less accessible and transcriptionally silent chromatin in vivo (Ascenzi and Gantt, 1999; Barra et al., 2000; Fan et al., 2005; Raghuram et al., 2009). Loss of H1 has been reported to cause disparate changes in genomic DNA methylation: mice with reduced H1 specifically lose DNA methylation at the regulatory regions of several imprinted genes (Fan et al., 2005), whereas loss of H1 leads to extensive hypermethylation in the fungus Ascobolus immersus (Barra et al., 2000) and apparently stochastic methylation gains and losses in Arabidopsis (Wierzbicki and Jerzmanowski, 2005).
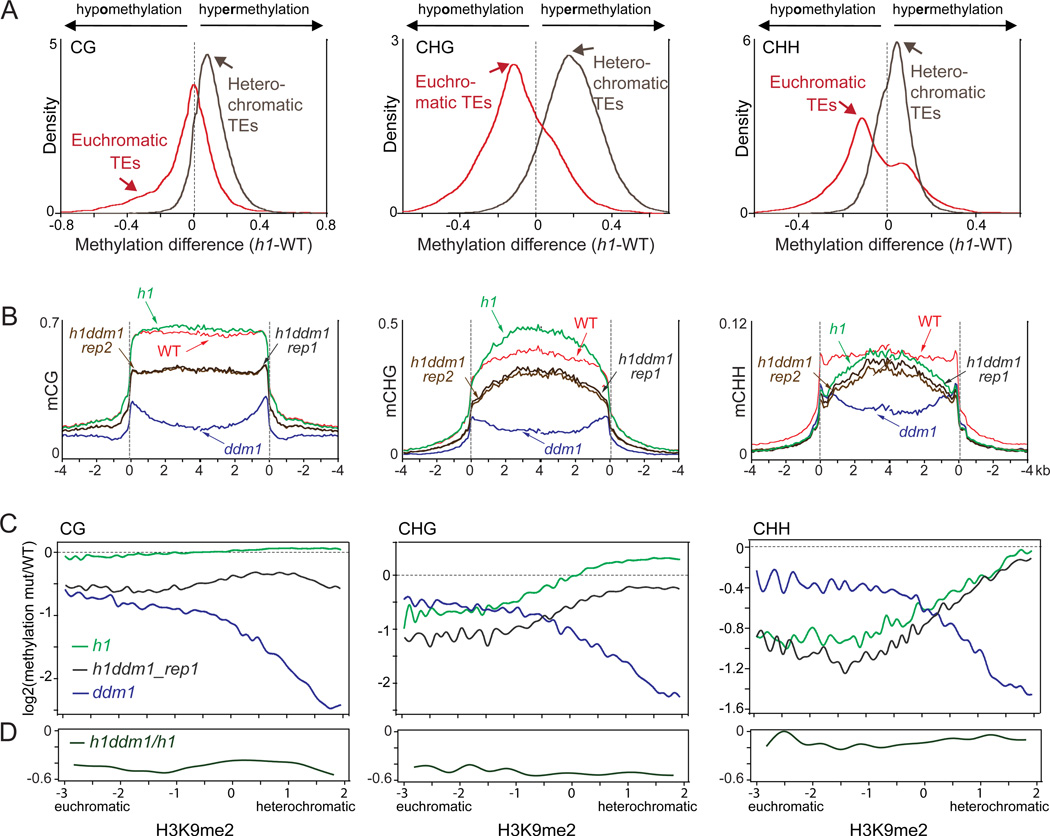
Plants with mutant alleles in the two canonical Arabidopsis H1 genes (Rea et al., 2012; Wierzbicki and Jerzmanowski, 2005) (h1 plants; Figures S3A–C) exhibit a complex DNA methylation phenotype. Euchromatic TEs (those with low H3K9me2) lose DNA methylation in h1 (Figure 3A), whereas H3K9me2-rich heterochromatic TEs exhibit a global increase of DNA methylation (Figures 3A–B), supporting our hypothesis that H1 impedes access of DNA methyltransferases to heterochromatin. Loss of H1 almost completely suppresses the reduction of TE CHH methylation in ddm1, and greatly ameliorates the reduction of TE CG and CHG methylation (Figures 3B–C and S3D–E). Most strikingly, H3K9me2-rich heterochromatic TEs are not preferentially hypomethylated in h1ddm1 plants, as they are in ddm1 (Figures 3C and S3D). Instead, h1ddm1 causes heterochromatic TEs to lose less DNA methylation than more euchromatic TEs (black traces in Figure 3C), similar to h1 (green traces in Figure 3C), indicating that the loss of DDM1 affects euchromatic and heterochromatic TEs similarly when H1 is not present (Figure 3D). Lack of H1 still destabilizes the methylation of euchromatic TEs when combined with ddm1, but heterochromatic TEs are methylated rather efficiently when both DDM1 and H1 are absent (Figures 3C and S3D–E). Our results indicate that the differential importance of DDM1 for the maintenance of DNA methylation in heterochromatic versus euchromatic TEs is governed by H1.
Figure 3. Lack of H1 ameliorates the loss of methylation in ddm1 plants.
(A) Kernel density plots of methylation differences between h1 and wild-type (positive numbers indicate greater methylation in h1). TEs with H3K9me2 log2 scores lower than -1 and higher than 1 are considered euchromatic and heterochromatic, respectively. The colored arrows emphasize global differences (a shifted peak) or extensive local differences (a broad shoulder). (B) Average methylation of TEs in sibling wild-type (WT), h1, ddm1, and h1ddm1 (two biological replicates) seedlings is plotted as in Figure 1. (C–D) M-spline curve fits of log2 DNA methylation ratios in 50-bp windows plotted against H3K9me2 level. See also Figure S3.
Methylation of TE families depends on sRNA abundance and chromatin features
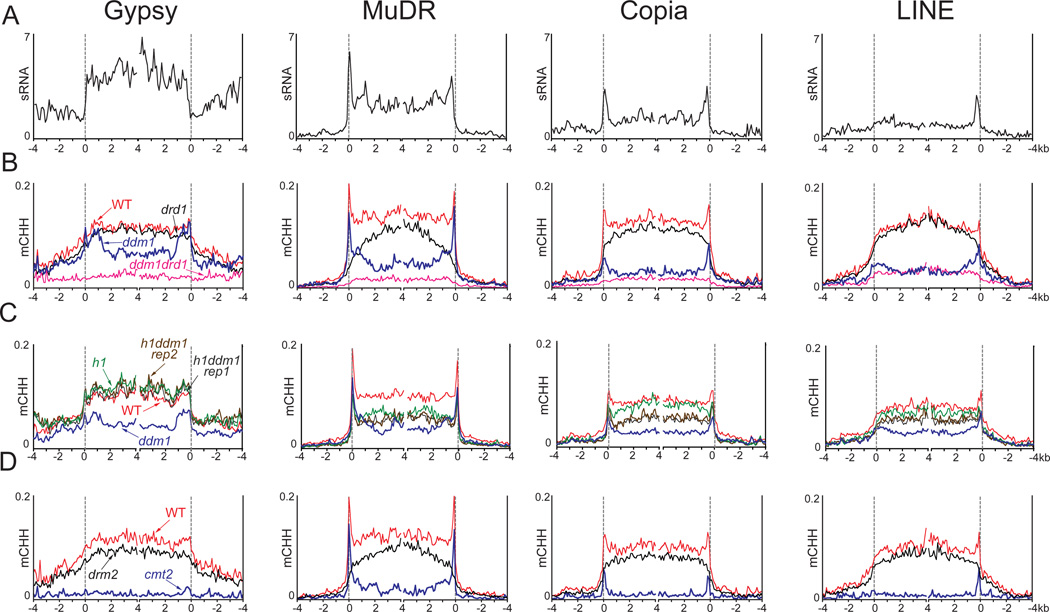
The Arabidopsis genome contains a variety of TE families that have different mechanisms of transposition, internal structure, and chromosomal localization (Ahmed et al., 2011; Buisine et al., 2008). We chose four such families to examine DNA methylation mediated by DDM1 and RdDM in more detail: Gypsy, Copia, MuDR and LINE elements. Gypsy elements are long terminal repeat (LTR)-flanked retrotransposons that are concentrated in pericentric heterochromatin (Figure S1C). Copia LTR elements are more evenly dispersed, as are LINE non-LTR retrotransposons and the terminal inverted repeat (TIR)-flanked MuDR DNA transposons (Figure S1C).
The four TE families have distinct sRNA distributions: Gypsy elements have high levels of sRNA across the entire sequence, Copia and MuDR elements preferentially accumulate sRNA at their 5' and 3' terminal repeats, and LINEs, which lack 5' repeats, have a spike in sRNA abundance at the 3' end (Figure 4A). DDM1-mediated DNA methylation – the residual methylation in the drd1 mutant – does not appear to be influenced by sRNA, as exemplified by efficient methylation of sRNA-poor Copia and LINE TE bodies as well as sRNA-rich Gypsy elements in drd1 (compare the black traces in Figures 4B and S4A with Figure 4A), supporting our family-independent TE analysis (Figure 2A). CHH RdDM – the residual methylation in the ddm1 mutant – resembles the distribution of sRNA in Copia, MuDR and LINE elements (compare the blue trace in Figure 4B with Figure 4A). Although Gypsy elements are evenly covered by sRNA (Figure 4A), RdDM is still preferentially localized at their edges (Figure 4B), supporting our conclusion that the heterochromatic environment of internal Gypsy sequences inhibits RdDM (Figure 2E). Lack of H1 ameliorates the CG, CHG and CHH methylation losses caused by ddm1 in all TE families (Figures 4C and S4B). In particular, CHH methylation in h1ddm1 is similar to that in h1 at Gypsy and MuDR elements, and to a lesser extent at Copia and LINE TEs (Figure 4C).
Figure 4. Methylation of TE families depends on sRNA abundance and chromatin features.
(A–D) Averaged sRNA abundance (A) and CHH methylation levels (B–D) are plotted as in Figure 1 for TEs belonging to four distinct families. The ddm1 trace in (C) represents siblings of the wild-type (WT), h1 and h1ddm1 seedlings analyzed in this panel, and is independent of the ddm1 roots analyzed in (B). See also Figure S4.
CHH methylation in drm2 plants closely resembles that in drd1 mutants at all four TE families (compare the black traces in Figures 4B and 4D), further substantiating the link between RdDM and DRM2. The cmt2 CHH methylation phenotype is similar to but stronger than that of ddm1 at MuDR, Copia, and LINE elements (compare the blue traces in Figures 4B and 4D) – virtually all CHH methylation is lost in cmt2 plants except at sRNA-targeted terminal repeat sequences (Figure 4D), supporting our conclusion that CMT2 mediates CHH methylation independently of sRNA. Surprisingly, lack of CMT2 essentially eliminates CHH methylation at Gypsy elements (Figure 4D), even though Gypsy CHH methylation can be maintained by RdDM (Figure 4B). This result suggests that CHH methylation feeds back on the levels of the sRNA molecules that mediate RdDM, and is consistent with the recent finding that sRNA production from short TEs requires pol V, but sRNA production from Gypsy elements does not (Lee et al., 2012). Lack of the pol V pathway, as exemplified by drd1, leads to major CHH hypomethylation of short TEs and TE edges (Figures 1E–F), and would thus be expected to reduce CHH methylation-dependent sRNA production from such sequences, whereas sRNA production from Gypsy elements that maintain RdDM-independent CHH methylation at sRNA-corresponding sequences (Figure 4B) would not require RdDM, but would require CMT2.
DDM1 and RdDM collaborate to repress TE transcription and transposition
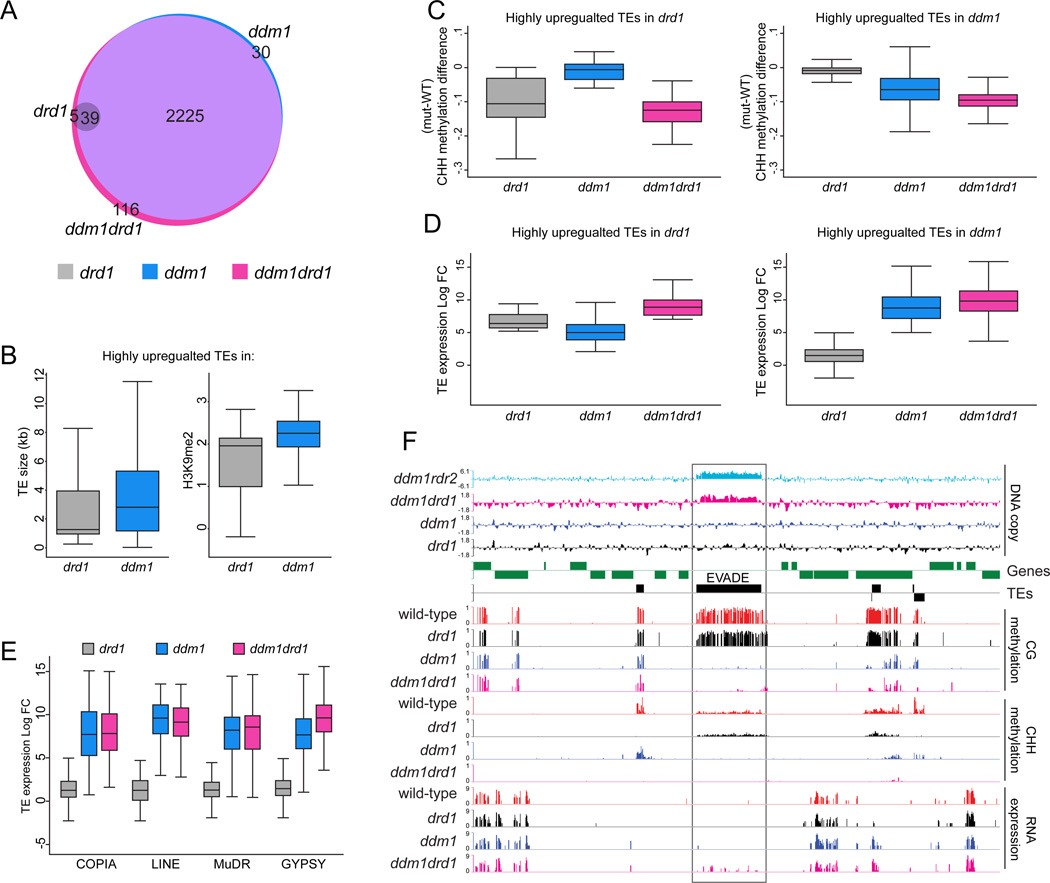
Consistent with the importance of DDM1 for the maintenance of DNA methylation in the bodies of long TEs, where TE-encoded genes required for transposition are located, our RNA-seq analysis revealed that many TEs (2294) are reactivated in ddm1 mutant plants (Figure 5A). In contrast, lack of DRD1, which primarily affects RdDM of non-coding short TEs and TE edges, caused the reactivation of just 44 TEs (Figure 5A). In agreement with our methylation analyses, TEs reactivated in ddm1 are longer and more heterochromatic than those reactivated in drd1 (Figure 5B). In both mutants, TE reactivation is associated with DNA hypomethylation (Figures 5C and S5A). The ddm1drd1 double mutant, which showed additive to synergistic hypomethylation (Figures 5C and S5A), led to stronger TE transcriptional reactivation than either of the single mutants (Figure 5D). This is particularly exemplified by Gypsy elements that require both DDM1 and RdDM for full methylation (Figures 4B and S4A), which are synergistically hyperactivated in ddm1drd1 (Figure 5E).
Figure 5. DDM1 and RdDM collaborate to repress TE expression and transposition.
(A) Venn diagram of significantly upregulated TEs in drd1, ddm1 and ddm1drd1 mutants. (B–D) Box plots of the sizes and H3K9me2 levels (B), absolute fractional CHH demethylation of 50-bp windows (C), and expression compared to wild-type (D) of TEs that are at least 32-fold overexpressed either in drd1 or in ddm1. (E) Box plots of TE family expression in the indicated mutants with respect to wild-type. (F) DNA sequencing coverage (log2(reads in mutant/reads in wild-type)), DNA methylation and RNA levels near the LTR retrotransposon EVADE (AT5TE20395). The sequence coverage is indicative of EVADE copy number relative to wild type (Tsukahara et al., 2009). See also Figure S5.
Mutations in DDM1 and MET1 have been shown to cause transposition of a few TEs, including CACTA and EVADE, but only after several generations of inbreeding (Mirouze et al., 2009; Tsukahara et al., 2009). We found that CACTA and EVADE transpose within the first homozygous generation of ddm1drd1 and ddm1rdr2 mutants (Figures 5F and S5B). This result emphasizes that the DDM1 and RdDM pathways collaborate to prevent TE expression and mobilization.
DDM1 mediates gene body DNA methylation
Plant genes, including those with presumed or demonstrated biological functions, can have methylation patterns that resemble TEs (You et al., 2012; Zemach et al., 2010a), and may therefore be regulated more like TEs than conventional genes. TEs are generally highly methylated at CG sites (Figure 6A, left panel), whereas most Arabidopsis genes have lower methylation levels, except for a distinct group that resembles TEs (Figure 6A). A cutoff of 60% overall CG methylation separates the two genic groups rather cleanly (Figure 6A). The 1284 highly methylated genes resemble TEs in many aspects – they are concentrated in pericentric heterochromatin (Figure S6A), and enriched for non-CG methylation (Figures 6B and S6B) and H3K9me2 (Figure 6C). CG methylation of such genes is lost in ddm1 but not drd1 mutants (Figure 6A), and non-CG methylation is lost partially in both mutants and synergistically in ddm1drd1 (Figures 6B and S6B). Because of these characteristics, we will refer to the highly methylated TE-like genes as heterochromatic genes, and to the rest as euchromatic genes.
Figure 6. DDM1 mediates genic DNA methylation.
(A) CG methylation was averaged in TEs (left panel) and genes for the indicated genotypes (right panels), and plotted against TE or gene size. Note the group of relatively short genes above the red line that are highly methylated in wild-type and significantly hypomethylated in ddm1 and ddm1drd1 mutants, similarly to TEs. (B) Box plots of averaged CHG methylation in TEs, euchromatic genes (mCG < 0.6), and heterochromatic genes (mCG > 0.6). (C) Box plots of H3K9me2 in euchromatic and heterochromatic genes. (D) Genes with average CG methylation between 20% and 60% (euchromatic genes) were aligned as described in Figure 1A. The Y-axis of the CHG plot was broken at 0.02 to improve visualization. (E) Distribution of CG methylation in representative genes AT1G04700, AT1G04750 and AT1G67220 (emphasized by horizontal black bars) that lose methylation in ddm1 but not in h1ddm1. (F) Box plots of wild-type CG methylation (left), absolute fractional CG demethylation in ddm1 (middle), and H3K9me2 (right) of 50-bp windows within long TEs (> 4 kb), short TEs (< 500 bp) and euchromatic genes. (G) Heat maps of CG (red) and CHG (yellow) DNA methylation in genes aligned at the 5' end (left half of each panel) and the 3' end (right half of each panel). More intense color indicates greater methylation. Genes without wild-type CG methylation (shown in the top half of each panel) were stacked from the top of chromosome 1 to the bottom of chromosome 5; genes containing CG methylation islands (shown at the bottom of each panel) were sorted based on the starting position (for 5' panels) or ending position (for 3' panels) of the wild-type CG methylation island. See also Figure S6.
Despite the reported requirement of DDM1 and its mouse homolog Lsh for maintenance of DNA methylation in TEs but not genes (Lippman et al., 2004; Tao et al., 2011), we find that CG methylation of at least 5,348 euchromatic gene bodies (50% of all methylated euchromatic genes) is significantly reduced in ddm1 plants (Fisher’s exact test p-value < 0.0005), whereas only 85 genes are significantly hypermethylated, leading to an overall methylation loss of 20% (Figures 6D and S6C). Lack of H1 also destabilizes genic methylation (Figures S3E and S6C), with at least 3,712 genes significantly hypomethylated and 2,003 genes hypermethylated (p-value < 0.0005). Nevertheless, h1 suppresses the genic hypomethylation caused by ddm1 (Figures 6D–E and S3E), with numbers of significantly hypomethylated (3,878) and hypermethylated (1,488) genes in h1ddm1 resembling h1. These results demonstrate that DDM1 is important for DNA methyltransferase access to all types of sequences, and that the DDM1 requirement depends on the extent of heterochromatin, from the most heterochromatic long TEs, to the less heterochromatic short TEs, to the least heterochromatic genes (Figure 6F).
Loss of DDM1 causes extensive genic CHG hypermethylation
Loss of DDM1 has been reported to cause CHG hypermethylation of a few genes (Lippman et al., 2004; Saze and Kakutani, 2007), and genic hypermethylation was also reported in mouse cells lacking Lsh (Tao et al., 2011), but whether ddm1 causes extensive genic CHG hypermethylation, as has been reported for Arabidopsis met1 and ibm1 mutants (Lister et al., 2008; Saze and Kakutani, 2011), is unknown. We find that lack of DDM1 leads to substantial CHG hypermethylation in euchromatic gene bodies (Figure 6D), but this hypermethylation differs from that caused by met1. Hypermethylation in ddm1 plants is higher toward the 3' end, whereas met1-mediated CHG hypermethylation is more prevalent near the 5' end (Figures 6D and S6D). CHG hypermethylation in ddm1 is restricted to genes that exhibit CG methylation, and is excluded from 5' and 3' genic sequences like wild-type CG methylation (Figure 6G). In contrast, met1 causes CHG methylation of genes that lack CG methylation in wild-type, and the hypermethylation extends to the 3', and to a lesser extent the 5' regions of genes that are normally not CG methylated (Figure 6G).
Like the ddm1 phenotype, CHG hypermethylation in plants lacking the H3K9me2 demethylase IBM1 (Figure 6D) is confined almost exclusively to genic regions that bear CG methylation (Figure 6G). Such genes also exhibit some CHG methylation in wild-type plants (Figure 6G), which may be due to imperfect activity of IBM1. The ibm1 mutation also causes substantial CHH hypermethylation of genes (Figure 6D) (Coleman-Derr and Zilberman, 2012) that is insensitive to drd1 (Figure S6E), and is thus likely mediated by CMT2. However, hypermethylation in ddm1 plants is unlikely to be primarily caused by IBM1 malfunction (Miura et al., 2009), because genic CHG methylation is 5'-biased in ibm1, unlike the 3'-biased methylation in ddm1 (Figures 6D and S6D). This interpretation is also supported by the normal expression of the IBM1 transcript in ddm1, which is disrupted in met1 (Figure S6F) (Rigal et al., 2012). Thus, lack of MET1, IBM1 and DDM1 leads to distinct genic hypermethylation phenotypes likely mediated by different, though potentially overlapping mechanisms.
DDM1 and RdDM synergistically regulate gene expression
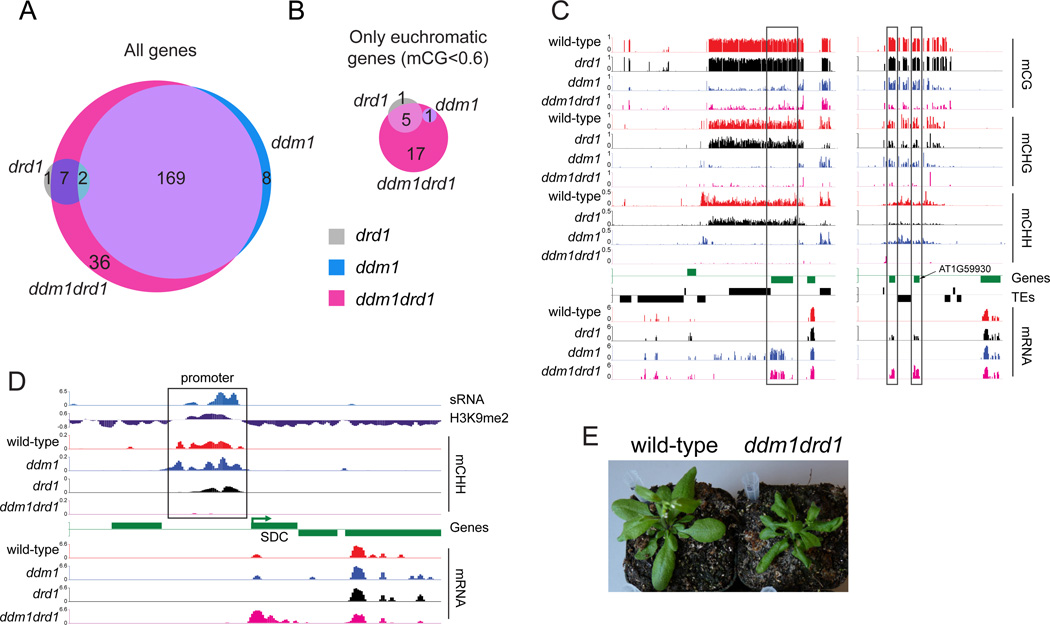
The importance of TE silencing by DNA methylation is not restricted to preventing transposition – demethylation and activation of TEs can also alter the expression of nearby genes (Henderson and Jacobsen, 2008; Hsieh et al., 2011). We found 179 genes upregulated in ddm1 plants (Figure 7A and S7A), all but one of which are heterochromatic (Figure 7B), consistent with the importance of DDM1 for the methylation of heterochromatin generally and heterochromatic genes specifically (Figures 6A–B, S6B and 7C). Only ten genes are upregulated in drd1 plants (Figure 7A), six of which are euchromatic (Figure 7B), and only two of which overlap with genes upregulated by ddm1. One of the heterochromatic genes upregulated in drd1 is AT1G59930 (Figure 7C), a maternally-expressed imprinted gene that is activated by DNA demethylation (Hsieh et al., 2011). All six drd1-upregulated euchromatic genes (AT1G21940, AT1G35730, AT4G01985, AT4G09350, AT4G16460, AT5G41830) have short TEs or TE edges that are hypomethylated in drd1 but not in ddm1 in close proximity to the transcriptional start sites, such as the 150 bp Copia fragment (META1) upstream of AT1G35730 (Figure S7B).
Figure 7. DDM1 and RdDM synergistically regulate gene expression.
(A–B) Venn diagram of significantly (p < 0.05) upregulated genes in drd1, ddm1 and ddm1drd1 mutants. (C) DNA methylation and RNA levels near AT1G46696, and the linked genes AT1G59920 and AT1G59930. (D) sRNA, H3K9me2, CHH methylation and RNA levels near SDC (AT2G17690). (E) Phenotypes of wild-type (flat leaves) and ddm1drd1 (leaves curled downward) plants. See also Figure S7.
More genes (214) are overexpressed in the ddm1drd1 double mutant, including 23 euchromatic genes and five of the six euchromatic genes upregulated in drd1 (Figures 7A–B). One such gene is SDC (Figure 7D), the overexpression of which confers the characteristic phenotype of the drm1drm2cmt3 methyltransferase mutant line (Henderson and Jacobsen, 2008) that is also exhibited by ddm1drd1 plants (Figure 7E). Similarly to Gypsy elements, the repetitive SDC promoter is targeted by sRNA and H3K9me2 (Figure 7D), and its methyation and silencing is mediated by DDM1 and RdDM (Figure 7D). SDC is also a maternally-expressed imprinted gene regulated by DNA demethylation (Hsieh et al., 2011), as is FWA, which is modestly upregulated in ddm1drd1 plants due to demethylation of its SINE-related promoter (Figure S7C). Overall, our results demonstrate that DDM1 and RdDM collaborate to methylate gene-adjacent repetitive elements, thereby maintaining appropriate patterns of gene expression.
Discussion
The targeting of plant DNA methylation has been carefully dissected at individual loci, and the methyltransferases that catalyze CG and CHG methylation throughout the genome are known (Law and Jacobsen, 2010), but the identity of the pathways that mediate the bulk of genomic methylation has remained a mystery. Here, we find that DDM1 and RdDM separately mediate nearly all DNA methylation in Arabidopsis TEs. DDM1 is required for methylation in all sequence contexts of highly heterochromatic TEs (Figures 1A and 2A). This requirement is reduced at less heterochromatic elements, is least in euchromatic genes (Figure 6F), and depends on histone H1 (Figures 3B–D). Together with the preferential demethylation of short euchromatic TEs during plant sexual development (Ibarra et al., 2012; Zemach et al., 2010a), our results demonstrate that a division of the genome into genes and TEs can only explain the biology of DNA methylation if chromatin configuration is also considered.
Lack of access to DNA is postulated to be a core property of heterochromatin that enforces gene silencing by preventing binding of transcription factors and RNA polymerase (Grewal and Jia, 2007). At the same time, stable maintenance of inaccessible heterochromatin requires DNA methylation and histone modifications like H3K9me2 that are catalyzed by enzymes that need to access chromatin. This conundrum is exemplified by the RdDM pathway that silences TE expression, yet requires TE transcripts to function (Haag and Pikaard, 2011). Our results indicate that H1 restricts access to nucleosomal DNA, and that DDM1 overcomes this restriction to enable the maintenance of DNA methylation and silencing of diverse TEs. Without DDM1, DNA methyltransferases cannot efficiently methylate inaccessible heterochromatic TEs (Figures 1A and 2A), leading to derepression and transposition (Figures 5 and S5). Without H1, less heterochromatic sequences lose methylation (Figures 3A and S3E), presumably because they become more accessible to enzymes that catalyze euchromatic histone modifications and antagonize DNA methylation (Ibarra et al., 2012; Probst et al., 2004; Qian et al., 2012). The balance between exclusion and access is thus essential for the stable propagation of chromatin states.
The influence of the chromatin environment on DNA methylation has important implications for how different classes of TEs are silenced. Short TEs, which are preferentially found near active genes (Ibarra et al., 2012; Zemach et al., 2010a), are generally relatively euchromatic (Figure 2B), and rely on RdDM for silencing (Figure 5B), whereas silencing of the more heterochromatic longer TEs (Figure 2B) that are usually found away from genes (Ibarra et al., 2012; Zemach et al., 2010a) relies primarily on DDM1 (Figure 5B). Despite these differences, DDM1 and RdDM contribute to the methylation and silencing of most TEs, leading to a synergistic loss of methylation (Figures 1A, 4B and 5C) and repression (Figures 5D–E), as well as to enhanced transposition (Figures 5F and S5B), when both pathways are mutated.
Our data suggest that RdDM operates primarily through DRM2, and is thus responsible for a relatively minor fraction of genomic methylation (Figures 1A–B) (Wierzbicki et al., 2012). We show that the majority of CHH methylation is mediated by CMT2 (Figure S1A) independently of RdDM (Figures 1A–C, 4D and Table S2), presumably by binding to H3K9me2 like its CMT3 homolog (Du et al., 2012). CMT2 forms a distinct family in monocots and dicots (Figure 1D), indicating that this enzyme catalyzes CHH methylation throughout flowering plants, including important crops such as rice. Curiously, we have not been able to identify a CMT2 homolog in maize, suggesting that CHH methylation may be entirely dependent on RdDM in this species.
RdDM, by definition, only targets loci that generate sRNA, and therefore affects TEs, which are at least somewhat heterochromatic compared to genes, almost exclusively (Figures 1, 4B and 6D) (Zhong et al., 2012). Production of sRNA can be influenced by many factors, including TE structure and copy number (Martienssen, 2003). Thus, the abundant Gypsy LTR retrotransposons that make up the bulk of pericentric heterochromatin generate sRNA differently from the more dispersed Copia, LINE and MuDR elements (Figure 4A), and RdDM is crucial for Gypsy silencing (Figure 5E) despite the highly heterochromatic nature of these elements. Nonetheless, RdDM is more efficient at less heterochromatic sRNA-producing loci (Figure 2E) (Schoft et al., 2009). This is a curious observation because RdDM, which functions to methylate and silence TEs, might be expected to work well in heterochromatin. Furthermore, why would DDM1, a protein apparently adapted to remodel heterochromatic nucleosomes, not facilitate RdDM?
Our observation that DRD1 is required for RdDM offers a potential explanation. DRD1 is associated with RNA polymerase V, a derivative of RNA polymerase II that shares most pol II subunits (Haag and Pikaard, 2011). Pol II has evolved to transcribe genes, and thus the machinery associated with pol V is likely adapted to function in euchromatin. Unlike DDM1, DRD1 may remodel heterochromatic nucleosomes inefficiently. Because RdDM is a branch of the RNA interference pathway that functions to cleave aberrant and viral mRNA, it likely evolved independently of heterochromatic DDM1-associated pathways. Thus, plants possess two separate mechanisms for methylating and silencing TEs that rely on distinct remodelers with differing nucleosome preferences.
Experimental Procedures
Biological materials
The ddm1-2 (Jeddeloh et al., 1999), met1-6 (Xiao et al., 2003), rdr2-1 (Xie et al., 2004) and ibm1-6 (Coleman-Derr and Zilberman, 2012) mutant lines were described previously. The drd1-7 (GABI_503F06) and h1.2-1 (AT2G30620; GABI_406H11) mutants in the Col-0 ecotype were obtained from the GABI-KAT collection (www.gabi-kat.de). T-DNA insertion lines for drm2–3 (SALK_150863), cmt3–12 (SALK_148381), cmt2–3 (SALK_012874), cmt2–4 (SALK_201637), cmt2–5 (SAIL_906_G03; CS863642), cmt2–6 (SAIL_1236_D12; CS863007) and h1.1-1 (AT1G06760, SALK_128430C) alleles in the Col-0 ecotype were obtained from TAIR (www.arabidopsis.org). T-DNA insertions were confirmed by PCR.
Genomic data acquisition
Bisulfite conversion, Illumina library construction, sequencing and data processing were performed exactly as described in (Ibarra et al., 2012). DNA for the MNase-seq library was prepared essentially as described in (Zilberman et al., 2008). RNA-seq was performed as described in (Zemach et al., 2010b).
Published genomic data
Data for sRNA were derived from (Lister et al., 2008), for ibm1–6 DNA methylation from (Coleman-Derr and Zilberman, 2012), for H3K9me2 from (Bernatavichute et al., 2008) and (Moissiard et al., 2012), for H3K27me3 from (Kim et al., 2012), and for all other histone modifications from (Roudier et al., 2011).
Phylogenetic analysis
Phylogenetic analysis was performed as described in (Zemach et al., 2010b).
Gene and TE DNA methylation and chromatin pattern analyses
Gene and TE meta-analysis was performed as described in (Coleman-Derr and Zilberman, 2012). Genes with CG methylation over 60% were excluded from the analysis because they behave like transposons. Genes with low CG methylation (<20%) were also excluded from methylation analyses because they decrease the dynamic range without substantively contributing to the analysis. To avoid complications in calculating TE size caused by serial TE insertions near the centromeres, only TEs located on the chromosome arms were included in analyses where TEs were filtered by size. GC content was calculated by averaging in 50-bp windows.
Kernel density plots
Density plots in Figure 3A were generated with the difference between h1 and wild-type root methylation in 50-bp windows with at least 10 informative sequenced cytosines and fractional methylation of at least 0.3 for CG and 0.1 for CHG and CHH in h1 or wild-type. Genes with over 60% and under 20% CG methylation were excluded from the analysis.
Expression analysis
RNA-seq datasets were mapped to the TAIR cDNA and TE annotations and analyzed using the Bioconductor package edgeR (Robinson et al., 2010).
Accession Numbers
Sequencing data have been deposited in Gene Expression Omnibus under accession number GSE41302.
Supplementary Material
The chromomethylase CMT2 is required for most asymmetric (CHH) methylation
Chromatin features determine the importance of DDM1 and the efficiency of RdDM
Lack of histone H1 suppresses the dependence of DNA methyltransferases on DDM1
DDM1 and RdDM regulate the DNA methylation and expression of genes
Acknowledgements
We thank Toshiro Nishimura for computational support, Minyong Chung for Illumina sequencing and Robert Fisher for critical reading of the manuscript. A.Z. was supported by a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. P-H. H. is supported by a Taiwan Ministry of Education Scholarship. D.Z. is a Young Investigator of the Arnold and Mabel Beckman Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ahmed I, Sarazin A, Bowler C, Colot V, Quesneville H. Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 2011;39:6919–6931. doi: 10.1093/nar/gkr324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascenzi R, Gantt JS. Subnuclear distribution of the entire complement of linker histone variants in Arabidopsis thaliana. Chromosoma. 1999;108:345–355. doi: 10.1007/s004120050386. [DOI] [PubMed] [Google Scholar]
- 3.Barra JL, Rhounim L, Rossignol JL, Faugeron G. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol Cell Biol. 2000;20:61–69. doi: 10.1128/mcb.20.1.61-69.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blevins T, Pontes O, Pikaard CS, Meins F., Jr Heterochromatic siRNAs and DDM1 independently silence aberrant 5S rDNA transcripts in Arabidopsis. PLoS One. 2009;4:e5932. doi: 10.1371/journal.pone.0005932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- 7.Buisine N, Quesneville H, Colot V. Improved detection and annotation of transposable elements in sequenced genomes using multiple reference sequence sets. Genomics. 2008;91:467–475. doi: 10.1016/j.ygeno.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman-Derr D, Zilberman D. Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. Dual Binding of Chromomethylase Domains to H3K9me2-Containing Nucleosomes Directs DNA Methylation in Plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 12.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 13.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 14.Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22:1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci U S A. 2011;108:1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibarra CA, Feng X, Schoft VK, Hsieh TH, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer VR. Nucleosome positioning: bringing order to the eukaryotic genome. Trends Cell Biol. 2012;22:250–256. doi: 10.1016/j.tcb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 19.Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. DNA Methylation Dynamics during Sexual Reproduction in Arabidopsis thaliana. Curr Biol. 2012;22:1825–1830. doi: 10.1016/j.cub.2012.07.061. [DOI] [PubMed] [Google Scholar]
- 20.Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 21.Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 2012;8:e1002512. doi: 10.1371/journal.pgen.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TF, Gurazada SG, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC. RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics. 2012;7:781–795. doi: 10.4161/epi.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 26.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martienssen RA. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat Genet. 2003;35:213–214. doi: 10.1038/ng1252. [DOI] [PubMed] [Google Scholar]
- 28.Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 29.Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pennings S, Meersseman G, Bradbury EM. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci U S A. 1994;91:10275–10279. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghuram N, Carrero G, Th'ng J, Hendzel MJ. Molecular dynamics of histone H1. Biochem Cell Biol. 2009;87:189–206. doi: 10.1139/O08-127. [DOI] [PubMed] [Google Scholar]
- 35.Rea M, Zheng W, Chen M, Braud C, Bhangu D, Rognan TN, Xiao W. Histone H1 affects gene imprinting and DNA methylation in Arabidopsis. Plant J. 2012;71:776–786. doi: 10.1111/j.1365-313X.2012.05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigal M, Kevei Z, Pelissier T, Mathieu O. DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 2012;31:2981–2993. doi: 10.1038/emboj.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson PJ, Rhodes D. Structure of the '30 nm' chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan DP, Owen-Hughes T. Snf2-family proteins: chromatin remodellers for any occasion. Curr Opin Chem Biol. 2011;15:649–656. doi: 10.1016/j.cbpa.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saze H, Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007;26:3641–3652. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saze H, Kakutani T. Differentiation of epigenetic modifications between transposons and genes. Curr Opin Plant Biol. 2011;14:81–87. doi: 10.1016/j.pbi.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Schoft VK, Chumak N, Mosiolek M, Slusarz L, Komnenovic V, Brownfield L, Twell D, Kakutani T, Tamaru H. Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep. 2009;10:1015–1021. doi: 10.1038/embor.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao Y, Xi S, Shan J, Maunakea A, Che A, Briones V, Lee EY, Geiman T, Huang J, Stephens R, et al. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc Natl Acad Sci U S A. 2011;108:5626–5631. doi: 10.1073/pnas.1017000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 46.Thomas JO. Histone H1: location and role. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 47.Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- 48.Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wierzbicki AT, Jerzmanowski A. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics. 2005;169:997–1008. doi: 10.1534/genetics.104.031997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Developmental Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- 51.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You W, Tyczewska A, Spencer M, Daxinger L, Schmid MW, Grossniklaus U, Simon SA, Meyers BC, Matzke AJ, Matzke M. Atypical DNA methylation of genes encoding cysteine-rich peptides in Arabidopsis thaliana. BMC Plant Biol. 2012;12:51. doi: 10.1186/1471-2229-12-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci U S A. 2010a;107:18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010b;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 55.Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–130. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.