Abstract
Preoperative breast pain in women with breast cancer may result from a number of causes. Previous work from our team found that breast pain occurred in 28.2% of women (n=398) who were about to undergo breast cancer surgery. The occurrence of preoperative breast pain was associated with a number of demographic and clinical characteristics, as well as variation in two cytokine genes. Given that ion channels regulate excitability of sensory neurons, we hypothesized that variations in potassium channel genes would be associated with preoperative breast pain in these patients. Therefore, in this study we evaluated for associations between single nucleotide polymorphisms and inferred haplotypes among 10 potassium channel genes and the occurrence of preoperative breast pain in patients scheduled to undergo breast cancer surgery. Multivariable logistic regression analyses were used to identify those genetic variations that were associated with the occurrence of preoperative breast pain while controlling for age and genomic estimates of and self-reported race/ethnicity. Variations in four potassium channel genes: 1) potassium voltage-gated channel, delayed rectifier, subfamily S, member 1 (KCNS1); 2) potassium inwardly-rectifying channel, subfamily J, member 3 (KCNJ3); 3) KCNJ6; and 4) potassium channel, subfamily K, member 9 (KCNK9) were associated with the occurrence of breast pain. Findings from this study warrant replication in an independent sample of women who report breast pain following one or more breast biopsies.
Keywords: breast pain, preoperative pain, potassium channel genes, breast cancer, candidate genes
INTRODUCTION
Breast pain prior to breast cancer surgery was noted by surgeons several decades ago (Corry, 1952; Lane-Claypon, 1926). Prevalence estimates for preoperative breast pain range from 14% to 53% (Corry, 1952; Poleshuck et al., 2006; Tasmuth, von Smitten, & Kalso, 1996). Before the advent of screening mammography and diagnostic biopsies, this localized pain was considered a somewhat reliable indicator of malignant disease (Corry, 1952). This preoperative pain was described as aching or stabbing (Corry, 1952) and reported to interfere with daily activities (Tasmuth, et al., 1996). Aside from these observations, very little information is available on the characteristics of and mechanisms that underlie this clinical condition.
In an attempt to address this gap, our group recently reported on the occurrence of preoperative breast pain as well as its severity, qualities, and impact on function (McCann et al., 2012). Consistent with published reports, 28% of women reported pain in the breast prior to surgery. Average and worst pain scores were 2.2 and 3.6, respectively, using a 0 to 10 numeric rating scale (NRS). This pain significantly interfered with activities of daily living an average of 6.2 hours per day for approximately 3 days a week. Using the Pain Qualities Assessment Scale (PQAS) (Jensen et al., 2006), the qualities with the highest ratings were tender, dull, and aching. In addition, preoperative breast pain interfered with patients’ sleep and mood. Compared to women without preoperative breast pain, women with pain were younger; more likely to be non-white; less likely to be post-menopausal; had lower functional status scores; and more breast biopsies in the past year (McCann et al., 2012). In addition, preoperative breast pain was associated with higher depressive symptom scores and poorer physical well-being (Kyranou et al., 2012). Moreover, women who reported preoperative breast pain were significantly more likely to report persistent pain for six months following breast cancer surgery (Miaskowski et al., 2012b).
We hypothesized that this preoperative breast pain would have an inflammatory component. Consistent with this hypothesis, the rare allele of a single nucleotide polymorphism (SNP) in interleukin (IL) receptor 2 (IL1R2; rs2110726) was associated with decreased risk for preoperative pain and the rare allele of a SNP in IL13 (rs1295686) was associated with increased risk for preoperative pain (McCann et al., 2012). However, given that pain is a complex trait, other genetic factors may contribute to the variability in the occurrence of preoperative breast pain.
This preoperative breast pain may be due to altered neuronal excitability. Potassium channels, the most ubiquitous type of ion channel (Miller, 2000), are distributed centrally and peripherally, and play a key role in the maintenance of resting membrane potential, the regulation of neuronal excitability (Dodson & Forsythe, 2004; Wickenden, 2002), and the transmission of nociceptive information to the central nervous system (Xie, 2007). Variations in a number of potassium channel genes are associated with thermal hyperalgesia (Alloui et al., 2006; Marker, Stoffel, & Wickman, 2004) and inflammatory pain (Marsh et al., 2012) in rodents, analgesic responses in mice and humans (Blednov et al., 2003; Marker et al., 2004; Nishizawa et al., 2009), and a number of chronic pain conditions in humans (Costigan et al., 2010).
Given their involvement in pain and analgesia, we hypothesized that variations in potassium channel genes would be associated with the occurrence of preoperative breast pain in women prior to breast cancer surgery. Specifically, we evaluated for associations between variations in 10 potassium channel genes and the occurrence of preoperative breast pain in a sample of patients scheduled to undergo breast cancer surgery. These candidate genes encode for three classes of potassium channels (KCN): voltage-gated potassium channels (i.e., KCNA1, KCND2, KCNS1), inward-rectifying potassium channels (i.e., KCNJ3, KCNJ5, KCNJ6, KCNJ9), and two-pore domain potassium channels (i.e., KCNK2, KCNK3, KCNK9).
METHODS
Patients and Settings
This analysis is part of a larger study of women undergoing breast cancer surgery (McCann et al., 2012; Miaskowski et al., 2012b; Miaskowski et al., 2013). Patients were recruited from seven breast care centers in the San Francisco Bay Area.
Eligible patients were adult women (>18 years) scheduled to undergo breast cancer surgery on one breast; were able to read, write, and understand English; and gave written informed consent. Exclusion criteria included having breast cancer surgery on both breasts and/or distant metastasis at the time of diagnosis. Of the 516 patients who were approached to participate, 410 were enrolled (response rate 79.4%), and 398 completed the baseline assessment.
Instruments
A demographic questionnaire obtained information on age, marital status, education, ethnicity, employment status, and living arrangements. The Karnofsky Performance Status (KPS) scale was used to evaluate functional status (Karnofsky et al., 1948). For this study, the KPS scale ranged from 30 (I feel severely disabled and need to be hospitalized) to 100 (I feel normal; I have no complaints or symptoms). The KPS scale has well established validity and reliability (Karnofsky, 1977).
The Self-Administered Comorbidity Questionnaire (SCQ) was used to measure the occurrence, severity, and functional limitations of 13 common medical conditions (Sangha et al., 2003). The SCQ has well-established validity and reliability and has been used in studies of patients with a variety of chronic conditions (Brunner et al., 2008; Sangha et al., 2003).
At the time of enrollment, patients were asked whether they currently had pain in their affected breast (yes/no). Responses to this question were used to dichotomize the sample into patients with (n=110) and without (n=280) breast pain prior to surgery.
Study Procedures
The study was approved by the Committee on Human Research at the University of California, San Francisco and by the Institutional Review Boards at each of the study sites. During the patient’s preoperative visit, a clinician explained the study and determined the patient’s willingness to participate. Women who were willing to participate met with the research nurse who determined eligibility and obtained written informed consent prior to surgery. Patients completed the enrollment questionnaires on average four days prior to surgery. Medical records were reviewed for disease and treatment information.
Genomic Analyses
Gene selection
Candidate genes were selected based on evidence in the literature of an association between the gene and various pain outcomes (e.g., pain severity). In addition to a literature search of potassium channel genes and pain in humans, the Pain Genes Database (Lacroix-Fralish, Ledoux, & Mogil, 2007) was used to identify potassium channel genes. In total, 10 potassium channel genes were selected. Three of the selected genes encode for voltage-gated potassium channels (i.e., KCNA1, KCND2, KCNS1); four encode for inward-rectifying potassium channels (i.e., KCNJ3, KCNJ5, KCNJ6, KCNJ9); and three encode for two-pore domain potassium channels (i.e., KCNK2, KCNK3, KCNK9).
Blood collection and genotyping
Of the 398 patients who completed the baseline questionnaires, 302 provided a blood sample. Genomic DNA was extracted from PBMCs using the PUREGene DNA Isolation System (Invitrogen, Carlsbad, CA). Samples were genotyped using the Golden Gate genotyping platform (Illumina, San Diego, CA) and processed using GenomeStudio (Illumina, San Diego, CA). Genotyping was performed blinded to pain group status and positive and negative controls were included.
SNP Selection
A combination of tag-SNPs and literature driven SNPs were selected for analysis. Tag-SNPs were common (minor allele frequency ≥0.05) in public databases. SNPs with call rates <95% or Hardy-Weinberg p<.001 were excluded. As shown in Table 1, a total of 155 SNPs among the 10 candidate genes (KCNA1: 1 SNP; KCND2: 9 SNPs; KCNS1: 4 SNPs; KCNJ3: 28 SNPs; KCNJ5: 8 SNPs; KCNJ6: 58 SNPs; KCNJ9: 2 SNPs; KCNK2: 22 SNPs, KCNK3: 6 SNPs; KCNK9: 17 SNPs) passed all quality control filters and were included in the genetic association analyses. Potential functional roles for SNPs associated with preoperative breast pain were examined using PUPASuite 3.1 (Conde et al., 2006).
Table 1.
Summary of Potassium Channel Gene Single Nucleotide Polymorphisms (SNPs) and Haplotypes Analyzed for Pain Versus No Pain in Women Prior to Breast Cancer Surgery
| Gene | SNP | Position | Chr | MAF | Alleles | Chi Square |
p-value | Model |
|---|---|---|---|---|---|---|---|---|
| VOLTAGE-GATED POTASSIUM CHANNELS | ||||||||
| KCNA1 | rs4766311 | 4892699 | 1 | 0.466 | C>T | 3.16 | 0.206 | A |
| KCND2 | rs17376373 | 119787721 | 7 | 0.197 | T>G | 4.36 | 0.113 | A |
| KCND2 | rs702414 | 119924204 | 7 | 0.249 | G>C | 3.52 | 0.172 | A |
| KCND2 | rs802340 | 119975021 | 7 | 0.293 | G>T | 1.87 | 0.393 | A |
| KCND2 | rs12706292 | 120012310 | 7 | 0.346 | A>G | 0.08 | 0.963 | A |
| KCND2 | rs4730967 | 120060462 | 7 | 0.320 | T>C | 4.52 | 0.104 | A |
| KCND2 | rs1072198 | 120114585 | 7 | 0.304 | A>G | 0.63 | 0.730 | A |
| KCND2 | rs11489533 | 120117902 | 7 | 0.268 | A>G | 0.70 | 0.704 | A |
| KCND2 | rs4727914 | 120122574 | 7 | 0.343 | A>G | 4.40 | 0.111 | A |
| KCND2 | rs12673992 | 120160059 | 7 | 0.319 | A>G | 0.01 | 0.996 | A |
| KCND2 | HapA1 | 0.77 | 0.682 | |||||
| KCND2 | HapA3 | 4.40 | 0.111 | |||||
| KCNS1 | rs4499491 | 43154833 | 20 | 0.432 | C>A | FE | 0.001 | R |
| KCNS1 | rs6124684 | 43154907 | 20 | 0.223 | C>T | FE | 0.025 | D |
| KCNS1 | rs734784 | 43157041 | 20 | 0.447 | A>G | 4.31 | 0.116 | A |
| KCNS1 | rs6073643 | 43161484 | 20 | 0.274 | T>C | 1.31 | 0.520 | A |
| KCNS1 | HapA1 | 12.71 | 0.002 | |||||
| KCNS1 | HapA2 | 3.96 | 0.138 | |||||
| KCNS1 | HapA3 | 5.36 | 0.069 | |||||
| KCNS1 | HapB1 | 4.31 | 0.116 | |||||
| KCNS1 | HapB2 | 5.43 | 0.066 | |||||
| KCNS1 | HapB3 | 1.38 | 0.501 | |||||
| INWARD-RECTIFYING POTASSIUM CHANNELS | ||||||||
| KCNJ3 | rs6435329 | 155265893 | 2 | 0.445 | G>T | FE | 0.014 | D |
| KCNJ3 | rs3111020 | 155275635 | 2 | 0.450 | T>C | FE | 0.025 | D |
| KCNJ3 | rs11895478 | 155279369 | 2 | 0.246 | C>T | FE | 0.027 | D |
| KCNJ3 | rs3106653 | 155283806 | 2 | 0.262 | A>C | FE | 0.014 | D |
| KCNJ3 | rs3111003 | 155300413 | 2 | 0.465 | C>T | 2.45 | 0.294 | A |
| KCNJ3 | rs3111006 | 155302345 | 2 | 0.375 | C>T | 4.59 | 0.101 | A |
| KCNJ3 | rs12471193 | 155304383 | 2 | 0.343 | A>G | 4.83 | 0.089 | A |
| KCNJ3 | rs6711727 | 155304684 | 2 | 0.485 | G>A | 1.29 | 0.524 | A |
| KCNJ3 | rs2652443 | 155313983 | 2 | 0.395 | G>A | 5.49 | 0.064 | A |
| KCNJ3 | rs7574878 | 155315394 | 2 | 0.429 | T>G | FE | <.0001 | D |
| KCNJ3 | rs2121085 | 155315711 | 2 | 0.447 | A>G | 9.04 | 0.011 | A |
| KCNJ3 | rs2121089 | 155317633 | 2 | 0.479 | C>A | FE | 0.041 | D |
| KCNJ3 | rs2961959 | 155326068 | 2 | 0.432 | C>G | FE | 0.045 | R |
| KCNJ3 | rs2591168 | 155326179 | 2 | 0.316 | A>G | 2.37 | 0.306 | A |
| KCNJ3 | rs2591172 | 155330423 | 2 | 0.333 | T>G | 9.29 | 0.010 | A |
| KCNJ3 | rs12995382 | 155340539 | 2 | 0.290 | T>C | 1.40 | 0.498 | A |
| KCNJ3 | rs13398937 | 155348593 | 2 | 0.362 | C>G | 2.16 | 0.339 | A |
| KCNJ3 | rs13390038 | 155351011 | 2 | 0.403 | G>A | 2.96 | 0.228 | A |
| KCNJ3 | rs12616121 | 155353928 | 2 | 0.469 | A>G | 2.00 | 0.367 | A |
| KCNJ3 | rs2591158 | 155355912 | 2 | 0.280 | A>C | 1.42 | 0.491 | A |
| KCNJ3 | rs2591157 | 155356612 | 2 | 0.330 | A>G | 1.25 | 0.536 | A |
| KCNJ3 | rs717175 | 155356841 | 2 | 0.332 | C>T | 5.27 | 0.072 | A |
| KCNJ3 | rs1037091 | 155360603 | 2 | 0.375 | G>A | 2.63 | 0.269 | A |
| KCNJ3 | rs17641121 | 155373998 | 2 | 0.259 | T>C | 0.39 | 0.824 | A |
| KCNJ3 | rs2591173 | 155395322 | 2 | 0.477 | C>A | 2.53 | 0.283 | A |
| KCNJ3 | rs2971902 | 155400624 | 2 | 0.220 | G>T | 0.87 | 0.649 | A |
| KCNJ3 | rs2937600 | 155411014 | 2 | 0.299 | A>G | 0.34 | 0.843 | A |
| KCNJ3 | rs4467223 | 155414657 | 2 | 0.479 | T>A | 0.28 | 0.871 | A |
| KCNJ3 | HapA1 | 0.20 | 0.907 | |||||
| KCNJ3 | HapA2 | 5.64 | 0.060 | |||||
| KCNJ3 | HapA3 | 6.08 | 0.048 | |||||
| KCNJ3 | HapB1 | 6.43 | 0.040 | |||||
| KCNJ3 | HapB4 | 5.75 | 0.056 | |||||
| KCNJ3 | HapC3 | 5.46 | 0.065 | |||||
| KCNJ3 | HapC5 | 5.87 | 0.053 | |||||
| KCNJ3 | HapD1 | 5.95 | 0.051 | |||||
| KCNJ3 | HapD4 | 5.69 | 0.058 | |||||
| KCNJ3 | HapE1 | 10.55 | 0.005 | |||||
| KCNJ3 | HapE2 | 3.69 | 0.158 | |||||
| KCNJ3 | HapE4 | 2.06 | 0.358 | |||||
| KCNJ3 | HapF1 | 1.49 | 0.475 | |||||
| KCNJ3 | HapF2 | 0.31 | 0.858 | |||||
| KCNJ3 | HapF4 | 1.35 | 0.509 | |||||
| KCNJ3 | HapG1 | 0.89 | 0.642 | |||||
| KCNJ3 | HapG3 | 0.87 | 0.649 | |||||
| KCNJ3 | HapG4 | 2.46 | 0.293 | |||||
| KCNJ5 | rs7941582 | 128266885 | 11 | 0.408 | A>G | 1.87 | 0.393 | A |
| KCNJ5 | rs2846700 | 128274148 | 11 | 0.172 | A>G | 1.51 | 0.469 | A |
| KCNJ5 | rs4937384 | 128285012 | 11 | 0.223 | T>C | 2.47 | 0.290 | A |
| KCNJ5 | rs11221503 | 128277662 | 11 | 0.184 | C>T | 0.45 | 0.800 | A |
| KCNJ5 | rs2604212 | 128278165 | 11 | 0.459 | C>G | 0.17 | 0.920 | A |
| KCNJ5 | rs4937387 | 128278623 | 11 | 0.257 | T>C | 0.29 | 0.865 | A |
| KCNJ5 | rs11221510 | 128285907 | 11 | 0.241 | A>T | 0.27 | 0.876 | A |
| KCNJ5 | rs6590357 | 128286549 | 11 | 0.163 | C>T | 0.03 | 0.987 | A |
| KCNJ5 | HapA1 | 0.38 | 0.826 | |||||
| KCNJ5 | HapA2 | 0.17 | 0.920 | |||||
| KCNJ5 | HapA5 | 0.33 | 0.846 | |||||
| KCNJ6 | rs860795 | 37937160 | 21 | 0.208 | G>C | 0.25 | 0.884 | A |
| KCNJ6 | rs1709838 | 37941983 | 21 | 0.431 | C>A | 0.68 | 0.710 | A |
| KCNJ6 | rs10483038 | 37946641 | 21 | 0.279 | T>C | 0.36 | 0.835 | A |
| KCNJ6 | rs857967 | 37954006 | 21 | 0.197 | T>A | 3.41 | 0.182 | A |
| KCNJ6 | rs2835885 | 37961436 | 21 | 0.432 | T>G | 0.67 | 0.714 | A |
| KCNJ6 | rs858010 | 37987109 | 21 | 0.166 | G>A | 0.03 | 0.983 | A |
| KCNJ6 | rs1005546 | 37990742 | 21 | 0.450 | C>T | 0.51 | 0.777 | A |
| KCNJ6 | rs858003 | 37994854 | 21 | 0.197 | C>T | 1.76 | 0.416 | A |
| KCNJ6 | rs1709816 | 37999129 | 21 | 0.390 | G>T | 1.20 | 0.548 | A |
| KCNJ6 | rs13049947 | 38002710 | 21 | 0.403 | C>T | 0.26 | 0.877 | A |
| KCNJ6 | rs2835914 | 38020720 | 21 | 0.347 | G>C | FE | 0.027 | D |
| KCNJ6 | rs858035 | 38021061 | 21 | 0.344 | T>C | FE | 0.010 | D |
| KCNJ6 | rs13048511 | 38037731 | 21 | 0.468 | A>G | 1.82 | 0.403 | A |
| KCNJ6 | rs2835925 | 38041173 | 21 | 0.176 | A>G | 1.40 | 0.497 | A |
| KCNJ6 | rs857989 | 38042001 | 21 | 0.115 | G>C | 0.20 | 0.906 | A |
| KCNJ6 | rs2835931 | 38043518 | 21 | 0.282 | C>T | 1.51 | 0.469 | A |
| KCNJ6 | rs1399596 | 38045382 | 21 | 0.260 | T>C | 2.44 | 0.295 | A |
| KCNJ6 | rs2835942 | 38052778 | 21 | 0.303 | C>T | 3.44 | 0.179 | A |
| KCNJ6 | rs2835945 | 38057170 | 21 | 0.398 | G>A | 2.43 | 0.297 | A |
| KCNJ6 | rs1160350 | 38065897 | 21 | 0.494 | G>C | 4.31 | 0.116 | A |
| KCNJ6 | rs762145 | 38068188 | 21 | 0.366 | C>T | 1.14 | 0.566 | A |
| KCNJ6 | rs2226356 | 38075902 | 21 | 0.427 | C>T | 1.25 | 0.535 | A |
| KCNJ6 | rs1787337 | 38077824 | 21 | 0.494 | A>G | 2.59 | 0.274 | A |
| KCNJ6 | rs2835961 | 38083028 | 21 | 0.482 | G>A | 2.62 | 0.269 | A |
| KCNJ6 | rs2835976 | 38103779 | 21 | 0.385 | C>T | 0.47 | 0.790 | A |
| KCNJ6 | rs2835977 | 38104067 | 21 | 0.224 | G>A | 0.07 | 0.967 | A |
| KCNJ6 | rs2211842 | 38105403 | 21 | 0.376 | C>A | 0.33 | 0.846 | A |
| KCNJ6 | rs2211843 | 38106055 | 21 | 0.234 | G>T | 1.78 | 0.410 | A |
| KCNJ6 | rs2211845 | 38106371 | 21 | 0.447 | T>C | 1.20 | 0.549 | A |
| KCNJ6 | rs2835982 | 38110247 | 21 | 0.368 | C>A | 0.84 | 0.658 | A |
| KCNJ6 | rs2835983 | 38110476 | 21 | 0.304 | G>A | 1.30 | 0.521 | A |
| KCNJ6 | rs2835984 | 38110657 | 21 | 0.497 | A>T | 0.40 | 0.818 | A |
| KCNJ6 | rs3787835 | 38111440 | 21 | 0.455 | C>T | 1.28 | 0.527 | A |
| KCNJ6 | rs6517435 | 38117092 | 21 | 0.422 | G>A | 0.30 | 0.859 | A |
| KCNJ6 | rs2154556 | 38120757 | 21 | 0.344 | T>C | 1.55 | 0.462 | A |
| KCNJ6 | rs4817896 | 38123831 | 21 | 0.248 | C>T | 0.23 | 0.892 | A |
| KCNJ6 | rs3787840 | 38124263 | 21 | 0.139 | C>T | 1.17 | 0.558 | A |
| KCNJ6 | rs991985 | 38128024 | 21 | 0.286 | C>A | 0.04 | 0.979 | A |
| KCNJ6 | rs2836007 | 38128761 | 21 | 0.194 | C>T | 0.11 | 0.947 | A |
| KCNJ6 | rs2836013 | 38132582 | 21 | 0.292 | C>T | 0.39 | 0.823 | A |
| KCNJ6 | rs2836016 | 38134890 | 21 | 0.411 | A>G | 0.49 | 0.785 | A |
| KCNJ6 | rs2836019 | 38136864 | 21 | 0.327 | C>T | 2.04 | 0.360 | A |
| KCNJ6 | rs915800 | 38138203 | 21 | 0.455 | C>T | 1.18 | 0.554 | A |
| KCNJ6 | rs2226741 | 38146803 | 21 | 0.147 | A>G | 1.40 | 0.496 | A |
| KCNJ6 | rs7276928 | 38147607 | 21 | 0.288 | G>A | 2.00 | 0.368 | A |
| KCNJ6 | rs3827199 | 38149472 | 21 | 0.408 | G>A | 3.27 | 0.195 | A |
| KCNJ6 | rs4816585 | 38151120 | 21 | 0.495 | G>A | 1.26 | 0.533 | A |
| KCNJ6 | rs9305628 | 38166861 | 21 | 0.227 | A>G | FE | 0.036 | D |
| KCNJ6 | rs9974219 | 38168568 | 21 | 0.277 | A>T | 4.10 | 0.129 | A |
| KCNJ6 | rs7277957 | 38168770 | 21 | 0.492 | A>G | 1.50 | 0.473 | A |
| KCNJ6 | rs1892682 | 38169935 | 21 | 0.265 | G>A | 0.61 | 0.738 | A |
| KCNJ6 | rs928765 | 38173472 | 21 | 0.292 | C>T | 1.77 | 0.413 | A |
| KCNJ6 | rs3787862 | 38174571 | 21 | 0.197 | G>A | 0.74 | 0.692 | A |
| KCNJ6 | rs10775660 | 38175388 | 21 | 0.415 | C>T | 0.90 | 0.637 | A |
| KCNJ6 | rs8129919 | 38176410 | 21 | 0.471 | G>A | 10.69 | 0.005 | A |
| KCNJ6 | rs2836039 | 38188930 | 21 | 0.195 | G>A | n/a | n/a | n/a |
| KCNJ6 | rs2836048 | 38206168 | 21 | 0.321 | G>A | 3.04 | 0.218 | A |
| KCNJ6 | rs2836050 | 38206705 | 21 | 0.227 | C>T | FE | 0.044 | R |
| KCNJ6 | rs3787870 | 38207323 | 21 | 0.463 | A>G | 0.07 | 0.966 | A |
| KCNJ6 | HapA1 | 0.25 | 0.882 | |||||
| KCNJ6 | HapA2 | 0.58 | 0.749 | |||||
| KCNJ6 | HapA3 | 0.22 | 0.895 | |||||
| KCNJ6 | HapB1 | 0.55 | 0.761 | |||||
| KCNJ6 | HapB2 | 3.41 | 0.182 | |||||
| KCNJ6 | HapB3 | 0.36 | 0.835 | |||||
| KCNJ6 | HapC1 | 0.51 | 0.777 | |||||
| KCNJ6 | HapC2 | 0.21 | 0.902 | |||||
| KCNJ6 | HapC3 | 0.03 | 0.983 | |||||
| KCNJ6 | HapD1 | 0.23 | 0.892 | |||||
| KCNJ6 | HapD4 | 1.27 | 0.529 | |||||
| KCNJ6 | HapD6 | 1.41 | 0.493 | |||||
| KCNJ6 | HapE1 | 2.47 | 0.290 | |||||
| KCNJ6 | HapE2 | 3.42 | 0.181 | |||||
| KCNJ6 | HapE5 | 1.77 | 0.413 | |||||
| KCNJ6 | HapE7 | 1.40 | 0.496 | |||||
| KCNJ6 | HapF1 | 2.77 | 0.251 | |||||
| KCNJ6 | HapF2 | 0.01 | 0.993 | |||||
| KCNJ6 | HapF4 | 3.04 | 0.219 | |||||
| KCNJ6 | HapG1 | 1.64 | 0.441 | |||||
| KCNJ6 | HapG5 | 0.98 | 0.613 | |||||
| KCNJ6 | HapG6 | 0.08 | 0.962 | |||||
| KCNJ6 | HapH1 | 0.68 | 0.713 | |||||
| KCNJ6 | HapH3 | 1.18 | 0.556 | |||||
| KCNJ6 | HapH5 | 1.74 | 0.418 | |||||
| KCNJ6 | HapI1 | 0.14 | 0.932 | |||||
| KCNJ6 | HapI5 | 0.28 | 0.869 | |||||
| KCNJ6 | HapJ1 | 1.01 | 0.605 | |||||
| KCNJ6 | HapJ2 | 0.39 | 0.823 | |||||
| KCNJ6 | HapJ3 | 0.09 | 0.955 | |||||
| KCNJ6 | HapK1 | 1.18 | 0.554 | |||||
| KCNJ6 | HapK4 | 2.05 | 0.358 | |||||
| KCNJ6 | HapL1 | 1.80 | 0.407 | |||||
| KCNJ6 | HapL4 | 3.15 | 0.207 | |||||
| KCNJ6 | HapL5 | 5.77 | 0.056 | |||||
| KCNJ6 | HapM1 | 0.71 | 0.702 | |||||
| KCNJ6 | HapM4 | 1.59 | 0.451 | |||||
| KCNJ6 | HapM6 | 1.73 | 0.421 | |||||
| KCNJ6 | HapN2 | 3.44 | 0.179 | |||||
| KCNJ6 | HapN3 | 4.66 | 0.097 | |||||
| KCNJ9 | rs6677510 | 158318743 | 1 | 0.442 | A>G | 0.93 | 0.629 | A |
| KCNJ9 | rs2753268 | 158324876 | 1 | 0.260 | C>T | 3.82 | 0.148 | A |
| TWO-PORE POTASSIUM CHANNELS | ||||||||
| KCNK2 | rs2601640 | 213253979 | 1 | 0.492 | A>G | 0.92 | 0.630 | A |
| KCNK2 | rs12141327 | 213273537 | 1 | 0.335 | G>A | 0.44 | 0.802 | A |
| KCNK2 | rs1452619 | 213280153 | 1 | 0.120 | A>G | 0.64 | 0.725 | A |
| KCNK2 | rs10494991 | 213287222 | 1 | 0.331 | T>C | 1.57 | 0.456 | A |
| KCNK2 | rs1584759 | 213289445 | 1 | 0.453 | A>C | 0.75 | 0.687 | A |
| KCNK2 | rs12064317 | 213293664 | 1 | 0.136 | G>T | 0.26 | 0.880 | A |
| KCNK2 | rs6665177 | 213298091 | 1 | 0.155 | G>A | 0.14 | 0.933 | A |
| KCNK2 | rs12028008 | 213298169 | 1 | 0.497 | A>G | 0.34 | 0.842 | A |
| KCNK2 | rs12038094 | 213302819 | 1 | 0.291 | C>T | 1.96 | 0.375 | A |
| KCNK2 | rs17024179 | 213304166 | 1 | 0.163 | T>C | 2.06 | 0.358 | A |
| KCNK2 | rs7528988 | 213315040 | 1 | 0.259 | C>T | 3.01 | 0.222 | A |
| KCNK2 | rs2363561 | 213321930 | 1 | 0.395 | C>T | 0.41 | 0.815 | A |
| KCNK2 | rs12133857 | 213331109 | 1 | 0.128 | G>T | 0.34 | 0.843 | A |
| KCNK2 | rs4411107 | 213355542 | 1 | 0.375 | T>C | 1.08 | 0.584 | A |
| KCNK2 | rs4303048 | 213385781 | 1 | 0.236 | G>A | 2.10 | 0.349 | A |
| KCNK2 | rs12757222 | 213391641 | 1 | 0.233 | A>G | 2.81 | 0.246 | A |
| KCNK2 | rs1556905 | 213428215 | 1 | 0.411 | C>A | 0.21 | 0.899 | A |
| KCNK2 | rs10494994 | 213428830 | 1 | 0.207 | G>A | 0.53 | 0.767 | A |
| KCNK2 | rs12038695 | 213444580 | 1 | 0.494 | A>C | 1.16 | 0.559 | A |
| KCNK2 | rs2027320 | 213446566 | 1 | 0.385 | G>A | 0.37 | 0.833 | A |
| KCNK2 | rs12143625 | 213458463 | 1 | 0.235 | T>C | 0.61 | 0.739 | A |
| KCNK2 | rs12080135 | 213463166 | 1 | 0.252 | T>G | 0.20 | 0.905 | A |
| KCNK2 | HapA1 | 0.44 | 0.802 | |||||
| KCNK2 | HapA4 | 0.92 | 0.630 | |||||
| KCNK2 | HapB1 | 1.45 | 0.485 | |||||
| KCNK2 | HapB4 | 0.97 | 0.615 | |||||
| KCNK2 | HapC1 | 0.45 | 0.799 | |||||
| KCNK2 | HapC4 | 1.59 | 0.451 | |||||
| KCNK2 | HapC5 | 0.34 | 0.842 | |||||
| KCNK2 | HapD1 | 0.01 | 0.994 | |||||
| KCNK2 | HapD3 | 0.41 | 0.815 | |||||
| KCNK2 | HapE1 | 0.14 | 0.935 | |||||
| KCNK2 | HapE3 | 1.18 | 0.554 | |||||
| KCNK2 | HapE4 | 0.62 | 0.733 | |||||
| KCNK2 | HapF2 | 0.37 | 0.833 | |||||
| KCNK2 | HapF3 | 1.16 | 0.559 | |||||
| KCNK3 | rs1275982 | 26772593 | 2 | 0.497 | C>T | 4.75 | 0.093 | A |
| KCNK3 | rs1275977 | 26776359 | 2 | 0.414 | A>G | 4.79 | 0.091 | A |
| KCNK3 | rs11126666 | 26782315 | 2 | 0.330 | G>A | 4.00 | 0.135 | A |
| KCNK3 | rs1662987 | 26791686 | 2 | 0.243 | A>G | 4.80 | 0.091 | A |
| KCNK3 | rs1662988 | 26793738 | 2 | 0.290 | C>T | 0.72 | 0.699 | A |
| KCNK3 | rs7584568 | 26798797 | 2 | 0.471 | G>A | 1.80 | 0.407 | A |
| KCNK3 | HapA1 | 4.00 | 0.135 | |||||
| KCNK3 | HapA4 | 4.75 | 0.093 | |||||
| KCNK3 | HapB1 | 1.51 | 0.470 | |||||
| KCNK3 | HapB2 | 4.22 | 0.121 | |||||
| KCNK3 | HapB4 | 0.50 | 0.780 | |||||
| KCNK9 | rs2542424 | 140701683 | 8 | 0.362 | A>G | 4.75 | 0.093 | A |
| KCNK9 | rs2542422 | 140706306 | 8 | 0.328 | C>A | 0.08 | 0.961 | A |
| KCNK9 | rs2014712 | 140709816 | 8 | 0.235 | C>T | 1.01 | 0.604 | A |
| KCNK9 | rs2545462 | 140714686 | 8 | 0.343 | C>A | 0.24 | 0.887 | A |
| KCNK9 | rs2542420 | 140714883 | 8 | 0.419 | C>G | 0.21 | 0.902 | A |
| KCNK9 | rs2545461 | 140717431 | 8 | 0.257 | A>G | n/a | n/a | n/a |
| KCNK9 | rs3780051 | 140727983 | 8 | 0.471 | A>G | 0.12 | 0.943 | A |
| KCNK9 | rs2545457 | 140730467 | 8 | 0.350 | T>C | 1.43 | 0.489 | A |
| KCNK9 | rs2005895 | 140738217 | 8 | 0.256 | T>C | 0.34 | 0.842 | A |
| KCNK9 | rs888349 | 140738927 | 8 | 0.197 | A>C | 0.45 | 0.801 | A |
| KCNK9 | rs759656 | 140739149 | 8 | 0.320 | T>C | n/a | n/a | n/a |
| KCNK9 | rs13277242 | 140739269 | 8 | 0.495 | G>A | 3.84 | 0.147 | A |
| KCNK9 | rs885724 | 140740112 | 8 | 0.380 | A>C | FE | 0.048 | D |
| KCNK9 | rs3780039 | 140745846 | 8 | 0.372 | T>G | FE | 0.003 | D |
| KCNK9 | rs10110946 | 140754803 | 8 | 0.333 | T>C | 1.08 | 0.583 | A |
| KCNK9 | rs7828107 | 140756023 | 8 | 0.409 | C>A | 4.05 | 0.132 | A |
| KCNK9 | rs983740 | 140762922 | 8 | 0.472 | T>G | 3.72 | 0.156 | A |
| KCNK9 | rs11166921 | 140776937 | 8 | 0.395 | C>A | FE | 0.020 | R |
| KCNK9 | rs13278664 | 140779544 | 8 | 0.455 | A>G | 3.52 | 0.173 | A |
| KCNK9 | HapA1 | 0.16 | 0.925 | |||||
| KCNK9 | HapA2 | 0.13 | 0.938 | |||||
| KCNK9 | HapA3 | 4.28 | 0.118 | |||||
| KCNK9 | HapB1 | 0.09 | 0.957 | |||||
| KCNK9 | HapB4 | 0.13 | 0.939 | |||||
| KCNK9 | HapC1 | 1.13 | 0.568 | |||||
| KCNK9 | HapC3 | 1.54 | 0.464 | |||||
| KCNK9 | HapC4 | 3.85 | 0.146 | |||||
| KCNK9 | HapD1 | 0.42 | 0.812 | |||||
| KCNK9 | HapD2 | 3.18 | 0.204 | |||||
| KCNK9 | HapD3 | 6.60 | 0.037 | |||||
Abbreviations: A = additive model, Chr = chromosome, D = dominant model, Hap = haplotype, KCNA = voltage-sensitive potassium channel, KCND = voltage-gated potassium channel, KCNJ = potassium inward-rectifying channel, KCNK = potassium channel, subfamily K, MAF = minor allele frequency, n/a = not assayed because SNP violated Hardy-Weinberg expectations (p<.001) or because MAF was <.05, R = recessive model, SNP= single nucleotide polymorphism
Statistical Analyses for the Phenotypic Data
Data were analyzed using SPSS version 19 (SPSS, 2010) and STATA Version 12 (StataCorp, 2005). Independent samples t-tests, Mann-Whitney U tests, and Chi-square analyses were used to evaluate for differences in demographic and clinical characteristics between the pain and no pain groups. All calculations used actual values. Adjustments were not made for missing data.
Statistical Analyses for the Genetic Data
Allele and genotype frequencies were determined by gene counting. Hardy-Weinberg equilibrium was assessed by the Chi-square or Fisher Exact tests. Measures of linkage disequilibrium ((LD); i.e., D’ and r2) were computed from the patients’ genotypes using Haploview 4.2. LD-based haplotype block definition was based on D’ confidence interval (Gabriel et al., 2002).
Haplotype analyses were conducted in order to localize the association signal within each gene and to determine if haplotypes improved the strength of the association with the phenotype. Haplotypes were constructed using the PHASE version 2.1 (Stephens, Smith, & Donnelly, 2001). Only haplotypes that were inferred with probability estimates of >.85, across five iterations, were retained for subsequent analyses.
One hundred and six ancestry informative markers (AIMs) were used to control for population stratification (i.e., race/ethnicity) (Halder et al., 2008; Hoggart et al., 2003; Tian, Gregersen, & Seldin, 2008). Using Helix Tree (Golden Helix, Bozeman, MT), homogeneity in ancestry among patients was verified by principal component (PC) analysis (Price et al., 2006) (data not shown). The first three PCs were selected to adjust for potential confounding due to population substructure by including the three covariates in all regression models.
For association tests, additive, dominant, and recessive genetic models were assessed for each SNP. Barring small improvements from the additive model (i.e., delta <10%), the model that best fit the data (by maximizing the significance of the p-value) was selected for each SNP. Logistic regression analysis that controlled for significant covariates, genomic estimates of and self-reported race/ethnicity, and variation in other SNPs/haplotypes within the same gene, was used to evaluate the association between genotype and pain group membership. A backwards stepwise approach was used to create a parsimonious model. Genetic model fit and covariate-adjusted odds ratios were estimated using STATA version 12.
As done previously (Dunn et al., 2013; Illi et al., 2012; McCann et al., 2012; Miaskowski et al., 2012a), based on recommendations in the literature (Hattersley & McCarthy, 2005; Rothman, 1990) as well as the implementation of rigorous quality controls, the non-independence of genetic markers in LD, and the exploratory nature of the analyses, adjustments were not made for multiple testing. Moreover, significant SNPs identified in the bivariate analyses were evaluated using regression analyses that controlled for differences in phenotypic characteristics, potential confounding due to population stratification, and variation in other SNPs/haplotypes within the same gene. Only those SNPs that remained statistically significant in the multivariable analyses were included in the final presentation of the results. Therefore, the identified significant genetic associations are unlikely to be due solely to chance. Unadjusted associations are reported for all SNPs passing quality control criteria in Table 1 to allow for subsequent comparisons and meta-analyses.
RESULTS
Differences in Demographic and Clinical Characteristics
A detailed description of the differences in demographic and clinical characteristics between our patients with and without preoperative breast pain is available elsewhere (McCann et al., 2012). Table 2 summarizes only those characteristics that differed significantly between the two groups. The characteristics associated with the occurrence of preoperative breast pain were younger age, lower functional status, being non-white, being pre-menopausal, and having more biopsies in the past year.
Table 2.
Significant Differences in Demographic and Clinical Characteristics Between Patients With (n = 110) and Without (n = 280) Preoperative Breast Pain*
| Characteristic | NO PAIN Mean (SD) |
PAIN Mean (SD) |
Statistic and p-value |
|---|---|---|---|
| Age (years) | 56.5 (11.8) | 50.9 (9.8) | t=4.81; p<0.001 |
| Karnofsky Performance Status score | 94.0 (10.3) | 90.9 (10.1) | t=2.66; p= 0.008 |
| Number of biopsies in past year | 1.5 (0.8) | 1.6 (0.8) | U=12887.0, p<0.01 |
| % (N) | % (N) | ||
| Non-white race/ethnicity | 31.9 (89) | 45.0 (49) | FE; p=0.018 |
| Post-menopausal | 67.9 (186) | 53.8 (57) | FE; p=0.012 |
Abbreviations: FE = Fisher’s Exact, SD = standard deviation
Modified from McCann, B., Miaskowski, C., Koetters, T., Baggott, C., West, C., Levine, J. D., et al. (2012). Associations between pro- and anti-inflammatory cytokine genes and breast pain in women prior to breast cancer surgery. J Pain, 13, 425-437.
Regression Analyses for KCNJ3, KCNJ6, KCNK9, and KCNS1 Genotypes and Haplotypes
In order to better estimate the magnitude (i.e., odds ratio, OR) and precision (95% confidence interval [CI]) of genotype on the odds of reporting preoperative breast pain, multivariate logistic regression models were fit. Using a backwards stepwise approach, age was the only phenotypic characteristic listed in Table 2 that remained significant in this initial logistic regression model. Age was included as a covariate in subsequent models that evaluated genotypic predictors. Each 5-year increase in age was associated with a 23% reduction in odds of reporting preoperative breast pain (OR: 0.77; 95% CI: 0.682, 0.878).
After controlling for age and genomic estimates of and self-reported race/ethnicity, and variation in other SNPs/haplotypes within the same gene, eight genetic associations (7 SNPs, 1 haplotype) among four candidate genes were associated with pain group membership: KCNS1 (rs4499491); KCNJ3 (rs7574878) and haplotype E1 (composed of rs2591168 and rs2591172); KCNJ6 (rs2835914, rs8129919, rs2836050); and KCNK9 (rs3780039, rs11166921; see Table 3).
Table 3.
Multiple Logistic Regression Analyses for Single Nucleotide Polymorphisms in KCNS1, KCNJ3, KCNJ6, and KCNK9 and the Occurrence of Preoperative Breast Pain (N=302)
| Pain Group Comparison |
Predictor | Odds Ratio |
Standard Error | 95% CI | Z | p-value |
|---|---|---|---|---|---|---|
| No pain (n=218) versus Pain (n=84) |
KCNS1 rs4499491 | 3.01 | 1.004 | 1.561, 5.785 | 3.29 | 0.001 |
| Age | 0.77 | 0.051 | 0.677, 0.878 | −3.92 | <0.001 | |
| Overall model fit: χ2 = 34.94, p <.0001; R2 = 0.0989 | ||||||
| No pain (n=218) versus Pain (n=84) |
KCNJ3 rs7574878 | 0.52 | 0.165 | 0.276, 0.966 | −2.07 | 0.038 |
| KCNJ3 Hap E1 | 1.69 | 0.408 | 1.049, 2.710 | 2.16 | 0.031 | |
| Age | 0.76 | 0.051 | 0.664, 0.863 | −4.16 | <0.001 | |
| Overall model fit: χ2 = 40.74, p <.0001; R2 = 0.1154 | ||||||
| No pain (n=218) versus Pain (n=84) |
KCNJ6 rs2835914 | 0.48 | 0.140 | 0.274, 0.850 | −2.52 | 0.012 |
| KCNJ6 rs8129919 | 2.10 | 0.459 | 1.367, 3.220 | 3.39 | 0.001 | |
| KCNJ6 rs2836050 | 3.58 | 2.001 | 1.199, 10.708 | 2.29 | 0.022 | |
| Age | 0.74 | 0.052 | 0.648, 0.854 | −4.20 | <0.001 | |
| Overall model fit: χ2 = 46.71, p <.0001; R2 = 0.1322 | ||||||
| No pain (n=218) versus Pain (n=84) |
KCNK9 rs3780039 | 1.90 | 0.594 | 1.027, 3.506 | 2.05 | 0.041 |
| KCNK9 rs11166921 | 2.40 | 0.830 | 1.218, 4.726 | 2.53 | 0.011 | |
| Age | 0.77 | 0.051 | 0.678, 0.878 | −3.92 | <0.001 | |
| Overall model fit: χ2 = 35.59, p <.0001; R2 = 0.1008 | ||||||
Multiple logistic regression analyses of candidate gene associations with no pain versus pain. The first three principal components identified from the analysis of ancestry informative markers as well as self-reported race/ethnicity were retained in all models to adjust for potential confounding due to population substructure (data not shown). Predictors evaluated in each model included genotype (KCNS1 rs4499491: CC+CA (no pain (n=189), pain (n=58)) versus AA (no pain (n=29), pain (n=26)); KCNJ3 rs7574878: TT (no pain (n=61), pain (n=42)) versus TG+GG (no pain (n=157), pain (n=42)); KCNJ3 haplotype E1 composed of rs2591168-rs2591172: zero, one, or two doses of the A-G haplotype; KCNJ6 rs2835914: GG (no pain (n=85), pain (n=45)) versus GC+CC (no pain (n=133), pain (n=39)); KCNJ6 rs8129919: GG (no pain (n=65), pain (n=15)) versus GA (no pain (n=121), pain (n=44)) versus AA (no pain (n=32), pain (n=25)); KCNJ6 rs2836050: CC+CT (no pain (n=207), pain (n=74)) versus TT (no pain (n=11), pain (n=10)); KCNK9 rs3780039: TT (no pain (n=103), pain (n=24)) versus TG+GG (no pain (n=114), pain (n=60)); KCNK9 rs11166921: CC+CA (no pain (n=185), pain (n=61)) versus AA (no pain (n=33), pain (n=23))), and age (in 5 year increments).
Abbreviations: CI =confidence interval; Hap = haplotype; KCNJ3 = potassium inwardly-rectifying channel, subfamily J, member 3; KCNJ6 = potassium inwardly-rectifying channel, subfamily J, member 6; KCNK9 = potassium channel subfamily K, member 9; KCNS1 = potassium voltagegated channel, delayed-rectifier, subfamily S, member 1.
For KCNS1 rs4499491, individuals homozygous for the rare A allele (CC+CA versus AA) had a 3.0-fold increase in the odds of reporting preoperative breast pain.
For KCNJ3 rs7574878, individuals who were heterozygous or homozygous for the rare G allele (TT versus TG+GG) had a 48% reduction in the odds of reporting preoperative breast pain. In addition, each dose of the KCNJ3 haplotype E1 (composed of the common A allele at rs2591168 and the rare G allele at rs2591172; Figure 1) was associated with a 1.7-fold increase in the odds of reporting preoperative breast pain.
Figure 1.
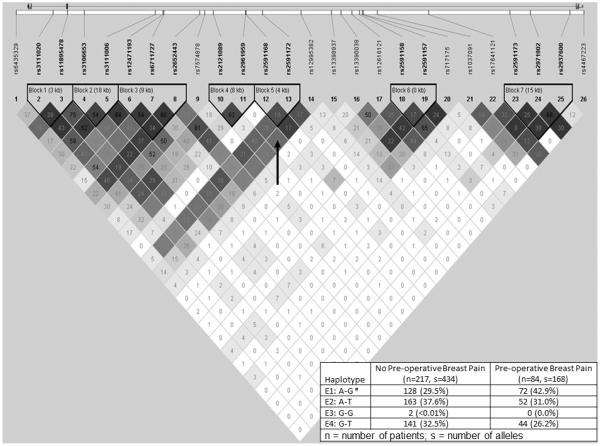
KCNJ3 linkage disequilibrium-based heatmap and haplotype analysis. An ideogram of potassium inwardly-rectifying channel, subfamily J, member 3 (KCNJ3, GIRK1, Kir3.1) is presented above the white bar that represents the physical distance along human (chromosome 2 position 155555093 to 155714864; genome build 37.10, NC_000002.11). Exons are represented as boxes. Gray lines connecting the exons represent introns. The direction of transcription is from left to right. Reference sequence identifiers (rsID) for each single nucleotide polymorphism (SNP) are plotted both in terms of their physical distance (i.e., the white bar at the top of the figure) and equidistantly in order to render the pairwise linkage disequilibrium (LD) estimates that were calculated and visualized with Haploview 4.2. The gene structure for KCNJ3 (i.e., hg18 NM_002239) was rendered with FancyGene 1.4. The correlation statistics (r2 and D’) are provided in the heatmap. LD-based haplotype block definition was based on D’ confidence interval (Conde et al., 2006a). The haploblock is outlined in a bolded triangle and its component SNPs are rendered in bold font. Pairwise D’ values (range: 0-1, inclusive) were rendered in greyscale, with dark grey diamonds representing D’ values approaching 1.0. When the r2 values (range of 0-100, inclusive) are not equal to 0 or 100, they are provided in a given diamond. The haplotypes i.e., HapE1-HapE4) observed in the haploblock 5 (i.e., “Block 5” indicated by the vertical black arrow in the figure) are listed in each row, starting with the nucleotide composition across the two SNPs that compose the haplotype (i.e., rs2591168, rs2591172) and the count frequency (%) of each haplotype observed in the no preoperative breast pain and preoperative breast pain groups.
#The haplotype E1, composed of the “A” common allele at rs2591168 and the “G” rare allele at rs2591172, identified in the bivariate analyses (Table 1) remained significant after controlling for relevant covariates.
For KCNJ6, three SNPs (i.e., rs2835914, rs8129919, rs2836050) were associated with the occurrence of preoperative breast pain. For KCNJ6 rs2835914, individuals who were heterozygous or homozygous for the rare C allele (GG versus GC+CC) had a 52% reduction in the odds of reporting preoperative breast pain. For KCNJ6 rs8129919, each dose of the rare A allele (GG versus GA versus AA) was associated with a 2.1-fold increase in the odds of reporting preoperative breast pain. For KCNJ6 rs2836050, individuals homozygous for the rare T allele (CC+CT versus TT) had a 3.6-fold increase in the odds of reporting preoperative breast pain.
For KCNK9, two SNPs (i.e., rs3780039, rs11166921) were associated with the occurrence of preoperative breast pain. For KCNK9 rs3780039, individuals who were heterozygous or homozygous for the rare G allele (TT versus TG+GG) had a 1.9-fold increase in the odds of reporting preoperative breast pain. For KCNK9 rs11166921, individuals homozygous for the rare A allele (CC+CA versus AA) had a 2.4-fold increase in the odds of reporting preoperative breast pain.
DISCUSSION
This study provides new evidence of associations between four potassium channel genes and the occurrence of breast pain prior to breast cancer surgery. These findings build on our previous work that identified associations between cytokine gene variations and preoperative breast pain (McCann et al., 2012). While our initial phenotypic and genotypic findings suggested that preoperative breast pain has an inflammatory component, the current findings suggest that potassium channel activity also contributes to preoperative breast pain. These findings are not discrepant, because interplay may exist between potassium channels and cytokines that presents an interesting avenue for future research.
Differences in demographic and clinical characteristics between women with and without breast pain prior to breast cancer surgery are discussed in detail elsewhere (McCann et al., 2012). However, it is interesting to note that age was the only phenotypic characteristic that remained significant in the final phenotypic regression model. Each 5-year increase in age was associated with a 23% reduction in the odds of reporting breast pain. This finding is consistent with previous reports of age-related differences in the occurrence of cancer pain (Gibson & Helme, 2001). In addition, it is consistent with work from our research group that found decreases in the occurrence rates for a number of common symptoms (e.g., depressive symptoms, fatigue, sleep disturbance) in older oncology patients (Dunn et al., 2012; Dunn et al., 2013; Illi et al., 2012; Linden et al., 2012). The effect of age on the occurrence of preoperative breast pain warrants additional investigation, as some reviews noted that a number of persistent pain conditions increase with age (Fillingim, 2005; Gibson & Helme, 2001).
Of the three voltage-gated potassium channel genes that were evaluated, only KCNS1, which encodes for the potassium channel, Kv9.1, demonstrated an association with the occurrence of preoperative breast pain. In this sample, patients who were homozygous for the rare “A” allele for KCNS1 rs4499491, located in the 3′ untranslated region of KCNS1, had a 3-fold increase in the odds of reporting preoperative breast pain. While no functional data are reported for this SNP, it is located in a conserved region of the gene. Therefore, it may be functional or may be in LD with an unmeasured functional SNPs. Interestingly, the minor allele of a nearby functional SNP rs734784 (isoleucine to valine missense mutation) was associated with an increased risk for a number of persistent pain conditions (Costigan et al., 2010). However, rs734784 was not associated with ratings of average pain in women at least one year after surgery for breast cancer (Costigan et al.). The estimates of LD between rs4499491 and rs734784 (D’ = 0.416, r2 = 0.106) in our study suggest that the association observed between rs4199491 and preoperative pain is not likely to be attributable to its LD with rs734784. Moreover, no association was found between KCNS1 rs734784 and preoperative pain in our study.
Located on sensory neurons, voltage-gated potassium channels play a key role in modulating resting membrane potentials as well as the shape and magnitude of action potentials (Takeda et al., 2011; Tsantoulas et al., 2012). Although non-functional on its own, Kv9.1 modifies the activity of co-expressed functional voltage-gated potassium channels. (Richardson & Kaczmarek, 2000). Taken together, these findings suggest that KCNS1 may play a role in the pathophysiology of pain. The specific SNP identified in this study (i.e., rs4499491) warrants additional investigation in terms of its role in preoperative pain as well as persistent postsurgical pain.
Four genes that encode for G-protein-gated inwardly-rectifying potassium (GIRK) channels were evaluated in our study. Only KCNJ3 (GIRK1) and KCNJ6 (GIRK2) were associated with the occurrence of preoperative breast pain. All four of the polymorphisms identified are intronic. However, with the exception of one SNP in haplotype E1, they are located in conserved regions of the gene. Therefore, they may be functional or in LD with nearby functional SNPs.
GIRK channels are involved in postsynaptic inhibition in response to a number of neurotransmitters (Luscher & Slesinger, 2010), including those implicated in pain transmission (e.g., dopamine, serotonin, gamma-aminobutyric acid, opioids) (Fields, Heinricher, & Mason, 1991; Luscher & Slesinger, 2010). In addition, GIRK1 and GIRK2 channels closely interact with opioid receptors to modulate neuronal transmission (Ulens, Daenens, & Tytgat, 1999). Evidence from animal models suggests that GIRK1 (KCNJ3) and GIRK2 (KCNJ6), but not GIRK3 (KCNJ9), subunits are expressed in the superficial layers of the dorsal horn; play a role in thermal nociception and analgesic responses to morphine; and are highly interactive (Marker et al., 2004). Consistent with findings of altered thermal nociception in transgenic mice (Marker et al., 2004), only variations in KCNJ3 and KCNJ6, but not KCNJ9, were associated with the occurrence of preoperative breast pain in our sample. Given the substantial evidence for the role of these channels in the modulation of nociceptive transmission, additional investigations are warranted on the role of GIRK channels in preoperative breast pain.
Of the three two-pore domain potassium leak channel genes evaluated in this study, only variations in KCNK9 (TWIK-related acid sensing potassium channel-3; TASK3) were associated with an increased risk for the occurrence of preoperative breast pain. While these two intronic SNPs have no known function, they are located in a conserved region of KCNK9 and may be in LD with an unmeasured functional SNPs.
Two-pore domain potassium leak channels establish resting membrane potentials, play a key role in the modulation of neuronal excitability (Lesage, 2003; Talley et al., 2003), and are expressed in sensory neurons of rat DRG (Rau, Cooper, & Johnson, 2006). Like GIRK channels, TASK channels are inhibited by several neurotransmitters (Talley et al., 2000). Recently, in a rodent model of cutaneous inflammation, TASK3 mRNA expression in DRG neurons was reduced bilaterally four days after unilateral inflammation compared to one day after inflammation (Marsh et al., 2012). In addition, reduced mRNA expression was associated with reduced ipsilateral spontaneous pain behavior (i.e., foot lifting) (Marsh, et al., 2012). Further investigation of KCNK9, including an evaluation of whether genetic variations (e.g., rs3780039, rs11166921) are associated with TASK3 expression levels, is warranted.
In light of our previous findings (McCann et al., 2012), which suggested that preoperative breast pain has an inflammatory component, it is possible that gene × gene interactions between cytokine and potassium channel genes may occur and result in a higher risk for preoperative breast pain. This hypothesis is supported by work that demonstrates that potassium channel activity can impact cytokine production in lymphocytes (Feske, Skolnik, & Prakriya, 2012) and THP-1 cells through toll-like receptor 4 inhibition (Jo et al., 2011). Larger samples are needed to test for such potential epistatic interactions in patients with preoperative breast pain.
Some study limitations should be noted. Firstly, in this study, preoperative breast pain was operationalized as pain occurring in the affected breast prior to breast cancer surgery. Because the timeframe of occurrence was not evaluated, it is not clear whether this preoperative breast pain was acute or chronic in nature. Secondly, as with any association study, it is important to note that the genetic associations identified herein are not necessarily causal. Replication in independent samples, followed by functional studies and/or deep sequencing may be required before a causal relationship between these SNPs and the occurrence of preoperative breast pain is established. Thirdly, with an increased sample size, additional genetic associations may be identified. Likewise, regression models would have sufficient power to fit interaction terms (e.g., gene × environment interactions). Finally, although our sample size of 302 is substantial, it is possible that these associations may not be replicated in an independent sample (Ioannidis et al., 2001).
In summary, this study identified eight genetic variations among four potassium channel genes (i.e., KCNS1, KCNJ3, KCNJ6, KCNK9) that were significantly associated with the occurrence of preoperative breast pain. Variation among these genes may constitute important risk factors for the occurrence of preoperative breast pain. Moreover, in our work preoperative breast pain was associated with more severe postoperative pain data in preparation), as well as with the development of persistent breast pain after breast cancer surgery (Miaskowski et al., 2012b). An evaluation of genetic associations may help to identify the underlying mechanisms for preoperative, postoperative, and persistent pain in patients who undergo breast cancer surgery.
Acknowledgements
This study was funded by grants from the National Cancer Institute (CA107091 and CA118658). Dr. Miaskowski is an American Cancer Society (ACS) Clinical Research Professor. Dr. Dhruva is funded through NIH Mentored Patient-Oriented Research Career Development Award (K23 AT005340). Dr. Langford is supported by a Department of Defense Breast Cancer Research Program Postdoctoral Fellowship. Dr. Merriman was supported by an NINR fellowship (F31 NR012604), an ACS Doctoral Degree Scholarship (DSCN-10-087), an Oncology Nursing Society Doctoral Scholarship, and a UCSF Nursing Alumni Association Scholarship. Dr. Baggott is funded by an American Cancer Society Mentored Research Scholar Award (MRSG 12-01-PCSM). Dr. Leutwyler is funded by the KL2 Program (RR624130). Ms. Ward Sullivan is supported by an NINR T32. This project is supported by NIH/NCRR UCSF-CTSI Grant Number UL1 RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
REFERENCES
- Alloui A, Zimmermann K, Mamet J, Duprat F, Noel J, Chemin J, et al. TREK-1, a K+ channel involved in polymodal pain perception. EMBO J. 2006;25:2368–2376. doi: 10.1038/sj.emboj.7601116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Alva H, Harris RA. A pervasive mechanism for analgesia: activation of GIRK2 channels. Proc Natl Acad Sci U S A. 2003;100:277–282. doi: 10.1073/pnas.012682399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F, Bachmann LM, Weber U, Kessels AG, Perez RS, Marinus J, et al. Complex regional pain syndrome 1--the Swiss cohort study. BMC Musculoskelet Disord. 2008;9:92. doi: 10.1186/1471-2474-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde L, Vaquerizas JM, Dopazo H, Arbiza L, Reumers J, Rousseau F, et al. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res. 2006;34:W621–625. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry DC. Pain in carcinoma of the breast. Lancet. 1952;1:274–276. doi: 10.1016/s0140-6736(52)90339-5. [DOI] [PubMed] [Google Scholar]
- Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133:2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson PD, Forsythe ID. Presynaptic K+ channels: electrifying regulators of synaptic terminal excitability. Trends Neurosci. 2004;27:210–217. doi: 10.1016/j.tins.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Dunn LB, Aouizerat BE, Cooper BA, Dodd M, Lee K, West C, et al. Trajectories of anxiety in oncology patients and family caregivers during and after radiation therapy. Eur J Oncol Nurs. 2012;16:1–9. doi: 10.1016/j.ejon.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LB, Aouizerat BE, Langford DJ, Cooper BA, Dhruva A, Cataldo JK, et al. Cytokine gene variation is associated with depressive symptom trajectories in oncology patients and family caregivers. Eur J Oncol Nurs. 2013;17:346–353. doi: 10.1016/j.ejon.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S, Skolnik EY, Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat Rev Immunol. 2012;12:532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields HL, Heinricher MM, Mason P. Neurotransmitters in nociceptive modulatory circuits. Annu Rev Neurosci. 1991;14:219–245. doi: 10.1146/annurev.ne.14.030191.001251. [DOI] [PubMed] [Google Scholar]
- Fillingim RB. Individual differences in pain responses. Curr Rheumatol Rep. 2005;7:342–347. doi: 10.1007/s11926-005-0018-7. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gibson SJ, Helme RD. Age-related differences in pain perception and report. Clin Geriatr Med. 2001;17:433–456. doi: 10.1016/s0749-0690(05)70079-3. [DOI] [PubMed] [Google Scholar]
- Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366:1315–1323. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illi J, Miaskowski C, Cooper B, Levine JD, Dunn L, West C, et al. Association between pro- and anti-inflammatory cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression. Cytokine. 2012;58:437–447. doi: 10.1016/j.cyto.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Jensen MP, Gammaitoni AR, Olaleye DO, Oleka N, Nalamachu SR, Galer BS. The pain quality assessment scale: assessment of pain quality in carpal tunnel syndrome. J Pain. 2006;7:823–832. doi: 10.1016/j.jpain.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Jo HY, Kim SY, Lee S, Jeong S, Kim SJ, Kang TM, et al. Kir3.1 channel is functionally involved in TLR4-mediated signaling. Biochem Biophys Res Commun. 2011;407:687–691. doi: 10.1016/j.bbrc.2011.03.076. [DOI] [PubMed] [Google Scholar]
- Karnofsky D. Performance Scale. Plenum Press; New York, NY: 1977. [Google Scholar]
- Karnofsky D, Abelmann WH, Craver LV, Burchenal JH. The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer. 1948;1:634–656. [Google Scholar]
- Kyranou M, Paul SM, Dunn LB, Puntillo K, Aouizerat BE, Abrams G, et al. Differences in depression, anxiety, and quality of life between women with and without breast pain prior to breast cancer surgery. Eur J Oncol Nurs. 2013;17:190–195. doi: 10.1016/j.ejon.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix-Fralish ML, Ledoux JB, Mogil JS. The Pain Genes Database: An interactive web browser of pain-related transgenic knockout studies. Pain. 2007;131:3.e1–3.e4. doi: 10.1016/j.pain.2007.04.041. [DOI] [PubMed] [Google Scholar]
- Lane-Claypon JE. Reports on Public Health and Medical Subjects. Ministry of Health; London: 1926. A further report on cancer of the breast with special reference to its associated antecedent conditions. [Google Scholar]
- Lesage F. Pharmacology of neuronal background potassium channels. Neuropharmacology. 2003;44:1–7. doi: 10.1016/s0028-3908(02)00339-8. [DOI] [PubMed] [Google Scholar]
- Linden W, Vodermaier A, Mackenzie R, Greig D. Anxiety and depression after cancer diagnosis: Prevalence rates by cancer type, gender, and age. J Affect Disord. 2012;141:343–351. doi: 10.1016/j.jad.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Luscher C, Slesinger PA. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat Rev Neurosci. 2010;11:301–315. doi: 10.1038/nrn2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker CL, Stoffel M, Wickman K. Spinal G-protein-gated K+ channels formed by GIRK1 and GIRK2 subunits modulate thermal nociception and contribute to morphine analgesia. J Neurosci. 2004;24:2806–2812. doi: 10.1523/JNEUROSCI.5251-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh B, Acosta C, Djouhri L, Lawson SN. Leak K(+) channel mRNAs in dorsal root ganglia: relation to inflammation and spontaneous pain behaviour. Mol Cell Neurosci. 2012;49:375–386. doi: 10.1016/j.mcn.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann B, Miaskowski C, Koetters T, Baggott C, West C, Levine JD, et al. Associations between pro- and anti-inflammatory cytokine genes and breast pain in women prior to breast cancer surgery. J Pain. 2012;13:425–437. doi: 10.1016/j.jpain.2011.02.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaskowski C, Cooper BA, Dhruva A, Dunn LB, Langford DJ, Cataldo JK, et al. Evidence of associations between cytokine genes and subjective reports of sleep disturbance in oncology patients and their family caregivers. PLoS One. 2012a;7:e40560. doi: 10.1371/journal.pone.0040560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaskowski C, Cooper B, Paul SM, West C, Langford D, Levine JD, et al. Identification of patient subgroups and risk factors for persistent breast pain following breast cancer surgery. J Pain. 2012b;13:1172–1187. doi: 10.1016/j.jpain.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaskowski C, Dodd M, Paul SM, West C, Hamolsky D, Abrams G, et al. Lymphatic and Angiogenic Candidate Genes Predict the Development of Secondary Lymphedema following Breast Cancer Surgery. PLoS One. 2013;8:e60164. doi: 10.1371/journal.pone.0060164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. An overview of the potassium channel family. Genome Biol. 2000;1:1–5. doi: 10.1186/gb-2000-1-4-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa D, Nagashima M, Katoh R, Satoh Y, Tagami M, Kasai S, et al. Association between KCNJ6 (GIRK2) gene polymorphisms and postoperative analgesic requirements after major abdominal surgery. PLoS One. 2009;4:e7060. doi: 10.1371/journal.pone.0007060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshuck EL, Katz J, Andrus CH, Hogan LA, Jung BF, Kulick DI, et al. Risk factors for chronic pain following breast cancer surgery: a prospective study. J Pain. 2006;7:626–634. doi: 10.1016/j.jpain.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rau KK, Cooper BY, Johnson RD. Expression of TWIK-related acid sensitive K+ channels in capsaicin sensitive and insensitive cells of rat dorsal root ganglia. Neuroscience. 2006;141:955–963. doi: 10.1016/j.neuroscience.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Richardson FC, Kaczmarek LK. Modification of delayed rectifier potassium currents by the Kv9.1 potassium channel subunit. Hear Res. 2000;147:21–30. doi: 10.1016/s0378-5955(00)00117-9. [DOI] [PubMed] [Google Scholar]
- Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–46. [PubMed] [Google Scholar]
- Sangha O, Stucki G, Liang MH, Fossel AH, Katz JN. The Self-Administered Comorbidity Questionnaire: a new method to assess comorbidity for clinical and health services research. Arthritis Rheum. 2003;49:156–163. doi: 10.1002/art.10993. [DOI] [PubMed] [Google Scholar]
- SPSS . IBM SPSS for Windows (Version 19) SPSS, Inc; Chicago, Illinois: 2010. [Google Scholar]
- StataCorp . Stata Statistical Software: Release 9. Stata Corporation; College Station, Texas: 2005. [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Tsuboi Y, Kitagawa J, Nakagawa K, Iwata K, Matsumoto S. Potassium channels as a potential therapeutic target for trigeminal neuropathic and inflammatory pain. Mol Pain. 2011;7:5. doi: 10.1186/1744-8069-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley EM, Lei Q, Sirois JE, Bayliss DA. TASK-1, a two-pore domain K+ channel, is modulated by multiple neurotransmitters in motoneurons. Neuron. 2000;25:399–410. doi: 10.1016/s0896-6273(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Talley EM, Sirois JE, Lei Q, Bayliss DA. Two-pore-Domain (KCNK) potassium channels: dynamic roles in neuronal function. Neuroscientist. 2003;9:46–56. doi: 10.1177/1073858402239590. [DOI] [PubMed] [Google Scholar]
- Tasmuth T, von Smitten K, Kalso E. Pain and other symptoms during the first year after radical and conservative surgery for breast cancer. Br J Cancer. 1996;74:2024–2031. doi: 10.1038/bjc.1996.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantoulas C, Zhu L, Shaifta Y, Grist J, Ward JP, Raouf R, et al. Sensory neuron downregulation of the Kv9.1 potassium channel subunit mediates neuropathic pain following nerve injury. J Neurosci. 2012;32:17502–17513. doi: 10.1523/JNEUROSCI.3561-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulens C, Daenens P, Tytgat J. The dual modulation of GIRK1/GIRK2 channels by opioid receptor ligands. Eur J Pharmacol. 1999;385:239–245. doi: 10.1016/s0014-2999(99)00736-0. [DOI] [PubMed] [Google Scholar]
- Wickenden A. K(+) channels as therapeutic drug targets. Pharmacol Ther. 2002;94:157–182. doi: 10.1016/s0163-7258(02)00201-2. [DOI] [PubMed] [Google Scholar]
- Xie W. Ion channels in pain transmission. Int Anesthesiol Clin. 2007;45:107–120. doi: 10.1097/AIA.0b013e31803419e7. [DOI] [PubMed] [Google Scholar]


