Abstract
The Magnetization-Prepared 2 Rapid Acquisition Gradient Echo (MP2RAGE) method achieves spatially uniform contrast across the entire brain between gray matter and surrounding white matter tissue and cerebrospinal fluid by rapidly acquiring data at two points during an inversion recovery, and then combining the two volumes so as to cancel out sources of intensity and contrast bias, making it useful for neuroimaging studies at ultrahigh field strengths (≥ 7 T).
To quantify the effectiveness of the MP2RAGE method for quantitative morphometric neuroimaging, we performed tissue segmentation and cerebral cortical surface reconstruction of the MP2RAGE data and compared the results with those generated from conventional Multi-Echo MPRAGE (MEMPRAGE) data across a group of healthy subjects.
To do so, we developed a preprocessing scheme for the MP2RAGE image data to allow for automatic cortical segmentation and surface reconstruction using FreeSurfer and analysis methods to compare positioning of the surface meshes.
Using image volumes with 1 mm isotropic voxels we found a scan-rescan reproducibility of cortical thickness estimates to be 0.15 mm (or 6%) for the MEMPRAGE data and a slightly lower reproducibility of 0.19 mm (or 8%) for the MP2RAGE data. We also found that the thickness estimates were systematically smaller in the MP2RAGE data, and that both the interior and exterior cortical boundaries estimated from the MP2RAGE data were consistently positioned within the corresponding boundaries estimated from the MEMPRAGE data. Therefore several measureable differences exist in the appearance of cortical gray matter and its effect on automatic segmentation methods that must be considered when choosing an acquisition or segmentation method for studies requiring cortical surface reconstructions.
We propose potential extensions to the MP2RAGE method that may help to reduce or eliminate these discrepancies.
Keywords: Ultra-high-field MRI, quantitative morphometry, brain segmentation, FreeSurfer, cortical thickness
1. INTRODUCTION
Accurate identification and segmentation of the cerebral cortical gray matter and subcortical gray matter structures from volumetric imaging data are critical for many anatomical and functional brain imaging studies. For anatomical studies, automatic cortical surface reconstructions enable quantitative morphometric analyses of brain anatomy that can be sensitive to sub-voxel changes in cortical thickness, and can be used to track or detect brain atrophy, plasticity, and development {Kuperberg et al., 2003; Rosas et al., 2002; Salat et al, 2009; Sowell et al, 2004}. For functional studies, cortical surface reconstructions provide a model of the folded cortical sheet, and many distributed features of neuronal processing have a distinct spatial pattern across the cortex that can only be identified with an anatomically-informed, surface-based analysis {Huang et al, 2012; Polimeni et al., 2010a}. As more clinical and scientific studies migrate to ultrahigh field strengths, such as 7 Tesla, techniques for cortical surface reconstruction must be made available, however challenges associated with high magnetic fields—such as tissue dielectric properties which cause a “central brightening” effect, and reduced T1 differences between tissues—must be overcome.
The magnetization-prepared rapid gradient echo, or MPRAGE, pulse sequence {Mugler & Brookeman, 1990} achieves excellent T1-weighted image contrast between the cortical gray matter (GM) and the adjacent white matter (WM) and cerebrospinal fluid (CSF) in a time-efficient acquisition at conventional field strengths, but at ultrahigh field strengths the dielectric effects in the human head lead to a spatially varying image contrast due to spatial non-uniformity in the transmit B1+ field {Collins et al, 2005}. Furthermore, spatial intensity biases can be caused by the rapid fall-off in receive coil sensitivity profiles, especially for the high-element-count arrays (employing smalldiameter coil detectors) that are becoming increasingly common {Keil et al, 2010; Ledden et al., 2007; Wiggins et al, 2009}. While receive coil biases can be corrected in post-processing, transmit-side biases are more challenging to address. Specialized adiabatic inversion pulses may be used to reduce B1+ field variations caused by dielectric effects and spatially varying transmit coil efficiency {Hurley et al, 2010; Ordidge et al., 1996; Wrede et al, 2012}, however these pulses tend to generate high levels of SAR and can be sensitive to off-resonance effects near susceptibility gradients around air-tissue interfaces. Other approaches to mitigate B1+ inhomogeneity make use of multiple transmit coil arrays by using parallel transmit techniques for tailored pulse design {Cloos et al., 2012; Setsompop et al., 2009}. While promising, the parallel transmit approach requires specialized hardware, subject-specific pulse design, and careful online SAR monitoring, and therefore is not yet suitable for routine clinical use.
Another approach to reduce both transmit-side and receive-side sources of bias involves acquiring a second, reference image volume containing similar biases, then uses this reference to normalize the MPRAGE volume {Van de Moortele et al., 2009}. The Magnetization-Prepared 2 Rapid Acquisition Gradient Echo (MP2RAGE) method {Marques et al., 2010} adopts this approach in a time-efficient manner by including two full image readouts during each inversion recovery of the MPRAGE acquisition— effectively acquiring two images each with its own inversion time—and employs an image reconstruction that produces a normalized image with dramatically reduced image biases and a more spatially uniform tissue contrast over the brain. The first inversion time (TI1) is chosen to produce a T1-weighted image with the cortical gray matter effectively nulled at the center of k-space, while the second inversion time (TI2) is long and therefore produces an approximately proton density-weighted contrast. A normalized combination of these two images provides a synthetic image with strong GM-WM-CSF contrast across the brain.
In this study, we have attempted to quantify both the precision and accuracy of cortical gray matter segmentation and the corresponding cortical surface reconstruction from the synthetic T1-weighted image data generated with the MP2RAGE method. Assessing the accuracy of the gray matter segmentation is extremely challenging in the absence of any ground-truth data identifying the gray matter borders. Furthermore, the exact anatomical position of the GM-WM and GM-CSF boundaries can be defined somewhat subjectively; and the sharpness of the GM-WM boundary appears to vary across brain areas and can change with normal ageing {Salat et al, 2009}. Therefore, to address the accuracy of the surfaces generated from the MP2RAGE method we characterized biases in cortical thickness measures and in cortical surface reconstruction placement using a well-validated surface reconstruction method, FreeSurfer {Fischl, 2012}, on both data acquired with the MP2RAGE method and conventional data acquired with the multi-echo MPRAGE (MEMPRAGE) method {van der Kouwe et al., 2008}. Because in many studies the exact placement of the cortical boundaries is not as critical as having a method that provides consistent boundaries over time or across subject groups, which provides the sensitivity to detect subtle changes in cortical morphometry. We also quantified the precision of the cortical segmentation for both the MP2RAGE and MEMPRAGE data by calculating the scanrescan reproducibility of the cortical thickness measures and in cortical surface reconstruction placement as has been reported previously for FreeSurfer using similar MPRAGE data {Han et al., 2006; Reuter et al., 2012}. For both characterizations, we have included analyses of the position of the reconstructed boundary surfaces of the cortical gray matter to better understand which boundary (the GM-WM boundary or the GM-CSF boundary) gives rise to any detected differences in cortical thickness measures seen between acquisition methods.
To perform these analyses, we developed a modified FreeSurfer pipeline to process MP2RAGE data, described below. While the MP2RAGE method does reduce the intensity and contrast biases at 7 T relative to the MEMPRAGE method, the generation of the MP2RAGE synthetic image amplifies noise in the CSF region immediately adjacent to the cortical GM which, if left unaddressed, causes local inaccuracies in the surface reconstruction (see Fig. 1). With our modified processing stream, we demonstrate surface reconstructions of 7 T MP2RAGE data, evaluate the precision and accuracy of the reconstructions at 3 T where reliable, automatic reconstructions of MEMPRAGE data are possible, and directly compare the 7 T and 3 T data. We find systematic differences between surface reconstructions based on the MP2RAGE and MEMPRAGE acquisition methods as well as a difference in the level of precision in the two methods. Preliminary accounts of these results have appeared in abstract form {Fujimoto et al., 2011a, 2011b}.
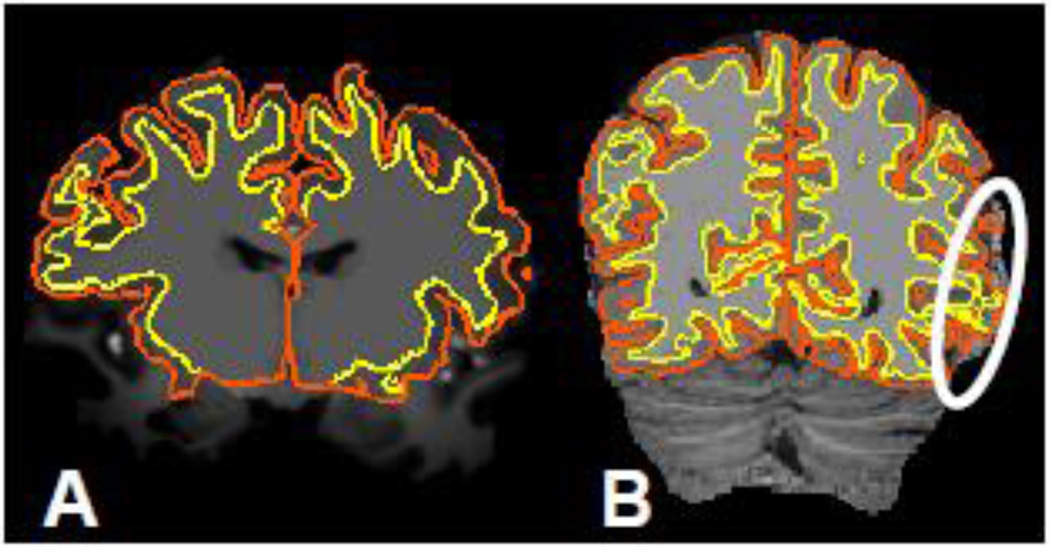
Fig. 1.
(A) Example of an unprocessed 7 T MEMPRAGE image slice with cross-sections of FreeSurfer surface reconstructions generated from this data overlaid. (Yellow contour represents intersection of slice with the white matter surface, while the orange contour represents the pial surface.) (B) Example of a synthetically generated 7 T MP2RAGE “flat image” slice with FreeSurfer surface reconstructions generated using the standard processing stream applied to this data overlaid. The marker indicates a region where the amplified noise in the CSF region causes errors in the cortical GM segmentation, which leads to inaccurate surface reconstructions.
2. METHODS
2.1 Participants
Eight healthy adults volunteered to participate in the study. Written informed consent was obtained from each participant prior to the experiment in accordance with our institution’s Human Research Committee. All subjects participated in a 7 T session, and four subjects returned on another day for a 3 T session.
2.2 Data Acquisition
All 7 T data were acquired with a Siemens whole-body scanner (Siemens Healthcare, Erlangen, Germany) equipped with body gradients using a custom-made 31-channel brain receive coil array and birdcage volume coil for transmit {Keil et al, 2010}. The MP2RAGE data acquisition and online image reconstruction were performed with a vendor-supplied MP2RAGE package (WIP #412). For each volunteer at 7 T we acquired a single whole-brain T1-weighted volume with the MP2RAGE method with 1 mm isotropic voxel size and the following protocol parameters: TI1/TI2/TE/TR/flip-1/flip2/BW/echo spacing=901 ms / 3201 ms / 2.82 ms / 5000 ms / 4° / 5° / 240 Hz/pix / 6.9 ms, a 256×256 encoding matrix and 192 partitions, with R=3 acceleration in the primary phase encoding direction (32 reference lines) and online GRAPPA image reconstruction, for a total acquisition time of 8 min 22 s per volume. (For all scans, accelerated parallel imaging was employed as it is now used routinely in clinical practice. Also, because the reduced acquisition times limit the vulnerability to subject head movement, motion artifacts are minimized.) After data acquisition, all volumes were corrected for gradient nonlinearity by applying the online 3D gradient nonlinearity geometric distortion correction {Jovicich et al., 2006}.
All 3 T data were acquired with a Siemens whole-body TIM Trio scanner using the vendor 32-channel brain receive coil array. The MP2RAGE data acquisition and online image reconstruction were performed with a vendor-supplied MP2RAGE package (WIP #602). During the 3 T sessions we acquired both MP2RAGE data and MEMPRAGE data to assess both the reproducibility of the cortical segmentations from both methods as well as to compare the results of the two acquisition methods. We acquired wholebrain T1-weighted MP2RAGE volumes with 1 mm isotropic voxel size and the following protocol parameters: TI1/TI2/TE/TR/flip1/flip2/BW/echo spacing=700 ms / 2500 ms / 2.96 ms / 5000 ms / 4° / 5° / 240 Hz/pix / 7.1 ms, a 256×256 encoding matrix and 192 partitions, with R=3 acceleration (32 reference lines) in the primary phase encoding direction and online GRAPPA image reconstruction, for a total acquisition time of 8 min 52 s per volume. The whole-brain T1-weighted MEMPRAGE volumes were also acquired with 1 mm isotropic voxel size and four echoes with the following protocol parameters: TI/TE1/TE2/TE3/TE4/TR/flip/BW/echo spacing = 1200 ms / 1.64 ms / 3.5 ms / 5.36 ms / 7.22 ms / 2510 ms / 7° / 651 Hz/pix / 10.3 ms, a 256×256 encoding matrix and 192 partitions, with R=2 acceleration in the primary phase encoding direction (32 reference lines) and online GRAPPA image reconstruction, for a total acquisition time of 6 min 02 s per volume. In each 3 T session, we acquired three MP2RAGE volumes and three MEMPRAGE volumes for a total of six volumes per session: we first acquired two MEMPRAGE volumes and two MP2RAGE volumes (termed “repeats”) in the first scanning block; then we repositioned the subject within the scanner and re-acquired an MEMPRAGE volume and an MP2RAGE volume (termed “rescans”) in a second scanning block (see Fig. 2). Comparing the “repeats” provides a baseline measure of precision while comparing the “rescans” provides a measure of across-session reproducibility. The subjects were instructed to remain still throughout the scanning. After data acquisition, all volumes were corrected for gradient nonlinearity {Jovicich et al., 2006} by applying the vendor-supplied online 3D gradient nonlinearity geometric distortion correction.
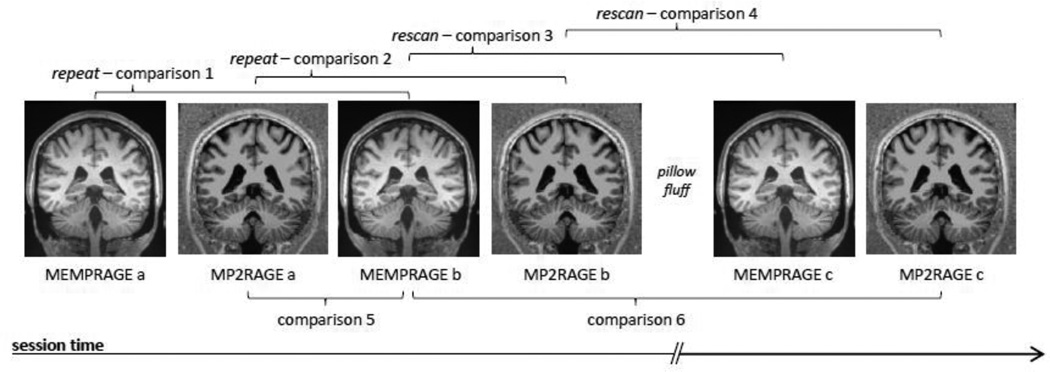
Fig. 2.
Schematic depicting the 6 comparisons performed between the image volumes acquired in a single 3 T session. During each session six volumes were acquired. First, two MEMPRAGE volumes and two MP2RAGE volumes were acquired. Then, the subject was briefly removed from the scanner and then immediately repositioned to simulate the effects of multiple scan sessions (i.e., to allow for the subject’s head to change positions). Then, one additional MEMPRAGE and one MP2RAGE volume was acquired. Across these six volumes we performed six comparisons. Volumes a and b were therefore acquired with the subject in the same position and volumes c were acquired with the subject in a slightly different position. The repeat and rescan comparisons were designed to address the reproducibility of the image volume segmentation and surface reconstruction for a within-scan and across-scan precision estimation, respectively. We also performed two cross-modality accuracy estimates by performing comparisons directly between the MEMPRAGE image volumes and the MP2RAGE image volumes. (Not shown is Comparison 7, which was performed between the 3 T MEMPRAGE and a 7 T MP2RAGE volume acquired from the same subject across different days.)
2.3 Surface reconstruction
Cortical surfaces were computed automatically from the MEMPRAGE image data using FreeSurfer {Dale et al., 1999; Fischl et al., 1999, 2001, 2002, 2004b; Ségonne et al., 2004},(http://surfer.nmr.mgh.harvard.edu/) Version 5.1 (2011). Additional steps were required to compute surfaces from the MP2RAGE data. The MP2RAGE image reconstruction combines images from two inversion times to generate a “Flat Image”, which produces spatially uniform tissue contrast but amplifies noise outside of the brain and adjacent to the cortical gray matter (Fig. 3a) that can introduce errors in the automatic segmentation (Fig. 3b). To mask this amplified noise, we multiplied the Flat Image by the proton density-weighted image acquired during the second half of the inversion recovery (Fig. 3c), which naturally has low voxel intensities in the image background regions. This multiplicative product image was then skull-stripped using FreeSurfer with the standard parameter settings. This robustly generated a brain mask including the brain but excluding the amplified noise regions. The resulting mask was then applied to the original Flat Image to segment the brain from the surrounding amplified noise (Fig. 3d), and this final masked Flat Image was input into the standard FreeSurfer segmentation and surface reconstruction pipeline for further segmentation and cortical surface reconstruction. Therefore, in practice the FreeSurfer “recon-all” command was invoked twice in two independent streams. First, recon-all was invoked in a primary stream with the default Flat Image as input and in a secondary stream with the derived multiplicative product image described above as input. Both streams were then paused immediately after the brain masking stage. The brain mask from the primary stream was then replaced by the brain mask produced from the secondary stream (calculated from the multiplicative product image). Each resulting brain mask was visually inspected to verify that the masked region did not include cortical gray matter. Finally, the primary stream was resumed with this new brain mask while the secondary stream was terminated and discarded. All invocations of “recon-all” were made without any additional flags, therefore the default options were used for all reconstructions.
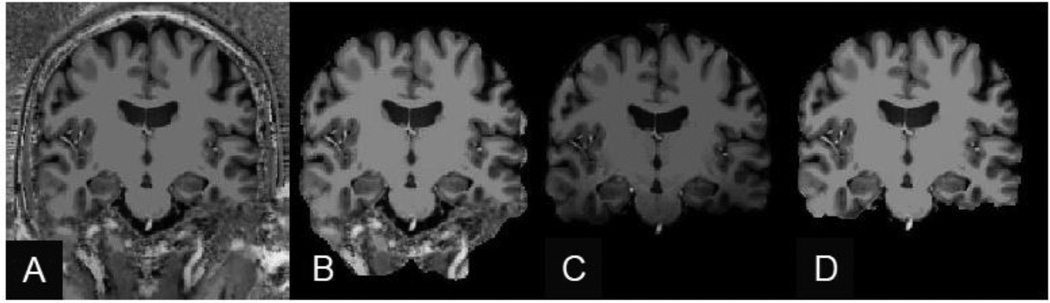
Fig. 3.
Segmenting the brain in MP2RAGE data. (A) Example “flat image” of 7 T MP2RAGE, in which the background noise due to the MP2RAGE image processing is seen both outside of the head and in the skull region adjacent to the cortical gray matter. (B) Example “skullstripping” produced by FreeSurfer on a 7 T MP2RAGE volume. Because the background noise in places has the same image intensity as the gray matter, segmenting the brain can be challenging. (C) Example of the mask created by multiplying the flat Image by the TI2 image. (D) Example of a brain segmentation from a 7 T MP2RAGE volume using the proposed masking procedure. This mask removes the background noise without removing the brain tissue regions.
2.4 Surface alignment and vertex correspondence calculation
To evaluate the differences between surfaces generated from image volume data acquired with the MP2RAGE method and surfaces generated from image volume data acquired with the MEMPRAGE method, we performed several comparisons to characterize both the reproducibility of the surface reconstructions derived from either method as well as the level of agreement between the surfaces derived from the two imaging methods. Because of the potential for head movement between acquisitions, and because surface meshes derived from different volumes are likely to have differing numbers and distributions of vertices across the cortex, first the surfaces generated from two volumes need to be brought into alignment with one another, then a vertex correspondence must be calculated to compare the surfaces on a vertex-by-vertex basis.
Thus to compare surface reconstructions generated from a set of image volumes for a particular subject, we used the Longitudinal Processing Stream framework {Reuter et al., 2012} as implemented in FreeSurfer v5.1; this provided a comparison that was not biased in favor of any one volume within any comparison set. The longitudinal processing was applied to comparing the surface reconstructions for the multiple volumes acquired with each volunteer within a single scanning session as follows. For each comparison, we computed a bias-free template volume {Reuter & Fischl, 2011} derived from only those volumes being considered in the comparison. This template was based on a robust registration method that automatically discounts outlier voxels to achieve improved registration accuracy {Reuter et al., 2010}. Initial surfaces were reconstructed from this template, and these surfaces were rigidly transformed to each individual volume in the comparison set. Finally, the surfaces were deformed to the gray matter boundaries based on local image intensities in the volume using the same optimization and cost function used in the conventional FreeSurfer surface reconstruction. This strategy forces each surface reconstruction in the comparison set to have the same mesh topology and the same number of vertices, and provides a natural vertex correspondence between all surface reconstructions in each comparison set.
Note that all comparisons were restricted to only those vertices in the mesh that were identified as cortex by FreeSurfer—there is a collection of vertices in the mesh along the medial wall in the location of the corpus callosum that are not cortex but are added by FreeSurfer to force the surface mesh to be topologically closed. (These noncortex vertices can be seen as a gap in the surface overlays in Figs. 6 and 8.)
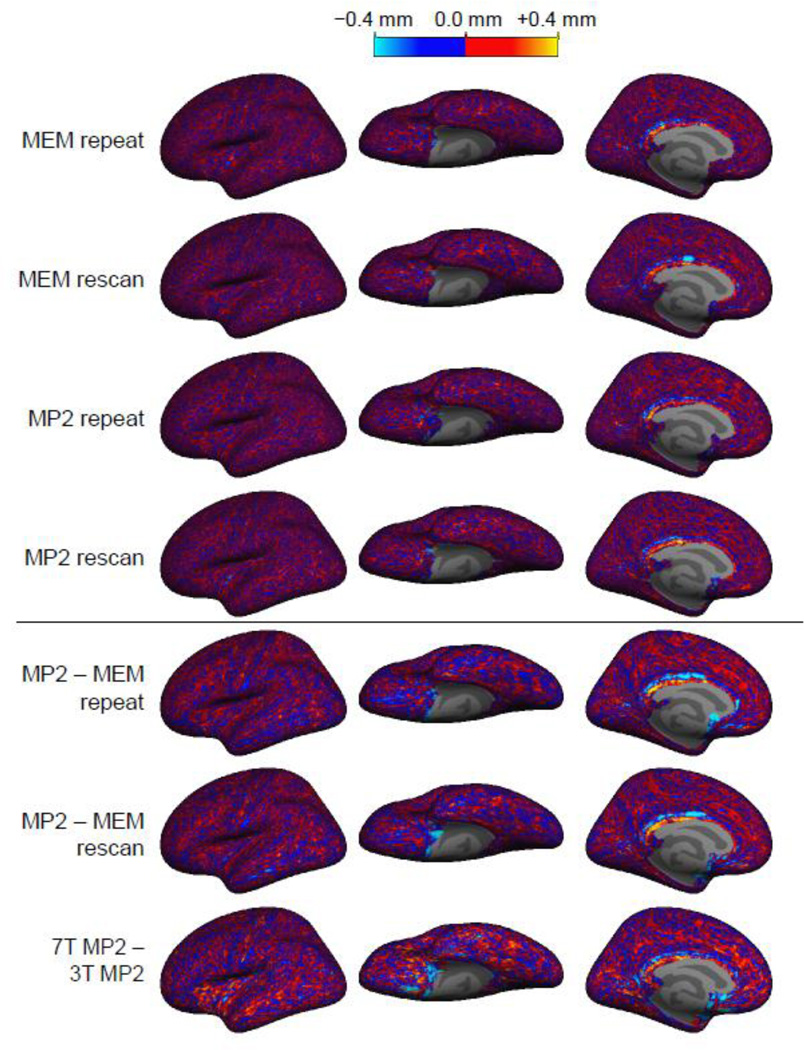
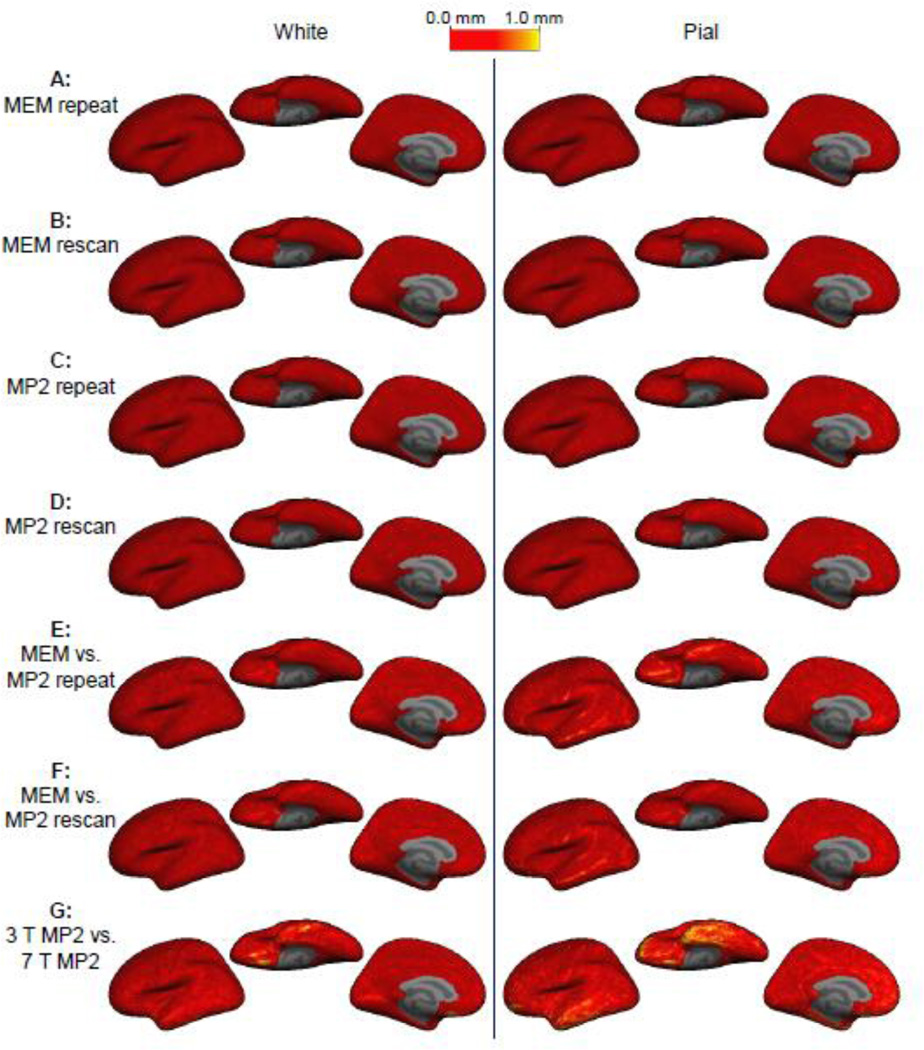
Fig. 6.
Signed cortical thickness differences between surface reconstructions. Cortical thickness was measured from the surface reconstructions generated from seven acquisitions on each subject, and the difference in the resulting thickness estimate was computed for each location on the surface and then averaged across the four subjects on the common surface space. No spatial structure is apparent in the distribution of thickness difference across the cortex for all comparisons except for the one comparison between 3 T and 7 T.
Fig. 8.
Gray-white and gray-pial boundaries absolute position difference. Boundaries were measured from the surface reconstructions generated from seven acquisitions on each subject, and the difference in the resulting position difference was computed for each location on the surface in each coordinate, calculated a distance and then averaged across the four subjects on the common surface space.
2.5 Quantitative comparison of surface reconstructions
To quantify the differences between surfaces reconstructed from data acquired with the MP2RAGE method and surfaces reconstructed from data acquired with the MEMPRAGE method, we performed a total of seven comparisons. Specifically, we compared surfaces reconstructed from: (1) the two repeated MEMPRAGE volumes acquired within the first scanning block; (2) the two repeated MP2RAGE volumes acquired within the first scanning block; (3) one MEMPRAGE volume acquired in the first block with the rescanned MEMPRAGE volume acquired in the second block; (4) one MP2RAGE volume acquired in the first block with the rescanned MP2RAGE volume acquired in the second block; (5) a MEMPRAGE volume and an MP2RAGE volume acquired in the first block; (6) a MEMPRAGE volume acquired in the first block and an MP2RAGE volume acquired in the second block; and (7) the 3 T MEMPRAGE and 7 T MP2RAGE volumes. (See Fig. 2 for a schematic describing these comparisons.) Therefore, under the Longitudinal Processing Stream framework described above, we generated seven templates per subject. For all comparisons we included only two volumes in the comparison set (e.g., only sequentially acquired volumes b and c were compared for rescan comparisons, see Fig. 2). For each comparison we evaluated the discrepancies between the surface reconstructions from the two volumes in terms of differences in cortical thickness as well as differences in the white matter surface placement and differences in the pial surface placement. Note that comparison 7 between 3 T MEMPRAGE and 7 T MP2RAGE was chosen because the 3 T MEMPRAGE is the conventional volume used for cortical surface reconstruction and the 7 T MP2RAGE is a potential viable replacement, therefore we wished to evaluate 7 T MP2RAGE here. Also, because 7 T MEMPRAGE surface reconstructions currently require some manual editing to represent the cortex accurately (especially around the temporal poles) due to the spatially varying contrast caused by dielectric effects at high fields, we did not include any 7 T MEMPRAGE comparisons in this study.
Given a vertex correspondence between surfaces generated from a pair of image volumes, the cortical thickness derived from the cortical gray matter segmentation can be compared between the two volumes on a point-by-point basis. For the seven comparisons performed for each subject, we computed the thickness difference between the reconstructions at each location of the cortex and compared the absolute value of this difference along the surface. To assess whether the spatial pattern of absolute cortical thickness difference was similar across subjects (which would suggest a systematic bias), we then mapped these absolute cortical thickness differences computed for each subject into a common surface space, i.e., the “fsaverage” atlas. This spatial normalization was performed via surface-based registration {Fischl et al., 1999} as implemented in FreeSurfer. Once each absolute cortical thickness difference map was in this atlas space, we were able to calculate an average of the absolute cortical thickness differences across subjects on each vertex of the common surface space.
Beyond assessing differences in measured cortical thickness amongst the cortical reconstructions being compared, we also sought to quantitatively compare the positioning of the two surface reconstructions—the surface representing the interface between the cortical gray matter and white matter and the surface representing the interface between the cortical gray matter and pia mater/CSF. Differences in cortical thickness between two segmentations could reflect either a discrepancy between the positioning of the gray-white surfaces or between the gray-pial surfaces, or both. To better understand the causes for discrepancies in cortical surface we developed a procedure for directly comparing the positions of the cortical surface meshes themselves.
The procedure for computing surface distance discrepancy began with the rigid alignment and vertex correspondence calculation described above. Then, for each vertex in the surface reconstructed from the reference volume we calculated the 3D Euclidean distance to the corresponding vertex in the surface reconstructed from the movable volume. This provided an absolute distance discrepancy for each vertex on the white matter surface reconstruction and for each vertex on the pial surface reconstruction, accounting for possible head motion between acquisitions. To further assess the direction of the displacement of any given vertex on the movable surface relative to the reference surface, we computed a signed vertex displacement by calculating the sign of the dot product of the displacement vector with the surface normal of the vertex on the reference surface, then multiplying the absolute distance discrepancy by the sign of this dot product. As with the thickness comparison described above, to assess any pattern in the distance discrepancy across subjects, the pervertex distance discrepancy map for each subject, surface, and comparison was projected into the common surface space for averaging across subjects and visualization.
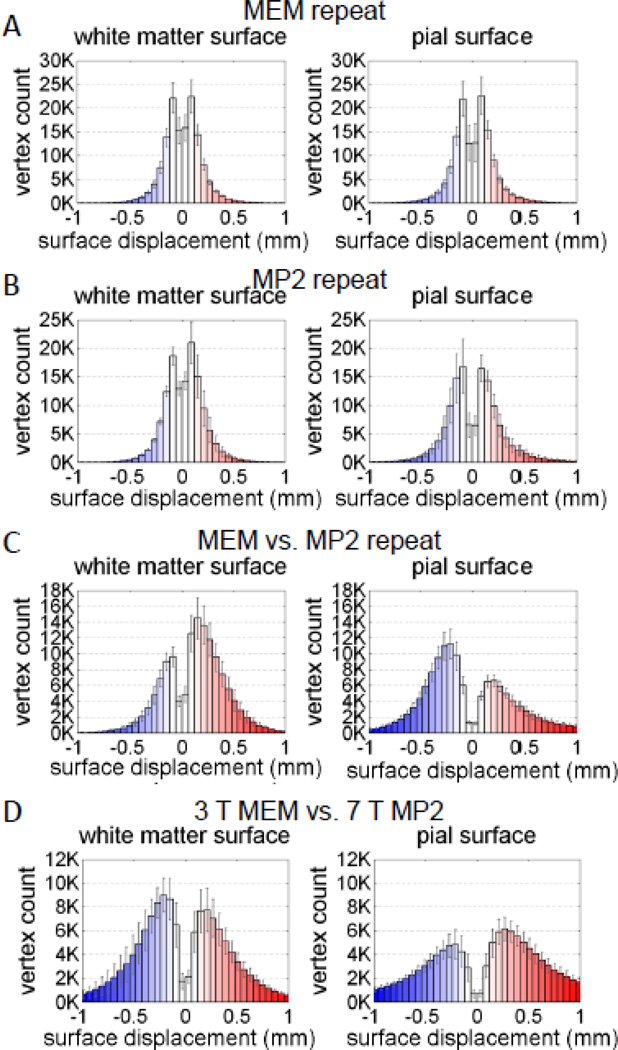
To detect and quantify any bias in the surface positioning between a pair of surfaces, for each comparison we generated a histogram of the signed displacement between the two surfaces across all vertices. Asymmetry in this histogram indicates that one surface tends to be positioned either inside or outside of a reference surface.
For all cortical thickness comparisons and boundary surface position comparisons, we excluded the portion along medial wall in each surface. Vertices within this medial wall region are labeled as “non-cortex” by FreeSurfer and thus are straightforward to remove from any analysis. This medial wall portion is included in the mesh only to close the surface, making it a topological sphere.
3. RESULTS
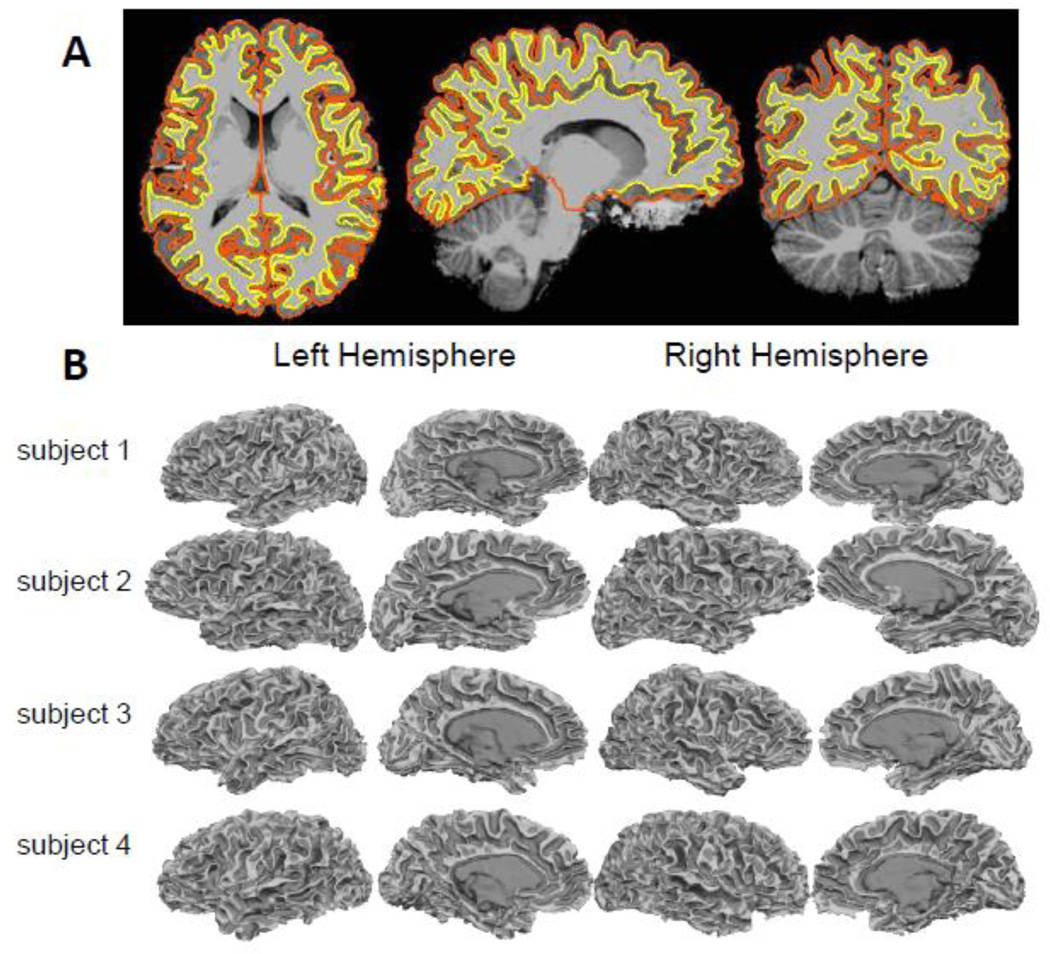
The modification to the FreeSurfer surface reconstruction stream for the T1-weighted images generated by the MP2RAGE method was successful in all subjects tested. The masking strategy described above successfully removed the noisy regions generated by the MP2RAGE processing and enabled robust cortical surface reconstruction. An example surface reconstruction of the interior and exterior boundaries of the cortical gray matter is shown in Fig. 4a overlaid on the image data; surface reconstructions for four representative 7 T data sets, including two conventional-bandwidth (one of which is shown in Fig. 3) and two high-bandwidth examples, are shown in Fig. 4b.
Fig. 4.
Example surface reconstructions from 7 T MP2RAGE data. (A) Masked MP2RAGE flat image with cross-sections of surface reconstructions representing the gray-white and gray-pial boundaries overlaid. (B) Final FreeSurfer white matter surface reconstructions of four subjects, including conventional-bandwidth (240 Hz/pix) and high-bandwidth (975 Hz/pix) examples. (Dark gray indicates sulcal regions and light gray indicates gyral regions.)
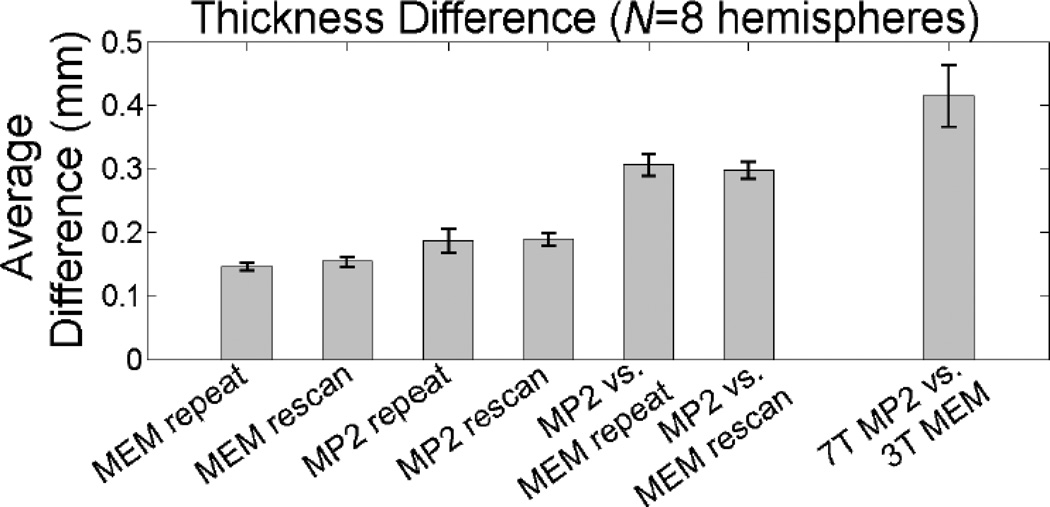
The quantitative comparison of cortical gray matter thickness measures taken from the four subjects scanned both at 3 T and 7 T is provided in Figs. 5 & 6. Of the seven comparisons made, the first four comparisons quantify the reproducibility of the cortical thickness measure seen in the two acquisitions (to address the precision provided by each acquisition method), and the last three comparisons quantify the agreement of the thickness measure between the acquisitions (to address the accuracy provided by each acquisition method). The average measured thickness difference over the entire cortical hemisphere—excluding the non-cortex portion along medial wall— calculated across the four subjects is provided in Fig. 5 and enumerated in Table 1 for each of the seven comparisons. The highest precision was 0.145 mm (± 0.006 mm), seen between the back-to-back repeated 3 T MEMPRAGE acquisitions (i.e., the “repeat” acquisitions). When the medial wall was included in the calculation, this value decreased slightly to 0.140 (± 0.005); this is in close agreement with a similar precision measurement made by Han et al. {Han et al., 2006} who reported a precision in the FreeSurfer thickness measurement of about 0.120 mm when including the medial wall, and is also in agreement with a subsequent precision measurement made by Reuter et al. {Reuter et al., 2012}. A slightly lower precision of 0.154 mm (± 0.008 mm) is seen between the two MEMPRAGE acquisitions that were separated by a repositioning of the subject (i.e., the “rescan” acquisitions). This also suggests that the robust registration used to align the two volumes acquired after repositioning the subject {Reuter et al., 2010} was highly effective. Overall lower precision is seen in the MP2RAGE data. The precision of the cortical thickness measure in the 3 T MP2RAGE data was 0.187 mm (± 0.019 mm) between the “repeat” acquisitions and was 0.189 mm (± 0.009 mm) between the “rescan” acquisitions.
Fig. 5.
Summary of the average absolute thickness difference comparison across four subjects (eight hemispheres). Error bars indicate population standard deviation across the hemispheres.
Table 1.
Comparison of average absolute thickness difference across four subjects (eight hemispheres) with population standard deviation across the hemispheres.
| comparison | average difference (mm) |
standard deviation of difference (mm) |
|---|---|---|
| repeat | 0.145 | 0.006 |
| MEM rescan | 0.154 | 0.008 |
| MP2 repeat | 0.187 | 0.019 |
| MP2 rescan | 0.189 | 0.009 |
| MP2 vs. MEM, repeat | 0.307 | 0.017 |
| MP2 vs. MEM, rescan | 0.298 | 0.013 |
| 7 T MP2 vs. 3 T MP2 | 0.414 | 0.049 |
For the comparisons between the MP2RAGE surface reconstructions and the MEMPRAGE surface reconstructions, the average absolute thickness difference was 0.307 mm (± 0.017 mm) for volumes acquired within the same block and 0.298 mm (± 0.013 mm) for volumes acquired in different blocks separated by a repositioning of the subject. The close agreement between the two measures again suggests that the robust registration was capable of accurately aligning data separated by a repositioning even with differing contrast in the MEMPRAGE and MP2RAGE data. Finally, the largest discrepancy in absolute cortical thickness difference was seen in the comparison between the 3 T MEMPRAGE and the 7 T MP2RAGE; an absolute thickness difference of 0.414 mm (± 0.049 mm) was observed across methods and field strengths. Given that the voxel size for both acquisitions was 1 mm3, this discrepancy is over one third of the linear dimension of a voxel, therefore the surfaces reconstructed from the 7 T MP2RAGE data would not seem able to substitute for surfaces reconstructed from the 3 T MEMPRAGE. However, in the absence of truth data it is not clear which reconstruction is more accurate—see Discussion.
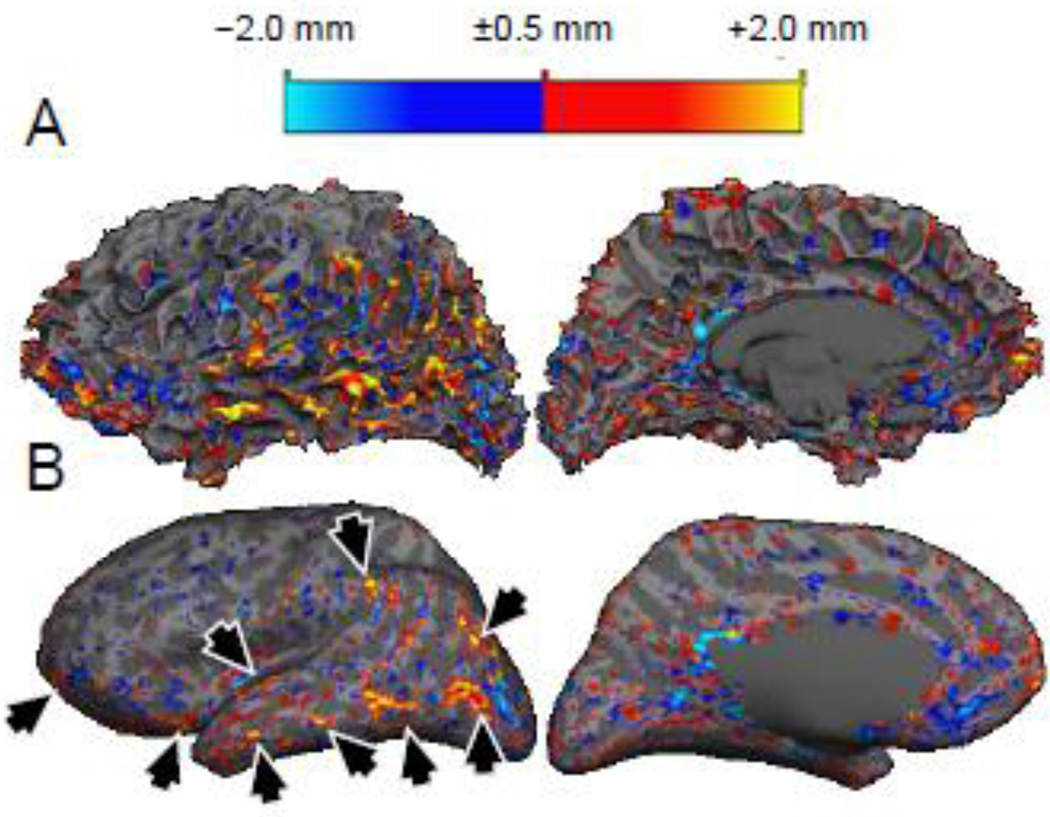
To assess whether any consistent patterns of thickness discrepancy between the acquisitions were present across our population of subjects, the signed thickness difference was averaged across subjects in the common surface atlas space. The results are presented in Fig. 6. For the first four comparisons (addressing the reproducibility of both acquisition methods), the spatial pattern of thickness difference is incoherent across the surface atlas indicating that there is little if any systematic discrepancies across subjects in any region of the brain. However, for the two comparisons between the two acquisition methods at 3 T a distinct spatial pattern is apparent. There is a clear bias for a thinner cortex over almost the entire hemisphere seen in the MP2RAGE data compared to the MEMPRAGE data. For the comparison between the 3 T MEMPRAGE and the 7 T MP2RAGE, there are regions where the 7 T MP2RAGE cortical segmentation is thicker than that of the 3 T MEMPRAGE, and regions where it is thinner. A large discrepancy is seen near the temporal pole, which is a region where tissue contrast is reduced in high field acquisitions due to the dielectric effects. Also a large, focal discrepancy is seen on the ventral aspect of the frontal lobe, which may be attributed to differential geometric distortion (or to poor inversion caused by narrow-bandwidth inversion pulses) due to susceptibility gradients around the frontal sinuses. To rule out the possibility that the averaging of the individuals thickness maps in the atlas space could cancel out effects seen at the single-subject level, we also computed the average cortical thickness in native subject space for all segmentations (N=24). The average thickness across the population for the MEMPRAGE was 2.42 mm ± 0.80 mm and for the MP2RAGE was 2.30 mm ± 0.83 mm.
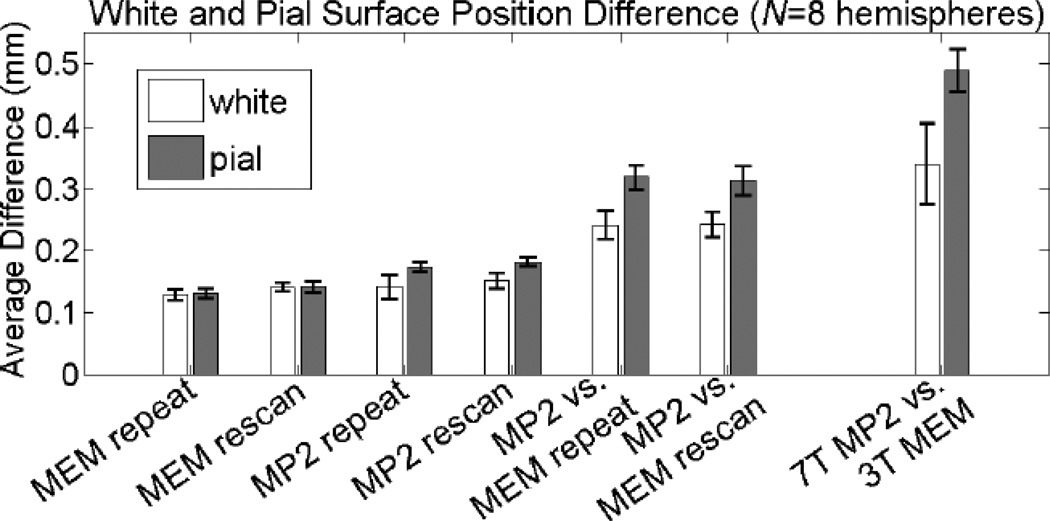
The results of the surface positioning discrepancy are summarized in Figs. 7 & 8 and Table 2, where the average distance between corresponding vertices across the white matter surfaces and the average distance between corresponding vertices across the pial surfaces across all subjects are reported. The reproducibility of the MEMPRAGE white and pial surfaces were high both for the repeat and rescan comparisons. Overall, the positioning of the white matter surface was more consistent between volumes for all comparisons conducted. This may be due to the potential of mis-classification of dura mater (and—when using a local transmit coil—blood vessels) as gray matter, which can displace the pial surface boundary (see Discussion). As expected, the positioning of the surfaces is more consistent between volumes acquired with the same acquisition method. In the first four comparisons addressing reproducibility, there is a trend towards a larger discrepancy seen in the MP2RAGE data relative to the MEMPRAGE data.
Fig. 7.
Summary of the gray-white and gray-pial boundary absolute position difference comparison across four subjects (eight hemispheres). Error bars indicate population standard deviation across the hemispheres.
Table 2.
Comparison of gray-white and gray-pial boundary position across four subjects with population standard deviation across the hemispheres. (All units are in mm.)
| comparison | Gray- White Boundary mean (mm) |
std. dev. (mm) |
Gray- Pial Boundary mean (mm) |
std. dev. (mm) |
|---|---|---|---|---|
| MEM repeat | 0.129 | 0.008 | 0.131 | 0.009 |
| MEM rescan | 0.141 | 0.009 | 0.142 | 0.007 |
| MP2 repeat | 0.142 | 0.009 | 0.174 | 0.020 |
| MP2 rescan | 0.152 | 0.007 | 0.182 | 0.013 |
| MP2 vs. MEM, repeat | 0.242 | 0.020 | 0.319 | 0.023 |
| MP2 vs. MEM, rescan | 0.243 | 0.024 | 0.314 | 0.020 |
| 7 T MP2 vs. 3 T MP2 | 0.340 | 0.034 | 0.489 | 0.064 |
A large discrepancy can be seen between the surfaces reconstructed with the MP2RAGE data compared to the surfaces reconstructed with the MEMPRAGE data. The average distance was 0.242 mm (± 0.020 mm) between white matter surfaces reconstructed from volumes acquired within the same block and 0.243 mm (± 0.024 mm) between white matter surfaces reconstructed from volumes acquired in different blocks separated by a repositioning of the subject; and the average distance was 0.319 mm (± 0.023 mm) between pial surfaces reconstructed from volumes acquired within the same block and 0.314 mm (± 0.020 mm) between pial surfaces reconstructed from volumes acquired in different blocks separated by a repositioning of the subject. Again the close agreement in the distance measures from data acquired within the same block with distance measures from data acquired across the two blocks indicates a successful robust registration between the MP2RAGE and MEMPRAGE data, and that measured differences can be attributed to differences in the tissue contrast and geometry. Finally, as with the thickness comparisons, the largest average distance was seen in the comparison between the 3 T MEMPRAGE and the 7 T MP2RAGE; an average distance of 0.340 mm (± 0.034 mm) was observed between white matter surfaces and an average distance of 0.489 mm (± 0.064 mm) was observed between pial surfaces.
To assess whether any consistent patterns of distance between corresponding surfaces generated from the different image volumes were present across our population of subjects, absolute distance between surfaces was averaged across subjects in the common surface atlas space. The results are presented in Fig. 8. For the first four comparisons (addressing the reproducibility of both acquisition methods), the spatial pattern of surface position difference is incoherent in the surface atlas space for both the white surfaces and for the pial surfaces indicating that there is little if any systematic discrepancies across subjects in any region of the brain. However, for the two comparisons between the two acquisition methods at 3 T a subtle spatial pattern is apparent only in the case of the pial surface comparisons. In these cross-method comparisons a larger distance is seen along the ridges of the gyri across subjects, suggesting that perhaps dura, which can cause errors in the pial surface placement in gyral regions where dura closely abuts the gray matter, is a primary cause of the discrepancy. For the comparison between the 3 T MEMPRAGE and the 7 T MP2RAGE, regions of large discrepancies consistent across subjects are again distinct near the temporal pole directly above the ear canals and in the low contrast region, and the near the ventral frontal lobe directly above the frontal sinuses.
To detect and quantify bias in the surface positioning we compiled a histogram of signed distance discrepancy—for both the white matter vertices and the pial surface vertices—for each of the comparisons in Fig. 8. The results are shown in Fig. 9 for four relevant comparisons. (The remaining three comparisons are shown in Supplementary Fig. 1.) The reproducibility of the surface placement for the MEMPRAGE method and the MP2RAGE method is presented in Figs. 9a and 9b, respectively. The histograms for these comparisons appear symmetric, indicating that there is no apparent bias in the placement of the surfaces between the repeated acquisitions, as expected. This indicates that any discrepancy between surfaces generated from repeated acquisitions is due to statistical errors (i.e., noise) as opposed to systematic errors (i.e., bias). The histograms characterizing the surface positioning discrepancy between back-to-back MEMPRAGE and MP2RAGE acquisitions, with the MEMPRAGE surfaces acting as the reference surfaces, are shown in Fig. 9c. Here the histograms for both the white matter surface placement and for the pial surface placement exhibit a clear asymmetry indicating that for the white matter the surfaces generated from the MP2RAGE data tend to be outside the surfaces generated from the MEMPRAGE data (i.e., the signed distances are more often positive with respect to the outward-pointing surface normal of the MEMPRAGE white surface); and for the pial surface the surfaces generated from the MP2RAGE data tend to be inside the surfaces generated from the MEMPRAGE data. While it may be expected that the MP2RAGE pial surfaces may lie outside the MEMPRAGE pial surface due to the possible inclusion of dura mater in the MP2RAGE gray matter segmentation (see Discussion), the finding that the MP2RAGE white surfaces are systematically placed inside of the gray matter compared to the MEMPRAGE white surfaces is less expected and may be due to subtle differences in the gray-white interface transition (or the spatial slope of the image intensities at this contrast boundary) seen in the MP2RAGE data compared to the MEMPRAGE data. Together, these two findings indicate that both MP2RAGE surfaces are shifted relative to the corresponding MEMPRAGE surfaces. Finally, the histogram characterizing the surface positioning discrepancy between the 3 T MEMPRAGE surfaces and the 7 T MP2RAGE surfaces, with the 3 T MEMPRAGE surfaces acting as the reference surfaces, is shown in Fig. 9d. While less asymmetry is apparent in the histogram comparing the two acquisition methods and the two field strengths, the tails of these histograms are quite heavy compared to those of the other comparisons (95% of the absolute displacements were less than 1.1 mm for the white surface and less than 2.0 mm for the pial surface), indicating that overall there is a much larger discrepancy between the surfaces generated from these volumes in agreement with the trend seen in Fig. 7.
Fig. 9.
Histograms of signed distance across four subjects (eight hemispheres) for test-retest pairs and 3 T MEMPRAGE vs. 7 T MP2RAGE. Both (A) MEMPRAGE repeat and (B) MP2RAGE repeat have symmetric histograms whereas the (C) MEMPRAGE– MP2RAGE repeat comparison shows that the MP2RAGE white matter surfaces are positioned outward (i.e., the signed distance is more often positive valued, with a positive-valued distance indicating a displacement in the direction of the outwardpointing surface normal of the MEMPRAGE white surface) and the MP2RAGE pial surfaces are positioned inward relative to the MEMPRAGE surfaces. (D) The 3 T MEMPRAGE vs. 7 T MP2RAGE rescan comparison is also symmetric, although a larger discrepancy between the 3 T and 7 T surfaces relative to the other comparisons is apparent from the heavy histogram tails. The number of surface vertices across the population was 145249 ± 15762; the number of vertices in the FreeSurfer white and pial surface meshes are identical by design. (Histograms have been restricted to ± 1 mm to aid visualization.)
Note that in each histogram there is a distinct dip in the center of the histogram corresponding to a small signed distance discrepancy between surfaces. While it might have been expected that the histogram would be zero-mean, this dip indicates that there are a small number of vertices in perfect agreement between each examined pair of surfaces. Given the resolution of the surface mesh, it is likely that in many cases the surfaces may be close to one another but the corresponding vertices might be displaced laterally in the direction tangential to the surface, which could partially account for this dip.
4. DISCUSSION
The proposed preprocessing steps designed to eliminate the noise enhancement in regions surrounding the brain that is induced by the MP2RAGE image reconstruction {Marques et al., 2010; Van de Moortele et al., 2009} enabled successful automatic cortical segmentation and surface reconstruction with FreeSurfer. Recently a straightforward regularization approach has been proposed to reduce this noise enhancement {O’Brien et al., 2013}, which warrants evaluation. Note that in some cases our preprocessing step was not needed for the MP2RAGE data—the conventional FreeSurfer segmentation and brain masking were able to successfully avoid the noise enhancement regions and properly segment the brain from the surrounding regions. However our preprocessing was required in other cases and so we recommend it as a default initial step when using MP2RAGE data with FreeSurfer. The proposed methodology can therefore be applied to generate surface models from 7 T data to allow for same-session morphometric analysis of anatomical data or surface-based analysis of functional data.
The greatest challenge in assessing accuracy in surface reconstructions is the lack of ground truth for the gray matter segmentation and associated surface mesh placement. Despite this, the cortical segmentation and surface reconstruction algorithms implemented in the FreeSurfer software have been validated and shown to capture the cortical boundary seen in the imaging data {Fischl & Dale, 2000; Kuperberg et al., 2003; Ly et al., 2012; Rosas et al., 2002}, agreeing with histology {Rosas et al., 2002}. Furthermore, FreeSurfer has been shown to provide a sub-voxel precision (i.e., scan-rescan reproducibility) of 120 µm in its cortical thickness measurements derived from standard 1×1×1 mm3 voxels {Han et al., 2006}. Given that the exact anatomical boundary between gray matter and white matter is partially indistinct {Hutsler & Avino, 2012} and that it has been shown to become blurry and ambiguous in some cortical regions during healthy aging {Salat et al, 2009}, the exact definition for the gray-white boundary is somewhat subjective. However, because cortical thickness is typically used as a tool to track or detect changes in gray matter either longitudinally (in studies of brain atrophy or plasticity) or cross-sectionally across groups (in studies of neurodegeneration or development for example), the exact definition of the cortical boundaries may not be as relevant as the precision with which the thickness is measured. The reproducibility of the cortical thickness estimate is therefore a measure of the sensitivity of the method to quantify gray matter differences.
We have observed a distinct bias in the positioning of the white matter surface between the MP2RAGE data and the MEMPRAGE data. Although we cannot determine which surface is more accurate at the gray-white boundary, the definition of the graypial boundary is less ambiguous, and we have detected errors in the MP2RAGE data segmentation due to mis-labeled dura mater (see below). The FreeSurfer method has been tested on and, to some extent, tailored to (ME)MPRAGE data, and it can robustly detect the gray-white boundary in images with the contrast achieved by standard MEMPRAGE protocols. Therefore it is possible that the segmentation algorithm could be adapted to be better suited to the MP2RAGE image data.
Previous studies have evaluated the reliability of cortical surface reconstructions by comparing estimates of cortical thickness in individuals across scans {Han et al., 2006; Reuter et al., 2012}, thereby assessing the precision of this morphometric measure. In the present study, when we performed the same evaluation on MEMPRAGE data and found a scan-rescan reproducibility of about 0.140 mm, which is comparable to the reproducibility of 0.120 mm reported by {Han et al., 2006}. We used this reproducibility measure to compare the reliability of MP2RAGE data compared with MEMPRAGE data and found that the reproducibility of the MP2RAGE data was slightly (but not significantly) worse than that of MEMPRAGE data.
The comparisons conducted in this study to assess the reproducibility of the cortical surface reconstructions and the agreement of the reconstructions between acquisitions were performed using the Longitudinal Processing Stream analysis implemented in FreeSurfer. The motivation behind our use of this longitudinal processing was to generate topologically equivalent surface meshes for any two volumes under comparison (which provides a natural vertex correspondence between the resulting surface reconstructions) in a way that was unbiased to favor either volume. The surface mesh generated from the unbiased template (which is an average of the two volumes, after robust registration) is used as an initialization for the re-positioning of the surface mesh relative to each of the two individual volumes, and this re-positioning is driven by optimizing the same cost function that is used in the conventional FreeSurfer surface reconstruction algorithm. If the surface placement optimization were guaranteed to find a global minimum to this cost function, the re-positioning procedure would provide surfaces with the same geometry as would be produced by the default, conventional FreeSurfer stream, but local minima can be encountered, and noise in the individual image volumes may influence the surface placement. This Longitudinal Processing Stream processing may therefore provide higher accuracy than the conventional approach due to the reduced noise present in the unbiased template that provides an initialization that is closer to the tissue boundaries. This suggests, however, that the results provided by the Longitudinal Processing Stream may not generalize to those provided by the conventional stream, which is based on the original image volumes that contain higher levels of noise than the unbiased template. It is possible that one could also detect these same thickness differences using the conventional stream if the measurements across a large population of subjects were pooled, which would help to average out the effects caused by the higher noise levels seen in the individual image volumes. In addition to the potential discrepancy caused by differing noise levels, the Longitudinal Processing Stream may also provide more accurate thickness difference measures by virtue of the vertex correspondence it provides. While surface-based registration (e.g., via a surface atlas) can generate a good correspondence of cortical areas and cortical folds based on large-scale geometric features of the cortical folding pattern, it is not intended to align surface meshes at the vertex level. Therefore for these two reasons the Longitudinal Processing Stream may provide a more accurate estimate of differences between surface reconstructions and the true differences in apparent cortical thickness derived from the various image volumes tested.
One potential drawback of the MP2RAGE approach is that the GM-WM boundary in the resulting image volumes may be strongly dependent on the interaction between the TI1 parameter and the T1 value of cortical gray matter. However, the distribution of GM T1 is known to vary across the brain {Fischl et al., 2004a} and even appears to vary between different cortical layers and across different cortical areas {Barazany & Assaf, 2012}. Thus, as with any T1-weighted anatomical imaging, biases will exist in the boundaries of the GM seen in the MP2RAGE data across the brain—and the biases in the MP2RAGE data are expected to differ from those same biases seen in other methods. It may be possible to reduce or remove this bias by generating surface reconstructions directly from a quantitative T1 map {Fischl et al., 2004a} rather than a T1-weighted image, such as the T1 map generated online from the two inversion recovery readouts acquired in the MP2RAGE method {Fujimoto et al., 2013}. However, because the derived T1 map, when treated as an image, has somewhat different noise properties than a native T1-weighted image {Fischl et al., 2004a} and can have lower gray-white contrast-to-noise ratio than an MPRAGE image volume {Fujimoto et al., 2013}, the precision of the gray matter segmentation and of the positions of the derived surfaces may be affected as a consequence.
To better understand the sources of discrepancy between cortical thickness estimates derived from MP2RAGE data and MEMPRAGE data, we developed a novel analysis approach to compare surface positioning and to calculate biases in the placement of the cortical surface meshes. This method provided insight into the consistently thinner estimates from the MP2RAGE data (despite the sporadic contamination of the cortical gray matter segmentation with abutting dura), which appear to be caused by a systematic relative displacement of the gray-white surfaces generated from MP2RAGE data outside of the gray-white surfaces generated from MEMPRAGE data. This type of approach should generally be used in future studies assessing cortical gray matter segmentation and surface reconstruction techniques. The quantification of the positioning of the surfaces is also relevant for studies investigating the anatomy, physiology, and function of cerebral cortical layers, since these studies typically estimate the positions of the layers based on the radial distance from the graywhite surface or based on the percent thickness between the gray-white and gray-pial surfaces {Polimeni et al., 2010a, 2010b; Waehnert et al., 2012}.
A recent study {Lüsebrink et al., 2013} investigated the effect of voxel size on the cortical thickness measures derived from 3 T and 7 T anatomical data, using an imaging technique similar to MP2RAGE {Van de Moortele et al., 2009}, and found consistently thinner cortical estimates from 0.5×0.5×0.5 mm3 voxels compared to conventional 1×1×1 mm3 voxels, as well as a close agreement between thickness measures derived from 1×1×1 mm3 3 T and 7 T surfaces. Although there were several differences in contrast and blurring effects between the 0.5×0.5×0.5 mm3 acquisition and the 1×1×1 mm3 acquisition, making the differences due solely to nominal voxel size difficult to quantify, future high-resolution comparisons between MP2RAGE and MEMPRAGE, perhaps with prospective motion correction {Tisdall et al., 2012, 2013}, could aid in identifying the observed differences in surface placement reported in the present study.
While the current report has focused on the quality of the cerebral cortical gray matter segmentation and associated morphometric measures, many studies require automatic parcellation of the cortex into named sulci and gyri as well as segmentation and identification of subcortical structures, both of which are provided by FreeSurfer {Fischl et al., 2002, Fischl et al., 2004b}. The preprocessing of the MP2RAGE data introduced here in order to generate cortical segmentations and surface reconstructions also provides a T1-weighted volume that is suitable for cortical parcellation and subcortical segmentation. A qualitative comparison of cortical parcellation and subcortical segmentation generated by FreeSurfer on the MEMPRAGE and MP2RAGE volumes is provided in Supplementary Fig. 2. Future work will be required to quantitatively compare and evaluate the cortical parcellation and subcortical segmentation using methods similar to those employed in this report.
The MP2RAGE method, by providing uniform tissue contrast across the brain, has already found uses in clinical applications {Tanner et al., 2012} and exhibits sufficient contrast in human cortical gray matter to distinguish adjacent cortical areas based on laminar differences in myelin content seen in high-resolution acquisitions {Geyer et al., 2011}. However our analysis shows some unexpected differences between the MP2RAGE tissue contrast and that of the MEMPRAGE method. There are unfortunately several differences between the MP2RAGE method and the MEMPRAGE method that confound our ability to isolate which factor contributes most to the observed discrepancies. One potential feature of the MP2RAGE method that could explain the observed reduction in cortical thickness relative to the MEMPRAGE method is the reduced T2* contrast achieved by the bias removal {Marques et al., 2010; Van de Moortele et al., 2009}. It is possible that the T2* signal that remains in the MEMPRAGE images could shift the apparent gray-white boundary, and because these effects grow with magnetic field strength this could be one source of discrepancy when comparing MEMPRAGE-based surface reconstructions acquired across field strengths. However, given that T2* weighted images do also provide a sharp gray-white boundary it is unclear to what extent the T2* content in the MEMPRAGE data would bias the localization of this boundary. Quantitative T2* mapping has also demonstrated that the orientation of the cortical gray matter relative to the direction of the B0 field causes local changes in T2* value {Cohen-Adad et al., 2012}, and this could potentially lead to a position-dependent effect. It is less clear how the residual T2* signal in the MEMPRAGE could bias the position of the pial surface boundary. In a post-hoc analysis we confirmed that the pial boundary displacement was not a result of our MP2RAGE masking procedure by carefully visually re-inspecting all masked volumes.
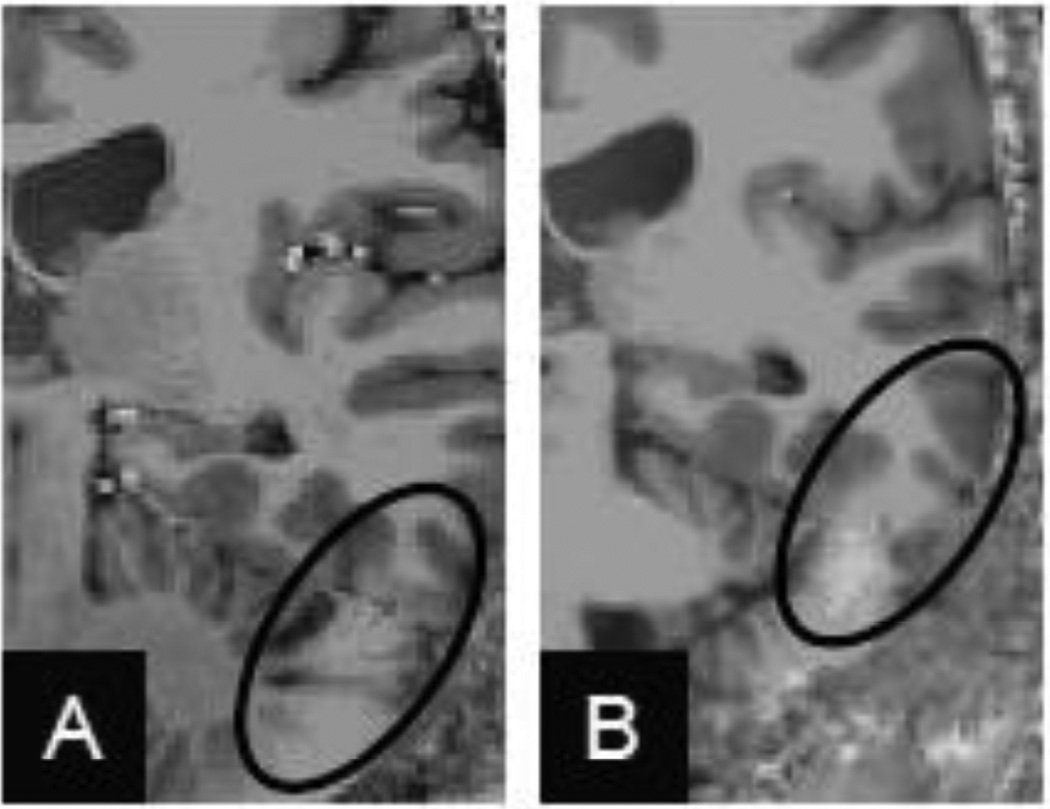
A major feature of the MEMPRAGE method that is not currently available in our MP2RAGE data is the partial suppression of the dura mater superjacent to the cerebral cortical gray matter. This is achieved through the acquisition of multiple echoes with progressively longer echo times, in which the intensity of the dura diminishes due to its relatively short T2* value {van der Kouwe et al., 2008}. While the dura contamination cannot explain the trend where the pial surface of the MP2RAGE data is positioned inside of the pial surface of the MEMPRAGE data (see Fig. 9C), it may partly explain the lower precision of the pial surface placement (see Fig. 7). However, it may be possible to exploit the short T2* feature of the dura to suppress it in an MP2RAGE acquisition using a similar multi-echo approach. To test this idea, in Fig. 10 we show a preliminary comparison of the pial surface placement between surface reconstructions generated from the first and fourth echoes of a prototype “MEMP2RAGE” acquisition in one subject, where the echo times are matched to our MEMPRAGE acquisition protocol. The discrepancy between the two sets of surface reconstructions is largest at the crowns of gyri within the folding pattern, exactly where contamination of the overlying dura mater occurs. This observation is consistent with the discrepancy across subjects in the placement of the pial surface reconstruction near the crowns of gyri, as seen in the population maps shown in Fig. 8E & F. This preliminary finding suggests that a multi-echo MP2RAGE method may be beneficial for removing this one potential source of error in the MP2RAGE method (and for removing potential variability due to geometric distortion in the readout direction due to susceptibility effects) and may thereby reduce the differences in cortical thickness seen between MP2RAGE and MEMPRAGE. However the utility of the multi-echo approach applied to MP2RAGE may be somewhat diminished by the reduced T2* contrast in the MP2RAGE images, described above. An alternative route to increasing the contrast to noise ratio would be to investigate strategies for reducing noise. Accelerated parallel imaging was employed in all anatomical protocols, thus there is increased noise relative to a longer, unaccelerated protocol. One feature of the original strategy of Van de Moortele et al., {2009} in which the MPRAGE volume is acquired separate from the subsequent protondensity-weighted gradient-recalled echo reference volume is that each volume can be acquired with different levels of acceleration, enabling the possibility of reducing the acceleration factor of the gradient-recalled echo reference to decrease noise and thereby increase contrast to noise ratio. However this increased contrast to noise ratio would require a longer acquisition time.
Fig. 10.
Thickness difference between surfaces independently reconstructed from the first and fourth echoes of an example four-echo “ME-MP2RAGE”, visualized on the white matter surface. (A) Original folded representation. Large thickness discrepancies can be seen on the crowns of the gyri. (B) Inflated representation. Arrowheads indicate example regions of large thickness discrepancy between the echoes concentrated on the tops of the gyri.
Care was taken to use the recommended imaging protocols for each technique to best capture the differences in image quality expected to be seen in practice. However, these protocols differ not only in terms of image contrast but also in terms of, for example: image distortion along the readout direction (due to macroscopic susceptibility gradients); T1 image blurring during the inversion recovery; motion artifact vulnerability due to the different acquisition durations; and contrast-to-noise ratio. We did not detect any systematic differences between the MP2RAGE data and the MEMPRAGE data in any of the image encoding directions (see Figs. 8E & F), but it is possible that some of the increased variability seen in the MP2RAGE data is due to the reduced bandwidth of this protocol {Fischl et al., 2004a} or to the longer duration and greater potential for motion-induced blurring {Tisdall et al., 2012, 2013}. Ideally all acquisition differences between the two acquisitions would be carefully controlled in order to directly assess the impact of image contrast on the location of the gray matter borders.
In this study we compared data acquired with the recommended MEMPRAGE protocol to data acquired with the recommended MP2RAGE protocol in order to provide a practical comparison of the segmentation results from the most commonly-used protocols. As a consequence, the two protocols have different readout bandwidth values which may lead to discrepancies that could be reduced by matching the bandwidth; however by increasing the bandwidth used in the MP2RAGE protocol we would also reduce the image CNR, which would add another confound. Given that we observed no systematic patterns of discrepancy between the two methods at 3 T, no clear discrepancy near the known regions of large susceptibility gradients, and a systematic reduction in cortical thickness seen in the MP2RAGE relative to the MEMPRAGE segmentation, the mismatch in readout bandwidth is unlikely to be the main cause for the observed discrepancies. Rather, differences in contrast and positions of the contrast boundaries are more likely the cause of the discrepancies observed at 3 T.
Susceptibility differences are also present between the 3 T and 7 T data, and the discrepancies seen in the comparison between 3 T MEMPRAGE and 7 T MP2RAGE may also be caused by effects other than differences in image contrast. Not only may differential geometric distortions cause discrepancies, but other factors such as noninverted spins near susceptibility regions due to the narrow bandwidth of the adiabatic inversion pulses {Wrede et al., 2012}, residual contrast nonuniformity due to dielectric effects {Marques & Gruetter, 2013; Marques et al., 2010}, and bright signal in blood vessels {Van de Moortele et al., 2009} may cause discrepancies that should not be attributed to differences in the two acquisition methods alone. Ongoing work seeking to improve the quality of the 7 T data by addressing these issues will help make the tissue segmentation to be consistent across field strengths.
The MP2RAGE method was initially introduced as a method to remove the spatially varying tissue contrast that occurs at ultrahigh field strengths such as 7 T due to the dielectric properties of the tissue that give rise to spatially varying transmit efficiency and flip angles {Marques et al., 2010; Van de Moortele et al., 2009}. Additionally, different transmit head coil designs can exhibit different levels of transmit uniformity across the brain, and therefore can affect the severity of the spatially varying loss of tissue contrast. A typical example of this is shown in Fig. 11 in which MP2RAGE data acquired with both a small bandpass birdcage volume coil (16 rungs, 27 cm inner diameter, 21 cm endring-to-endring length) built for an insert head gradient coil system and with a large bandpass birdcage volume coil (16 rungs, 30 cm inner diameter, 33 cm endring-to-endring length) built for a body gradient coil system are presented. Transmit efficiency varies with coil design and geometry {Wald et al., 2005}, and in this case the two volume transmit coils exhibit different spatial efficiency patterns especially near the temporal lobe and cerebellum, which noticeably impact the image contrast in these regions. Therefore, as this example shows, the extent to which the MP2RAGE method can successfully remove all of the lost tissue contrast at 7 T can depend somewhat on the transmit coil design, and so the generalizability of our 7 T results may be influenced by the specific transmit coil in use.
Fig. 11.
Example of “flat image” with (A) a small birdcage transmit coil and (B) a large birdcage transmit coil, shown on the same subject. The small transmit coil provides better tissue contrast than the large birdcage coil. The ovals show a region with reduced tissue contrast in both examples.
As expected, the MP2RAGE method provided high-quality T1-weighted anatomical volumes that we were able to process automatically with the FreeSurfer software tools. Despite the abovementioned focal image artifacts seen in the 7 T MP2RAGE data at the temporal pole, the FreeSurfer method is robust to a sharp, focal loss of contrast. The resulting surface reconstructions are valid elsewhere in the brain even when these image artifacts are present, although care must be taken in interpreting cortical surface placement and thickness measures in the immediate vicinity of these artifacts. These artifacts were far larger in extent in the 7 T MEMPRAGE data, and covered a large portion of the temporal lobe bilaterally. The image artifacts included both a loss of tissue contrast between gray matter and white matter, as well as an overall loss of image intensity that lead to a loss of contrast between cortical gray matter and the surrounding CSF. Because 7 T MEMPRAGE surface reconstructions currently require some manual editing to accurately reconstruct the entire surface (especially around the temporal poles) due to the spatially varying contrast and intensity, we conservatively did not include any 7 T MEMPRAGE comparisons in this study.
5. CONCLUSIONS
A method for preprocessing 3 T and 7 T MP2RAGE image data for subsequent automatic tissue segmentation and cortical surface reconstruction with FreeSurfer was presented. Using these surfaces, we were able to quantitatively compare the segmentation results between data acquired with the MP2RAGE method to those generated from the MEMPRAGE method. The reproducibility of the cortical thickness measure and the cortical surface placement was shown to be lower for the MP2RAGE data relative to the MEMPRAGE data, and the consistent observation of a thinner cortex in MP2RAGE data relative to the MEMPRAGE data. These results suggest that tissue segmentation methods may need to be tailored to the MP2RAGE data, or that further developments of the MP2RAGE method, such as a multi-echo MP2RAGE, may be required to reduce or to better understand this discrepancy.
Supplementary Material
HIGHLIGHTS.
-
-
FreeSurfer automatic cortical surface reconstruction is extended to MP2RAGE data.
-
-
Reproducibility is quantified for 3T MP2RAGE and ME-MPRAGE surface reconstructions.
-
-
A novel analysis of surface discrepancies across imaging methods is introduced.
-
-
Systematic differences are found between MP2RAGE and ME-MPRAGE surfaces.
-
-
Potential extensions to MP2RAGE are suggested to reduce these discrepancies.
ACKNOWLEDGEMENTS
Support for this research was provided in part by the NIH National Institute for Biomedical Imaging and Bioengineering (R01-EB006758, P41-EB015896, K01-EB011498 and R01-EB006847), the National Institute on Aging (R01-AG022381, R01-AG008122), the National Center for Alternative Medicine (RC1-AT005728), the National Institute for Neurological Disorders and Stroke (R01-NS0525851, R21-NS072652, R01-NS070963), and the National Center for Research Resources (P41-RR14075, and the NCRR BIRN Morphometric Project BIRN002, U24-RR021382), and was made possible by the resources provided by Shared Instrumentation Grants S10-RR023401, S10-RR019307, S10-RR023043, S10-RR019371, and S10-RR020948. Additional support was provided by The Autism & Dyslexia Project funded by the Ellison Medical Foundation, and by the NIH Blueprint for Neuroscience Research (U01-MH093765), part of the multi-institutional Human Connectome Project. We thank Allison Player, Nick Schmansky, Thomas Witzel, Bastien Guerin, Dylan Tisdall, and Boris Keil for helpful discussions. We thank an anonymous reviewer for suggesting the novel idea of a MEMPRAGE acquisition with a GRE reference.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barazany D, Assaf Y. Visualization of cortical lamination patterns with magnetic resonance imaging. Cerebral Cortex. 2012;22(9):2016–2023. doi: 10.1093/cercor/bhr277. [DOI] [PubMed] [Google Scholar]
- Cloos MA, Boulant N, Luong M, Ferrand G, Giacomini E, Hang M-F, Wiggins CJ, Le Bihan D, Amadon A. Parallel-transmission-enabled magnetization-prepared rapid gradient-echo T1-weighted imaging of the human brain at 7 T. NeuroImage. 2012;62(3):2140–2150. doi: 10.1016/j.neuroimage.2012.05.068. [DOI] [PubMed] [Google Scholar]
- Cohen-Adad J, Polimeni JR, Helmer KG, Benner T, McNab JA, Wald LL, Rosen BR, Mainero C. T2* mapping and B0 orientation-dependence at 7 T reveal cytoand myeloarchitecture organization of the human cortex. NeuroImage. 2012;60(2):1006–1114. doi: 10.1016/j.neuroimage.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CM, Liu W, Schreiber W, Yang QX, Smith MB. Central brightening due to constructive interference with, without, and despite dielectric resonance. Journal of Magnetic Resonance Imaging. 2005;21(2):192–196. doi: 10.1002/jmri.20245. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Transactions on Medical Imaging. 2001;20(1):70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Ségonne F, Quinn BT, Dale AM. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004a;23(Suppl 1):S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human Brain Mapping. 1999;8(4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004b;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Polimeni JR, van der Kouwe AJ, Kober T, Benner T, Fischl B, Wald LL, Reuter M. Comparison of cortical surface reconstructions from MP2RAGE data at 3T and 7T. Proc Intl Soc Mag Reson Med. 2011a;19:130. doi: 10.1016/j.neuroimage.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K, Polimeni JR, van der Kouwe AJ, Reuter M, Kober T, Benner T, Fischl B, Wald LL. Comparison of cortical surface reconstructions from MP2RAGE data at 3T and 7T; 17th Annual Meeting of the Organization for Human Brain Mapping; 2011b. [Google Scholar]
- Fujimoto K, Wald LL, Polimeni JR. Comparison of cortical surface reconstructions between quantitative T1 and T1-weighted volumetric data. Proc Intl Soc Mag Reson Med. 2013;21:950. [Google Scholar]
- Geyer S, Weiss M, Reimann K, Lohmann G, Turner R. Microstructural parcellation of the human cerebral cortex - from Brodmann’s post-mortem map to in vivo mapping with high-field magnetic resonance imaging. Frontiers in Human Neuroscience. 2011 Feb;5:19. doi: 10.3389/fnhum.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Busa E, Pacheco J, Albert M, Killiany R, Maguire P, Rosas D, Makris N, Dale A, Dickerson B, Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. NeuroImage. 2006;32(1):180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Huang R-S, Chen C, Tran AT, Holstein KL, Sereno MI. Mapping multisensory parietal face and body areas in humans. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(44):18114–18119. doi: 10.1073/pnas.1207946109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley AC, Al-Radaideh A, Bai L, Aickelin U, Coxon R, Glover P, Gowland PA. Tailored RF pulse for magnetization inversion at ultrahigh field. Magnetic Resonance in Medicine. 2010;63(1):51–58. doi: 10.1002/mrm.22167. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Avino TA. Sigmoid fits to locate and characterize cortical boundaries in human cerebral cortex. Journal of Neuroscience Methods. 2012;212(2):242–246. doi: 10.1016/j.jneumeth.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006;30(2):436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Keil B, Triantafyllou C, Hamm M, Wald LL. Design optimization of a 32-channel head coil at 7T. Proc Intl Soc Mag Reson Med. 2010;18:1493. [Google Scholar]
- Kuperberg GR, Broome MR, McGuire PK, David AS, Eddy M, Ozawa F, Goff D, West WC, Williams SCR, van der Kouwe AJW, Salat DH, Dale AM, Fischl B. Regionally localized thinning of the cerebral cortex in schizophrenia. Archives of General Psychiatry. 2003;60(9):878–888. doi: 10.1001/archpsyc.60.9.878. [DOI] [PubMed] [Google Scholar]
- Ledden PJ, Mareyam A, Wang S, Van Gelderen P, Duyn J. 32 channel receive-only SENSE array for brain imaging at 7T. Proc Intl Soc Mag Reson Med. 2007;15:242. [Google Scholar]
- Lüsebrink F, Wollrab A, Speck O. Cortical thickness determination of the human brain using high resolution 3T and 7T MRI data. NeuroImage. 2013;70:122–131. doi: 10.1016/j.neuroimage.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Ly M, Motzkin JC, Philippi CL, Kirk GR, Newman JP, Kiehl KA, Koenigs M. Cortical thinning in psychopathy. American Journal of Psychiatry. 2012;169(7):743–749. doi: 10.1176/appi.ajp.2012.11111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JP, Gruetter R. Yacoub E. PloS one. 7. Vol. 8. 2013. New developments and applications of the MP2RAGE sequence--focusing the contrast and high spatial resolution R1 mapping; p. e69294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele P-F, Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage. 2010;49(2):1271–1281. doi: 10.1016/j.neuroimage.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Mugler JP, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magnetic Resonance in Medicine. 1990;15(1):152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- O’Brien K, Krueger G, Lazeyras F, Gruetter R, Roche A. A simple method to denoise MP2RAGE. Proc Intl Soc Mag Reson Med. 2013;21:269. [Google Scholar]
- Ordidge RJ, Wylezinska M, Hugg JW, Butterworth E, Franconi F. Frequency offset corrected inversion (FOCI) pulses for use in localized spectroscopy. Magnetic Resonance in Medicine. 1996;36(4):562–566. doi: 10.1002/mrm.1910360410. [DOI] [PubMed] [Google Scholar]
- Polimeni JR, Fischl B, Greve DN, Wald LL. Laminar analysis of 7T BOLD using an imposed spatial activation pattern in human V1. NeuroImage. 2010a;52(4):1334–1346. doi: 10.1016/j.neuroimage.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimeni JR, Fischl BR, Greve DN, Wald LL. Laminar-specific functional connectivity: distinguishing directionality in cortical networks; 16th Annual Meeting of the Organization for Human Brain Mapping; 2010b. [Google Scholar]
- Reuter M, Fischl B. Avoiding asymmetry-induced bias in longitudinal image processing. NeuroImage. 2011;57(1):19–21. doi: 10.1016/j.neuroimage.2011.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. NeuroImage. 2010;53(4):1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. NeuroImage. 2012;61(4):1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Liu AK, Hersch S, Glessner M, Ferrante RJ, Salat DH, van der Kouwe A, Jenkins BG, Dale AM, Fischl B. Regional and progressive thinning of the cortical ribbon in Huntington’s disease. Neurology. 2002;58(5):695–701. doi: 10.1212/wnl.58.5.695. [DOI] [PubMed] [Google Scholar]
- Salat DH, Lee SY, van der Kouwe a J, Greve DN, Fischl B, Rosas HD. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. NeuroImage. 2009;48(1):21–28. doi: 10.1016/j.neuroimage.2009.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. A hybrid approach to the skull stripping problem in MRI. NeuroImage. 2004;22(3):1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Setsompop K, Alagappan V, Gagoski BA, Potthast A, Hebrank F, Fontius U, Schmitt F, Wald LL, Adalsteinsson E. Broadband slab selection with B1+ mitigation at 7T via parallel spectral-spatial excitation. Magnetic Resonance in Medicine. 2009;61(2):493–500. doi: 10.1002/mrm.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. Journal of Neuroscience. 2004;24(38):8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdall MD, Hess AT, Reuter M, Meintjes EM, Fischl B, van der Kouwe AJW. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magnetic Resonance in Medicine. 2012;68(2):389–399. doi: 10.1002/mrm.23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdall MD, Polimeni JR, Augustinack J, van der Kouwe AJW. 350 µm isotropic, high-contrast, low-blur, low-distortion MPRAGE morphometry acquisition at 3T; 19th Annual Meeting of the Organization for Human Brain Mapping; 2013. [Google Scholar]
- Van de Moortele P-F, Auerbach EJ, Olman C, Yacoub E, Uğurbil K, Moeller S. T1 weighted brain images at 7 Tesla unbiased for Proton Density, T2* contrast and RF coil receive B1 sensitivity with simultaneous vessel visualization. NeuroImage. 2009;46(2):432–446. doi: 10.1016/j.neuroimage.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kouwe AJW, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. NeuroImage. 2008;40(2):559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waehnert M, Weiss M, Streicher M, Bazin P-L, Geyer S, Turner R. Do cortical layers conform to the Laplace equation. 18th Annual Meeting of the Organization for Human Brain Mapping. 2012;18:MT898. [Google Scholar]
- Wald LL, Wiggins GC, Potthast a, Wiggins CJ, Triantafyllou C. Design considerations and coil comparisons for 7 T brain imaging. Applied Magnetic Resonance. 2005;29(1):19–37. [Google Scholar]
- Wiggins GC, Polimeni JR, Potthast A, Schmitt M, Alagappan V, Wald LLL. 96-Channel receive-only head coil for 3 Tesla: design optimization and evaluation. Magnetic Resonance in Medicine. 2009;62(3):754–762. doi: 10.1002/mrm.22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede KH, Johst S, Dammann P, Umutlu L, Schlamann MU, Sandalcioglu IE, Sure U, Ladd ME, Maderwald S. Caudal image contrast inversion in MPRAGE at 7 Tesla: problem and solution. Academic Radiology. 2012;19(2):172–178. doi: 10.1016/j.acra.2011.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.