Abstract
The eukaryotic genome exists in vivo at an equimolar ratio with histones, thus forming a polymer composed of DNA and histone proteins. Each nucleosomal unit in this polymer provides versatile capabilities and dynamic range. Substitutions of the individual components of the histone core with structurally distinct histone variants and covalent modifications alter the local fabric of the chromatin fiber, resulting in epigenetic changes that can be regulated by the cell. In this review, we highlight recent advances in the study of histone variant structure, assembly, and inheritance, their influence on nucleosome positioning, and their cumulative effect upon gene expression, DNA repair and the progression of disease. We also highlight fundamental questions that remain unanswered regarding the behavior of histone variants and their influence on cellular function in the normal and diseased states.
Introduction
In Norse mythology, the trickster Loki plays the role of “Stirrer of strife, mischief-monger, Maker of laughter and bringer of change, Friend and foeman, order and chaos” [1]. Akin to Loki, tiny, positively charged proteins called histones impose different chromatin states and encode epigenetic changes in an otherwise staid genome. These proteins date back to the dawn of eukaryotic evolution, spanning protozoans, fungi, animals, and plants. Indeed, prokaryal and archaeal species are the earliest genomes known to have evolved histone-like proteins [2,3]. Bacterial genomes contain histone-like HU proteins, which bind and bend DNA, stabilize higher order chromosomal folding during replication, and regulate transcription (Figure 1a) [2]. Histone-like proteins are also present in the archaea [3]. For example, in the extremophile Methanothermus fervidus, archaeal histones (Figure 1b,c) form tetrameric complexes, which wrap ~70 bp of DNA in a right-handed toroid, into which histone subunits are exchanged in response to environmental stressors such as salt concentration or temperature [4]. Another archaeal organism, Methanopyrus kandleri, contains a fused “doublet” histone fold protein, wherein one of the histone folds shares homology with the histone folds of eukaryotic H2A and H4 (Figure 1d), suggesting that the eukaryotic histone genes for H2A, H2B, H3, and H4 probably arose from duplication of primitive archaeal histone genes [5].
Figure 1.
Histone-like proteins are found in every kingdom of life and serve to regulate access to the genome, paritally through formation of tertiary structure. In panel A, the co-crystal structure of the HU protein from Anabena and DNA (PDB ID 1P78) shows the HU protein dimer binding the DNA duplex. Panel B shows the crystal structure of the HmfA homodimer from Methanothermus fervidus (PDB ID 1B67) while panel C shows the crystal structure of HmfB (PDB ID 1A7W), also from Methanothermus fervidus. The crystal structure of the fused doublet HmfA from Methanopyrus kandleri (PBD ID 1F1E) is shown in panel D. In panel E, the crystal structure of the canonical eukaryotic histones (PDB ID 1AOI) shows the assembly of the histone octamer, the protein core of the nucleosome. Nucleosomes were previously thought to form 30 nm solenoid fibers as shown in panel F, through recent evidence suggests the genome exists primarily as a 10 nm “beads-on-a-string” fiber, as shown in panel G, with some regions of higher order organization like those shown in panel H.
In eukaryotes, 147 bp of DNA wrap in a left-handed torus around an octameric complex composed of two copies each of the invariant histones H3, H2A, H2B and H4 (Figure 1e) [6]. Since the discovery that the vast majority (>70%) of DNA in eukaryotes is packaged into nucleosomes, and the landmark X-ray diffraction study by Finch and Klug showing chromatin was organized into highly compacted 30 nm wide solenoidal coils (Figure 1f), histones were proposed to function primarily as packaging material for ever-growing eukaryotic genomes [7]. However, a serious challenge to the existence of the 30 nm chromatin fiber comes from recent theoretical and experimental analyses. Computational modeling of the chromatin fiber suggests that the nucleoprotein polymer is theoretically far less efficient for packaging than was previously assumed [8,9], and a series of experimental studies provide support for these computational models. Using cryo-electron microscopy (EM) coupled with careful measurements, 30 nm fibers were not detected in interphase nuclei, or even in metaphase chromosomes [10•,11•]. Using small-angle X ray scattering (SAXS), another group likewise reported it was unable to detect 30 nm fibers in vivo, but rather raised the startling possibility that the data which first reported 30 nm fibers might instead have been periodic reiterations of ribosomes, which are 30 nm in width and were found to coat the chromatin under certain preparative procedures [12•]. Despite the ongoing debate on this issue [13], it does appear that much of the chromatin fiber exists in the 10 nm fiber state (beads on a string) (Figure 1g), with a few locally folded areas comprising 5-10 nucleosomes and with 3D “fractal globule” arrangements of chromatin fibers stabilized in a cross-array format (Figure 1h), the density of which is possibly coordinated by linker histone H1 and networks of non-histone proteins [14]. These results, along with the evolutionary evidence that archaeal histones do not function as a packaging molecule, lend themselves to the possibility that histones may have evolved primarily as a means of regulating local access to genes [15,16]. Thus, if canonical histones generally serve to regulate access to the DNA, what additional roles do specialized histone variants play in regulating the various cellular processes that occur throughout the genome?
All eukaryotes studied thus far contain the histone variant H3.3 and the centromere-specific histone variant CENP-A/CENH3, even when they lack other H3 types [17]. Additional variants include H2A.Z/HTZ, H2A.X, H2Av, H2A.Bbd, macroH2A [17], the primate-specific histones H3.X and H3.Y [18] (Table 1), and a plethora of histone H1 variants. Remarkably, while these proteins were discovered decades ago, their precise function, the mechanisms by which they effect change on the chromatin fiber, how they are inherited in vivo, and their contributions to the progression of disease states remain open questions in biology.
Table 1.
A list of known functions for histone variants in the eukaryotic genome
| Histone variant | Function | Conserved? |
|---|---|---|
| CENP-A/CID/cse4 | Epigenetic marker of the centromere | Yes |
| H3.3 | Transcription | Yes |
| H2A.Z/H2AV | Transcription/double strand break repair | Yes |
| H2A.X | Double strand break repair/meiotic remodeling of sex chromosomes | Yes |
| macroH2A | Gene silencing/X chromosome inactivation | Yes |
| H2A.Bbd | Epigenetic mark of active chromatin | Yes |
| H3.Z | Regulation of cellular response to outside stimuli | No |
| H3.Y | Regulation of cellular response to outside stimuli | No |
In this review, we highlight recent advances and yet to be answered fundamental questions regarding the behavior of histone variants and their influence on cellular function in the normal and diseased states.
An ancient foe of the polymerase: the role of histone variants in regulating transcription
The histone variants H3.3 and H2A.Z have both been individually linked to a role in regulating transcription, but biochemical purification suggests that these two variants may come together in a single nucleosome. Using HeLa cells expressing a Flag-tagged H3.3 histone, single nucleosomes were isolated and subjected to immunoprecipitation (IP) followed by sequencing to determine their location relative to the transcription start sites (TSS) of three separate classifications of genes: highly expressed, intermediately expressed, and silent [19]. H3.3/H2A.Z hybrid nucleosomes localized to the TSS of active genes, at sites that have previously been characterized as nucleosome depleted regions (NDRs). Upon modulating the salt concentration used in the nucleosome isolation, it was discovered that H3.3/H2A.Z nucleosomes are unstable in vivo, causing them to dissociate from the DNA during extraction, leaving behind a NDR. Although a crystal structure is not available for this double hybrid, in vitro characterization of the H3.3/H2A.Z nucleosome's stability by salt induced dissociation revealed only very small differences compared to the stability of the canonical nucleosome, resulting in a puzzling discrepancy between in vivo and in vitro results [20]. However, a recent investigation into a post-translational modification (PTM) found not on the histone tail, but at H3K122, in the center of the nucleosome core, suggests a plausible explanation that could neatly resolve this discrepancy [21••]. Acetylation at H3K122 disrupts the interaction between the histone core and DNA, destabilizing the nucleosome [22••]. Furthermore, it co-localizes with H3.3 and H2A.Z in vivo, leading to the compelling hypothesis that K122 acetylation on H3.3, which is absent in the in vitro studies, may be responsible for the destabilized H3.3/H2A.Z nucleosome in vivo [21••]. An alternative attractive explanation for the instability of the H2A.Z/H3.3 hybrid nucleosome may lie with a newly characterized H2A.Z splice variant, H2A.Z.2.2 [23]. Due to its unique docking domain, this particular histone physically destabilizes the octameric core of the nucleosome. While it is unknown whether H2A.Z.2.2 co-localizes with H3.3 in the cell, the decreased stability observed in H2A.Z/H3.3 hybrid nucleosomes could be attributed to the splice variants. An additional key example of nucleosome conformation variability has also been documented for native CENP-A nucleosomes in vivo, which exhibit a surprising bi-stability across the human cell cycle, concurrent with cell-cycle regulated acetylation on K124, in the center of the CENP-A octameric core [24,25]. Thus, it is feasible that other histone variants display modification-dependent conformational oscillations that impact their inheritance and function in vivo.
While nucleosomes have been shown to associate with specific locations within the genome, such as the localization of H3.3 and H2A.Z to TSS, the mechanisms underlying nucleosome positioning in the cell are still being debated. Both experimental and theoretical research have uncovered subtle structural motifs embedded within the primary sequence of DNA as a key component driving preferential nucleosome formation, albeit at subsaturating levels of histones [26,27]. Many of these motifs turn out to have a venerable lineage: a recent study demonstrates that archaeal tetramers are positioned relative to specific motifs in DNA sequences, tending to prefer bendable GC-containing DNA motifs to stiff AT-containing DNA motifs, similar to their eukaryotic counterparts [28•]. However, in eukaryotes, genome-wide nucleosome positioning does not appear to be dictated solely by DNA sequence, as the addition of ATP to chromatin incubated in whole cell extracts is necessary to recapitulate nucleosome phasing in vitro, indicating that ATP-dependent chromatin remodelers play an important role in defining nucleosome positions within the cell [29]. Yet, other studies have highlighted the importance of AT-rich DNA sequences in maintaining NDRs in vivo [30,31]. Thus, while the primary sequence of DNA does position nucleosomes in select locations in the genome, trans-acting factors play an equally significant role in over-ruling intrinsic DNA-sequence based nucleosome positioning. Together, evolutionary conserved nucleosome positioning coupled to ATP-driven chromatin remodelers provide a powerful one-two punch, permitting chromatin structure to be flexible and responsive to changing environmental cues from the cell.
Despite decades of nucleosome positioning research, surprisingly little information is available on the interplay between key histone variants and nucleosome positioning. Using a 208 bp fragment of DNA, it is apparent simply from monitoring the migration of the nucleosomes through a native gel that the histone variants H3.3 and H2A.Z both modify the position of the nucleosome upon the DNA in vitro [20]. However, no extant study has yet undertaken the difficult yet exciting task of investigating whether individual histone variants, which are all at subsaturating levels in vivo, manipulate structural motifs within DNA sequences to potentially out-compete other histone variants for certain positions in the genome, or to create specialized chromatin structures that are co-dependent on the presence of the histone variant and the sequence of the underlying DNA.
Epigenetic inheritance and histone variants
While histone variants play an important role in regulating gene expression, they may also participate in their own epigenetic inheritance, maintaining correct localization on the newly synthesized daughter strands following DNA replication. Using a SILAC-based (stable isotope labeling by amino acids in cell culture) approach, it was recently determined that after two cell cycles, ~20% of the core (H3.3/H4)2 tetramer within nucleosomes were split into H3.3/H4 dimers, assembled with newly synthesized H3.3/H4 [32]. These data support a model in which segregated deposition of parental H3.3/H4 after DNA synthesis is responsible for maintaining the local epigenetic state (Figure 2a) [33]. The splitting process appears to be primarily replication-dependent, as treatment with hydroxyurea or aphidicolin significantly reduced splitting events. In contrast, the remaining (H3.3/H4)2 tetramers, along with the canonical (H3.1/H4)2 tetramers, were not split during replication, providing support instead for a model in which epigenetic information is regenerated after DNA synthesis using the past epigenetic state of neighboring nucleosomes as a template (Figure 2b). These data indicate that epigenetic inheritance of modified histones may proceed via more than one pathway. Another example of templating comes from Drosophila, in which the centromeric histone variant CID derived from the sperm is used to template CID deposition at the centromere during embryogenesis [34•]. While fertilization can occur with sperm that lack CID, the embryos do not develop normally, and paternal chromosomes lose the ability to recruit maternal CID and re-establish functional centromeres. Thus CID deposition during embryogenesis also appears to depend on a templating mechanism, although it is unclear whether it proceeds via direct or indirect recruitment. Interestingly, several epigenetic marks on the H3 histones appear to be important for proper recycling of old histones to the newly replicated DNA, and these marks have been shown to change under conditions of replication stress [35]. However, the mechanism by which nucleosome inheritance is regulated still remains unexplored. Investigations into the influence of transcription rate, histone availability, and timing of replication may all provide important insights into how histones provide the genome with a molecular memory.
Figure 2.
Histone variant H3.3 is inherited after DNA replication by at least two different mechanisms. In panel A, old (H3.3/H4)2 tetramers (orange) can split in a replication dependent manner into H3.3/H4 dimers, which are deposited on either daughter strand with newly synthesized H3.3/4 dimers (green). In panel B, the (H3.3/H4)2 tetramers are deposited intact onto either daughter strand, where it serves as a template for re-establishment of the chromatin state by filling in gaps with nucleosomes containing newly synthesized (H3.3/H4)2 tetramers. The centromeric histone variant found in Drosophila, CID, and canonical (H3.1/H4)2 tetramers are also re-established via a templating mechanism.
Friend and foe: versatile roles of histones in DNA damage repair
The ability of chromatin to protect DNA from ionizing radiation was established in a seminal study over 20 years ago. When DNA was completely stripped of its nucleosomes and exposed to 20 Gy of gamma-radiation, the occurrence of double strand breaks (DSBs) was 10 times greater than that of intact cells [36]. However the discovery that histone variants are intimately tied to proper DNA damage response (DDR) progression is relatively recent. In particular, work has focused on the role played by variants of the H2A family: (γ)H2A.X, H2A.Z and macroH2A.
While the localized phosphorylation of H2A.X has been implicated in the response to DSBs for some time, it is only recently that the behavior of H2A.X in response to clustered DNA lesions has been elucidated. Interestingly, when clustered DSBs were induced by ionizing radiation in skin fibroblasts, H2A.X phosphorylation, monitored by immunostaining, was not limited to the region directly surrounding the break, but occurred throughout the genome in a dose dependent manner [37]. This response, catalyzed by two kinases, ATM and DNA-PK, was transient and not linked to apoptosis. Recently, using ChIP at a defined DSB, a second H2A variant usually involved in transcriptional regulation, H2A.Z, was found at the break site [38]. H2A.Z is deposited at the DSB by the ATP-dependent chromatin remodeler p400, and is thought to re-organize the chromatin surrounding the DSB into a more fluid conformation by promoting H4 acetylation (Figure 3). Surprisingly, macroH2A, normally accumulated at repetitive LINE elements on the inactive X chromosome, is also found at DSBs, though it is not necessarily incorporated into chromatin [39]. While macroH2A produces a signal by ChIP after analysis of formaldehyde crosslinked chromatin surrounding the DSB, it does not produce a signal by ChIP after analysis of uncrosslinked chromatin, suggesting only a transient interaction as part of the DDR pathway. Thus, histone variants represent a crucial player in the proper repair of double strand breaks and maintenance of the genome. While histone variants generally aid the DNA repair process, there are examples where histones can serve as an obstacle. In vitro experiments demonstrate that when an oxidized abasic site, one of the most common lesions resulting from oxidative damage, is present in the nucleosome, the lesion is not merely removed from the DNA, but can be transferred to the closest histone tail, usually the lysine rich tails of H3 or H4, creating a DNA/protein crosslink [40]. By monitoring the length of 32P-labeled substrates before and after incorporation into a nucleosome, the formation of single strand breaks (SSBs) was determined to increase between 130 and 550 fold, depending on the location of the lesion within the nucleosome, with lesions positioned near the entry/exit site of the DNA displaying the highest rates of SSB formation. While these experiments were conducted using recombinant, canonical histones, the effect of histone variants on the rate of SSB and DNA/protein crosslink formation is completely unknown.
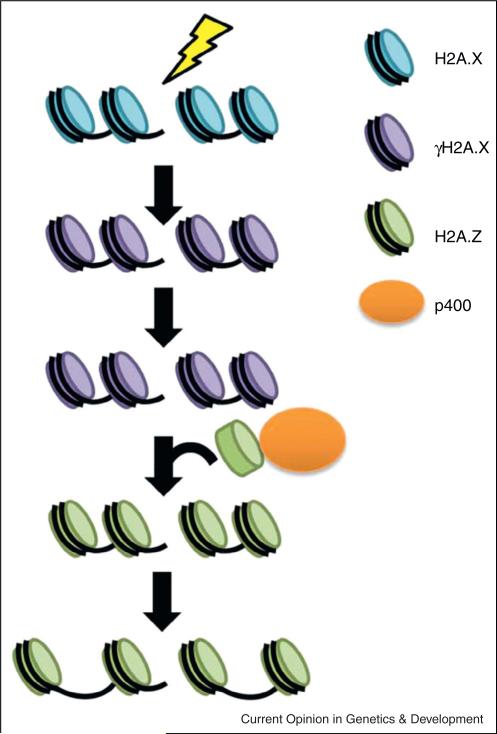
Figure 3.
The histone variants H2A.X and H2A.Z are involved in re-arrangement of chromatin around the DSB site. After a DSB occurs, H2A.X (blue) is phosphorylated (γH2A.X, purple), which leads to remodeling of the chromatin on either side of the DSB such that γH2A.X is exchanged for H2A.Z (green) by p400 (orange). H2A.Z then promotes the formation of more open, relaxed chromatin, which is poised for repair. It has been proposed that the incorporation of H2A.Z at DSBs creates a structure reminiscent of a TSS, and that H2A.Z may limit the spreading of nucleosome free regions at the break site.
Stirrers of strife: histone variants in aging and disease
Histones play an important role in cellular aging; histone levels decrease as part of the natural aging process in yeast [41]. Upon inactivation of the Hir histone chaperone complex or overexpression of histone proteins in S. cerevisiae, lifespan can be artificially increased, indicating that regeneration of cellular chromatin is vital for extending lifespan [42]. Histone variants are also implicated in cancer. A recent study has shown that specific splice variants of macroH2A are correlated with the known invasiveness of cancer cell lines [43]. While the total macroH2A content is consistent between the cell lines studied, when a cell has a greater amount of macroH2A1.1 as compared to macroH2A1.2, the cell is less invasive, as measured by migration through a porous membrane. Conversely, when the cell has a greater amount of macroH2A1.2, the cell tends to be more invasive. Mechanistically, it is not known if this correlation reflects an increase in fragile chromatin structure imparted by macroH2A1.2 versus macroH2A1.1, or whether the increase in macroH2A1.2 is an indirect downstream effect of other factors. Indeed, the potential for interaction of upregulated macroH2A1.2 with other histone variants remains a completely unexplored arena in the study of cancer invasiveness.
Interestingly, alterations in histone genes are not just associated with diseases of age. In pediatric glioblastomas, mutations such as K27M and G34R/V are found clustered on the tail of histone variant H3.3, causing a gain of function mutation [44••]. These mutations cause a reduction in the overall levels of methylation on H3K27 by targeting the active site of SET-domain containing methyl transferases [45••]. The loss of H3K27 methylation is predicted to disrupt a feedback loop that regulates the polycomb repressor complex 2 (PCR2), which then promotes the cancer state. Thus, histones can play a pivotal role in the progression of the disease state, making them potential candidates to consider for therapeutic targeting.
Conclusions and future directions
As is evident from the large body of literature on histones and their variants, nucleosomes and their structure, and chromatin organization in vitro and in vivo, this topic is a continuously evolving chapter in the study of genomes. Despite almost 40 years of steady progress on understanding chromatin, profound open questions persist that make this field one of the most exciting to investigate. Do histone variants have different preferences for particular DNA sequences? Do histones re-associate with the same DNA sequence after being disrupted? Is there true molecular memory at sites that are to be marked for the next cell cycle? How is such memory over-ridden when cells embark on different developmental programs? How does the vigorous compression in the mitotic chromosome physically affect the position and stability of various types of nucleosomes? When cells age or transit into resting phase, how does the proportion of histone variants and nucleosome positions change, and how do such phenomena affect the rate of gene expression, DNA repair, remodeling and replication? All these questions await answers, which will eventually bring a more complete conceptual framework of the behaviors used to regulate genetic accessibility by these tiny, but crucial proteins, the tricksters of the genome.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•of special interest
••of outstanding interest
- 1.Karlsdóttir A. Call to Loki. 1979 [Google Scholar]
- 2.Grove A. Functional evolution of bacterial histone-like HU proteins. Curr Issues Mol Biol. 2011;13:1–12. [PubMed] [Google Scholar]
- 3.Sandman K, Reeve JN. Archaeal histones and the origin of the histone fold. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Decanniere K, Babu AM, Sandman K, Reeve JN, Heinemann U. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J Mol Biol. 2000;303:35–47. doi: 10.1006/jmbi.2000.4104. [DOI] [PubMed] [Google Scholar]
- 5.Fahrner RL, Cascio D, Lake JA, Slesarev A. An ancestral nuclear protein assembly: crystal structure of the Methanopyrus kandleri histone. Protein Sci. 2001;10:2002–2007. doi: 10.1110/ps.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleo some core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokuda N, Terada T, Sasai M. Dynamical modeling of three-dimensional genome organization in interphase budding yeast. Biophys J. 2012;102:296–304. doi: 10.1016/j.bpj.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput Biol. 2008;4:e1531000. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Fussner E, Strauss M, Djuric U, Li R, Ahmed K, Hart M, Ellis J, Bazett-Jones DP. Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 2012;13:992–996. doi: 10.1038/embor.2012.139. [Along with Refs [11•,12•], these papers provide a robust challenge to the organization of chromatin as a regular invariant reiteration of 30nm in width. These papers use careful cryo-EM and SAXS measurements to demonstrate unequivocally that the bulk of chromatin in vivo exists mostly in the “beads on a string” format.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Eltsov M, MacLellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci U S A. 2008;105:19732–19740. doi: 10.1073/pnas.0810057105. [Along with Refs [12•], these papers provide a robust challenge to the organization of chromatin as a regular invariant reiteration of 30nm in width. These papers use careful cryo-EM and SAXS measurements to demonstrate unequivocally that the bulk of chromatin in vivo exists mostly in the “beads on a string” format.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Joti Y, Hikima T, Nishino Y, Kamada F, Hihara S, Takata H, Ishikawa T, Maeshima K. Chromosomes without a 30-nm chromatin fiber. Nucleus. 2012;3:404–410. doi: 10.4161/nucl.21222. [Along with Refs [11•], these papers provide a robust challenge to the organization of chromatin as a regular invariant reiteration of 30nm in width. These papers use careful cryo-EM and SAXS measurements to demonstrate unequivocally that the bulk of chromatin in vivo exists mostly in the “beads on a string” format.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Curr Opin Cell Biol. 2012;24:359–366. doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman-Aiden E, Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fussner E, Ching RW, Bazett-Jones DP. Living without 30 nm chromatin fibers. Trends Biochem Sci. 2011;36:1–6. doi: 10.1016/j.tibs.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J Biol Chem. 2008;283:34532–34540. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 18.Wiedemann SM, Mildner SN, Bönisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J Cell Biol. 2010;190:777–791. doi: 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/ H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nature. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, Fukui K, Tomschik M, Ausio J, Zlatanova J. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48:10852–10860. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- 21••.Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Bühler M, et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152:859–872. doi: 10.1016/j.cell.2013.01.032. [Together, Refs [22••] provide direct structural evidence for alteration of pseudodyad access deep within the nucleosomal core as a consequence of acetylation of H3K122. These findings are profound because they point to a novel mechanism to destabilize the four-helix bundle with the octameric core of the conventional nucleosome. Furthermore, Ref. [21••] demonstrates a functional consequence upon transcription in the presence of this modification.] [DOI] [PubMed] [Google Scholar]
- 22••.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A. 2011;108:12711–12720. doi: 10.1073/pnas.1106264108. [Together, Refs [21] provide direct structural evidence for alteration of pseudodyad access deep within the nucleosomal core as a consequence of acetylation of H3K122. These findings are profound because they point to a novel mechanism to destabilize the four-helix bundle with the octameric core of the conventional nucleosome. Furthermore, Ref. [21••] demonstrates a functional consequence upon transcription in the presence of this modification.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bönisch C, Schneider K, Pünzeler S, Wiedemann SM, Bielmeier C, Bocola M, Eberl HC, Kuegel W, Neumann J, Kremmer E, et al. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Research. 2012;40:5951–5964. doi: 10.1093/nar/gks267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bui M, Dimitriadis EK, Hoischen C, An E, Quénet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walkiewicz M, Dimitriadis EK, Dalal Y. CENP-A octamers do not confer a reduction in nucleosome height by AFM. Nature Struct Mol Biol. 2014;21:2–3. doi: 10.1038/nsmb.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 27.Hunter C, Moore I, Widom J. Structure-based identification of new high-affinity nucleosome binding sequences. J Mol Biol. 2012;420:8–16. doi: 10.1016/j.jmb.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 28•.Nalabothula N, Xi L, Bhattacharyya S, Widom J, Wang J, Reeve J, Santangelo T, Fondufe-Mittendorf Y. Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs. BMC Genomics. 2013;14:391. doi: 10.1186/1471-2164-14-391. [This paper by the late Jon Widom provides the first clue to the evolution of DNA motifs which favor preferential nucleosome assembly, showing that early archaea use the same motifs as those used by later eukaryotes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes AL, Jin Y, Rando OJ, Struhl K. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol Cell. 2012;48:5–15. doi: 10.1016/j.molcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet. 2012;44:743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 32.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 33.Groudine M, Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982;30:131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- 34•.Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in drosophila sperm. PLoS Biol. 2012;10:e4341001. doi: 10.1371/journal.pbio.1001434. [This paper provides evidence that templating of embryonic CENP-A on paternal centromeres relies on the presence of CENP-A that persists upon the chromatin fiber in sperms, thus yielding a wonderful example of epigenetic inheritance and its direct consequence on biology.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jasencakova Z, Scharf AND, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Molecular Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 36.Elia MC, Bradley MO. Influence of chromatin structure on the induction of DNA double strand breaks by ionizing radiation. Cancer Res. 1992;52:1580–1590. [PubMed] [Google Scholar]
- 37.Meyer B, Voss K, Tobias F, Jakob B, Durante M, Taucher-Scholz G. Clustered DNA damage induces pan-nuclear H2AX phosphorylation mediated by ATM and DNA-PK. Nucleic Acids Res. 2013;41:6109–6110. doi: 10.1093/nar/gkt304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD. The histone variant macroH2A1. 1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 2012;586:3920–3930. doi: 10.1016/j.febslet.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou C, Sczepanski JT, Greenberg MM. Histone modification via rapid cleavage of C40-oxidized abasic sites in nucleosome core particles. J Am Chem Soc. 2013;135:5274–5280. doi: 10.1021/ja400915w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585:2041–2050. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feser J, Truong D, Chandrima, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dardenne E, Pierredon S, Driouch K, Gratadou L, Lacroix-Triki M, Espinoza MP, Zonta E, Germann S, Mortada H, et al. Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nat Struct Mol Biol. 2012;19:1139–1146. doi: 10.1038/nsmb.2390. [DOI] [PubMed] [Google Scholar]
- 44••.Schwartzentruber J, Korshunov A, Liu X, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DK, Tönjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [Along with Ref. [45••], these studies demonstrate that gain of function mutations in H3 which mimic certain modifications have drastic consequences for downstream function of repressor complexes, thus resulting in promotion of the cancer state.] [DOI] [PubMed] [Google Scholar]
- 45••.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [Along with Ref. [45••], these studies demonstrate that gain of function mutations in H3 which mimic certain modifications have drastic consequences for downstream function of repressor complexes, thus resulting in promotion of the cancer state.] [DOI] [PMC free article] [PubMed] [Google Scholar]