Summary
Retinoid X receptor-alpha (RXRα), an intriguing and unique drug target, can serve as an intracellular target mediating the anti-cancer effects of certain non-steroidal anti-inflammatory drugs (NSAIDs), including Sulindac. We report the synthesis and characterization of two new Sulindac analogs, K-8008 and K-8012, which exert improved anti-cancer activities over Sulindac in a RXRα- dependent manner. The new analogs inhibit the interaction of the N-terminally truncated RXRα (tRXRα) with the p85α subunit of PI3K, leading to suppression of AKT activation and induction of apoptosis. Crystal structures of the RXRα ligand-binding domain (LBD) with K-8008 or K-8012 reveal that both compounds bind to tetrameric RXRα LBD at a site different from the classical ligand-binding pocket. Thus, these results identify K-8008 and K-8012 as new tRXRα modulators and define a new binding mechanism for regulating the nongenomic action of tRXRα.
Keywords: RXRα, tRXRα, Sulindac, Sulindac analogs, Nuclear receptor, AKT activation, Ligand-binding domain, Novel binding site, nongenomic action
Introduction
Retinoid X receptor alpha (RXRα), a unique member of the nuclear receptor superfamily, regulates a broad spectrum of physiological functions including cell differentiation, growth and apoptosis (Germain et al., 2006; Szanto et al., 2004). Like other nuclear receptors, RXRα acts as a ligand-dependent transcription factor (Germain et al., 2006; Szanto et al., 2004). Recent accumulating evidence indicates that RXRα also has extranuclear actions. RXRα resides in the cytoplasm at certain stages during development (Dufour and Kim, 1999; Fukunaka et al., 2001) and migrates from the nucleus to the cytoplasm in response to differentiation, apoptosis, and inflammation (Cao et al., 2004; Casas et al., 2003; Zimmerman et al., 2006). RXRα exhibits a modular organization structurally consisting of three main functional domains: an N-terminal region, a DNA-binding domain and a ligandbinding domain (LBD). The LBD possesses a ligand-binding pocket (LBP) for the binding of small molecule ligands, a transactivation function domain termed AF-2 composed of Helix 12 (H12) of the LBD, a coregulator binding surface, and a dimerization surface (Germain et al., 2006; Szanto et al., 2004). The ligand-dependent transcription regulation is predominately mediated through H12 that is highly mobile. Agonist ligand binds to the LBP and helps the H12 to adopt the active conformation that forms a surface to facilitate the binding of coactivators and subsequent transactivation. In contrast, in the absence of an agonist ligand or in the presence of an antagonist ligand, the H12 adopt an inactive conformation that favors the binding of corepressors to inhibit target gene transcription. Natural RXRα ligand 9-cis-retinoic acid (9-cis-RA) and synthetic ligands have been effective in preventing tumorigenesis in animals and RXRα has been a drug target for therapeutic applications, especially in the treatment of cancer (Bushue and Wan, 2010; Yen and Lamph, 2006). Targretin, a synthetic RXR-selective retinoid (rexinoid), was approved for treating cutaneous T-cell lymphoma (Dawson and Zhang, 2002). RXRα can bind to DNA and activate transcription of target genes either as a homodimer or a heterodimer with its heterodimerization partners including retinoic acid receptor (RAR), vitamin D receptor (VDR), thyroid hormone receptor (TR), and peroxisome-proliferator-activated receptor (Germain et al., 2006; Szanto et al., 2004). In addition to homodimer and heterodimer, RXRα could also self-associate into homotetramers in solution, which rapidly dissociate into active dimers upon binding of a cognate ligand (Chen et al., 1998; Kersten et al., 1995). Tetramer formation of RXRα might serve to sequester the receptor's active species, dimers and monomers, into a transcriptionally inactive tetramer complex (Gampe et al., 2000).
Efforts on discovery of small molecules targeting RXRα for therapeutic application have been primarily focused on the optimization of the molecules that bind to its classical LBP (de Lera et al., 2007; Germain et al., 2006; Szanto et al., 2004). However, various studies have recently identified small molecule modulators of nuclear receptors that function via unknown sites and undefined mechanisms of action (Buzon et al., 2012; Moore et al., 2010). The recent report of the structure of estrogen receptor-β (ERβ) with a second molecule of 4-hydroxytamoxifen bound in its coactivator-binding surface represents the first direct example of a second estrogen receptor ligand-binding site and provides insight into the possible pharmacological effects of the drug (Wang et al., 2006). Compounds that bind to the surface binding sites of LBD have been demonstrated for other nuclear receptors, including androgen receptor (AR), VDR and TR (Buzon et al., 2012; Moore et al., 2010). However, compounds that bind to RXRα at the sites other than the classical LBP have not been reported.
We recently showed that certain non-steroidal anti-inflammatory drugs (NSAIDs), including Etodolac (Kolluri et al., 2005) and Sulindac (Zhou et al., 2010), could bind to RXRα and modulate its biological activities. Interestingly, Sulindac but not 9-cis-RA could inhibit the binding of an N-terminally-truncated RXRα protein (tRXRα) to the p85α regulatory subunit of phosphatidylinositol-3-OH kinase (PI3K), leading to inhibition of tumor necrosis factor-α (TNFα)-activated PI3K/AKT pathway (Zhou et al., 2010). We also demonstrated, through a designed Sulindac analog K-80003, the feasibility of developing a new generation of RXRα-specific molecules for therapeutic application and mechanistic studies of RXRα (Wang et al., 2013; Zhou et al., 2010). These results identify Sulindac and related analogs as unique regulators of tRXRα activity through an undefined binding mechanism. Here we report our synthesis and characterization of K-80003-based analogs, K-8008 and K-8012, which exhibited improved activity in inhibiting tRXRα-mediated PI3K/AKT signaling pathway. Moreover, our X-ray crystallographic studies of the LBD of RXRα in complex with K-8008 or K-8012 revealed that both compounds bound to the RXRα LBD in its tetrameric form via a novel site outside of the classical RXRα LBP, providing a new strategy for developing RXRα-based agents for cancer therapy.
RESULTS
K-8008 and K-8012 are new antagonists of RXRα
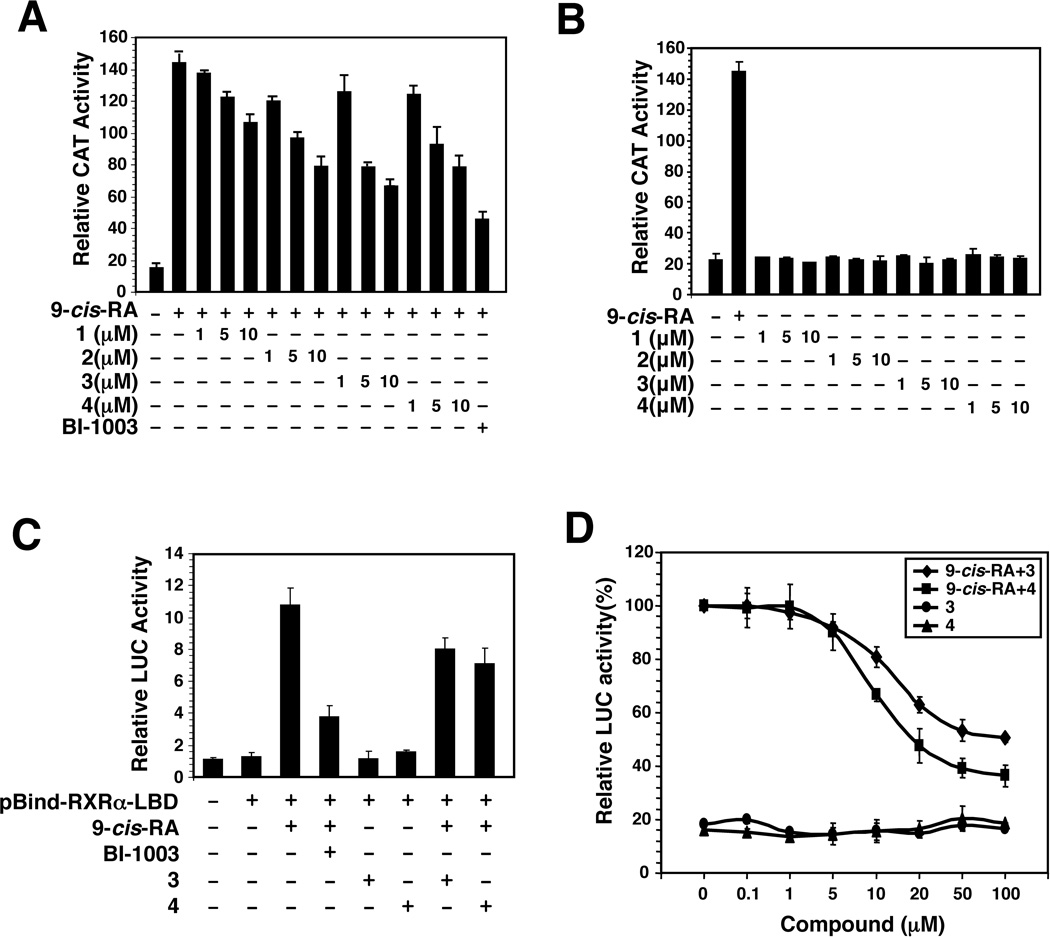
In an effort to identify improved Sulindac analogs for cancer therapy, we designed and synthesized a series of analogs around K-80003 (Wang et al., 2013; Zhou et al., 2010). Analogs shown in Figure 1 were initially evaluated by the reporter assay using a CAT reporter containing TREpal that known to bind to RXRα homodimer (Zhang et al., 1992). 9-cis-RA strongly induced the TREpal reporter activity, which was inhibited by BI-1003, a known RXRα antagonist (Lu et al., 2009). K-8008 and K-8012 also exhibited inhibitory effect on 9-cis-RA-induced TREpal reporter activity in a concentration dependent manner (Figure 2A), while they did not show any agonist activity at the concentrations used (Figure 2B). The antagonist effect of K-8008 and K-8012 was much better than Sulindac and comparable to K-80003 (Figure 2A). We also used the Gal4-RXRα-LBD chimera and Gal4 reporter system to evaluate the inhibitory effect of K-8008 and K-8012 on 9-cis-RA-induced reporter activity. Cotransfection of Gal4-RXRα-LBD strongly activated the Gal4 reporter in the presence of 9-cis-RA, which was inhibited by BI-1003 as well as K-8008 and K-8012 (Figure 2C). Dose response experiments showed that the IC50 values for K-8008 and K-8012 to inhibit 9-cis-RA-induced Gal4-RXRα-LBD transactivation were about 13.2 µM and 9.2 µM, respectively (Figure 2D). Thus, K-8008 and K-8012 are new antagonists of RXRα.
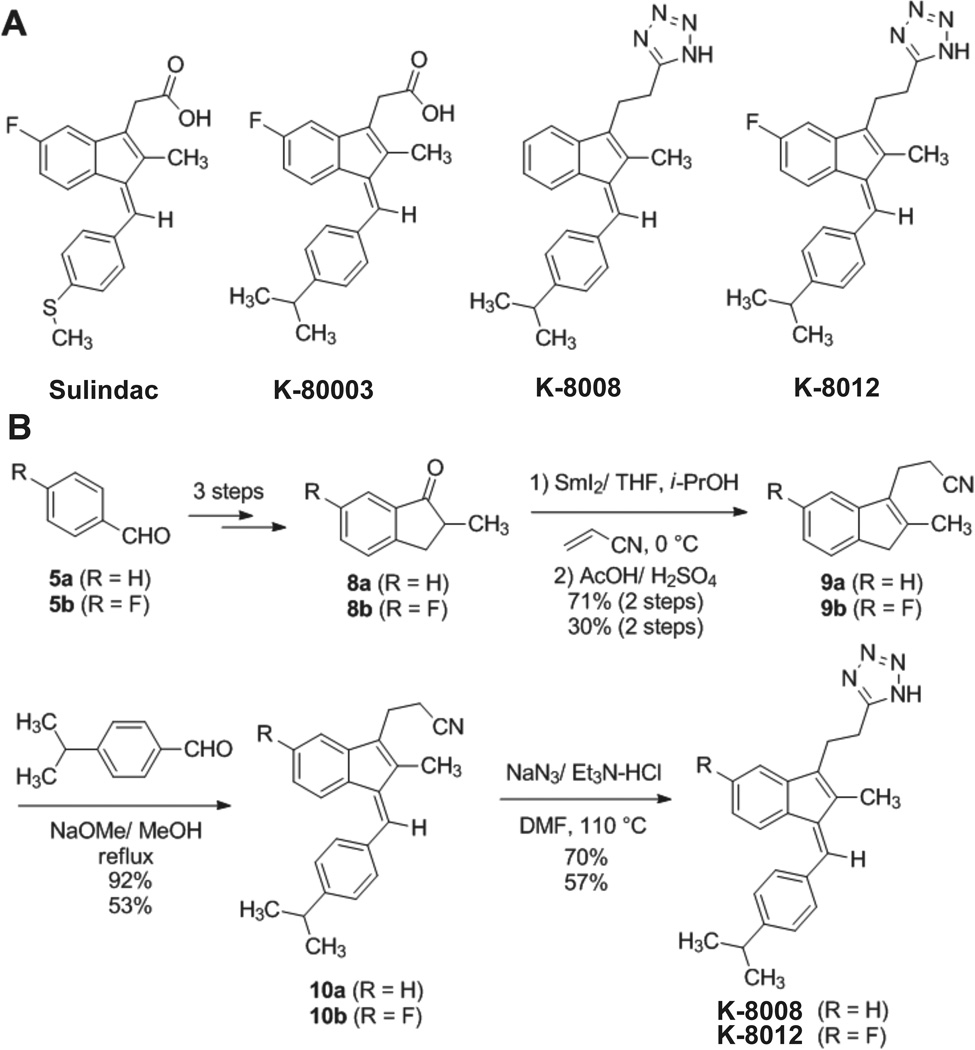
Figure 1.
Chemical structures and Synthesis. (A) The structures of Sulindac and its analogs K-80003, K-8008, and K-8012. (B) Synthesis Scheme of K-8008 and K-8012.
Figure 2.
Antagonist effect of Sulindac analogs. (A and B) Inhibition of RXRα transactivation. (TREpal)2-tk-CAT and RXRα were transiently transfected into CV-1 cells. Cells were treated with or without 9-cis-RA (10−7 M) in the presence or absence of the indicated concentration of Sulindac and its analogs. CAT activity was determined. BI-1003 (1 µM) was used for comparison. Error bars represent SEM. (C) Inhibition of 9-cis-RA-induced Gal4 reporter activity. pBind-RXRα-LBD and pG5luc were transiently transfected into HCT-116 cells. Cells were treated with or without 9-cis-RA (10−7 M) in the presence or absence of BI-1003 (1 µM), K-8008 (50 µM) and K-8012 (50 µM). Luciferase activity was determined. (D) Dose dependent effect of K-8008 and K-8012. HCT-116 cells transfected with pBind-RXRα-LBD and pG5luc were treated with indicated concentrations of K-8008 and K-8012 in the presence or absence of 9-cis-RA (10−7 M).
K-8008 and K-8012 induce apoptosis and inhibit AKT activation by preventing tRXRα from binding to p85α
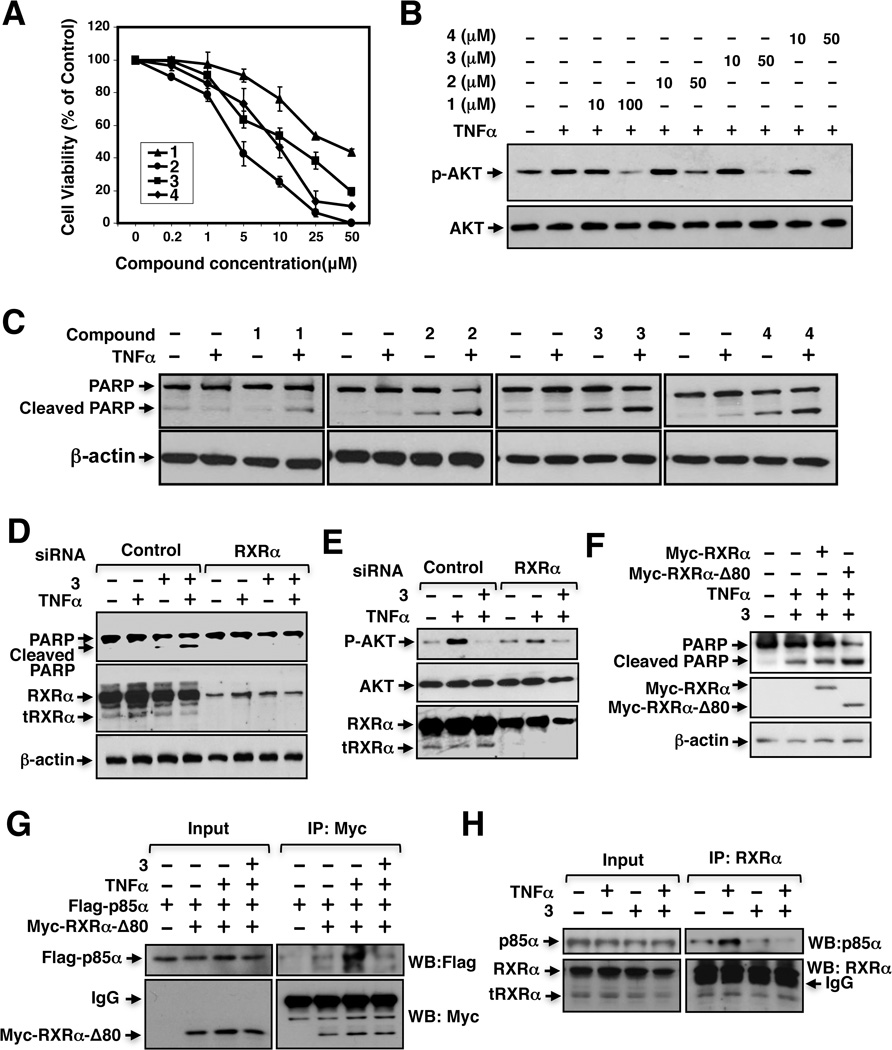
We next evaluated K-8008 and K-8012 for their effect on the growth of cancer cells. Compared to Sulindac, K-8008 and K-8012 were much more effective in inhibiting the growth of various cancer cells, including A549 lung cancer (Figure 3A), PC3 prostate cancer, ZR-75-1 and MB231 breast cancer cells (Figure S1). One unique property of Sulindac and analogs is their ability to inhibit TNFα-induced AKT activation (Zhou et al., 2010). Thus, A549 lung cancer cells were treated with TNFα in the absence or presence of K-8008 or K-8012. Treatment of cells with TNFα enhanced AKT activation as revealed by Western blotting (Figure 3B). However, when cells were cotreated with either K-8008 or K-8012, the TNFα-induced AKT activation was suppressed in a dose dependent manner (Figure 3B). Similar results were obtained in other cancer cell lines (Figure S2).
Figure 3.
Biological effects of K-8008 and K-8012. (A) Growth inhibition by Sulindac and its analogs. A549 lung cancer cells were treated with various concentrations of the indicated compounds. Cell viability was measured by the MTT colorimetric assay. (B) Inhibition of TNFα-induced AKT activation. A549 cells were pretreated with Sulindac or analogs for 1 h before exposed to TNFα (10 ng/mL) for 30 min. Phosphorylated AKT and total AKT were analyzed by immunoblotting. (C) Induction of apoptosis by Sulindac and analogs in the presence of TNFα. Cells cultured in medium with 1% FBS were treated with TNFα (10 ng/mL) and/or compound (40 µM) for 4 h, and analyzed for PARP cleavage by immunoblotting. (D) RXRα siRNA transfection inhibits the apoptotic effect of K-8008. HeLa cells transfected with control or RXRα siRNA for 48 h were treated with K-8008 (40 µM) and/or TNFα (10 ng/ml) for 6 h and analyzed by immunoblotting. (E) RXRα siRNA transfection antagonizes the inhibitory effect of K-8008 on AKT activation. HeLa cells transfected with control or RXRα siRNA for 48 h were treated with K-8008 (40 µM) for 1.5 h before exposed to TNFα (10 ng/ml) for 30min and analyzed by immunoblotting. (F) Myc-RXRα-Δ80 transfection enhances the apoptotic effect of K-8008. HeLa cells transfected with Myc-RXRα or Myc-RXRα-Δ80 were treated with K-8008 (20 µM) and/or TNFα for 12 h. PARP cleavage and transfected RXRα expression were analyzed by immunoblotting. (G) K-8008 inhibits the interaction of transfected tRXRα and p85α. HeLa cells transfected with Myc-RXRα-Δ80 and/or Flag-p85α for 24 h were treated with vehicle or 20 µM K-8008 in the presence of absence of 10 ng/ml TNFα for 1 h. Cell lysates were immunoprecipitated using anti-Myc antibody and analyzed by Western blotting (WB) analysis using the indicated antibody. (H) K-8008 inhibits the interaction of endogenous tRXRα and p85α. A549 cells were pretreated with vehicle or 40 µM K-8008 for 1 h before exposed to 10 ng/mL TNFα for 30 min. Cell lysates were immunoprecipitated with ΔN197 anti-RXRα antibody and analyzed by Western blotting. See also Figure S1, S2, S3 and S4.
TNFα is a multifunctional cytokine that controls diverse cellular events such as cell survival and death (Balkwill, 2009; Wang and Lin, 2008). Inhibition of TNFα-induced AKT activation by Sulindac and analogs in cancer cells led to a shift of TNFα signaling from survival to death (Zhou et al., 2010). We therefore evaluated the effect of K-8008 and K-8012 alone or in combination with TNFα on the cleavage of PARP, an indication of apoptosis in cancer cells (Lazebnik et al., 1994). Treatment of A549 cells with TNFα did not have effect on PARP cleavage, whereas treatment with Sulindac or analogs slightly induced PARP cleavage. Combination of Sulindac or analogs with TNFα, however, caused a significant induction of PARP cleavage (Figure 3C and Figure S3). Thus, K-8008 and K-8012, like Sulindac, could convert TNFα signaling from survival to death in cancer cells.
The growth inhibitory effect of K-8008 and K-8012 and the induction of apoptosis by K-8008 occurred at low micromolar concentrations, suggesting that they might exert their anti-cancer effects through RXRα binding. To address the issues, cancer cells were transfected with RXRα siRNA and evaluated for the effect of K-8008 on inducing PARP cleavage and inhibiting AKT activation. Knocking down RXRα expression by RXRα siRNA transfection significantly diminished the effect of K-8008 on inducing PARP cleavage (Figure 3D) and inhibiting TNFα-induced AKT activation (Figure 3E). To address the role of tRXRα, RXRα-Δ80, a RXRα mutant lacking its N-terminal 80 amino acids and mimicking tRXRα (Wang et al., 2013; Zhou et al., 2010), was transfected into HeLa cells. Transfection of RXRα-Δ80 but not the full-length RXRα enhanced the effect of K-8008 on inducing PARP cleavage in the presence of TNFα (Figure 3F). Together, these results demonstrate that tRXRα plays a crucial role in mediating the biological effects of K-8008.
We next determined whether K-8008 could affect tRXRα interaction with p85α, an interaction known to activate AKT (Zhou et al., 2010). HeLa cells were transfected with Myc-tagged RXRα-Δ80 and Flag-tagged p85α expression vectors and treated with or without TNFα and/or K-8008. Co-immunoprecipitation assays using anti-Myc antibody showed that Flag-p85α was coimmunoprecipitated together with Myc-RXRα-Δ80 in cells treated with TNFα (Figure 3G). However, when cells were cotreated with K-8008, TNFα-induced interaction of Myc-RXRα-Δ80 with Flag-p85α was almost completely inhibited. We also examined the effect of K-8008 on interaction of endogenous tRXRα with p85α in A549 cells. Cell lysates prepared from A549 cells treated with TNFα in the presence or absence of K-8008 were analyzed by co-immunoprecipitation using Δ197 anti-RXRα antibody that recognizes both tRXRα and RXRα (Zhou et al., 2010). Figure 3H showed that treatment of cells with TNFα promoted the interaction of endogenous tRXRα with p85α, consistent with previous finding (Zhou et al., 2010). When cells were co-treated with K-8008, the interaction was largely inhibited. Such an effect of K-8008 on inhibiting TNFα-induced p85α interaction with tRXRα was also observed in other cancer cell lines, including PC3 and HepG2 cells (Figure S4). Together, these results demonstrate that K-8008 can induce TNFα-dependent apoptosis by suppressing the tRXRα-mediated activation of AKT through its inhibition of tRXRα interaction with p85α.
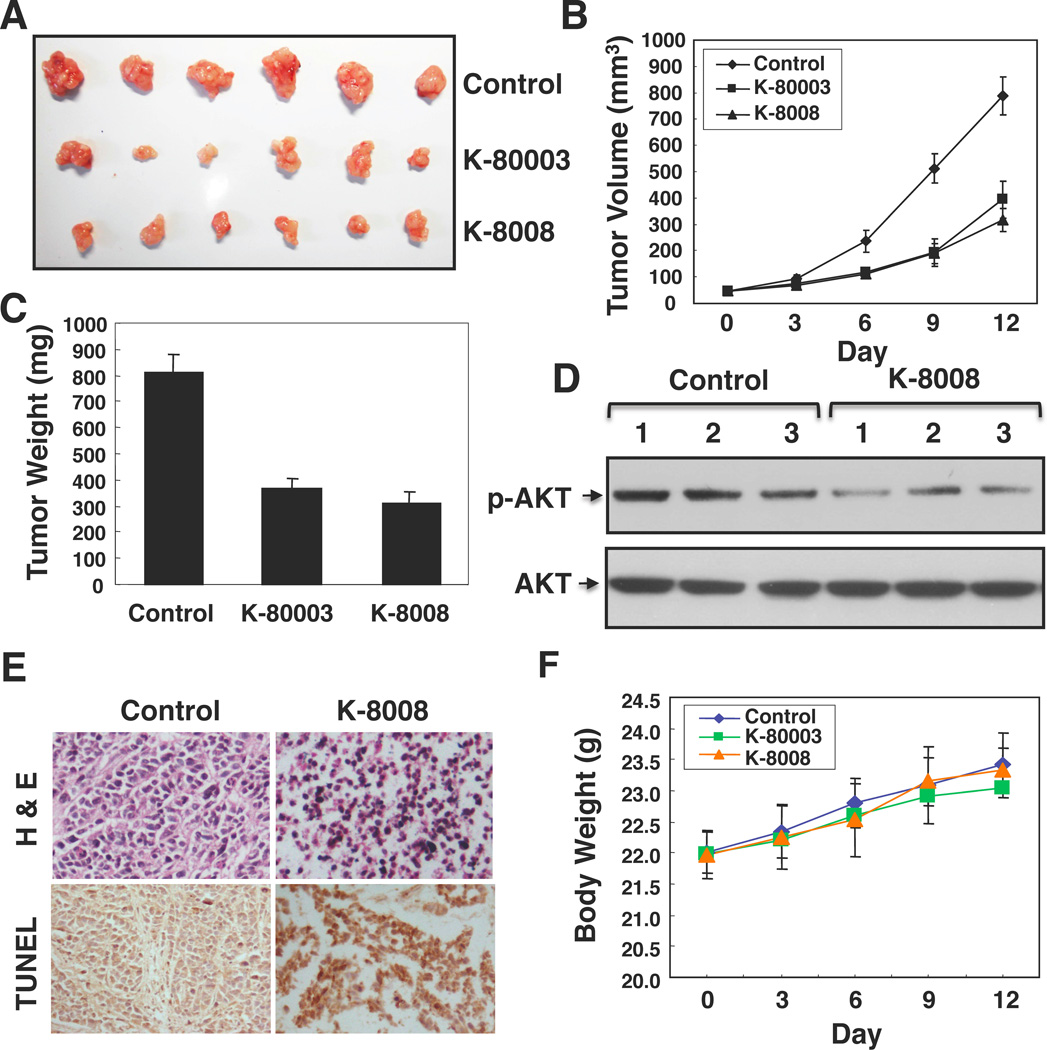
To further evaluate the anti-cancer effect of K-8008, mice with HepG2 tumor xenografts were treated with 20 mg/kg K-8008 or K-80003. Administration of K-8008 inhibited the growth of HepG2 tumor in a time dependent manner (Figure 4A), resulting in a 61.23% reduction of tumor weight after a 12-day treatment (Figures 4B–C), which was comparable with the inhibitory effect of K-80003 (54.84% reduction). Consistent with our in vitro observation, examination of three tumors treated with or without K-8008 showed reduction of AKT activation by K-8008 (Figure 4D). Moreover TUNEL staining revealed induction of apoptosis by K-8008 (Figure 4E). Significantly, administration of either K-80003 or K-8008 did not show any apparent toxic effects such as loss of body weight (Figure 4F).
Figure 4. Inhibition of HepG2 tumor growth in animals.
(A, B and C) Nude mice with HepG2 heptoma xenografts were intraperitoneally injected daily with vehicle, K-8008 (20 mg/kg) or K-80003 (20 mg/kg) for 12 days. Tumors were removed and measured. Tumor sizes and weights in control, K-80003 and K-8008-treated mice were compared. (D) Lysates prepared from three tumors treated with vehicle or K-8008 were analyzed by Western blotting assay for p-AKT expression. (E) H&E staining and TUNEL assay. Tumor sections were stained for H&E or TUNEL by immunohistochemistry. Increased apoptotic tumor cells were observed in tumor from K-8008 treated mice. (F) K-8008 does not exhibit apparent toxicity. Body weight was measured every three days. Each point represents the mean ± standard deviation of six mice. The differences between the compound treated group and control group are not significant (P>5%).
K-8008 and K-8012 do not bind to the classical LBP of RXRα
Although Sulindac and analogs can bind tRXRα and induce tRXRα-dependent apoptosis of cancer cells, how they bind to tRXRα to regulate tRXRα functions remains undefined. According to current understanding of the mechanism by which ligands regulate the transcriptional activity of nuclear receptors, K-8008 and K-8012 might bind to the canonical binding site, the LBP of RXRα, acting as conventional antagonists. Thus, we evaluated their binding to the LBP of RXRα using the classical radioligand competition binding assay (Zhou et al., 2010). Unlike 9-cis-RA and K-80003 that competed well with [3H]9-cis-RA for binding to the LBP of RXRα, K-8008 and K-8012 failed to replace [3H]9- cis-RA for its binding to the RXRα LBP (Figure 5A).
Figure 5.
Unique binding of K-8008 and K-8012 to RXRα. (A) K-8008 and K-8012 fail to compete with the binding of 9-cis-RA to RXRα. RXRα-LBD protein was incubated with [3H]9-cis-RA in the presence or absence of Sulindac analogs K-80003, K-8008, K-8012, or unlabeled 9-cis-RA. Bound [3H]9-cis-RA was quantitated by liquid scintillation counting. (B) K-8008 and K-8012 reduce 9-cis-RAinduced FRET signal. GST-RXRα-LBD was incubated with K-8008 or K-8012 in the presence or absence of 9-cis-RA (10−8 M). BI-1003 (1 µM) was used as a control. (C) Dose dependent effect of K-8008 and K-8012 on 9-cis-RA-induced FRET signal. GST-RXRα-LBD was incubated with K-8008 or K-8012 in the presence of 9-cis-RA (10−8 M).
Results of the [3H]9-cis-RA binding competition assay demonstrated that K-8008 and K-8012 did not bind to the canonical binding site, suggesting a different binding mechanism. Other than the classical LBP, recent structural and functional studies have revealed the existence of distinct small molecule binding sites on the surface of the LBD of nuclear receptors (Buzon et al., 2012; Moore et al., 2010). One of the potential alternative sites that small molecules could bind and function as antagonists is the coregulator-binding site in the LBD. Antagonistic property displayed by ligands through binding to the coregulator site has been reported for several members of the nuclear receptor family. For example, small molecules mimicking the structure of the coactivator LXXLL alpha-helical motif have been developed for modulating the activities of AR, ER and VDR (Mita et al., 2010; Rodriguez et al., 2003) (Arnold et al., 2005; Moore et al., 2010). We therefore tested if K-8008 and K-8012 could bind to an alternative surface binding site by using the time-resolved fluorescence resonance energy transfer (TRFRET) RXRα co-activator peptide competition assay. Our results showed that both compounds could inhibit 9-cis-RA-induced interaction of RXRα LBD with its coactivator peptide (Figure 5B). The inhibitory effect of K-8008 and K-8012 was much stronger than Sulindac, with IC50 values of 16.8 µM and 14.5 µM, respectively (Figure 5C), which correlated well with their inhibition of 9-cis-RA-induced RXRα transactivation (Figure 2D). Taken together, K-8008 and K-8012 might act as RXRα antagonists by binding to a novel RXRα surface site, leading to inhibition of coactivator binding.
K-8008 and K-8012 bind to a tetrameric structure of the RXRα LBD
To gain direct and structural understanding of the binding of K-8008 or K-8012 to RXRα, we performed crystallographic studies of these ligands bound to the RXRα LBD. Crystals of protein-ligand complexes were obtained using co-crystallization method. The structures of RXRα LBD in complex with K-8008 and K-8012 were determined to the resolution of 2.0 Å and 2.2 Å, respectively. Both protein/ligand complexes crystallized as tetrameric oligomers in the space group of P21 with similar unit cell parameters and the molecular replacement method was used to obtain the initial phasing by using the published RXRα structure, PDB code 1G1U. Statistics of structure refinement and data collection is summarized in Table S1.
The crystal structure of the RXRα LBD in complex with the K-8008 exists as noncrystallograohic homotetramer similar to the reported apo homotetramer (Gampe et al., 2000), in which 2 homodimers pack in a bottom-to-bottom manner (Figure 6A and Figure S5A). Superposition of our crystal structure with the published apo structure (PDB code 1G1U) shows that the corresponding monomers have almost identical fold with small shift found in the orientation of H12 in the monomer where a K-8008 molecule is bound (Figure 6B). N-terminal residues, from 223 to 260, were found to be disordered and undetermined in the complex structures, though residues from 231 to 260 were defined in the Apo structure. In a tetramer, 2 modulator molecules were found to bind to one homotetramer, with a binding stoichiometric ratio of 1:2 between ligand and protein, as one ligand molecule binds only to one monomer within a dimer (Figure 6A). Observation of the same stoichiometric ratio between ligand and protein has been reported for other RXRα-LBD/antagonist(Gampe et al., 2000). K-8008 binds to a region that is close to the dimer-dimer interface, making interaction primarily with one monomer of the dimer and some interaction with one monomer of the other dimer. The structure of RXRα LBD in complex with K-8012 is very similar to that of RXRα LBD in complex with K-8008 (Figure S5B). Therefore we only describe and discuss about the protein/K-8008 complex structure here.
Figure 6.
Crystal structure of the RXRα LBD in complex with K-8008. (A) The tetramer structure of RXRα LBD in complex with K-8008. The two bound K-8008 molecules are shown as magenta sticks surrounded by an electron density mesh (see Figure 6D footnotes for details). The two biological dimers (A1–B1 and A2–B2) are shown as green/cyan and yellow/brown pairs respectively. The N- and C-termini of four subunits are marked by the corresponding residue numbers. (B) Superposition of the RXRα LBD monomers from the K-8008-binding structure (brown ribbons) and the apo protein structure (purple ribbons, from PDB 1G1U). K-8008 is shown as sticks (carbon/nitrogen atoms are in magenta/blue). The classic ligand binding site is also marked by a VDW ball model (in cyan/red) of 9-cis-RA taken from a superimposed PDB entry 1FBY. (C) The hydrophobicity of the K-8008 binding site presented as a surface fragment on top of the RXRα LBD monomer (brown ribbon). The hydrophobic side chains that contribute to the region are shown in teal and K-8008 is shown in the same fashion as in Figure 6B. For clarity, residues contributing to this region are not labeled. (D) The protein side chains (in sticks) that make VDW interaction with K-8008. The displayed region is an enlargement of the black box in Figure 6A. The view is slightly rotated, and fragments of the green subunit that do not interact with K-8008 are removed. The protein surface is shown as semitransparent envelope. The (Fo-Fc) electron density is shown around the ligands as a black mesh. It was calculated at a 3-σ level with omitted ligand atoms. The positive end of the H11 helix dipole is highlighted in orange. (E) Side Chains around K-8008 that make significant changes in comparison with the apo protein (PDB 1G1U). Side chains of the apo protein are shown in green sticks and the protein/K-8008 complex in orange sticks. The complex structure is shown in brown ribbon and the apo structure in purple. K-8008 is presented in the same fashion as in Figure 6B. See also Figure S5 and S6
Both K-8008 and K-8012 bind to a hydrophobic region of LBD near the entry and the edge of the cognate LBP. This region does not overlap with the binding region of 9-cis-RA (Figure 6B), which explains why both compounds failed to compete with the binding of 9-cis-RA (Figure 5A). To get a sense of how far the K-8008 binding region is away from the LBP, we calculated the distance between the centroid of the bound 9-cis-RA and the centroid of the bound K-8008 molecule using the tools available in Maestro. In the superimposed structures of the K-8008 bound LBD structure with the 9-cis-RA bound LBD structure, the 2 centroids are about 7 Å apart (Figure S6A). The K-8008 hydrophobic binding region is made of side chains primarily from one monomer: Ala271 and Ala272 from H3, Trp305 and Leu309 from H5, Leu326 and Leu330 from the beta-turn, Leu433 from H10, Leu436 from L10–11, Phe437, Phe438, Ile442 and Gly443 from H11 of chain B2, and Leu436 from L10–11 of chain A1 (Figure 6C). With respect to the monomer of RXRα LBD, this region is located on the surface of the RXRα monomer molecule. However, in the tetramer structure, this region is buried. K-8008 makes both hydrophobic interaction and polar interaction with the protein. The negatively charged tetrazole of the ligand sits on the top of the N-terminal end of H11 with a distance of 3 Å from the backbone N of residue Phe438 (Figure S6B), making charge-diploe interaction with H11(Figure 6B). At 4.0 Å cutoff, the lipophilic part of the ligand makes Van Der Waals (VDW) contacts with side chains of Ile268, Ala271, Trp305, Leu436, Phe438, Phe439 and Ile442 from chain B2 and Leu436 from chain A1 (Figure6D and 6A). The overall structure of the K-8008 bound RXRα LBD is identical to the apo structure of RXRα LBD tetramer. Binding of K-8008 does not induce much significant changes in the surrounding side chains except for the side chains of Phe439 and Leu309. Side chain of Ph439 swings out to make room for the ligand to bind and side chain of Leu309 rearranges to make better VDW contact with the ligand (Figure 6E). Compared to the structure of the RXRα LBD bound to 9-cis-RA (Egea et al., 2000), K-8008 binding does not result in change in the shape of the LBP, whereas 9-cis-RA binding induces a substantial change in the shape of the LBP that include the movement of H3 and H11 and the reorientation of H12 (Figure 7A). 9-cis-RA binding results in the formation of the coactivator-binding site, however, K-8008 binding doesn’t promote the formation of the coactivator-binding site, instead it leaves the receptor protein in an auto-repression state. Thus K-8008 acts as an antagonist. Similar to 9-cis-RA ligand, K-8008 is made of largely a lipophilic body linked to a charged group. The carboxylate group of 9-cis-RA acts as an anchor via forming charge-charge interaction with Arg316, however the bioisostere tetrazole of the carboxylate in K-8008 makes no interaction with Arg316. The tetrazole group in the RXRα LBD/K-8008 structure is about 15 Å away from Arg316. Both of 9-cis-RA and K-8008 make extensive hydrophobic interactions with the protein. Some of the residues contributing to the hydrophobic interactions between ligand (9-cis-RA or K-8008) and the protein are shared, such as Ile268, Ala271 and Leu326. But, because of the LBP shape change induced by the binding of 9-cis-RA, these shared residues are located in different regions of the structures of RXRα LBD/K-8008 and RXRα LBD/9-cis-RA (Figure S6C).
Figure 7.
Structural comparison and mutagenesis analysis of the K-8008 binding site. (A) Structural superposition of the protein/K-8008 complex and the protein/9-cis-RA complex. The 9-cis-RA bound structure (PDB code 1FBY) is in pink cartoon and 9-cis-RA in cyan (C atoms) and red (O atoms) sticks. The K-8008 bound structure is in light orange cartoon and K-8008 is in grey (C atoms) and blue (N atoms) sticks. Side chain Arg316 is displayed for distance comparison between distances to –COOH and to tetrazole. (B–D) Mutational analysis of the K-8008 binding site. The LBD of RXRα or mutants cloned into pBind vector and pG5luc were transiently co-transfected into HCT-116 cells. Cells were treated with or without 9-cis-RA (10−7 M) in the presence or absence of BI-1003 (1 µM) or K-8008 (50 µM). Luciferase activity was determined. See also Figure S6.
To further support and cross-validate the identification of this novel binding site on RXRα, we designed a couple of mutants that could potentially impact the binding of K-8008 but not 9-cis-RA. Comparison of the binding nature of the K-8008 molecule to that of the 9-cis-RA reveals that Leu433 is close to the tetrazole group and replacing Leu433 with Glu could weaken the binding of K-8008 due to the repulsive interaction between the deprotonated Glu and the negatively charged tetrazole. However, Leu433Glu is not likely to impact the binding of 9-cis-RA as in the 9-cis-RA bound RXRα structure (PDB: 1FBY) Leu433 is not close to the ligand and is also partially solvent-exposed. This prediction is supported by our reporter assays showing that 9-cis-RA could similarly activate the wild-type RXRα (Figure 7B) and the Leu433Glu RXRα mutant, RXRα-L433E (Figure 7C). By contrast, K-8008 inhibited 9-cis-RA-induced transactivation of RXRα but not RXRα-L433E. Structural comparison also suggests that mutating both Phe438 and Phe439 into Ala could affect the binding of K-8008 but not 9-cis-RA. Indeed, simultaneous substitution of Phe438 and Phe439 with Ala resulted in a mutant, RXRα-F438, 9A, whose transactivation could be strongly induced by 9-cis-RA, but K-8008 failed to inhibit the induced transcriptional activity (Figure 7D). Together, the mutagenesis studies confirm the K-8008 binding site identified by the crystal structure.
DISCUSSION
We report here our identification of two new Sulindac analogs, K-8008 and K-8012, which showed potent tRXRα inhibitory effects through a novel and unique binding mechanism. Our results demonstrated that K-8008 and K-8012 were more effective than Sulindac in inhibiting RXRα transactivation (Figure 2A). Sulindac binds to RXRα with an IC50 of 82.9 µM based on the classical ligand competition assays (Zhou et al., 2010). K-8008 and K-8012 could antagonize 9-cis-RA-induced transactivation and inhibit coactivator peptide binding to RXRα with IC50 value of around 10 µM. Consistently, K-8008 and K-8012 showed improved activity than Sulindac in inhibiting AKT activation and inducing apoptosis. About 100 µM of Sulindac is normally used to achieve its anti-cancer effects (Weggen et al., 2001; Yamamoto et al., 1999; Zhang et al., 2000), whereas 10 to 50 µM of K-8008 and K-8012 were able to inhibit AKT activation and induce apoptosis of cancer cells. Furthermore, K-8008 showed potent inhibitory effect on the growth of tumor cells in animals without apparent toxicity. Similar to Sulindac, inhibition of AKT activation and induction of apoptosis by K-8008 and K-8012 were RXRα dependent, likely due to their inhibition of the interaction between tRXRα and p85α.
K-8008 and K-8012 were synthesized as part of the structure-activity relationship studies of Sulindac, by replacing the carboxylate group in K-80003 with its bioisotere tetrazole (Herr, 2002). Based on the principle of bioisosteric replacement (Matta et al., 2010), we anticipated that tetrazole group acted like the carboxylate group, a common motif found in most of the cognate RXR ligands, which interacts with Arg316 in the LBP, and therefore both K-8008 and K-8012 would compete 9-cis-RA for binding. To our surprise, both compounds, unlike Sulindac and K-80003, failed to compete with 9-cis-RA for binding to the LBP, demonstrating that they exert their antagonist effect through a different binding mechanism from Sulindac and K-80003. Our structure analysis confirmed that the tetrazole group of K-8008 and K-8012 binds to a region away from Arg316 and it anchors to the RXRα protein by sitting atop the N-terminus of helix 11, forming the charge-helix dipole interaction. This charge-dipole interaction may also function to stabilize the orientation and conformation of H11 as in most of the cases ligand binding to the cognate LBP induces the conformation change and reorientation of H11(Egea et al., 2000; Sato et al., 2010). Tetrazole is a non-classical isotere of –COOH moiety (Herr, 2002; Matta et al., 2010). It has a similar PKa to –COOH, but has a different steric characteristics and different number of atoms from –COOH. Spatially tetrazole moiety is bulkier than –COOH and it seems that some spatial clash could prevent the tetrazole group of K-8008 from making an anchoring chargecharge interaction with Arg316. As a result, K-8008 and K-8012 bind to a novel region. The structural results show that the compounds bind to a region that doesn’t overlap with the 9-cis-RA binding space, offering a structural explanation for the inability of K-8008 and K-8012 to compete with 9-cis-RA for RXRα binding. Furthermore, our mutagenesis data (Figures 7B–D) support the existence of this novel K-8008 binding site.
Our crystal structures revealed that K-8008 and K-8012 bind to a RXRα LBD tetramer structure through a novel hydrophobic region that is located on the surface of a monomer and near the dimerdimer interface in the tetramer. Unlike the binding of other published ligands, the binding of K-8008 doesn’t change the shape of the apo RXRα LBP. In addition, K-8008 interacts with monomers of each dimer in the tetramer, contributing to the dimer-dimer interaction. Taken together, K-8008 or K-8012 binding may help to stabilize the tetramer. Stabilizing the tetrameric state of RXRα through ligand binding may have important implication for the regulation of the nongenomic biological activities of RXRα. Previous studies have demonstrated that tetramer formation of RXRα serves as a mechanism to suppress its transactivation (Gampe et al., 2000; Kersten et al., 1998). The fact that inhibition of tRXRα interaction with p85α by K-8008 and K-8012 was associated with their binding to the RXRα tetramer raises an intriguing question that the formation of RXRα homotetramer may also represent a mechanism to suppress its interaction with cytoplasmic p85α and activation of tRXRα-dependent PI3K/AKT signaling.
Unlike classical nuclear receptor antagonists that bind to the RXRα LBP with high affinity, weak antagonism is commonly observed for antagonists that target the receptor surface binding sites (Caboni et al., 2012b; Gunther et al., 2008; Hwang et al., 2009; Rodriguez et al., 2003). Similarly, K-8008 and K-8012 bind to a surface hydrophobic site and display weak antagonist effect. However, the therapeutic relevance of targeting the RXRα through this new binding site is evidenced by our observation that both K-8008 and K-8012 could inhibit tRXRα activities in cancer cells in vitro and tumor growth in animals (Figure 4). In fact, the existence and therapeutic relevance of the novel sites other than the LBP have been described for several nuclear receptors, including estrogen receptor, androgen receptor, and vitamin D receptor (Caboni et al., 2012a; Kojetin et al., 2008; Mizwicki et al., 2004; Moore et al., 2010; Sun et al., 2011). Furthermore, in the case of Sulindac and analogs, weak antagonism may be clinic relevant as conventional administration of Sulindac can result in about 10–15 µM Sulindac in the serum of patients (Davies and Watson, 1997; Yamamoto et al., 1999). Up to approximately 50 µM of Sulindac could be detected in the plasma of humans depending on dose and schedule (Davies and Watson, 1997). And Sulindac can be concentrated in epithelial cells at concentrations that are at least 20-fold higher than those seen in the serum (Duggan et al., 1980; Yamamoto et al., 1999). Consistently, we observed that administration of K-8008 at the dose that effectively inhibited the growth of tumor cells did not show any apparent toxicity to animals (Figure 4). Thus, although showing a relatively weak binding to RXRα, our new compounds could be clinically relevant.
EXPERIMENTAL PROCEDURES
Compound Synthesis
K-8008 and K-8012 were synthesized using scheme of Figure 1B. See Supporting Information for details.
Cell Culture and Transfection
PC3 prostate cancer, ZR-75-1 breast cancer and HeLa cervical cancer cells were grown in RPMI1640, CV-1 African green monkey kidney cells, HCT-116 colon cancer, A549 lung cancer cells were cultured in DMEM containing 10% fetal bovine serum. The cells were maintained at 5% CO2 at 37°C. Subconfluent cells with exponential growth were used throughout the experiments. Cell transfections were carried out by using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. Myc-RXRα-Δ80 and Flag-p85α expression vectors as well as RXRα siRNA were described (Zhou et al., 2010). RXRα-L433E and RXRα-F438, 9A mutants were constructed by standard procedure and their LBDs were cloned into pBind expression vector (Wang et al., 2013).
CAT Assay
(TREpal)2-tk-CAT (100 ng), β-galactosidase (100 ng) and RXRα (20 ng) were transiently transfected into CV-1 cells (Zhang et al., 1992). Cells were then treated with or without 9-cis-RA (10−7 M) in the presence or absence of increasing concentrations of compounds for an additional 24 h. Cells were harvested and assayed for CAT and β-gal activity. To normalize for transfection efficiency, CAT activities were corrected to β-gal activities.
Mammalian One Hybrid
HCT-116 cells seeded in 24-well plates were transiently transfected with 50 ng pG5luc, 25 ng pBind-RXRα-LBD or mutant. Twenty-four hours after transfection, the medium was replaced by medium containing the Sulindac analogs and/or 9-cis-RA. Cells were washed, lysed and assayed by using the Dual-Luciferase Reporter Assay System (Promega). Transfection efficiency was normalized to Renilla luciferase activity.
Protein Expression and Purification
The human RXRα LBD (residues Thr223 to Thr462) was cloned as an N-terminal histidine-tagged fusion protein in pET15b expression vector and overproduced in Escherichia coli BL21 strain. Briefly, cells were harvested and sonicated, and the extract was incubated with the His60 Ni Superflow resin. The protein-resin complexes were washed and eluted. The eluent was collected and concentrated to 5 mg/mL for subsequent trails(Bourguet et al., 1995; Peet et al., 1998). For crystallization experiment, the His tag was cleaved by bovine thrombin (Sigma) and removed on the Resource-Q column (GE), using 0.1–1.0 M NaCL gradient and the TrisCl pH 8.0 buffer. The additional purification was done by the gel filtration on a Superdex-200 2660 column (GE) pre-equilibrated with the 75 mM NaCl, 20 mM Tris-Cl buffer (pH 8.0).
Ligand-binding Competition Assay
The His-tagged human RXRα-LBD(223–462) was incubated in tubes with unlabeled 9-cis-RA or different concentrations of compounds in 200 µL binding buffer [0.15 M KCl, 10 mM Tris·HCl (pH7.4), 8% glycerol, and 0.5% CHAPS detergent] at 4°C for 1 h. [3H]-9-cis-RA was added to the tubes to final concentration of 7.5 nM and final volume of 300 µL and incubated overnight at 4°C. The RXRα-LBD was captured by nickel-coated beads. Bound [3H]-9-cis-RA was quantitated by liquid scintillation counting.
TR-FRET Retinoic X Receptor alpha Coactivator Assay
Invitrogen’s LanthsScreen TR-FRET RXRα Coactivator Assay was conducted according to the manufacture’s protocol. The TR-FRET ratio was calculated by dividing the emission signal at 520 nm by the emission signal at 495 nm.
MTT assay
Confluent cells cultured in 96-welll dishes were treated with various concentrations of compounds for 48 h. The cells were then incubated with 2 mg/mL (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) for 4 h at 37°C. MTT solution was then aspirated and formazan in cells was instantly dissolved by addition of 150 µL DMSO each well. Absorbance was measured at 570 nm.
Western Blotting
Cells were lysed and equal amounts of the lysates were electrophoresed on 10% SDS-PAGE gels and transferred onto PVDF membranes (Millipore). The membranes were blocked with 5% skimmed milk in TBST [50 mM Tris-HCl (pH7.4),150 mM NaCl and 0.1% Tween20] for 1 h, then incubated with primary antibodies and secondary antibodies and detected using ECL system (Thermo). The dilutions of the primary antibodies were anti-RXRα (ΔN197, Santa Cruz) in 1:1000, anti-PARP(H-250, Santa Cruz) in 1:3000, anti-p85α (Millipore) in 1:1000, anti-p-AKT (D9E, Cell Signaling Technology) in 1:1000, anti-AKT1/2/3 (H-136, Santa Cruz) in 1:1000, anti-β-actin (Sigma) in 1:5000, anti-c-myc (9E10, Santa Cruz), anti-Flag (F1804, Sigma).
Co-immunoprecipitation assay
Cells were harvested and lysed in buffer containing 50 mM Hepes-NaOH (pH7.5), 2.5 mM EDTA, 100 mM NaCl, 0.5% NP40, and 10% glycerol, with 1 mM DTT and proteinase inhibitor cocktail. Immunoprecipitation was performed as described (Zhou et al., 2010).
HepG2 Xenografts
Nude mice (BALB/c, SPF grade, 16–18 g, 4–5-week old) were housed at 28°C in a laminar flow under sterilized conditions. Mice were injected subcutaneously with 100 µL HepG2 cells (2×10−6). For drug treatment, mice (n = 6) were treated intraperitoneally after 7 days of transplantation with K-8008 (20 mg/kg), K-80003 (20 mg/kg) or vehicle (tween-80) once a day. Body weight and tumor size were measured every 3 days. Mice were scarified after 12-day drug treatment and the tumors removed for various assessments. All experimentations and animal usage were performed and approved by the Animal Care and Use Committee of Xiamen University.
Histology and Apoptosis analysis
Paraffin wax embedded tumors were cut into 5 µM-thick sections. These sections were deparaffinized and stained with hematoxylin and eosin (H&E) according to the standard protocol. Tumor sections of HepG2 xenografts were also stained with TUNEL for assessing spontaneous apoptosis according to the manufacturer’s instructions (In situCell Death Detection Kit; Roche). The images were taken under a fluorescent microscope (Carl Zeiss).
Crystallization and structure solution of the RXR LBD-ligand complexes
The initial crystallization conditions were determined using the sitting-drop vapor-diffusion method and the crystallization screens Index and PEG-Ion (Hampton research). Other crystallization chemicals were from Hampton research and Sigma. The data were collected from crystals grown in sitting drops of the 96-well Intelli-Plates (ARI) by the vapor diffusion method. 0.2 µl of the protein-ligand complex containing 0.37 mM of RXR LBD, 0.5–0.7 mM of a ligand, 100 mM NaCl and 20 mM Tris-Cl buffer (pH 8.0) were mixed with 0.2 µl of the well solution (20% PEG3330 and 0.2M Magnesium Formate for the K-8008 complex or 0.2 M Na Acetate for the K-8012 complex) and incubated at 20° C. The first crystals appeared in 5–10 days and grew within same amount of time into 0.2×0.2×0.05 mm plates. The crystals were flesh-frozen against the well solution containing 20% PEG400 as a cryoprotectant. The diffraction data were collected from the cryo-cooled crystals (@100° K) at the beamline BL11-2 of SSRL and processed using the program suits XDS (Kabsch, 2010) and ccp4i (Collaborative Computational Project, 1994).
The structures were solved by the molecular replacement program Phaser (McCoy et al., 2007) using pdb entry 1G1U as an initial model. The model rebuilding and refinement were done with Coot (Emsley and Cowtan, 2004) and the program suit Phenix (Adams et al., 2010). The initial models and parameter files for the ligands were prepared by eLBOW of Phenix. The data collection and refinement statistics are presented in the Table S1.
Data Analyses
Data were expressed as means ± SD from three or more experiments. Statistical analysis was performed using Student’s t test. Differences were considered statistically significant with P <0.05.
Supplementary Material
Highlights.
Two new Sulindac analogs with improved RXRα-dependent anti-cancer efficacy
New Sulindac analogs regulate tRXRα activity by a unique binding mechanism
New Sulindac analogs bind to a tetramer of RXRα ligand-binding domain
Crystal structures reveal a new small molecule binding site of RXRα
SIGNIFICANCE.
RXRα represents an intriguing and unique target for pharmacologic intervention. Unfortunately, the development of RXRα-based drugs has been hampered by the side effects associated with targeting its cognate LBP. Thus, discovery of new strategies for targeting RXRα is significant. We report here our identification of two Sulindac-derived analogs that exert their anticancer effects by binding to a new binding site of RXRα, which is different from the classical LBP. These molecules could serve as chemical probes for studying the role of tRXRα in cancer. Furthermore, our results provide new opportunity to target specifically this surface site and thus may circumvent side effects associated with binding to the classical RXRα LBP. The development of tRXRα-selective inhibitors targeting a novel binding site may support a departure from the traditional approach of targeting the LBP and lead to a new paradigm targeting a functionally important surface site, which may lead to more effective and specific therapeutics.
ACKNOWLEDGEMENTS
This work was supported by Grants from the U.S. Army Medical Research and Material Command (W81XWH-11-1-0677), the National Institutes of Health (CA140980, GM089927, CA179379), the National Natural Science Foundation of China (NSFC-91129302), the Ministry of Education of China (IRT1037), and Science and Technology Bureau of Xiamen (3502Z20133008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The coordinates for the crystal structures of RXRα LBD in complex with K-8008 or K-8012 were deposited with the Protein Data Bank under ID codes 4N8R and 4N5G, respectively.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 6 figures and one table and can be found with this article online at http://dx.doi.org/xxx
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold LA, Estébanez-Perpiñá E, Togashi M, Jouravel N, Shelat A, McReynolds AC, Mar E, Nguyen P, Baxter JD, Fletterick RJ, et al. Discovery of Small Molecule Inhibitors of the Interaction of the Thyroid Hormone Receptor with Transcriptional Coregulators. Journal of Biological Chemistry. 2005;280:43048–43055. doi: 10.1074/jbc.M506693200. [DOI] [PubMed] [Google Scholar]
- Balkwill F. Tumour necrosis factor and cancer. Nature reviews Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- Bourguet W, Ruff M, Bonnier D, Granger F, Boeglin M, Chambon P, Moras D, Gronemeyer H. Purification, functional characterization, and crystallization of the ligand binding domain of the retinoid X receptor. Protein expression and purification. 1995;6:604–608. doi: 10.1006/prep.1995.1079. [DOI] [PubMed] [Google Scholar]
- Bushue N, Wan Y-JY. Retinoid pathway and cancer therapeutics. Advanced Drug Delivery Reviews. 2010;62:1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon V, Carbo LR, Estruch SB, Fletterick RJ, Estebanez-Perpina E. A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Molecular and cellular endocrinology. 2012;348:394–402. doi: 10.1016/j.mce.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Caboni L, Kinsella GK, Blanco F, Fayne D, Jagoe WN, Carr M, Williams DC, Meegan MJ, Lloyd DG. "True" antiandrogens-selective non-ligand-binding pocket disruptors of androgen receptor-coactivator interactions: novel tools for prostate cancer. J Med Chem. 2012a;55:1635–1644. doi: 10.1021/jm201438f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboni L, Kinsella GK, Blanco F, Fayne D, Jagoe WN, Carr M, Williams DC, Meegan MJ, Lloyd DG. “True” Antiandrogens—Selective Non-Ligand-Binding Pocket Disruptors of Androgen Receptor–Coactivator Interactions: Novel Tools for Prostate Cancer. Journal of medicinal chemistry. 2012b;55:1635–1644. doi: 10.1021/jm201438f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Liu W, Lin F, Li H, Kolluri SK, Lin B, Han Y-H, Dawson MI, Zhang X-k. Retinoid X receptor regulates Nur77/TR3-dependent apoptosis by modulating its nuclear export and mitochondrial targeting. Mol Cell Biol. 2004;24:9705–9725. doi: 10.1128/MCB.24.22.9705-9725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas F, Daury L, Grandemange S, Busson M, Seyer P, Hatier R, Carazo A, Cabello G, Wrutniak-Cabello C. Endocrine regulation of mitochondrial activity: involvement of truncated RXRalpha and c-Erb Aalpha1 proteins. FASEB J. 2003;17:426–436. doi: 10.1096/fj.02-0732com. [DOI] [PubMed] [Google Scholar]
- Chen Z-P, Iyer J, Bourguet W, Held P, Mioskowski C, Lebeau L, Noy N, Chambon P, Gronemeyer H. Ligand- and DNA-induced dissociation of RXR tetramers. Journal of molecular biology. 1998;275:55–65. doi: 10.1006/jmbi.1997.1413. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta crystallographica Section D, Biological crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davies NM, Watson MS. Clinical Pharmacokinetics of Sulindac: A Dynamic Old Drug. Clinical Pharmacokinetics. 1997;32:437–459. doi: 10.2165/00003088-199732060-00002. [DOI] [PubMed] [Google Scholar]
- Dawson MI, Zhang XK. Discovery and design of retinoic acid receptor and retinoid X receptor class- and subtype-selective synthetic analogs of all-trans-retinoic acid and 9-cis-retinoic acid. Current medicinal chemistry. 2002;9:623–637. doi: 10.2174/0929867023370789. [DOI] [PubMed] [Google Scholar]
- de Lera AR, Bourguet W, Altucci L, Gronemeyer H. Design of selective nuclear receptor modulators: RAR and RXR as a case study. Nature reviews Drug discovery. 2007;6:811–820. doi: 10.1038/nrd2398. [DOI] [PubMed] [Google Scholar]
- Dufour JM, Kim KH. Cellular and subcellular localization of six retinoid receptors in rat testis during postnatal development: identification of potential heterodimeric receptors. Biol Reprod. 1999;61:1300–1308. doi: 10.1095/biolreprod61.5.1300. [DOI] [PubMed] [Google Scholar]
- Duggan DE, Hooke KF, Hwang SS. Kinetics of the tissue distributions of sulindac and metabolites. Relevance to sites and rates of bioactivation. Drug Metabolism and Disposition. 1980;8:241–246. [PubMed] [Google Scholar]
- Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. Crystal structure of the human RXR[alpha] ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. The EMBO journal. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fukunaka K, Saito T, Wataba K, Ashihara K, Ito E, Kudo R. Changes in expression and subcellular localization of nuclear retinoic acid receptors in human endometrial epithelium during the menstrual cycle. Mol Hum Reprod. 2001;7:437–446. doi: 10.1093/molehr/7.5.437. [DOI] [PubMed] [Google Scholar]
- Gampe RT, Montana VG, Lambert MH, Wisely GB, Milburn MV, Xu HE. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 2000;14:2229–2241. doi: 10.1101/gad.802300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LXIII. Retinoid X Receptors. Pharmacological Reviews. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- Gunther JR, Moore TW, Collins ML, Katzenellenbogen JA. Amphipathic Benzenes Are Designed Inhibitors of the Estrogen Receptor α/Steroid Receptor Coactivator Interaction. ACS chemical biology. 2008;3:282–286. doi: 10.1021/cb800056r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr RJ. 5-Substituted-1H-tetrazoles as carboxylic acid isosteres: medicinal chemistry and synthetic methods. Bioorganic & medicinal chemistry. 2002;10:3379–3393. doi: 10.1016/s0968-0896(02)00239-0. [DOI] [PubMed] [Google Scholar]
- Hwang JY, Arnold LA, Zhu F, Kosinski A, Mangano TJ, Setola V, Roth BL, Guy RK. Improvement of Pharmacological Properties of Irreversible Thyroid Receptor Coactivator Binding Inhibitors. Journal of medicinal chemistry. 2009;52:3892–3901. doi: 10.1021/jm9002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Dong D, Lee W, Reczek PR, Noy N. Auto-silencing by the retinoid X receptor. J Mol Biol. 1998;284:21–32. doi: 10.1006/jmbi.1998.2168. [DOI] [PubMed] [Google Scholar]
- Kersten S, Kelleher D, Chambon P, Gronemeyer H, Noy N. Retinoid X receptor alpha forms tetramers in solution. Proceedings of the National Academy of Sciences. 1995;92:8645–8649. doi: 10.1073/pnas.92.19.8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojetin DJ, Burris TP, Jensen EV, Khan SA. Implications of the binding of tamoxifen to the coactivator recognition site of the estrogen receptor. Endocrine-Related Cancer. 2008;15:851–870. doi: 10.1677/ERC-07-0281. [DOI] [PubMed] [Google Scholar]
- Kolluri SK, Corr M, James SY, Bernasconi M, Lu D, Liu W, Cottam HB, Leoni LM, Carson DA, Zhang X-k. The R-enantiomer of the nonsteroidal antiinflammatory drug etodolac binds retinoid X receptor and induces tumor-selective apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2525–2530. doi: 10.1073/pnas.0409721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- Lu J, Dawson MI, Hu QY, Xia Z, Dambacher JD, Ye M, Zhang X-K, Li E. The effect of antagonists on the conformational exchange of the retinoid X receptor alpha ligand-binding domain. Magnetic Resonance in Chemistry. 2009;47:1071–1080. doi: 10.1002/mrc.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta CF, Arabi AA, Weaver DF. The bioisosteric similarity of the tetrazole and carboxylate anions: Clues from the topologies of the electrostatic potential and of the electron density. European journal of medicinal chemistry. 2010;45:1868–1872. doi: 10.1016/j.ejmech.2010.01.025. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita Y, Dodo K, Noguchi-Yachide T, Miyachi H, Makishima M, Hashimoto Y, Ishikawa M. LXXLL peptide mimetics as inhibitors of the interaction of vitamin D receptor with coactivators. Bioorganic & Medicinal Chemistry Letters. 2010;20:1712–1717. doi: 10.1016/j.bmcl.2010.01.079. [DOI] [PubMed] [Google Scholar]
- Mizwicki MT, Keidel D, Bula CM, Bishop JE, Zanello LP, Wurtz JM, Moras D, Norman AW. Identification of an alternative ligand-binding pocket in the nuclear vitamin D receptor and its functional importance in 1alpha,25(OH)2-vitamin D3 signaling. Proc Natl Acad Sci U S A. 2004;101:12876–12881. doi: 10.1073/pnas.0403606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TW, Mayne CG, Katzenellenbogen JA. Minireview: Not Picking Pockets: Nuclear Receptor Alternate-Site Modulators (NRAMs) Mol Endocrinol. 2010;24:683–695. doi: 10.1210/me.2009-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet DJ, Doyle DF, Corey DR, Mangelsdorf DJ. Engineering novel specificities for ligand-activated transcription in the nuclear hormone receptor RXR. Chemistry & biology. 1998;5:13–21. doi: 10.1016/s1074-5521(98)90083-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez AL, Tamrazi A, Collins ML, Katzenellenbogen JA. Design, Synthesis, and in Vitro Biological Evaluation of Small Molecule Inhibitors of Estrogen Receptor α Coactivator Binding. Journal of medicinal chemistry. 2003;47:600–611. doi: 10.1021/jm030404c. [DOI] [PubMed] [Google Scholar]
- Sato Y, Ramalanjaona N, Huet T, Potier N, Osz J, Antony P, Peluso-Iltis C, Poussin-Courmontagne P, Ennifar E, Mely Y, et al. The "Phantom Effect" of the Rexinoid LG100754: structural and functional insights. PloS one. 2010;5:e15119. doi: 10.1371/journal.pone.0015119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A, Moore TW, Gunther JR, Kim MS, Rhoden E, Du Y, Fu H, Snyder JP, Katzenellenbogen JA. Discovering small-molecule estrogen receptor alpha/coactivator binding inhibitors: high-throughput screening, ligand development, and models for enhanced potency. ChemMedChem. 2011;6:654–666. doi: 10.1002/cmdc.201000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto A, Narkar V, Shen Q, Uray IP, Davies PJA, Nagy L. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ. 2004;11:S126–S143. doi: 10.1038/sj.cdd.4401533. [DOI] [PubMed] [Google Scholar]
- Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta pharmacologica Sinica. 2008;29:1275–1288. doi: 10.1111/j.1745-7254.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chirgadze NY, Briggs SL, Khan S, Jensen EV, Burris TP. A second binding site for hydroxytamoxifen within the coactivator-binding groove of estrogen receptor β. Proceedings of the National Academy of Sciences. 2006;103:9908–9911. doi: 10.1073/pnas.0510596103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-G, Chen L, Chen J, Zheng J-F, Gao W, Zeng Z, Zhou H, Zhang X-k, Huang P-Q, Su Y. Synthesis and SAR study of modulators inhibiting tRXRα-dependent AKT activation. European journal of medicinal chemistry. 2013;62:632–648. doi: 10.1016/j.ejmech.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weggen S, Eriksen JL, Das P, Sagi SA, Wang R, Pietrzik CU, Findlay KA, Smith TE, Murphy MP, Bulter T, et al. A subset of NSAIDs lower amyloidogenic A[beta]42 independently of cyclooxygenase activity. Nature. 2001;414:212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Yin M-J, Lin K-M, Gaynor RB. Sulindac Inhibits Activation of the NF-κB Pathway. Journal of Biological Chemistry. 1999;274:27307–27314. doi: 10.1074/jbc.274.38.27307. [DOI] [PubMed] [Google Scholar]
- Yen WC, Lamph WW. A selective retinoid X receptor agonist bexarotene (LGD1069, Targretin) prevents and overcomes multidrug resistance in advanced prostate cancer. The Prostate. 2006;66:305–316. doi: 10.1002/pros.20347. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu J, Park BH, Kinzler KW, Vogelstein B. Role of BAX in the Apoptotic Response to Anticancer Agents. Science. 2000;290:989–992. doi: 10.1126/science.290.5493.989. [DOI] [PubMed] [Google Scholar]
- Zhang X-k, Lehmann J, Hoffmann B, Dawson MI, Cameron J, Graupner G, Hermann T, Tran P, Pfahl M. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature. 1992;358:587–591. doi: 10.1038/358587a0. [DOI] [PubMed] [Google Scholar]
- Zhou H, Liu W, Su Y, Wei Z, Liu J, Kolluri SK, Wu H, Cao Y, Chen J, Wu Y, et al. NSAID sulindac and its analog bind RXRalpha and inhibit RXRalpha-dependent AKT signaling. Cancer Cell. 2010;17:560–573. doi: 10.1016/j.ccr.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman TL, Thevananther S, Ghose R, Burns AR, Karpen SJ. Nuclear export of retinoid X receptor alpha in response to interleukin-1beta-mediated cell signaling: roles for JNK and SER260. J Biol Chem. 2006;281:15434–15440. doi: 10.1074/jbc.M508277200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.