Abstract
The transition between transcriptional initiation and elongation by RNA polymerase (Pol) II is associated with phosphorylation of its C-terminal tail (CTD). Depletion of Kin28, the TFIIH subunit that phosphorylates the CTD, does not affect elongation but causes Pol II occupancy profiles to shift upstream in a FACT-independent manner indicative of a defect in promoter escape. Stronger defects in promoter escape are linked to stronger effects on preinitiation complex formation and transcription, suggesting that impairment in promoter escape results in premature dissociation of general factors and Pol II near the promoter. Kin28 has a stronger effect on genes whose transcription is dependent on SAGA as opposed to TFIID. Strikingly, Kin28 depletion causes a dramatic increase in Mediator at the core promoter. These observations suggest that TFIIH phosphorylation of the CTD causes Mediator dissociation, thereby permitting rapid promoter escape of Pol II from the preinitiation complex.
INTRODUCTION
Transcription by RNA polymerase II (Pol II) requires the association of the TATA-binding protein (TBP) and general transcription factors to form a pre-initiation complex (PIC) at core promoters. PIC formation is the rate-limiting step for transcription at the vast majority of yeast promoters (Kuras and Struhl, 1999; Li et al., 1999). It can be stimulated by activator proteins via co-activator complexes (SAGA, Swi/Snf nucleosome remodeling complex, Mediator) or inhibited by repressor proteins via co-repressor complexes (Cyc8-Tup1 or Rpd3 histone deacetylase complex). PIC composition appears to be essentially identical at all yeast promoters (Rhee and Pugh, 2012).
After PIC formation, Pol II initiates mRNA synthesis, but productive transcription requires Pol II to escape from the PIC and transit into transcription elongation. The transition between initiation and elongation is associated with phosphorylation at the serine 5 (Ser5) residues within the hepta-peptide repeats in the C-terminal domain (CTD) of the largest Pol II subunit. Ser5 phosphorylation is mediated primarily by Kin28, the kinase subunit of the general transcription factor TFIIH (Feaver et al., 1994).
The role of Kin28 and its kinase activity in Pol II transcription has been unclear. From early in vitro studies, it was suggested that Kin28 stimulates Pol II escape from the PIC, and therefore, is important for transcription (Akoulitchev et al., 1995; Svejstrup et al., 1997; Liu et al., 2004). In accord with such a general function, loss of Kin28 activity in vivo via temperature-sensitive or degron mutations results in a loss of mRNA comparable to that observed upon loss of Pol II itself (Cismowski et al., 1995; Valay et al., 1995; Holstege et al., 1998). However, under these conditions, there is only a very modest effect on TBP occupancy at promoters, and presumably PIC formation (Kuras and Struhl, 1999), or transcription mediated by strong activator proteins (Lee and Lis, 1998; McNeil et al., 1998). Specific inactivation of Kin28 kinase activity via a kin28-analog-sensitive (as) mutant has led to conflicting results with respect to whether Kin28-mediated phosphorylation is (Hong et al., 2009) or is not (Kanin et al., 2007) important for global mRNA synthesis. It is important to note that these studies analyzed mRNA levels upon loss of Kin28 function, and hence are unable to distinguish effects on transcription per se as opposed to post-transcriptional events relevant for mRNA stability.
Analysis of transcription in vivo is best done by measurements of Pol II occupancy at promoters and coding regions. Genome-wide analysis of Pol II occupancy in a kin28-as mutant shows that Kin28-mediated phosphorylation of Ser5 has only a modest effect on transcription in vivo (Hong et al., 2009; Kim et al., 2010; Bataille et al., 2012). Thus, the strong effect of Kin28 on mRNA production presumably reflects its role in recruiting factors for chromatin modification, transcription termination, and mRNA processing.
It was reported that, in the kin28-as mutant strain, Pol II occupancy increases at the 5′ ends of long genes, suggesting a potential role of Kin28 in transcriptional elongation (Kim et al., 2010). As Kin28 associates with promoters and not coding regions, any effect on elongation is presumably due to recruitment of “elongation factors” via Kin28-mediated Ser5 phosphorylation. Based on biochemical experiments (Akoulitchev et al., 1995; Liu et al., 2004; Sogaard and Svejstrup, 2007), the Pol II occupancy profile in the kin28-as strain was also interpreted in terms of Kin28 stimulating the transition between initiation and elongation (Bataille et al., 2012). However, due to the limited resolution of the analysis, it was not possible to distinguish effects of Kin28 on elongation or promoter escape. Thus, it is unclear whether Kin28 mediates promoter escape in vivo.
Mediator, a 24-subunit complex, associates with Pol II via the CTD, and it is recruited to enhancer regions by activator proteins under appropriate environmental conditions (Boube et al., 2002; Bourbon et al., 2004). Mediator stimulates basal Pol II transcription in vitro (Kim et al., 1994; Takagi and Kornberg, 2006), and several subunits of Mediator are essential for general Pol II transcription in yeast cells (Thompson and Young, 1995; Holstege et al., 1998), leading to the view that Mediator is a general and essential component of the Pol II machinery. Furthermore, Mediator association with Pol II in vitro is strongly inhibited by phosphorylation of the CTD by Kin28 (Sogaard and Svejstrup, 2007), suggesting that Mediator is a component of the PIC and that its phosphorylation is linked to promoter escape (Guidi et al., 2004). However, in yeast cells, Mediator has never been found to associate with core promoters, and hence PICs, whereas it shows robust association with enhancers (Fan et al., 2006). One proposed explanation for these apparently discordant results is that Mediator presence in the PIC, though critical for transcription in vivo, is very transient, thereby precluding detection in standard chromatin immunoprecipitation experiments (Fan et al., 2006). Evidence for this suggestion is lacking.
Here, we use the anchor-away technique (Haruki et al., 2008) to effectively remove Kin28 from the nucleus, and examine the role of Kin28 on Pol II occupancy using ChIP-sequencing (ChIP-seq). This approach differs from previous work that employed a kin28-as mutant and tiling arrays that afforded less resolution than ChIP-seq (Kim et al., 2010; Bataille et al., 2012). Our results provide strong evidence that Kin28 stimulates promoter escape in vivo and differentially affects SAGA- and TFIID-dependent transcription. Strikingly, depletion of Kin28 results in a dramatic increase in Mediator occupancy at the core promoter, suggesting Kin28 stimulates rapid promoter escape via dissociation of Mediator from the PIC.
RESULTS
Kin28 function can be effectively removed by the anchor-away technique
We used the anchor-away (AA) method (Haruki et al., 2008) to conditionally remove Kin28 from the nucleus. In an AA-strain, rapamycin, which causes a strong tri-partite interaction with the FRB and FKBP12 domains, induces rapid export of the FRB-fused target protein from the nucleus to the cytoplasm, where the target protein is anchored to ribosome via Rpl13A-FKBP12. As expected from the essential role of Kin28, a Kin28-AA strain is unable to grow in media containing rapamycin (Figure 1A). Chromatin immunoprecipitation analysis shows that Kin28-FRB binding at all highly active promoters analyzed is reduced to background level after rapamycin treatment (Figure 1B). Thus, the anchor-away method can rapidly and effectively remove Kin28 from the nucleus, thereby permitting an analysis of the in vivo role of Kin28 on transcription.
Figure 1. Kin28 is important, but not essential for transcription.
(A) Growth spotting tests for wild-type (WT) and Kin28 anchor-away (AA) strains on rich YPD media with or without rapamycin at 30 °C for 2 days.
(B) Kin28-FRB occupancy at the indicated promoters in the Kin28-AA and control strains before and after 1 hour of rapamycin treatment. Averages and standard errors of three individual experiments are shown.
(C) Percentage of Pol II occupancy remained at the indicated coding regions in the WT, Spt15-AA and Kin28-AA strains after 1 hour of rapamycin treatment. Averages and standard errors of three individual experiments are presented.
(D) Changes of Pol II occupancy (measured by Pol II ChIP-seq) at 3′ end of active genes in the Kin28-AA and Spt15-AA strains upon rapamycin treatment. Numbers of genes are plotted against the log2 ratios of Pol II levels after rapamycin treatment (+Rap) over Pol II levels before rapamycin treatment (−Rap).
See also Figure S1.
Kin28 function is important, but not essential, for Pol II transcription
To assess the role of Kin28 in transcription, we measured Pol II occupancy upon rapid depletion of Kin28. Effects observed upon rapid depletion are highly likely to be directly due to Kin28, and measurements of Pol II occupancy avoid problems related to RNA stability. Upon Kin28 anchor-away, Pol II occupancy at coding regions of the three highly active genes analyzed are reduced. However, the reduction of Pol II occupancy occurs to a much lesser extent than observed upon depletion of TBP (Spt15; Figure 1C).
To analyze the level of Pol II transcription throughout the entire gene on a genome-wide basis, we performed ChIP-seq using antibodies against the Rpb3 subunit or un-phosphorylated CTD of the Rpb1 subunit of Pol II (8WG16). Consistent with a previous report (Bataille et al., 2012), Pol II ChIP-seq profiles measured by the two antibodies are highly comparable (Figure S1A), indicating that the 8WG16 antibody can be used to reliably measure Pol II occupancy and that not all of the 26 heptad-repeats within the Rpb1 CTD are phosphorylated. As the antibody against Rpb1 CTD gives much stronger ChIP-seq signal than the Rpb3 antibody, we focused on the Rpb1 CTD dataset for more accurate measurement of transcription in subsequent analyses.
We determined the level of Pol II transcription by measuring Pol II ChIP-seq signal at the last 200 bp window of coding region for each gene. Before Kin28 removal, Pol II occupancy is indistinguishable between a parental control strain and the Kin28-AA strain (correlation coefficient, r = 0.96), suggesting that the FRB domain does not affect Kin28 function. For all subsequent analyses, we focused on 460 genes whose Pol II levels before Kin28 depletion are at least 5-fold higher than the background (see Experimental Procedures), thereby avoiding sensitivity issues related to sequencing depth. While rapamycin does not affect transcription in the control strain, anchor-away of Kin28 reduces genome-wide Pol II occupancy about 2-fold on average, although some genes appear unaffected (Figure 1D; see below). In contrast and as expected, depletion of TBP strongly reduces transcription of essentially all genes (Figure 1D and S1B). Anchor-away of an essential nuclear replication factor, Mcm2, has minimal effect on Pol II transcription (Figure S1B), indicating that the transcription effects in the Kin28 and TBP anchor-away strains are not caused indirectly by the loss of an essential cellular function. These results are in accord with previous studies (Kanin et al., 2007; Hong et al., 2009; Kim et al., 2010; Bataille et al., 2012), and they confirm that Kin28, unlike other general transcription factors, is not essential for Pol II transcription.
Kin28 depletion causes a defect in promoter escape, but not elongation
On a genome-wide basis, depletion of Kin28 does not simply reduce transcription, but rather causes an altered pattern of Pol II occupancy throughout transcribed regions (Figure 2A). First, Pol II occupancy near the promoter shows an upstream shift in the Kin28-depleted strain as compared to the control strain. Second, a small peak of Pol II occupancy appears in the vicinity of the promoter; i.e. when proceeding downstream from the promoter, there is a noticeable increase in Pol II occupancy followed by a decrease near the beginning of the coding region. This peak is not observed in the control strain, in which Pol II occupancy progressively increases from the promoter through the beginning of the coding region. Furthermore, this altered pattern is specific to depletion of Kin28, as depletion of other TFIIH subunits (Rad3, Ssl1, and Ssl2) via anchor-away results in a general decrease in transcription (Figure S2A). In addition, considerable, but somewhat reduced, levels of TFIIH subunits remain associated with the core promoter upon loss of Kin28, whereas depletion of another TFIIH subunit (Ssl2) results in loss of all TFIIH subunits tested (Figure S2B, C). These and other observations to be discussed below strongly suggest that the loss of Kin28 kinase, rather than the TFIIH complex, results in a defect in promoter escape, such that Pol II spends relatively more time at or near the PIC in the Kin28-depleted strain than in the control strain.
Figure 2. Kin28 depletion causes Pol II re-distribution to 5′ ends of genes, but does not affect Pol II processivity.
(A) Overall Pol II ChIP-seq binding profiles of active genes in WT, Kin28-AA, and Spt15-AA strains before and after 1 hour of rapamycin treatment.
(B) Changes of Pol II processivity (measured between +500–1000) of active genes greater than 1 kb in length in the indicated strains after 1 hour of rapamycin treatment. Changes of Pol II processivity are expressed as log2 ratios of gradients of the linear regression lines after and before rapamycin treatment.
See also Figure S2.
To investigate the effect of Kin28 depletion on Pol II elongation, we measured Pol II processivity, as defined by relative Pol II levels throughout coding regions. Reduced Pol II processivity is a hallmark of a defect in transcription elongation, which can be caused by an intrinsically slow Pol II, a reduced level of nucleotide precursors, or reduced function of an “elongation factor” (Mason and Struhl, 2005). To avoid complications due to effects at or near promoters, we analyzed downstream portions of coding regions (0.5 to 1 kb downstream of the ATG codon) of genes longer than 1 kb. As shown in Figure 2B, Pol II processivity in the Kin28-depleted strain is not significantly different (p-value = 0.88) from that occurring before Kin28 removal or in the wild-type control strain. Taken together, the observed Pol II profile suggests that, in the absence of Kin28 function, Pol II is less efficiently released from PIC, but once released is not defective for elongation.
Kin28 affects PIC levels and transcriptional initiation
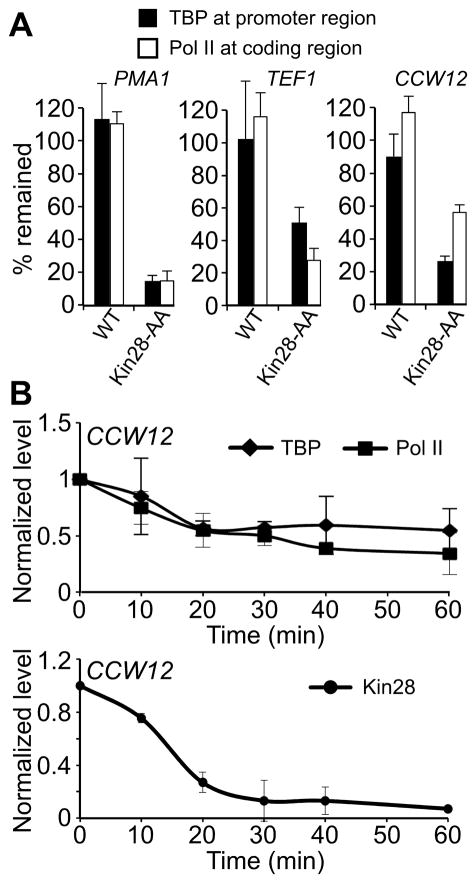
Given that transcription initiation and Pol II promoter escape are coupled, one might predict that a slower release of Pol II from PIC in the absence of Kin28 might increase PIC association with promoters. In contrast to this prediction, Kin28 depletion results in concomitant reduction of TBP (Figure 3A), TFIIA, and TFIIB (Figure S3) occupancies, and hence PIC levels, at all promoters analyzed. The kinetics of Kin28 depletion and the effects on TBP and Pol II occupancies are comparable (Figure 3B), strongly indicating that Kin28 directly affects PIC levels and Pol II transcription. The reduction in TBP occupancy is roughly comparable to the reduction in Pol II occupancy at individual genes (Figure 3A), in accord with the strong relationship between TBP occupancy and transcription observed in wild-type cells (Kuras and Struhl, 1999; Li et al., 1999).
Figure 3. Kin28 depletion affects PIC formation.
(A) TBP and Pol II occupancies at the promoter and the coding regions of three representative genes were analyzed before and after addition of rapamycin for 1 hour in the WT and Kin28-AA strains. Results are expressed as percentage of TBP and Pol II occupancies remained after the rapamycin treatment compared to those without the treatment.
(B) Time course analysis of TBP, Pol II and Kin28-FRB occupancies at the promoter region of highly expressed CCW12 upon rapamycin treatment. Values shown are occupancy levels relative to those before rapamycin treatment (i.e. Time = 0 min). Averages and standard errors of three individual experiments are presented.
See also Figure S3.
Differential transcriptional effects upon loss of Kin28
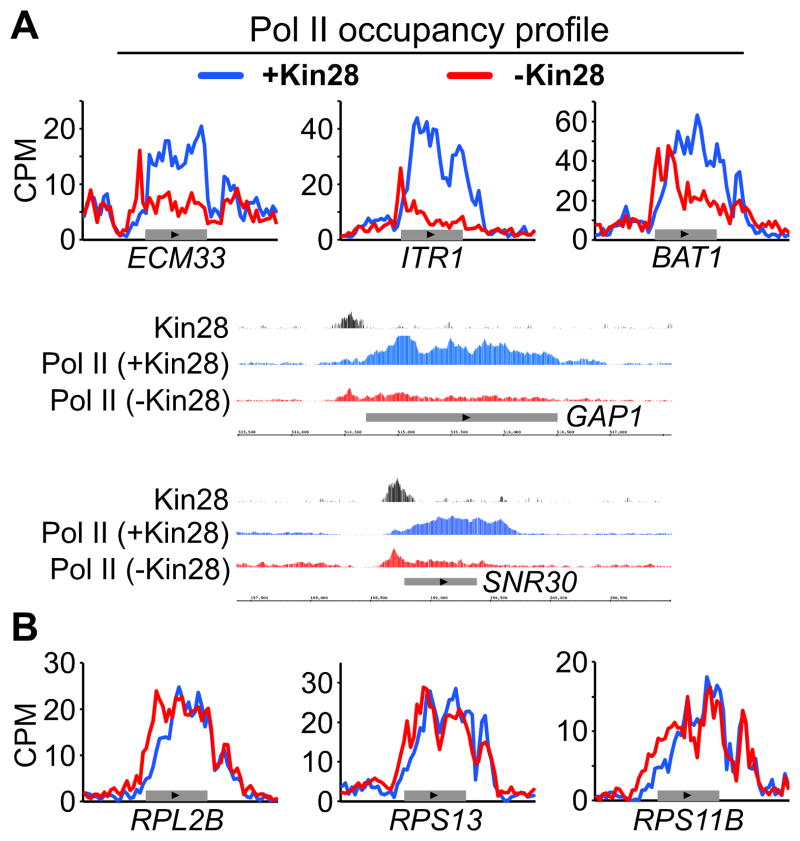
Although, Kin28 is a component of the general transcription factor TFIIH, depletion of Kin28 has diverse effects on Pol II occupancy of individual genes. For some genes, Pol II accumulates at the PIC (as defined by Kin28-FRB binding) with a substantial loss of Pol II at the coding region (Figure 4A), indicating a prominent role of Kin28 in the release of Pol II from PIC at these genes and subsequent transcription. In contrast, other genes display little or no effect on Pol II occupancy, and hence transcription, throughout the coding region (Figure 4B). At such genes, there is an upstream shift of Pol II in the vicinity of the promoter and the adjacent part of the coding region, suggesting a mild defect in promoter escape.
Figure 4. Kin28 depletion causes heterogeneous transcription effects.
(A, B) Pol II ChIP-seq binding profiles and genome-browser display of the indicated genes in the Kin28-AA strain before (+Kin28) and after (−Kin28) 1 hour of rapamycin treatment. See also Figure S4.
Previous studies suggested that Kin28 function is more important for mRNA levels of strongly transcribed genes (Hong et al., 2009) and elongation through long (>2 kb) genes (Kim et al., 2010). However, we found no relationship between the levels of Kin28 occupancy before its removal and effects on Pol II occupancy after Kin28 depletion (data not shown), and Kin28 depletion had comparable effects on the Pol II profiles of genes with high, medium, and low transcriptional activity (Figure S4A). In addition, the composite Pol II profile of long genes (> 2kb) shows no sign of a Pol II processivity defect (Figure S4B), and there is no correlation between gene length and Kin28 effect on Pol II occupancy at promoter and gene body (Figure S4B). While the transcriptional effects of Kin28 depletion appear slightly less pronounced for genes < 1 kb, this is due to the predominance of ribosomal protein genes that are less significantly affected (see below). Thus, differences in transcriptional activity and gene length do not account for diverse gene-specific effects that occur upon depletion of Kin28.
Kin28 is more important for PIC levels, promoter escape, and transcription of SAGA-dependent genes than TFIID-dependent genes
To address the basis of differential effects mediated by Kin28, we performed Gene Ontology analysis of genes that are rank-ordered with respect to their effects on Pol II occupancy within the coding region upon Kin28 depletion. Interestingly, genes encoding ribosomal proteins (RP) are highly enriched among genes with increased occupancy at 5′ ends and minimal effect at 3′ ends. As transcription of RP genes is TFIID-dependent, we asked whether the observed enrichment is specific to RP genes or to TFIID-dependent genes in general.
Indeed, there is a striking pattern that distinguishes TFIID-dependent genes from SAGA-dependent genes (Figure 5A). When analyzed separately, transcription (Pol II occupancy at the 3′ end of coding region) of SAGA-dependent genes is significantly (p-value = 10−15) more affected by Kin28 depletion as compared to TFIID-dependent genes. Analysis of published Pol II ChIP-chip data in the kin28-as strain under conditions in which Kin28 kinase activity is specifically inactivated (Kim et al., 2010), also shows this differential effect (Figure 5B), suggesting that the effect is caused by the lack of Kin28 kinase activity rather than the loss of Kin28 protein. Additionally, the difference is independent of Kin28 levels at promoters (Figure S5A), indicating that SAGA-mediated transcription is more strongly dependent on Kin28 function than TFIID-dependent transcription. Lastly, upon Kin28 depletion, SAGA-dependent promoters show a subtle but statistically significant (p-value = 0.003) stronger reduction in TFIIB occupancy, and hence PIC levels as compared to TFIID-dependent promoters (Figure 5C), suggesting that the differential transcription effect is partially mediated by Kin28’s role in transcription initiation.
Figure 5. Kin28 depletion affects SAGA-dependent transcription more than TFIID-dependent transcription.
(A, B) Cumulative frequency plots of transcription (Pol II occupancy) changes at SAGA- and TFIID-dependent genes (Huisinga and Pugh, 2004) upon Kin28 depletion in the Kin28-AA strain or inactivation of Kin28 kinase activity in the kin28-as strain.
(C) Cumulative frequency plots of TFIIB occupancy changes at SAGA- and TFIID-dependent genes upon Kin28 depletion by anchor-away.
(D) Pol II (blue and red lines) and Kin28-FRB (black line) ChIP-seq binding profiles across promoter regions (+/− 500 bp from ATG) of active SAGA- and TFIID-dependent genes in the Kin28-AA strain before and after Kin28 depletion.
(E) Spt16 ChIP-seq binding profiles were plotted for the same regions, as in (D).
See also Figures S5 and S6.
Given that Kin28 is required for the release of Pol II from the PIC, we addressed whether Kin28 anchor-away differentially affects the transition of Pol II at these two classes of promoters. Indeed, the averaged Pol II profiles show that Kin28 depletion causes distinct Pol II accumulation at SAGA-regulated PICs (defined by the Kin28 profile) with significant reductions of Pol II at coding regions (Figure 5D). By contrast, although Kin28 depletion also causes Pol II to shift upstream near the 5′ end of TFIID-dependent genes, which is indicative of a defect in promoter escape, distinct Pol II accumulation is not observed at the PIC and high levels of Pol II are still observed at coding regions (Figure 5D). Moreover, it is worthy of note that Kin28 depletion does not cause Spt16, an essential subunit of FACT that travels with elongating Pol II (Mason and Struhl, 2003), to shift upstream near the 5′ end of either SAGA or TFIID-dependent genes (Figure 5E), suggesting that observed upstream-shifted Pol II is not in the elongating form.
We used a theoretical treatment of Pol II occupancy profiles (Ehrensberger et al., 2013) to estimate the extent of the promoter escape defect in SAGA- and TFIID-dependent genes. Specifically, we varied the escape rate (i.e. “elongation rate” for the initial 20 bp region of the promoter) and examined the predicted Pol II occupancy profile (Figure S5B). Interestingly, a >70% reduction of the escape rate results in a Pol II profile similar to the observed profile of SAGA-dependent genes. A smaller reduction of 30% results in a profile similar to TFIID-dependent genes. Taken together, these results indicate that Kin28 function is relatively more important for PIC levels, promoter escape, and subsequent transcription at SAGA-regulated genes than at TFIID-dependent genes.
Pol II Ser5 phosphorylation are equally affected at SAGA- and TFIID-dependent genes upon Kin28 depletion
The differential effect of Kin28 depletion on SAGA- and TFIID-dependent transcription raises the question of whether Kin28 acts differently at these promoters or if it performs the same molecular function (i.e. Pol II Ser5 phosphorylation) but somehow differentially affects the two classes of promoters. It is also possible that Pol II promoter escape at the TFIID-dependent promoters might be facilitated via Pol II Ser5 phosphorylation by a redundant kinase in the absence of Kin28 function. However, while the overall level of Pol II Ser5 phosphorylation near the promoter is dramatically reduced when Kin28 is removed, the extent of decrease is indistinguishable for SAGA- and TFIID-dependent genes (Figure S6). Downstream Pol II Ser2 phosphorylation is also reduced (Figure S6), presumably due to impaired Ser5-dependent recruitment of the Ser2 kinases Ctk1 and Bur1/Bur2 (Qiu et al., 2009; Bataille et al., 2012). These observations not only indicate a common function of Kin28 at all promoters, but also eliminate the possibility that a redundant kinase is responsible for the TFIID-dependent transcription in the absence of Kin28 function.
Kin28 depletion causes dramatic accumulation of Mediator at pre-initiation complexes of active genes
Mediator is recruited much more strongly by activators of stress-responsive genes (typically SAGA-dependent) than the activator of ribosomal protein genes (typically TFIID-dependent) (Fan et al., 2006). In addition, the tail module of the Mediator complex is functionally more important at SAGA-dependent genes than TFIID-dependent genes (Ansari et al., 2012). Moreover, Kin28 phosphorylation of the CTD facilitates Pol II dissociation from Mediator in vitro (Sogaard and Svejstrup, 2007). These findings lead us to hypothesize that the observed difference between TFIID- and SAGA-dependent genes upon Kin28 depletion is related to different levels of Mediator function at these two classes of promoters.
To test this hypothesis, we compared Mediator binding (Rgr1, Med7, and Srb6 subunits) at core promoters of SAGA- and TFIID-dependent genes before and after Kin28 depletion. Consistent with our previous studies (Fan et al., 2006; Fan and Struhl, 2009), we could not detect significant Mediator association at the core promoters of highly expressed SAGA- and TFIID-dependent genes before Kin28 depletion. Strikingly, upon Kin28 depletion, Mediator very strongly associates with the core promoter region of several active genes analyzed (Figures 6A, S7A). Genome-wide analysis shows that Mediator occupancy increases at core promoters of all active genes irrespective of whether they are SAGA- or TFIID-dependent (Figure 6B). The Mediator binding profile upon Kin28 depletion is indistinguishable from that of TFIIB at core promoters (Figure 6C), indicating that Mediator associates with the PIC under these conditions. Increased Mediator association is strongly correlated with increased Pol II occupancy at the core promoter (Figure S7B), suggesting a mechanistic connection between Mediator dissociation and Pol II promoter escape. Interestingly, we noted a modest but statistically significant higher Mediator-to-Pol II ratio for SAGA-dependent genes as compared to TFIID-dependent genes (Figure 6D), which may be related to the differential Kin28 effect between the two classes of genes.
Figure 6. Kin28 depletion causes dramatic accumulation of Mediator at core promoters of all active genes.

(A) Mediator (Rgr1) occupancy at the core promoters of indicated genes in the Kin28-AA strain before and after Kin28 depletion. Averages and standard errors of three individual experiments are presented.
(B) Comparison between TFIIB levels and Mediator (Rgr1) binding changes in the Kin28-AA strain upon Kin28 depletion. Promoter TFIIB level was determined by the total count of TFIIB ChIP-seq reads within the 100 bp window centered on the transcription start site. For Mediator (Rgr1) binding changes, normalized read counts of analyzed promoters after Kin28 depletion were divided by those before Kin28 depletion and expressed as log2 ratios. Only promoters with more than 1000 TFIIB raw counts before Kin28 depletion were included in the analysis.
(C) Mediator (Rgr1) and TFIIB ChIP-seq binding profiles of active genes in the Kin28-AA strain before (blue line) and after (red line) Kin28 depletion.
(D) Boxplots of Mediator (Rgr1) to Pol II binding ratios at core promoter regions of active SAGA- and TFIID-dependent genes in the Kin28-AA strain before (+Kin28) and after (−Kin28) Kin28 depletion.
(E) Mediator (Rgr1) occupancy at the core promoters of indicated genes in the kin28-as strain upon inactivation of Kin28 kinase activity by the inhibitor NA-PP1. Averages and standard errors of three individual experiments are presented.
See also Figure S7.
Mediator association with the core promoter is strongly increased upon specific inactivation of Kin28 kinase activity (via the analog-sensitive allele; Figure 6E, S7C). We note that Mediator occupancy in the kin28-as strain is lower than in the Kin28-depleted strain; this might reflect differences in strain backgrounds or growth conditions, extent of Kin28 inactivation/loss, or an indirect effect on TFIIH function. Although a non-kinase function of Kin28 on Mediator association cannot be excluded, any such function has minimal effect on transcription, because the Pol II occupancy profile in Kin28-depleted cells is very similar to that inkin28 -as cells.
Taken together, these observations indicate that Mediator dissociation from PIC is severely inhibited in the absence of Kin28-mediated phosphorylation. Conversely, the fact that Mediator cannot be detected at the PIC under normal conditions suggests that Mediator association with the PIC is extremely transient. Thus, Kin28 facilitates efficient Mediator dissociation and Pol II release from the PIC in vivo.
DISCUSSION
Kin28 stimulates promoter escape, but not elongation, in vivo
In vitro, Kin28-mediated phosphorylation of Ser5 can stimulate the escape of Pol II from the PIC into an elongation competent form (Akoulitchev et al., 1995; Liu et al., 2004). However, it is unclear whether these in vitro experiments are physiologically relevant, particularly given the artificial conditions (i.e. no nucleotide triphosphate precursors and no Mediator) used to form a stable PIC. Here, we provide strong evidence that Kin28 stimulates promoter escape in vivo. Due to the mapping resolution afforded by chromatin immunoprecipitation, our definition of promoter escape requires sufficient movement of Pol II from the location of PIC (estimated to be between 10 and 20 bp). As such, our results do not address intermediate biochemical steps between PIC formation and promoter escape such as true initiation (synthesis of the first few nucleotides).
Most definitively, in the absence of Kin28, many genes show a peak of Pol II occupancy that coincides with the location of Kin28 in non-depleted cells. This peak of Pol II is not observed in wild-type cells, indicating that loss of Kin28 function results in Pol II being “stuck” at or very close (< 10 bp) to the position of the PIC. More generally, loss of Kin28 causes the Pol II profile to shift upstream of the profile observed in wild-type cells. This upstream shift is the predicted profile of a modest defect in promoter escape, and it cannot be due to a defect in Pol II processivity, which by definition occurs after the transition to elongation. Furthermore, the upstream-shifted Pol II is not associated with FACT, an H2A/H2B chaperone that travels with elongating Pol II but does not associate with the PIC (Mason and Struhl, 2003). In addition, and in contrast to a previous study that did not distinguish between processivity and promoter escape (Kim et al., 2010), we did not detect a defect in Pol II processivity in the absence of Kin28. As reduced processivity is a hallmark of an elongation defect (Mason and Struhl, 2005), our results indicate that Kin28 does not play a significant role in Pol II elongation. Thus, while less dramatic than the “stuck” Pol II at some promoters, the upstream shift at most promoters indicates increased Pol II dwell time in the immediate vicinity of the PIC, strongly indicating a broad defect in promoter escape.
Kin28 stimulates rapid promoter escape via dissociation of Mediator from the PIC
The prevailing view of Mediator is that it is generally required for transcriptional initiation in vivo (Thompson and Young, 1995; Holstege et al., 1998) and in vitro (Kim et al., 1994; Takagi and Kornberg, 2006), presumably involving a direct interaction with the unphosphorylated CTD (Svejstrup et al., 1997; Sogaard and Svejstrup, 2007) at the PIC. To account for the apparently conflicting observation that Mediator was not detected at core promoters by ChIP, we suggested that Mediator association with core promoters is very transient in vivo (Fan et al., 2006; Fan and Struhl, 2009). A prediction of this suggestion is that Mediator should be detected at core promoters if it is trapped there by conditions that inhibit its dissociation. Indeed, depletion of Kin28 results in robust Mediator association at essentially all active core promoters at a position coincident with the PIC. This observation indicates that, in vivo, 1) Kin28-mediated phosphorylation is important for Mediator dissociation, 2) Mediator is a component of the PIC, 3) Mediator association with the PIC is very transient under normal conditions, 4) Mediator dissociation is mechanistically linked to promoter escape, and 5) the transition between PIC formation and Pol II escape is very rapid.
Our results in vivo and previous findings in vitro strongly suggest a model in which Kin28 phosphorylation of the CTD causes dissociation of Mediator from the PIC, thereby facilitating promoter escape of Pol II (Figure 7). In the absence of Kin28-mediated phosphorylation, Mediator dissociation and promoter escape are severely affected. The transition between the PIC and the process of promoter escape must be rapid under normal physiological conditions, because Mediator and a peak of Pol II is not easily detected at core promoters. As CTD phosphorylation at Ser5 severely weakens the interaction of Pol II with Mediator in vitro, it is highly likely that this is the major (and perhaps exclusive) role of Kin28 in mediating promoter escape. However, other potential substrates of Kin28 may play a role, and in this regard, Kin28 can phosphorylate the Med4 and Rgr1 subunits of Mediator (Liu et al., 2004).
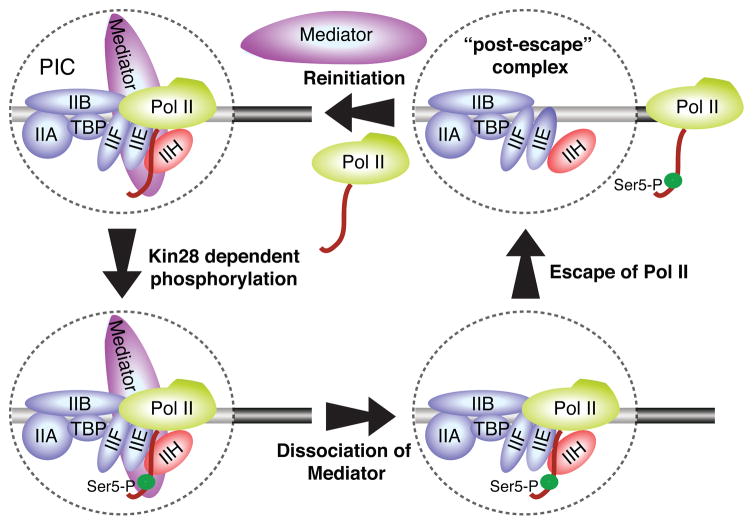
Figure 7. Model for the transition between initiation and elongation.
The general transcription factors and Mediator assemble at a core promoter to form a pre-initiation complex (PIC). Upon PIC formation, Kin28-dependent phosphorylation of Pol II CTD inhibits the interaction between Pol II and Mediator, leading to a rapid dissociation of Mediator from the PIC, facilitating Pol II escape from the PIC for productive elongation. Although it is formally possible that Mediator dissociation and Pol II escape from the PIC occur simultaneously or in the reverse order, we disfavor this possibility as CTD phosphorylation destabilizes the direct Mediator-Pol II interaction in vitro, and there is no evidence in vivo for direct stable Mediator-PIC association in the absence of Pol II. After Pol II promoter escape, a “post-escape” transcription initiation complex, which is relatively stable (minute time scale), remains bound at the core promoter such that transcription re-initiation can occur upon reloading of Mediator and Pol II.
The model in Figure 7 is of interest with respect to a transcriptional reinitiation intermediate that includes Mediator and is stable in vitro (Yudkovsky et al., 2000). In general, it is difficult to assess the physiological significance or stability of complexes that are formed under non-physiological conditions. However, a key feature of this intermediate is that it is stabilized by a transcriptional activator protein, and presumably involves activator-mediated recruitment of Mediator to activator binding sites. As the model of Figure 7 pertains to events at the core promoter, it is likely that the proposed reinitiation intermediate (Yudkovsky et al., 2000) reflects a different stage of the overall transcription process.
Promoter escape is differentially limiting at yeast genes and can lead to premature dissociation of Pol II
Kin28-mediated promoter escape is not essential for Pol II transcription, because we never observe a gene in which Pol II remains associated at the position of the PIC and does not traverse the coding region. As such, there must be a Kin28-independent mechanism that contributes to, but is not sufficient for, the efficient promoter escape that occurs in wild-type cells. Although Kin28 is presumably a general component of the PIC that phosphorylates Ser5 of the CTD at all core promoters, Kin28-mediated promoter escape is differentially limiting among genes. In the absence of Kin28, promoter escape is more limiting at genes showing a peak of Pol II occupancy at the position of the PIC than at genes with an upstream shift of Pol II. Interestingly, stronger defects in promoter escape are linked to lower levels of Pol II association throughout the entire coding region. These observations suggest that some Pol II molecules delayed in promoter escape prematurely dissociate from the template, thereby resulting in reduced transcription. This putative premature dissociation could occur at the level of the PIC and/or at an early stage of initiation.
Kin28 affects PIC formation/stability
One might predict that a defect in promoter escape would result in a longer-lived PIC and hence an increase in TBP occupancy. In contrast to this prediction, TBP occupancy decreases at many promoters upon Kin28 depletion, suggesting a defect in PIC formation/stability and hence transcriptional initiation. The presence of Kin28 at promoters, the rapid reduction in Pol II occupancy upon Kin28 depletion, and the strong relationship between TBP occupancy and transcriptional activity (Kuras and Struhl, 1999; Li et al., 1999) suggests that Kin28 plays an important role in transcriptional initiation. Any role in transcriptional initiation would involve the kinase activity of Kin28, because a similar transcriptional defect is observed in the kin28-as mutant strain (Hong et al., 2009; Kim et al., 2010).
There are two potential mechanisms by which Kin28 could affect PIC formation/stability. One possibility is that Kin28 directly stabilizes the PIC, presumably by phosphorylating a component of the basal Pol II machinery. Alternatively, Kin28 might indirectly affect PIC stability by virtue of its effects on promoter escape and premature Pol II dissociation. Specifically, premature dissociation of Pol II might result in rapid destabilization of the PIC, and hence a decrease in TBP occupancy. We favor this second model for two reasons. First, depletion of Pol II (via anchor-away) results in a rapid and drastic reduction in TBP occupancy (unpublished observations), indicating that Pol II is required for a stable PIC in vivo. Second, the linkage of strong defects in promoter escape to strong effects on TBP occupancy and Pol II transcription is easily explained by the model that invokes premature Pol II dissociation due to Kin28 depletion. In contrast, if Kin28 directly stabilizes the PIC, it is not obvious why Kin28 depletion would lead to gene-specific effects on promoter escape that are linked to transcriptional activity.
Kin28 differentially affects SAGA- and TFIID-dependent transcription
Several observations indicate that Kin28 is more important for promoter escape at SAGA-dependent genes as compared to TFIID-dependent genes. First, on an overall basis, SAGA-dependent genes show a peak of Pol II occupancy at the promoter, whereas TFIID-dependent genes display a milder upstream shift of the Pol II occupancy profile. Second, in comparison to TFIID-dependent genes, SAGA-dependent genes show a greater reduction of Pol II occupancy across coding regions, and hence transcription. Third, as Ser5 and Ser2 phosphorylation levels are comparable between the two classes of genes upon Kin28 depletion, the differential affect is unlikely to reflect a redundant kinase functioning at TFIID-dependent genes. Thus, while promoter escape is affected for both classes of genes in the absence of Kin28, escape appears less efficient at SAGA-regulated promoters than at TFIID-dependent promoters. As a consequence and as discussed above, Pol II would spend more time at SAGA-dependent promoters, which would increase premature Pol II dissociation and cause a more significant transcriptional effect.
Why does Kin28 differentially affect promoter escape and transcription at SAGA- and TFIID-dependent genes? Although the mechanistic basis is unknown, we suggest that Mediator-dependent inhibition of promoter escape is more pronounced at SAGA-dependent genes than at TFIID-dependent genes. In this regard, Mediator is recruited much more strongly by activators of stress-responsive genes (typically SAGA-dependent) than activators of ribosomal protein genes (typically TFIID-dependent)(Fan et al., 2006), and the Mediator tail module is functionally more important at SAGA-dependent genes (Ansari et al., 2012). In addition, the ratio of Mediator to Pol II is higher at SAGA-dependent promoters as opposed to TFIID-dependent promoters. Thus, the Kin28-mediated differences between TFIID- and SAGA-dependent genes may simply reflect higher levels of Mediator at SAGA-dependent genes, perhaps due to the stabilizing effects of activators that preferentially recruit Mediator. Such relatively higher levels of Mediator would lead to preferential inhibition of promoter escape at SAGA-dependent genes.
The transition between initiation and elongation in vivo
Based on our results and the assumption that formaldehyde crosslinking of an individual factor is similar at all stages of the transcription process, we can estimate kinetic parameters involved in the transition between initiation and elongation in vivo. As discussed previously (Reppas et al., 2006), the apparent Pol II ChIP occupancy at a given location within the coding region actually represents the sum of Pol II occupancies for ~150 different positions, which corresponds to the mean fragment size in the ChIP samples (Fan et al., 2008). As the Pol II elongation rate is ~30 nt/second (O’Brien and Lis, 1993; Iyer and Struhl, 1996; Mason and Struhl, 2005), a given level of Pol II occupancy corresponds to ~5 seconds of transcription. Under conditions with the most severe defect in promoter escape (SAGA-dependent genes in Kin28-depleted cells), Pol II occupancy at the PIC is roughly comparable to that observed in the downstream coding region. Thus, under these conditions of defective promoter escape, we estimate that Pol II spends ~5 seconds at or very near the position of the PIC.
Our results provide the first measurements of PIC levels in vivo, and they indicate that the PIC is a very short-lived entity in vivo. We define the PIC as the entity that contains all the general factors and Mediator, a definition supported by the facts that Mediator is generally required for Pol II transcription in vivo (Thompson and Young, 1995; Holstege et al., 1998), physically associates with the initiating form of Pol II (Svejstrup et al., 1997; Sogaard and Svejstrup, 2007), and stimulates basal transcription in vitro (Kim et al., 1994; Takagi and Kornberg, 2006). As such, measurements of Mediator occupancy at the core promoter represent measurements of PIC levels. At highly active promoters where Mediator occupancy at core promoters can be measured in wild-type cells, Mediator occupancy increases ~40-fold upon loss of Kin28. As Pol II spends ~5 seconds at promoters that are defective in promoter escape and have high levels of Mediator, we estimate that Mediator spends ~1/8 second in the PIC in wild-type cells (i.e. 40-fold less time to account for the 40-fold lower observed occupancy). Although we don’t have specific information about the time it takes between Mediator dissociation and Pol II escape, we infer that it must be < 1 second, because we would expect to see a small peak of Pol II occupancy at the PIC in wild-type cells if promoter escape were slower. Taken together, the PIC exists for considerably less than 1 second in vivo in wild-type cells, and promoter escape is very rapid. We note that Mediator is readily detected at core promoters in mammalian cells (Kagey et al., 2010), indicating that the PIC is longer-lived than in yeast cells.
In Kin28-depleted cells with high Mediator occupancy at the core promoter, the PIC is much longer-lived than in wild-type cells. Nevertheless, under these conditions, TBP and TFIIB occupancy levels actually decrease ~2 fold, such that the TBP/TFIIB:Mediator ratio is ~80-fold higher in wild-type cells than in Kin28-depleted cells. As the relative occupancy values of the various PIC components should be independent of PIC levels, this observation indicates that TBP and TFIIB association in wild-type cells occurs primarily in the absence of Mediator and Pol II. As a consequence, measurements of TBP and TFIIB occupancy in wild-type cells largely reflect the amount of a complex in which Pol II has already escaped but which is suitable for association of a new Pol II molecule and subsequent reinitiation (Figure 7). As the half-life of TBP occupancy at Pol II promoters in yeast is estimated to be 1–5 minutes (Poorey et al., 2013), a “post-escape” complex is sufficiently stable to permit multiple rounds of reinitiation (Struhl, 1996). By analogy to a biochemically defined reinitiation intermediate (Yudkovsky et al., 2000), it is possible that this post-escape complex could be stabilized by activator, thereby allowing increased activator-dependent reinitiation. Taken together, our results suggest that although the PIC can be a stable structure in vitro, it is very transient in vivo, in part because Kin28-dependent phosphorylation of the CTD causes dissociation of Mediator from the PIC, leading to rapid escape of Pol II from the promoter.
EXPERIMENTAL PROCEDURES
Yeast strains and growth conditions
Strains used in this study are listed in Table S1. Anchor-away strains were constructed as previously described (Fan et al., 2011). For the spotting assay, yeast cells were grown to mid-log phase, and 5-fold serial dilutions of cells were spotted on Synthetic Complete (SC) solid media with or without 1 μg/ml rapamycin. Cells were grown at 30°C for 48 hours. For anchor-away, strains were grown in SC liquid media to an OD600nm of 0.4, and rapamycin was then added at a final concentration of 1 μg/ml for 1 hr. The analog-sensitive mutant, kin28-as, and its isogenic WT strain were grown in SC liquid media to an OD600nm of 0.4, and NA-PP1 (or DMSO as mock treatment) was then added at a final concentration of 5 μM for 1 hour.
Chromatin immunoprecipitation (ChIP)
Chromatin, prepared as described previously (Fan et al., 2008), from ~4 ml of cells (OD600nm ~0.5) was immunoprecipitated with antibodies against Pol II unphosphorylated CTD (8WG16, Covance), Rpb3 (W0012, Neoclone), CTD Ser5-P (3E8, Millipore), CTD Ser2-P (3E10, Millipore), or FRB (Alexis), c-Myc (9E10, Santa Cruz), TFIIA, TFIIB, TBP, Tfb1, or Tfb3. Quantitative PCR in real time was performed using primers listed in Table S1.
ChIP-seq and data analyses
Barcoded sequencing libraries from ChIP DNA were constructed as described previously (Wong and Struhl, 2011; Wong et al., 2013). Sequence reads were mapped using Bowtie with the following options: -k 1 --best. The Integrated Genome Browser (Nicol et al., 2009) was used for visualizing ChIP-seq data, and for the screenshots in Figure 4. Transcription activity (Pol II level) of a gene was calculated by summing the number of Pol II ChIP-seq reads within full coding region or the last 200 bp of coding region normalized to the respective surveyed window size, and is expressed as counts per million mapped reads (CPM).
To define the active-gene set, genes were first sorted according to their transcription activities, and the median Pol II level of the bottom 5% of genes is set as the background level. Genes with Pol II level at least 5-fold higher than the background level in the Kin28 anchor-away strain without rapamycin treatment were selected for subsequent analyses. Pol II occupancy was calculated by first dividing the Pol II level at a given region over the background level and then subtracting one from the ratio. For Pol II binding profiles, Pol II occupancies for each gene were measured by read counts either in windows of 20 bp across the respective promoter or in 20 windows across its entire coding region. For each window, the median read count over all active genes was plotted in the overall Pol II binding profiles. Gene clustering was performed using the Hierarchical Clustering algorithm with Pearson Uncentered metric (Eisen et al., 1998; Bar-Joseph et al., 2001) from the MeV program (Saeed et al., 2003). TFIID- and SAGA-dependent genes were defined as in a previous study (Huisinga and Pugh, 2004). Pol II processivity change for each gene was determined by comparing the gradient of a linear regression line of Pol II occupancy across 10 sliding windows of 50 bp covering the coding region between +500 bp to +1 kb relative to the ATG of the gene before and after rapamycin treatment.
Supplementary Material
HIGHLIGHTS.
Kin28, the TFIIH kinase subunit, important for promoter escape but not elongation
Kin28 depletion dramatically increases Mediator occupancy at the core promoter
Stronger defects in promoter escape are linked to stronger transcriptional effects
TFIIH phosphorylation of the CTD causes Mediator dissociation and promoter escape
Acknowledgments
We thank Zarmik Moqtaderi for providing bioinformatics programs, Steven Hahn and Sun woo Hong for the kin28-as yeast mutants, Stephen Buratowski for antibodies against Tfb1 and Tfb3, Joseph Geisberg for helpful comments, and all members of the Struhl laboratory for insightful discussions. This work was supported by a grant to K.S. from the National Institutes of Health (GM30186). K.H.W was supported by the Croucher Foundation Fellowship.
Footnotes
Accession number
The ChIP-seq datasets discussed in this report have been deposited in the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) and are accessible through BioProject ID (PRJNA237486).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akoulitchev S, Makela TP, Weinberg RA, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- Ansari SA, Ganapathi M, Benschop JJ, Holstege FC, Wade JT, Morse RH. Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J. 2012;31:44–57. doi: 10.1038/emboj.2011.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Gifford DK, Jaakkola TS. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics. 2001;17:S22–29. doi: 10.1093/bioinformatics/17.suppl_1.s22. [DOI] [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Cismowski M, Laff G, Soloman M, Reed S. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrensberger AH, Kelly GP, Svejstrup JQ. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell. 2013;154:713–715. doi: 10.1016/j.cell.2013.07.032. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Chou D, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- Fan X, Lamarre-Vincent N, Wang Q, Struhl K. Extensive chromatin fragmentation improves enrichment of protein binding sites in chromatin immunoprecipitation experiments. Nucl Acids Res. 2008;36:e125. doi: 10.1093/nar/gkn535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Struhl K. Where does Mediator bind in vivo? PloS one. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Geisberg JV, Wong KH, Jin Y. Conditional depletion of nuclear proteins by the Anchor Away system. Curr Protoc Mol Biol. 2011;Chapter 13(Unit 13.10B) doi: 10.1002/0471142727.mb1310bs93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup J, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Guidi BW, Bjornsdottir G, Hopkins DC, Lacomis L, Erdjument-Bromage H, Tempst P, Myers LC. Mutual targeting of mediator and the TFIIH kinase Kin28. J Biol Chem. 2004;279:29114–29120. doi: 10.1074/jbc.M404426200. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Hong SW, Hong SM, Yoo JW, Lee YC, Kim S, Lis JT, Lee DK. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc Natl Acad Sci USA. 2009;106:14276–14280. doi: 10.1073/pnas.0903642106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–584. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci USA. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollack DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- Li XY, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- McNeil JB, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes & Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SGJ, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien T, Lis JT. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol Cell Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorey K, Viswanathan R, Carver MN, Karpova TS, Cirimotich SM, McNally JG, Bekiranov S, Auble DT. Measuring chromatin interaction dynamics on the second time scale at single copy genes. Science. 2013;342:369–372. doi: 10.1126/science.1242369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppas NB, Wade JT, Church G, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate-limiting. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sogaard TM, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282:14113–14120. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- Struhl K. Chromatin structure and RNA polymerase II connection: Implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ, Li Y, Fellows J, Gnatt A, Bjorkland S, Kornberg RD. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G. The KIN28 gene is required for RNA polymerase II-mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- Wong KH, Struhl K. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 2011;25:2525–2539. doi: 10.1101/gad.179275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K-H, Jin Y, Moqtaderi Z. Multiplex Illumina sequencing using DNA barcoding. Curr Protoc Mol Biol. 2013;Chapter 7(Unit 7.11) doi: 10.1002/0471142727.mb0711s101. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.