Abstract
Calcium/calmodulin-dependent protein kinase II (CaMKII) is essential for synaptic plasticity underlying memory formation. Some functions of CaMKII are mediated by interactions with synaptic proteins, and activity-triggered translocation of CaMKII to synapses has been heavily studied. However, CaMKII actions away from the postsynaptic density (PSD) remain poorly understood, in part because of the difficulty in discerning where CaMKII binds in live cells. We used photoactivated localization microscopy (PALM) in rat hippocampal neurons to track single molecules of CaMKIIα, mapping its spatial and kinetic heterogeneity at high resolution. We found that CaMKIIα exhibits at least three kinetic subpopulations, even within individual spines. Latrunculin application or coexpression of CaMKIIβ carrying its actin-binding domain strongly modulated CaMKII diffusion, indicating that a major subpopulation is regulated by the actin cytoskeleton. CaMKII in spines was typically more slowly mobile than in dendrites, consistent with presence of a higher density of binding partners or obstacles. Importantly, NMDA receptor stimulation that triggered CaMKII activation prompted the immobilization and presumed binding of CaMKII in spines not only at PSDs but also at other points up to several hundred nanometers away, suggesting that activated kinase does not target only the PSD. Consistent with this, single endogenous activated CaMKII molecules detected via STORM immunocytochemistry were concentrated in spines both at the PSD and at points quite distant from the synapse. Together, these results indicate that CaMKII mobility within spines is determined by association with multiple interacting proteins, even outside the PSD, suggesting diverse mechanisms by which CaMKII may regulate synaptic transmission.
Keywords: kinase, long-term synaptic plasticity, NMDA receptors, postsynaptic density, super-resolution imaging, synapse
Introduction
Cytosolic signaling molecules, such as kinases, are critically regulated by their ability to diffuse within the cell interior. Diffusion of kinases governs exposure to upstream activators and determines how quickly and how far they disperse to bind substrate targets and phosphatases. A key example in neurons is the regulated mobility of calcium/calmodulin-dependent protein kinase II (CaMKII), an abundant kinase that plays an essential role in learning and memory by modifying synaptic function (Coultrap and Bayer, 2012; Lisman et al., 2012). In neurons at rest, most CaMKII is inactive and abundant in dendritic shafts, but after its activation following NMDA receptor-mediated Ca2+ influx, CaMKII rapidly accumulates in dendritic spines via diffusion and binding (Shen and Meyer, 1999). This enrichment of CaMKII in the spine is critical for the induction of synaptic plasticity (Fink and Meyer, 2002; Lisman et al., 2002), and so understanding how and where CaMKII acts within spines is of great importance.
A variety of CaMKII actions, including regulating the number of synaptic AMPARs (Hayashi et al., 2000; Poncer et al., 2002; Opazo et al., 2010; Sumioka et al., 2010), may occur directly at the postsynaptic density (PSD). NMDAR stimulation drives enrichment of CaMKII in the PSD (Petersen et al., 2003; Otmakhov et al., 2004) where it docks to proteins, including the NMDAR GluN2B subunit (Bayer et al., 2001; Leonard et al., 2002; Merrill et al., 2005; Halt et al., 2012). However, it is also clear that CaMKII can regulate synaptic function through diverse mechanisms unlikely to occur at the PSD. For instance, CaMKII can regulate trafficking of AMPARs, NMDARs, and GABAA receptors through effects both in spines and dendrites (Guillaud et al., 2008; Marsden et al., 2010; Opazo et al., 2010; Lemieux et al., 2012). Critically, although spines contain numerous potential CaMKII targets known to regulate AMPAR trafficking or synapse function (Colbran, 2004), little is known about CaMKII targeting to substrates within the spine that are not at the PSD.
Our understanding of subspine targeting is limited in part because biochemical methods suited for identifying kinase–substrate interactions are difficult to confirm in living cells. However, measurement of protein mobility can reveal protein–protein interactions in cells (Sprague and McNally, 2005). Fluorescence recovery after photobleaching and photoactivation has identified mobile and immobile fractions of the total population of CaMKII, and following NMDAR stimulation, more CaMKII is immobilized, presumably through increased binding interactions (Otmakhov et al., 2004; Sharma et al., 2006; Zhang et al., 2008; Lee et al., 2009). Unfortunately, dissecting mobility of CaMKII at specific regions within individual spines is difficult because diffraction limits the spatial resolution of confocal microscopy. Equally important, interactions of interest may be transient or involve molecular subpopulations difficult to detect while observing the average behavior of the total population.
To overcome these obstacles, we studied CaMKII mobility in living neurons using single-molecule tracking photoactivated localization microscopy (PALM) (Manley et al., 2008; Frost et al., 2010), which provided methods to map molecular dynamics of CaMKII within spines. Using this approach, we find a distinctive and substantial regulation of CaMKII mobility both at and surrounding synapses following NMDAR stimulation.
Materials and Methods
Neuron culture, cDNA, and transfection.
Dissociated hippocampal neuron cultures were prepared from embryonic day 18 rats of either sex as described previously (Frost et al., 2010). Glass coverslips (Warner Instruments) were boiled in filtered water, ammonium hydroxide, and hydrogen peroxide (5:1:1), rinsed with filtered water, ethanol, and methanol, and flamed. Before plating, coverslips were coated with poly-L-lysine (Sigma) overnight. Cells were transfected with Lipofectamine 2000 (Invitrogen) at DIV12–16 and imaged 36–48 h later. cDNAs were obtained or constructed as follows: mEos2-CaMKIIα was constructed by subcloning CaMKIIα from GFP-CaMKIIα (a gift from T. Meyer, Stanford University, Stanford, CA) into mEos2–C1 (a gift from S. McKinney, Stowers Institute for Medical Research, Kansas City, MO). CaMKIIβ-Cerulean3 was constructed by subcloning CaMKIIβ from GFP-CaMKIIβ (a gift from T. Meyer, Stanford University, Stanford, CA) into Cerulean3-C1 (a gift from M. Rizzo, University of Maryland, Baltimore, Baltimore). PSD-95-mCherry (Kerr and Blanpied, 2012) and shrPSD-95-mEos2 (MacGillavry et al., 2013) were described previously. PSD-95-Cerulean3 was constructed by subcloning PSD-95 into Cerulean3-N1.
PALM imaging.
Cells expressing the indicated constructs were imaged at room temperature in extracellular imaging buffer containing 120 mm NaCl, 3 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 10 mm glucose, and 10 mm HEPES, pH 7.35. Imaging was conducted on an Olympus IX81 inverted microscope with a 100×/1.45 TIRF oil-immersion objective. Output from a set of diode lasers (Coherent) was directed to the rear of the microscope using a custom optical path. Molecules were simultaneously photoconverted and excited using 405-nm (<100 μW) and 561 nm (20 mW) illumination through oblique illumination (near TIR) to reduce background fluorescence. The AOTF was controlled separately by a TTL timing source (AMPI Master-8) so that excitation pulse length could be set independently from the exposure time, reducing noise caused by molecular motion during the acquisition of each frame (Frost et al., 2012). As a molecule diffuses even during the 20 ms of our exposure, its photons are dispersed, effectively broadening the molecule's image. This increases error in the localization estimate, which in turn is expected to degrade estimates of D (Michalet, 2010) as well as directionality, bounded region size, and other parameters (Frost et al., 2012). Imaging was conducted at 50 Hz, with 10 ms laser pulses for 10,000–20,000 frames. Fluorescence was detected by an iXon+ 897 EM-CCD camera (Andor Technology) placed after a 1.6× magnifying optic, resulting in a pixel size of 100 nm. The Olympus ZDC2 feedback positioning system was used to maintain Z stability.
Single-molecule localization and tracking analysis.
Raw images were analyzed by algorithms written in MATLAB (MathWorks). Images were bandpass filtered to identify candidate peaks. Molecules were then localized by fitting a 2D elliptical Gaussian function to a 9 × 9 pixel array surrounding the peak as previously described (Frost et al., 2010). PALM images were rendered by constructing a density map by binning molecules within 25 × 25 nm subpixel. Based on available algorithms (http://physics.georgetown.edu/matlab/), localized molecules that appeared in consecutive frames were considered as the same track if the frame-to-frame displacement (tracking radius) was within 500 nm; gaps (e.g., due to fluorophore blinking) were not permitted. The tracking radius was determined considering the maximum displacement CaMKII could have within 20 ms (<r2> = 4DΔt) based on estimated CaMKII Deff (< 5 μm2/s) (Sanabria et al., 2008; Khan et al., 2012). Using a 500 nm tracking radius permits monitoring molecules with diffusion coefficients up to 3 μm2/s. The histogram of Deff was fitted with a two- or three-component Guassian function by “peak analyzer” in Origin (OriginLab), and the proportion of each subpopulation was quantified. Tracks with at least 4 frames were plotted and color-coded according to its Deff to construct a diffusion map. To estimate the relative enrichment of CaMKII at the PSD, CaMKII single-molecule localization were overlaid with widefield images of PSD-95-Cerulean3, and the density of CaMKII localizations at the PSD (marked by PSD-95-Cerulean3) was divided by the density of CaMKII in the rest of the spine. The border of the PSD was defined as follows: widefield images at three z positions centered on the focal plane of the PALM experiment were acquired; they were deconvolved in ImageJ, and their maximum projection was thresholded at 50% of the resulting peak intensity. To study the relation between the distance of CaMKII within the spine to the border of the PSD and its mobility, regions were drawn that included the spine head and neck but excluded the dendrite shaft. The distance was calculated of each tracked molecule to the border of the PSD. The mean Deff was calculated within 100 nm distance bins. To simulate molecule movement in three dimensions, random 3D walks with fixed diffusion coefficient were simulated in MATLAB. The simulated trajectories were then projected onto 2D to calculate the Deff as previously described.
Diffusion coefficient calculation for trajectories.
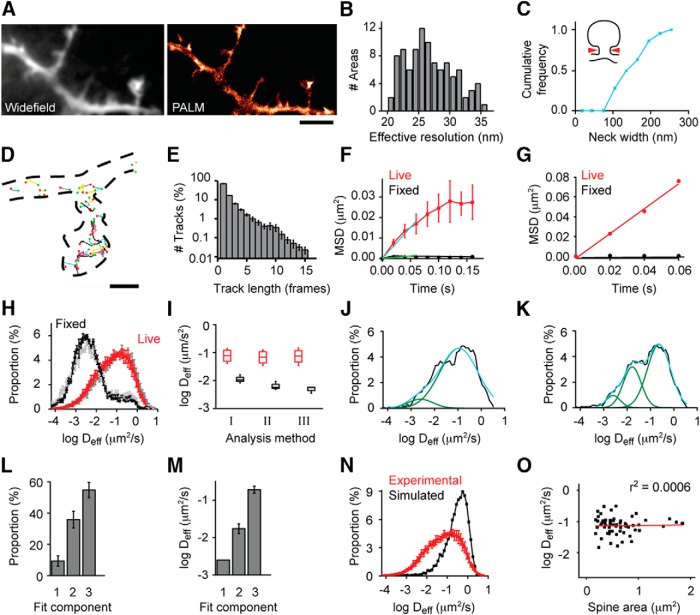
Tracks with at least 4 frames were used to calculate Deff based on a linear fit of the first 3 points of their MSD versus elapsed time. Because it was impractical to use the variance at each Δt because of the low number of samples per molecule in the short tracks typical of mEos, we weighted each by the square root of the number of sample measurements (i.e., in a 4-frame track, at Δt = 1 the weight was √3, and at Δt = 3, the weight was 1). A problem inherent to this approach is that the localization error inherent in each molecule's MSD versus time plot results in some best fits having a negative slope (and thus an uninterpretable Deff). Most problematic is that influence of this effect will be much greater for those points with low Deff, and thus present difficulties interpreting the effect of stimuli that prompt a shift in D. One approach to dealing with the molecules lost because of the negative slopes would be to assign them to an “immobile” group. However, this presumes that they are immobile and still undermines the advantages of monitoring the distribution; because it is sometimes quite a high fraction, removing these data would also lower the resolution of the resulting maps of Deff. We thus asked whether we could rescue some of the negative-slope fits without distorting the population by including an additional value of 0 at MSD(0), weighted lightly (as an n of 1), and then fit freely. This reduced the number of negative-slope results substantially, as expected (live vs fixed 3.0 ± 0.2%, n = 13 vs 6.8 ± 0.4%, n = 4; before vs after stimulation 2.5 ± 0.5 vs 4.8 ± 0.1%, n = 5). This confirms the risk that excluding these molecules will undermine our ability to interpret differences in the population between conditions. The overall distribution was not substantially different these two analyses (see Fig. 1H).
Figure 1.
Super-resolved CaMKII distribution and dynamic subpopulations in living neurons. A, Cells expressing mEos2-CaMKIIα were imaged by single-molecule tracking PALM at 50 Hz. The intensity of the images acquired was averaged to construct a diffraction-limited image (left), and all localizations were binned to create a density map (right). Scale bar, 2.5 μm. B, Histogram of effective resolution calculated from Reff = , where rnn is the mean nearest neighbor distance of molecules within the region analyzed and σ the mean SD of the localizations (Gould et al., 2009). C, Cumulative frequency of the width of spine necks and filopodia. D, Example tracks of mEos2-CaMKIIα, with green and red points indicating the first and last localized position in the track. Scale bar, 500 nm. E, Histogram of track length (n = >20,000 tracks per cell for 4 neurons). F, Mean MSD from different groups (red represents live cells; black represents fixed cells) were plotted as a function of elapsed time to construct MSD versus time plot. A linear fit of the first three points of the MSD versus plot was used to calculate Deff (n = 4 neurons). G, Example MSD versus time plots from molecules tracked for 4 frames. H, Histogram of Deff derived from tracks with at least 4 frames, using a weighted fit of the MSD versus time, including zero as described in Materials and Methods (red and black) or a fit excluding zero (grays) (n = 4 neurons). I, Median Deff from F and H. J, Single example of histogram of Deff fitted with a two-component Gaussian: black represents original data; green represents Gaussian peak fitted; blue represents sum of individual Gaussians. K, Single example of histogram of Deff fitted with a three-component Gaussian. L, The proportion of different subpopulations was quantified based on the characteristics of the fit. Area1: 9.3%; Area2: 35.9%; Area3: 54.8% (n = 11). M, Mean Deff of the subpopulations. Deff1: 0.0025 μm2/s; Deff2: 0.0173 μm2/s; Deff3: 0.1903 μm2/s. N, Histogram of Deff from simulated molecules (black) and experimental results (red). O, Correlation between spine size and mean Deff (n = 61 spines, 11 neurons).
In principle, the value of the fit at MSD = 0 reflects the error of the trajectory (Savin and Doyle, 2005), providing a means to exclude tracks of unacceptably high error. However, as a molecule diffuses even during the 10 ms of our exposure, its photons are dispersed, effectively broadening the molecule's image. This increases error in the localization estimate, which in turn degrades estimates of D (Michalet, 2010) as well as directionality, bounded region size, and other parameters (Frost et al., 2012), including cell morphology (Renner et al., 2011). The most troublesome aspect of this effect for our experiments is that the magnitude of the error is strongly dependent on the true D of the molecule. Immobilized molecules in live or fixed samples will be unaffected, whereas quickly moving molecules (D>∼0.1 μm2/s) are expected to be more poorly localized and subject to greater error. This D-dependent error was apparent in our experiments, as indicated by calculating the error (from linear fits excluding the point at Δt = 0) as ε = √(MSD(0)/8) (Savin and Doyle, 2005) under conditions where D was expected to vary: ε = 18.3 ± 1.4 nm (live cells) versus ε = 7.5 ± 0.5 nm (fixed cells); ε = 21.7 ± 3.3 nm (before Glu/Gly stimulation) versus ε = 11.39 ± 0.8 nm (after Glu/Gly stimulation). Thus, using MSD(0) as a way to select tracks thus preferentially eliminates the faster-moving molecules. Because we are centrally interested in the change of CaMKII mobility both within the cell and across stimulus conditions, this presents a confound that would make it difficult to interpret changes in the distribution of D, so we did not exclude tracks based on this criterion.
Live-cell confocal microscopy.
Live-cell imaging experiments were conducted on a spinning disc confocal system. This system consists of an Olympus IX-81 inverted microscope with a CSU-22 confocal (Yokagawa) and an Orca-ER CCD (Hamamatsu) mounted on the side-port, and excitation laser (Coherent) and emission filters (Semrock). Cells expressing the indicated constructs were imaged at room temperature in extracellular imaging buffer with a 60×1.42 NA oil objective and an extra 1.6× and 1.2× magnification in the light path, resulting in pixel size of 56 nm. Acquisition was controlled by IQ software (Andor Technology).
Immunostaining.
Cells were pretreated with 100 μm dl-APV and 10 μm KN-93 for 2 d and stimulated with 100 μm glutamate/10 μm glycine (Glu/Gly) for 30–60 s. Cells were then immediately fixed with 4% PFA and 4% sucrose supplemented with a phosphatase inhibitor mixture (PhosSTOP; Roche) in PBS, pH 7.4, for 10 min at room temperature. Cells were washed 3 times with PBS containing 0.1 m glycine (PBS/Gly) and blocked in PBS/Gly with 10% goat serum, 0.3% Tween 20 and PhosSTOP (Dil experiment) or with 10% goat serum, 0.3% Triton X-100, and phosSTOP for 1 h. Cells were incubated with rabbit anti-phospho-CaMKII (Thr286) (1:200, from Cell Signaling Technology or Promega) for 2 h at room temperature. Cells were washed 3 times with PBS/Gly and incubated with Alexa-647-conjugated goat anti-rabbit secondary antibodies (1:200, Invitrogen) for 1 h at room temperature. Cells were washed 3 times and postfixed with 4% PFA and 4% sucrose in PBS for 5 min, and washed 3 times with PBS prior imaging.
Two-color single-molecule imaging.
Cells expressing shrPSD-95-mEos2 were pretreated with 100 μm dl-APV and 10 μm KN-93 for 2 d and stimulated with Glu/Gly and stained with anti-phospho-CaMKII. Cells were imaged in STORM imaging buffer containing 50 mm Tris, 10 mm NaCl, 10% glucose, 0.5 mg/ml glucose oxidase (Sigma), 40 μg/ml catalase (Sigma), and 0.1 m cysteamine (Sigma). Emission light was passed through a Photometrics DV2 to split the emission light at 565 nm and separate the red and far-red light through emission filters (590/50 and 655 long pass) onto the EM-CCD. To overlay the red and far red channels, TetraSpeck beads (100 nm, Invitrogen) were dried on a glass coverslip and imaged (50 frames) for calibration at least once per experiment. Based on the average bead localizations, the “projective” transformation between the two channels was calculated using the MATLAB command “cp2tform.” Images were analyzed as described under “Single-molecule localization and analysis.”
Results
PALM reveals super-resolved CaMKII distribution in living neurons
To investigate CaMKIIα distribution at high resolution in living neurons, we used PALM to measure the location of single molecules (Betzig et al., 2006; Hess et al., 2006; Jones et al., 2011). Cultured rat hippocampal neurons were transfected with CaMKIIα tagged with mEos2, which undergoes photoconversion from green to red fluorescence upon UV absorbance (McKinney et al., 2009). Live-cell experiments were carried out using weak 405 nm illumination to photoconvert mEos2-CaMKIIα at a very low spatial density, enabling clear discrimination of single molecules. Red fluorescence was detected in 5000–10,000 frames at 50 Hz with 10 ms 561 nm excitation laser pulses. We used a shorter excitation pulse (10 ms) than camera exposure time (20 ms) because molecular motion during exposure deteriorates localization precision (Frost et al., 2012). Because CaMKIIα under basal conditions is substantially cytosolic (Michalet, 2010; Frost et al., 2012), the distribution of all localizations outlined cell morphology. Compared with a diffraction-limited view, this PALM image revealed subspine morphological details at extremely high resolution (Fig. 1A). We estimated the effective map resolution (Gould et al., 2009) to be <30 nm, considering the localization precision (mean = 12 nm) (Thompson et al., 2002) and the density of localizations (Fig. 1B). Under these conditions, small morphological structures, such as spine necks, were detected with widths as small as 80 nm (Fig. 1C), highlighting the ability of PALM to exploit a single molecular species to extract multiple modalities of information from the same dataset, including cell morphology, molecule distribution, and molecular mobility.
Multiple, distinct modes of CaMKII mobility resolved by single-molecule tracking
To examine CaMKII mobility, we noted that photobleaching or photoactivation is restricted by diffraction to a region not much smaller than a spine, so that only the “bulk” average motion of many molecules into and out of this region is measured (Kim et al., 2010). In addition, protein exchange rate between spines and dendrites is highly influenced by heterogeneous spine morphology (Bloodgood and Sabatini, 2005; Sheng and Hoogenraad, 2007). In combination, these factors may contribute to widely varying reports of time constants for the most mobile fraction of CaMKII exchange in spines (Sharma et al., 2006; Zhang et al., 2008; Lee et al., 2009; Khan et al., 2012; Dupuis et al., 2014). In contrast, single-molecule tracking PALM is not influenced by these factors, as it measures the motion of individually tracked molecules, and for each directly derives an effective diffusion coefficient (Hoze et al., 2012; Manley et al., 2008; Frost et al., 2010). Although the accuracy of the approach is degraded by noise in each measurement and a limited photon budget of the label, the greater spatial resolution of single-molecule tracking results in greater precision of measures, particularly for quickly moving molecules (Saxton and Jacobson, 1997). Given the brief track duration typical of mEos2 (Fig. 1D,E), we considered three methods for analyzing the effective diffusion coefficient, Deff, of CaMKII. First, to use the large number of the shortest, two-frame tracks, we calculated Deff based on the displacement between all consecutive measurements of all molecules tracked for at least two frames. Assuming that photobleaching or photoblinking are the predominant reasons for loss of molecules after more than one frame, and considering that consecutive steps in a random walk are independent, two-frame tracks should not sample a different population of molecules than those tracked over larger numbers of frames. We used the mean squared displacement MSD(Δt) = 4DeffΔt to calculate Deff for every molecule localized in two consecutive frames separated by time Δt. However, the distribution of Deff in fixed cells was broad (data not shown), resulting in a poor detection limit of the analysis. This limited its further usage for CaMKII tracking in living cells. As a second approach, we calculated Deff from plots of MSD versus time that included tracks with different lengths. Assuming that tracks with different lengths do not represent different populations, we grouped together tracks with the same length and calculated the mean MSD for each group. The mean MSD from different groups was plotted as a function of the elapsed time to construct an MSD versus time plot. Linear fitting was performed to the first 3 points of the MSD versus time plot. With this approach, the Deff of whole cells was 0.076 ± 0.02 μm2/s for live cells and 0.006 ± 0.001 μm2/s for fixed cells (Fig. 1F; n = 4 neurons in each group).
As a third approach, we sought to obtain both spatial and dynamic information of individual tracks. We constructed plots of MSD versus time for every track that consisted of at least 4 frames and calculated Deff as the best weighted linear fit of this plot (see Materials and Methods). Using this approach, we estimated the Deff of molecules in live cells to be 0.084 ± 0.024 μm2/s and for that in fixed cells 0.005 ± 0.001 μm2/s (Fig. 1G,H; n = 4 neurons). Nevertheless, in fixed samples, there was still some measured displacement between frames, potentially resulting from localization error from each molecule, incomplete immobilization of molecules after paraformaldehyde fixation (Tanaka et al., 2010), or the occasional misclassification of two molecules as the same track. With this method, we obtained a better separation between the distributions of Deff in live and fixed cells, and the Deff of fixed cells was lower (Fig. 1I).
Comparison of these three approaches is useful in several ways. First, it is clear that the information derived from the smaller number of long tracks cannot be replaced by averaging the much more numerous short tracks. This is presumably because shorter tracks are affected more by the localization error at each frame (Michalet, 2010; Frost et al., 2012), whereas longer tracks permit a greater time for diffusion and a resulting increase in displacement of unfixed molecules that dominates the error. Nevertheless, the distribution of 2-frame jump distances contains important information about molecular mobility, and the benefit of longer tracks will depend on the relationship between the localization error, D, and acquisition rate. The second approach, by accumulating all tracks, achieves high sensitivity but sacrifices spatial information. Notably, it provides high temporal resolution; this analysis of the mobility of the molecule population can be obtained at about once per seconds depending on the molecular density and size of the imaged area. For our further analysis of CaMKII mobility here, we used the third method because it provides sensitive measurements of CaMKII dynamics with spatial information, although the resulting maps of high spatial resolution are accumulated over a time frame of 200 s.
Using this approach, the distribution of Deff clearly appeared to consist of more than one subpopulation. A two-component Gaussian function fit, including one subpopulation whose mean was held constant at the mean Deff measured in fixed cells, resulted in fits that were clearly insufficient (R2 = 0.940; Fig. 1J). However, a three-component fit appeared adequate in all cases (R2 = 0.992; Fig. 1K). Thus, we continued to use a three-component analysis to characterize the distribution of Deff (Fig. 1L,M). Because CaMKII moves in three dimensions within cells, it is possible that the different Deff subpopulations we observed resulted from tracking this motion in 2D. To test this possibility, we simulated 3D random walks of molecules with a single diffusion coefficient and projected their displacement onto 2D for Deff calculation. The distribution of Deff from such simulations only had one population (Fig. 1N), indicating that tracking CaMKII movement in 2D is unlikely to result in artificial dynamic subpopulations. Alternatively, within small spines, the confined motion of CaMKII might result in reduced apparent mobility. However, we found no correlation between CaMKII mobility and spine size (r2 = 0.0006; Fig. 1O), suggesting that encounters between CaMKII molecules and the spine plasma membrane during the track were rare and of little influence on our measurements. Overall, these measurements indicate that CaMKII diffusion within cells is 10- to 100-fold slower than that measured in free solution (Sanabria et al., 2008), confirming the importance of understanding the mechanisms that restrict this mobility.
Actin cytoskeleton regulates CaMKII mobility
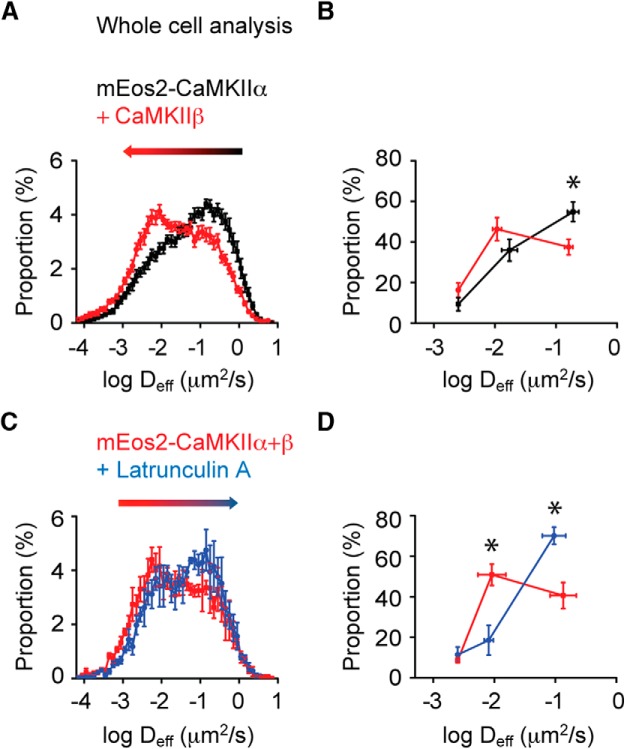
We inferred that the least mobile subpopulation of CaMKII contains those molecules that are immobilized via binding. Likewise, the fast population likely contains molecules that are the most free to diffuse unencumbered by interactions with effectors. However, the intermediate component suggests the presence of a third subpopulation of molecules not immediately interpretable in classical biochemical terms of bound and free. To investigate the mechanisms that result in this pool of molecules with intermediate mobility, we first considered that CaMKII holoenzymes are homomers or heteromers comprising both CaMKIIα and CaMKIIβ subunits (Lisman et al., 2012). CaMKIIβ interacts with actin filaments and could through this interaction slow the mobility of CaMKII holoenzymes (Shen et al., 1998; Lin and Redmond, 2008). To study the effect of CaMKIIβ on CaMKIIα mobility, we transfected neurons with mEos2-CaMKIIα alone or in combination with CaMKIIβ tagged with the photostable CFP variant Cerulean3 (Markwardt et al., 2011), and compared the Deff the two groups of cells (including dendrites and spines). Overexpression of CaMKIIβ significantly decreased CaMKIIα mobility (Fig. 2A; K-S test; p < 0.001). Three-component fitting showed that overexpression of CaMKIIβ decreased the proportion of the fast population from 54.8% to 37.4% of measured molecules while increasing the intermediate population from 35.9% to 46.3% (Fig. 2B). We next tested whether we could reverse the effect of CaMKIIβ overexpression on CaMKIIα mobility by depolymerizing actin filaments. To test the effect of reducing the number of actin filaments, we measured Deff in the same cells before and after treating the cells with the actin depolymerizing agent Latrunculin A (Lat A). For this, we chose dendrites with few spines to avoid the expected complication of Lat A reducing spine size. This treatment increased CaMKIIα mobility (Fig. 2C; K-S test; p < 0.001). Three-component fitting showed that Lat A treatment profoundly decreased the size of the intermediate population (from 50.8% to 18.5% of all measured molecules) while concomitantly increasing the fast population (from 40.6% to 70.1%; Fig. 2D). To assess whether this extended imaging (20,000 instead of 10,000 frames) exhausted the slowest population of molecules, we collected twice the number of frames and examined the proportion of slow molecules in the first and second cohort of frames. This showed no reduction in the size of the slow population (paired sample sign test for slow population; p > 0.5; intermediate populations; p > 0.2), suggesting that the population of slowly moving molecules was not depleted during prolonged but low-intensity photoactivation. Thus, the CaMKIIβ effect on the Deff distribution was abolished by Lat A. Consistent with the idea that the intermediate mobility arises from CaMKII interactions with the actin cytoskeleton, the intermediate subpopulation was most strongly regulated by these manipulations. Interestingly, the decrease of CaMKIIα mobility upon CaMKIIβ coexpression was not observed exclusively in spines, where F-actin is most dense, but also in dendrite shafts. This suggests that even the relatively sparse actin filaments in the dendrite (Xu et al., 2013) may regulate CaMKII mobility.
Figure 2.
Actin cytoskeleton regulates CaMKII mobility. A, Histogram of Deff of cells transfected with mEos2-CaMKIIα or mEos2-CaMKIIα/Cer3-CaMKIIβ (1:1) (n = 13 neurons for mEos2-CaMKIIα and n = 17 neurons for mEos2-CaMKIIα/Cer3-CaMKIIβ). B, Histograms of Deff were fit as in Figure 1K, and the proportion of each kinetic subpopulation was quantified. Deff1: 0.0025 μm2/s (9.3 ± 3.3%); Deff2: 0.0173 μm2/s (35.9 ± 5.4%); Deff3: 0.1903 ± 0.045 μm2/s (54.8 ± 4.9%) in the mEos2-CaMKIIα expressing cells (n = 11 neurons). Deff1: 0.0025 μm2/s (16.3 ± 3.6%); Deff2: 0.011 μm2/s (46.3 ± 5.7%); Deff3: 0.161 μm2/s (37.4 ± 3.8%) in the cells transfected with mEos2-CaMKIIα/Cer3-CaMKIIβ (n = 15 neurons). *p < 0.05 (two-way ANOVA repeated measures with one factor repeated). C, Histogram of Deff of cells transfected with mEos2-CaMKIIα/Cer3-CaMKIIβ before and after Lat A (n = 3 neurons). D, Deff distributions before or after Lat A treatment (n = 3 neurons). *p < 0.01 before versus after lat A (two-way ANOVA repeated measures with two factors repeated).
The spatial distribution of CaMKII mobility is heterogeneous within dendrites and spines
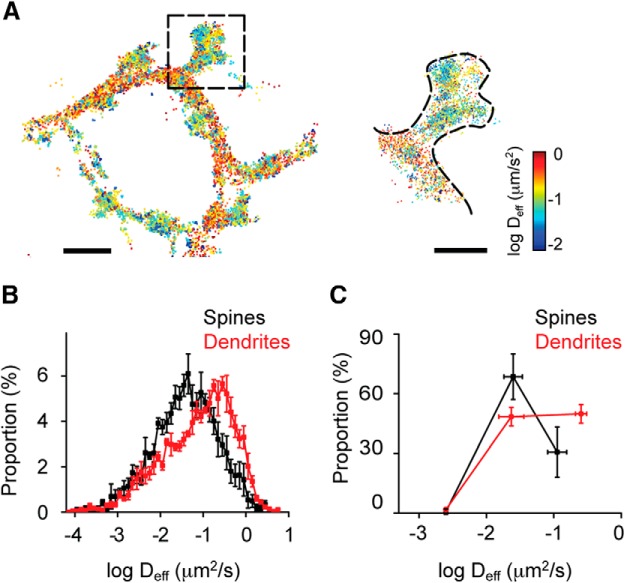
Because there are numerous CaMKII substrates within spines, it is tempting to speculate that CaMKII regulates synaptic function by interacting with other proteins at multiple locations, not exclusively at the PSD. However, it remains unclear where and when these interactions occur within spines. Reasoning that CaMKII mobility will decrease as a result of these interactions, we evaluated CaMKII Deff within spines. We plotted all tracked CaMKII molecules, color-coded according to their mobility to construct spatial diffusion maps. Interestingly, these maps indicated that the mobility of CaMKII was strongly heterogeneous across the cell, revealing dendritic subregions where the local Deff differed by 10- to 100-fold from closely adjacent areas (Fig. 3A). Notably, CaMKII mobility in spines was significantly lower than that in dendrites (Fig. 3B; K-S test, p < 0.001), consistent with the idea that there are more CaMKII binding partners within spines than within dendrites. Interestingly, three-component fitting of the mobility distribution in spines showed that most CaMKII belonged to the intermediate subpopulation (Fig. 3C; Deff1: 0.0025 μm2/s (0.76 ± 1.35%); Deff2: 0.025 μm2/s (68.5 ± 11.5%); Deff3: 0.112 μm2/s (30.7 ± 12.5%). By contrast, in dendritic shafts, a higher proportion of molecules occupied the fast subpopulation (Fig. 3C; Deff1: 0.0025 μm2/s (1.79 ± 0.82%); Deff2: 0.023 μm2/s (48.4 ± 4.7%); Deff3: 0.254 μm2/s (49.8 ± 4.7%). Because of the high density of polymerized actin in spines, this difference further supports the notion that a key regulator of CaMKII mobility is its interaction with the actin cytoskeleton.
Figure 3.
The spatial distribution of CaMKII mobility is heterogeneous within cells. A, Deff was calculated and color-coded to construct a spatial map of molecular mobility. Scale bar: left top, 1.5 μm. Enlarged view of the spine marked from the left. Scale bar: right top, 1 μm. B, Histogram of Deff from spines or dendrites (K-S test, p < 0.001, n = 5). C, Deff in spines or dendrites was fit as in Figure 1K, and the proportion of each kinetic subpopulation was quantified (n = 4).
NMDA receptor stimulation concentrates CaMKII in spines both at the PSD and away from the PSD
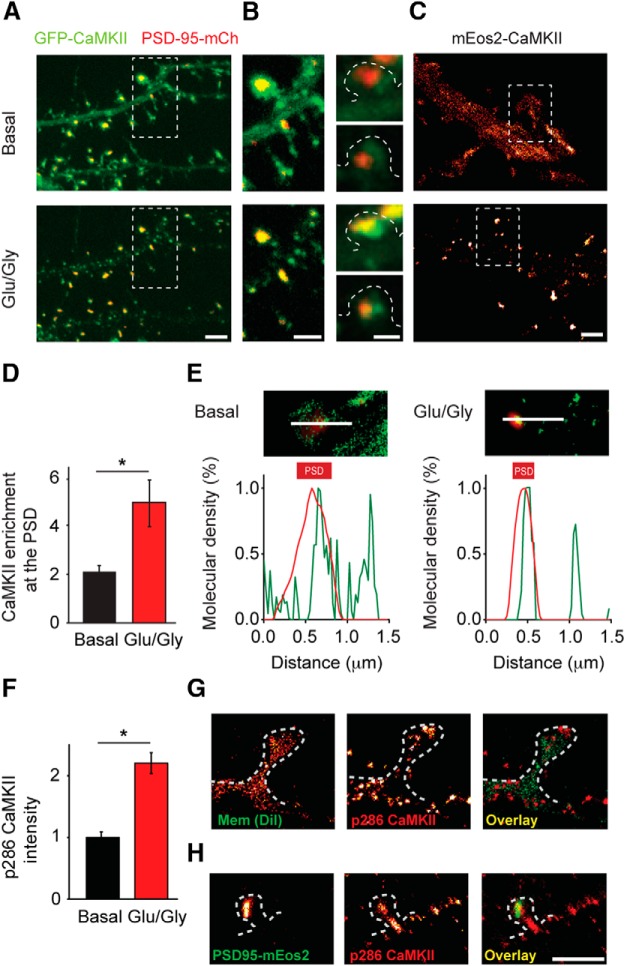
To investigate the activation-dependent dynamics of CaMKII within spines, we first used confocal microscopy to study CaMKII distribution before and after NMDAR stimulation. Cells were cotransfected with GFP-CaMKIIα and PSD-95-mCherry, then imaged before and after a brief (1 min) Glu/Gly stimulation. As expected, GFP-CaMKIIα was primarily diffusely distributed in neurons at rest, with some enrichment in spines, but was rapidly concentrated in spines following stimulation (Fig. 4A). Close examination of these spines revealed that CaMKII clearly colocalized with the PSD marked by PSD-95-mCherry but that typically the degree of overlap was rough and GFP-CaMKIIα was also found adjacent to the PSD or even in other parts of the spine (Fig. 4B). To explore this in greater detail, we used PALM to examine mEos2-CaMKIIα distribution in live cells coexpressing PSD-95-Cerulean3. To ensure enough CaMKII molecules are collected in each condition, results of basal or after Glu/Gly were collected from different cells. After Glu/Gly stimulation, PALM revealed the stimulation-driven concentration of CaMKII in spines (Fig. 4C). Quantitatively, we found that the proportion of CaMKII within the spine that colocalized with the PSD (marked with PSD-95-Cerulean3) approximately doubled after stimulation, compared with CaMKII enrichment under basal conditions (Fig. 4D). Notably, however, PALM images also confirmed that CaMKII concentrated at points away from the PSD. Line-scan analysis along the length of the spine showed clear intensities of mEos2-CaMKIIα not associated with PSD-95-Cerulean3 (Fig. 4E).
Figure 4.
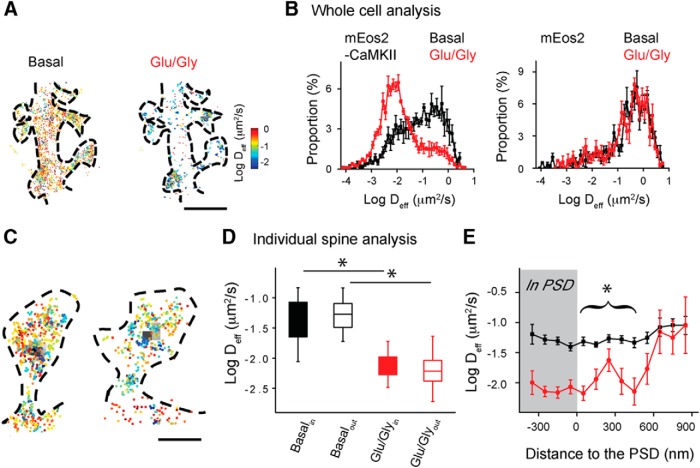
Activated CaMKII concentrates in spines both at the PSD and away from the PSD after NMDA receptor stimulation. A, Cells transfected with GFP-CaMKIIα and PSD-95-mCherry were imaged by confocal microscopy before and after NMDAR stimulation. Scale bar, 2 μm. B, Enlarged view from A (left) and other experiments before (right top) and after 0 Mg2+/Gly stimulation (right bottom). Scale bar, 1 μm. C, CaMKII localizations were binned (25 nm) to construct a map of CaMKII density under basal conditions and after Glu/Gly stimulation. Scale bar, 1 μm. D, CaMKII enrichment at the PSD was quantified in different cells under basal conditions and after Glu/Gly stimulation. *p < 0.05 (K-S test, n = 52 synapses for basal, n = 22 synapses for Glu/Gly). E, For basal condition and after Glu/Gly stimulation, image (top) shows density map of the spine from C (green) superimposed on deconvolved widefield image of PSD95-Cerulean3 (red). Lines indicate profiles along which intensity was measured, as shown in the graph (bottom). F, Quantification of the intensity of phospho-T286 immunostaining under basal conditions and after Glu/Gly stimulation. *p < 0.001 (K-S test, n = 11 cells). G, Example of phospho-T286 CaMKII (red) with the plasma membrane labeled with DiI (green) resolved by two-color localization microscopy. H, Example of phospho-T286 CaMKII as in G (red) with the PSD resolved by PALM of PSD-95-mEos2 (green). Scale bar, 1 μm.
CaMKII translocation following stimulation does not necessarily predict the kinase activation state (Lee et al., 2009). To test whether CaMKII that concentrated away from the PSD was activated, we stained cells with a phospho-T286-CaMKIIα-specific antibody, which specifically labels CaMKII in its activated state (Coultrap and Bayer, 2012). The amount of phospho-T286 is widely used to evaluate the status of CaMKII activation (Lemieux et al., 2012). Indeed, phospho-T286-CaMKII labeling intensity increased dramatically following Glu/Gly stimulation (Fig. 4F). However, it is important to note that CaMKII is activated by Ca2+/CaM also under conditions that do not lead to T286 autophosphorylation, and even more critically, T286-phosphorylated CaMKII is not fully active in absence of Ca2+/CaM (Coultrap et al., 2010). Accordingly, the phospho-T286 staining does show that the NMDAR stimulation (as used in the live tracking) indeed caused an activation of CaMKII, but there is not a way to direct assess the activity state of the kinase populations that were tracked live. To achieve nm resolution, we turned to two-color single-molecule imaging. First, after Glu/Gly stimulation, we labeled cells with anti-phospho-T286-CaMKII to mark activated CaMKII and DiI to outline morphology and imaged both simultaneously via dSTORM (van de Linde et al., 2011; Shim et al., 2012). Phosphorylated CaMKII was found distributed broadly throughout the cell but was largely concentrated in small clusters (Fig. 4G) that were difficult to distinguish in confocal microscopy (data not shown). Spines visualized with DiI clearly contained small clusters of CaMKII at multiple locations, including points typically not at the sites of synapses, such as the spine neck (Fig. 4G). To test this in more detail, we expressed shrPSD-95-mEos2 to visualize the PSD by PALM (MacGillavry et al., 2013), stimulated cells with Glu/Gly, and then stained them for phospho-T286-CaMKII for visualization by dSTORM. By visualizing both the PSD and activated CaMKII with super-resolution, we showed that phospho-T286 CaMKII clustered at locations both at and away from the PSD (Fig. 4H). Together, these observations indicate that NMDA receptor stimulation prompts concentration of activated CaMKII both at the PSD and elsewhere in spines, consistent with potential interaction of CaMKII with downstream effectors away from the synapse.
NMDAR stimulation immobilizes spine CaMKII both at and away from the PSD
To further evaluate whether CaMKII can interact with binding partners at locations away from the PSD, we measured CaMKII mobility before and after Glu/Gly stimulation. Overall, CaMKII mobility was significantly decreased after stimulation (Fig. 5A,B, left; K-S test, p < 0.001). In contrast, mobility of free mEos2 was not altered by stimulation (Fig. 5B, right; K-S test, p > 0.1). To quantify CaMKII mobility in relation to the synapse, we cotransfected PSD-95-Cerulean3. Under basal conditions, CaMKII mobility within the thresholded borders of the PSD was not different from that elsewhere in the spine (Fig. 5C,D; n = 52 spines). The results are in agreement with the previous observation that under basal conditions small or variable amounts of CaMKII are bound at the PSD (Dani et al., 2010; Feng et al., 2011), although the absolute amount is expected to correlate with the size and strength of the synapse (Asrican et al., 2007). After Glu/Gly stimulation, CaMKII was immobilized at the PSD but strikingly also elsewhere in the cell. This was clear at other points in the spine outside of the PSD (Fig. 5D), but a reduction was also seen in the dendrite shaft (median Deff basal 0.193 ± 0.012 μm2/s, stimulated 0.009 ± 0.002 μm2/s). To examine whether the reduction within the spine was restricted to the synapse, we grouped CaMKII localizations in the spine at 100 nm intervals relative to the border of the PSD. This revealed that the slowing or immobilization CaMKII molecules occurred found at distances even several hundred nanometers away from the border of the PSD (Fig. 5E). This suggests that NMDAR stimulation not only induces CaMKII binding to substrates at the PSD, but also to binding partners away even outside the PSD. Our results suggest that the mobility and binding of CaMKII within spines are determined by association with multiple interacting proteins that are broadly distributed within the cellular environment near the synapse.
Figure 5.
CaMKII is immobilized away from the PSD following NMDAR activation. A, Deff map of mEos2-CaMKIIα in the same neuron before (left) and after (right) Glu/Gly stimulation. Scale bar, 2 μm. B, Histograms of Deff of mEos2-CaMKIIα (left) or cytosolic mEos2 (right) before and after Glu/Gly stimulation (mEos2-CaMKIIα: K-S test, p < 0.001, n = 5 cells; mEos2: K-S test, p > 0.1, n = 5 cells). C, CaMKII mobility map superimposed on deconvolved widefield image of PSD-95-Cerulean3 (black). Scale bar, 500 nm. D, Mean Deff either within or outside of the PSD border (n = 54 synapses for basal condition, n = 22 synapses for Glu/Gly, two groups were from different cells). *p < 0.001 (two-way ANOVA repeated measures with one factor repeated). E, Mean Deff calculated in spatial bins either within or every 100 nm outside the PSD (n = 52 spines for basal condition, n = 22 spines for Glu/Gly). *p < 0.001 (two-way ANOVA).
Discussion
Discriminating protein distribution and sites of action within cells is critical for studying many kinases and other signaling molecules because even a few molecules of active kinase positioned away from the bulk of the population may have unique and important functions. Direct measurement of kinase mobility within spines will also be critical for developing accurate models of neuronal signaling cascades (Byrne et al., 2011; Faas et al., 2011; Khan et al., 2011). Using single-molecule tracking to achieve high spatial and kinetic resolution of protein motion within living neurons, we found that CaMKII mobility was strikingly heterogeneous. Mobility was on average lower in spines than in dendrite shafts, and even within single spines there were subregions of elevated or diminished mobility. Activation of NMDARs greatly slowed and redistributed the population of CaMKII molecules. In individual spines, immobilized molecules of potentially active, bound CaMKII were abundant at the synapse but also away from the PSD at other points of the spine. Our results are consistent with the idea that CaMKII exerts its function on multiple substrates at different locations within neuronal spines and dendrites, regulating diverse mechanisms of synaptic plasticity.
Multiple dynamic subpopulations
The measured diffusion coefficient of CaMKII molecules spanned several orders of magnitude, even in single spines of unstimulated cells under basal conditions. For convenience, we analyzed this as three subpopulations (fast, intermediate, and immobile), although it is likely that there are more than these, or indeed a broadly varying diversity. The slowest subpopulation of molecules (kinetically indistinguishable from chemically fixed molecules) we take to be bound to essentially immobile substrates. On the other hand, the subpopulation with the highest diffusion rates likely dominates the “mobile fraction” exchanging most quickly between the spine and dendrite as observed with bulk methods of FRAP and photoactivation. A FRAP exchange time of ∼1 min (Sharma et al., 2006; Zhang et al., 2008; Lee et al., 2009) can be accounted for in rough geometric models by an average CaMKII D of ∼0.17 μm2/s (Lee and Yasuda, 2009; Lee et al., 2009).This is in good accord with the fast population here (mean Deff = 0.19 μm2/s), suggesting that the mobility we observe may explain single-spine FRAP and photoactivation results. However, in spines we found that this fraction comprised only 30% of CaMKII molecules, much smaller than the FRAP mobile fraction of ∼80%. This suggests that the bulk “mobile fraction” comprises molecules with both fast and intermediate diffusion coefficients spanning at least two orders of magnitude, and which may represent multiple states and be subject to quite diverse regulatory influences.
Mechanisms regulating basal CaMKII mobility
The high concentration of actin filaments in the spine is thought to modulate CaMKII mobility, as binding between CaMKIIβ and filaments slows the exchange rate of CaMKII into and out of spines (Shen et al., 1998; Okamoto et al., 2009). In addition, modeling suggests that obstruction by actin filaments in the spine neck may reduce the rate of CaMKII exchange (Byrne et al., 2011). However, how actin filaments control CaMKII motion within the spine head has not been clear. Here, we show directly that CaMKII mobility within the spine head is strongly influenced by the cytoskeleton. This regulation was exerted most clearly on CaMKII molecules that moved with an intermediate diffusion coefficient, as this subpopulation was enlarged in the presence of overexpressed CaMKIIβ (along with the slowest population) and strongly reduced by latrunculin A treatment. Restricted mobility is likely mediated at least in part by direct binding of CaMKII oligomers to the sides of actin filaments (Okamoto et al., 2007). However, filament-bound CaMKII would most likely display a motion velocity similar to polymerized actin monomers or other filament-binding proteins, which move within the cell only at extremely slow speeds generated by polymerization-dependent flow along the filament (<50 nm/s, Deff < 10−3 μm2/s) (Danuser and Waterman-Storer, 2006; Frost et al., 2010). Motion at these rates would be classified in our current analysis as immobile rather than intermediate.
An intermediate Deff could also arise from transition of holoenzymes between bound and free (or vice versa) during their tracking. However, the unbinding rate is likely very slow (Khan et al., 2012), suggesting that these transitions occur rarely during the brief periods we track individual molecules, and so the effect may not contribute substantially. Further, even in high or low CaMKIIα:β ratios (expected to minimize the frequency of these transitions), a substantial population retained the intermediate mobility. Thus, filament-binding alone does not easily explain the characteristics of this population.
Several additional mechanisms could underlie actin-sensitive kinase mobility. Actin filaments in spines are highly branched and complex (Landis and Reese, 1983; Korobova and Svitkina, 2010; Hotulainen and Hoogenraad, 2010; Burette et al., 2012), and this dense array could act as a sieve for the bulky CaMKII holoenzyme (Khan et al., 2011). Even though kinase might still move freely during periods between interaction with filaments, its mobility averaged over longer time periods would be reduced because it would often encounter impassable obstructions. In addition, binding to other actin-associated targets, such as α-actinin2 (Walikonis et al., 2001; Robison et al., 2005), may reduce diffusion even of holoenzymes that do not contain CaMKIIβ. Overall, although the details are still unclear, it is likely that multiple actin-related mechanisms reduce CaMKII diffusion.
Spatial regulation of CaMKII mobility
We observed substantial but localized variations in the CaMKII effective diffusion coefficient. This makes clear that, as the kinase diffuses within the cell, it encounters a complex and varied environment where it likely interacts with diverse proteins. Overall, CaMKII mobility was higher in dendrites than spines, in line with earlier FRAP studies showing faster recovery after targeted photobleaching in dendrites than spines (Sharma et al., 2006). However, we expand these findings by demonstrating marked heterogeneity of mobility across the interior of individual spines even under basal conditions. This likely arises from various protein interactions with CaMKII. Some interactors may simply be obstacles that produce no binding during elastic collisions, whereas many interactions are likely to involve binding over a broad range of affinities. Notably, very low-affinity interactions of the type most difficult to identify via traditional biochemistry would be expected to slow or immobilize the kinase for only very brief periods.
Following glutamate receptor stimulation, the fraction of CaMKII molecules that were immobilized and presumably bound increased dramatically. A substantial proportion of these immobile molecules likely maintain at least partial activity because such brief, global stimulation elevates kinase activity for up to several minutes (Bayer et al., 2006; Lemieux et al., 2012). Consistent with this, staining of phosphorylated CaMKII increased dramatically following stimulation, as seen previously (Redondo et al., 2010; Lemieux et al., 2012), and this distribution within the cell was similar to immobilized mEos2-CaMKII. Activated immobilization of CaMKII occurred at the PSD, in the spine away from the PSD, and in the dendrite. Within the dendrite, we currently cannot define the mechanism of this effect, although several influences are likely. Actin filaments in the shaft are far less dense than in spines (Korobova and Svitkina, 2010; Xu et al., 2013), but CaMKIIα holoenzymes can self-associate to form macrooligomers (Hudmon et al., 1996, 2005). Although the chance of CaMKII self-association should be rare under basal conditions because Ca2+/CaM binding initiates the process (Hudmon et al., 2005), larger complexes would be highly sensitive to the sieving effect by the actin cytoskeleton. Additionally, recent work suggests that dendritic microtubules are a target of activated CaMKII (Lemieux et al., 2012).
At the synapse, the key CaMKII substrate GluN2B (Strack and Colbran, 1998; Bayer et al., 2001; Leonard et al., 2002; Dupuis et al., 2014) is clustered within subdomains of the PSD (MacGillavry et al., 2013). These subdomains might serve as CaMKII accumulation points to enrich phosphorylation of its targets in this local subregion of the PSD. There are many synaptic CaMKII substrates, including GKAP (Shin et al., 2012), PSD95 (Steiner et al., 2008), AMPARs (Lu and Roche, 2012; Coultrap et al., 2014), and TARPs (Opazo et al., 2010; Sumioka et al., 2010), each of which individually has been proposed to regulate synaptic strength. Clustering phosphorylated substrates by docking CaMKII at specific PSD sites (Petersen et al., 2003; Robison et al., 2005) may provide a means to accentuate or coordinate these effects.
Outside synapses, activated CaMKII could serve numerous functions. Autonomous CaMKII can phosphorylate a set of factors that control the actin regulator Rac1 (Penzes et al., 2008). Notably, CaMKII interaction with microtubules also increases synaptic AMPAR via regulating receptor trafficking to spines (Lemieux et al., 2012). Prior work has demonstrated the facilitation of NMDAR traffic by the CaMKII-triggered release of GluN2B-containing transport vesicles following phosphorylation of the kinesin-2 KIF17 (Guillaud et al., 2008). It is tempting to speculate that CaMKII action perisynaptically or at points further distributed away from the PSD could represent sites of phosphorylation controlling traffic of vesicles containing AMPARs or NMDARs. Defining molecular dynamics through single-molecule analysis of kinetic subpopulations will provide an important tool for deciphering the multiple CaMKII–substrate interactions that regulate synapse function.
Footnotes
This work was supported by the National Institutes of Health Grants MH080046 and MH096376, the Dana Foundation, the Broad Foundation, and the Katherine D. and Theodore J. Carski Fund. We thank Scott Thompson, Sri Raghavachari, Hiroaki Misono, Yasunori Hayashi, and members of the T.A.B. laboratory for helpful discussions and critical evaluation of the manuscript; and Tamar Davis and Minerva Contreras for excellent technical support.
The authors declare no competing financial interests.
References
- Asrican B, Lisman J, Otmakhov N. Synaptic strength of individual spines correlates with bound Ca2+-calmodulin-dependent kinase II. J Neurosci. 2007;27:14007–14011. doi: 10.1523/JNEUROSCI.3587-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- Bayer KU, LeBel E, McDonald GL, O'Leary H, Schulman H, De Koninck P. Transition from reversible to persistent binding of CaMKII to postsynaptic sites and NR2B. J Neurosci. 2006;26:1164–1174. doi: 10.1523/JNEUROSCI.3116-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 2005;310:866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
- Burette AC, Lesperance T, Crum J, Martone M, Volkmann N, Ellisman MH, Weinberg RJ. Electron tomographic analysis of synaptic ultrastructure. J Comp Neurol. 2012;520:2697–2711. doi: 10.1002/cne.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne MJ, Waxham MN, Kubota Y. The impacts of geometry and binding on CaMKII diffusion and retention in dendritic spines. J Comput Neurosci. 2011;31:1–12. doi: 10.1007/s10827-010-0293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbran RJ. Targeting of calcium/calmodulin-dependent protein kinase II. Biochem J. 2004;378:1–16. doi: 10.1042/BJ20031547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coultrap SJ, Bayer KU. CaMKII regulation in information processing and storage. Trends Neurosci. 2012;35:607–618. doi: 10.1016/j.tins.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coultrap SJ, Buard I, Kulbe JR, Dell'Acqua ML, Bayer KU. CaMKII autonomy is substrate-dependent and further stimulated by Ca2+/calmodulin. J Biol Chem. 2010;285:17930–17937. doi: 10.1074/jbc.M109.069351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coultrap SJ, Freund RK, O'Leary H, Sanderson JL, Roche KW, Dell'Acqua ML, Bayer KU. Autonomous CaMKII mediates both LTP and LTD using a mechanism for differential substrate site selection. Cell Rep. 2014;6:431–437. doi: 10.1016/j.celrep.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:361–387. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- Dupuis JP, Ladépêche L, Seth H, Bard L, Varela J, Mikasova L, Bouchet D, Rogemond V, Honnorat J, Hanse E, Groc L. Surface dynamics of GluN2B-NMDA receptors controls plasticity of maturing glutamate synapses. EMBO J. 2014;33:842–861. doi: 10.1002/embj.201386356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faas GC, Raghavachari S, Lisman JE, Mody I. Calmodulin as a direct detector of Ca2+ signals. Nat Neurosci. 2011;14:301–304. doi: 10.1038/nn.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Raghavachari S, Lisman J. Quantitative estimates of the cytoplasmic, PSD, and NMDAR-bound pools of CaMKII in dendritic spines. Brain Res. 2011;1419:46–52. doi: 10.1016/j.brainres.2011.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink CC, Meyer T. Molecular mechanisms of CaMKII activation in neuronal plasticity. Curr Opin Neurobiol. 2002;12:293–299. doi: 10.1016/S0959-4388(02)00327-6. [DOI] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Lu HE, Blanpied TA. Optimization of cell morphology measurement via single-molecule tracking PALM. PLoS One. 2012;7:e36751. doi: 10.1371/journal.pone.0036751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TJ, Verkhusha VV, Hess ST. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat Protoc. 2009;4:291–308. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaud L, Wong R, Hirokawa N. Disruption of KIF17-Mint1 interaction by CaMKII-dependent phosphorylation: a molecular model of kinesin-cargo release. Nat Cell Biol. 2008;10:19–29. doi: 10.1038/ncb1665. [DOI] [PubMed] [Google Scholar]
- Halt AR, Dallapiazza RF, Zhou Y, Stein IS, Qian H, Juntti S, Wojcik S, Brose N, Silva AJ, Hell JW. CaMKII binding to GluN2B is critical during memory consolidation. EMBO J. 2012;31:1203–1216. doi: 10.1038/emboj.2011.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R. Driving AMPA receptors into synapses by LTP and CaMKII: requirement for GluR1 and PDZ domain interaction. Science. 2000;287:2262–2267. doi: 10.1126/science.287.5461.2262. [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoze N, Nair D, Hosy E, Sieben C, Manley S, Herrmann A, Sibarita JB, Choquet D, Holcman D. Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proc Natl Acad Sci U S A. 2012;109:17052–17057. doi: 10.1073/pnas.1204589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudmon A, Aronowski J, Kolb SJ, Waxham MN. Inactivation and self-association of Ca2+/calmodulin-dependent protein kinase II during autophosphorylation. J Biol Chem. 1996;271:8800–8808. doi: 10.1074/jbc.271.15.8800. [DOI] [PubMed] [Google Scholar]
- Hudmon A, Lebel E, Roy H, Sik A, Schulman H, Waxham MN, De Koninck P. A mechanism for Ca2+/calmodulin-dependent protein kinase II clustering at synaptic and nonsynaptic sites based on self-association. J Neurosci. 2005;25:6971–6983. doi: 10.1523/JNEUROSCI.4698-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JM, Blanpied TA. Subsynaptic AMPA receptor distribution is acutely regulated by actin-driven reorganization of the postsynaptic density. J Neurosci. 2012;32:658–673. doi: 10.1523/JNEUROSCI.2927-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Zou Y, Amjad A, Gardezi A, Smith CL, Winters C, Reese TS. Sequestration of CaMKII in dendritic spines in silico. J Comput Neurosci. 2011;31:581–594. doi: 10.1007/s10827-011-0323-2. [DOI] [PubMed] [Google Scholar]
- Khan S, Reese TS, Rajpoot N, Shabbir A. Spatiotemporal maps of CaMKII in dendritic spines. J Comput Neurosci. 2012;33:123–139. doi: 10.1007/s10827-011-0377-1. [DOI] [PubMed] [Google Scholar]
- Kim SA, Sanabria H, Digman MA, Gratton E, Schwille P, Zipfel WR, Waxham MN. Quantifying translational mobility in neurons: comparison between current optical techniques. J Neurosci. 2010;30:16409–16416. doi: 10.1523/JNEUROSCI.3063-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis DM, Reese TS. Cytoplasmic organization in cerebellar dendritic spines. J Cell Biol. 1983;97:1169–1178. doi: 10.1083/jcb.97.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Yasuda R. Spatiotemporal regulation of signaling in and out of dendritic spines: CaMKII and Ras. Open Neurosci J. 2009;3:117–127. doi: 10.2174/1874082000903020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux M, Labrecque S, Tardif C, Labrie-Dion É, Lebel É, De Koninck P. Translocation of CaMKII to dendritic microtubules supports the plasticity of local synapses. J Cell Biol. 2012;198:1055–1073. doi: 10.1083/jcb.201202058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AS, Bayer KU, Merrill MA, Lim IA, Shea MA, Schulman H, Hell JW. Regulation of calcium/calmodulin-dependent protein kinase II docking to N-methyl-D-aspartate receptors by calcium/calmodulin and alpha-actinin. J Biol Chem. 2002;277:48441–48448. doi: 10.1074/jbc.M205164200. [DOI] [PubMed] [Google Scholar]
- Lin YC, Redmond L. CaMKIIbeta binding to stable F-actin in vivo regulates F-actin filament stability. Proc Natl Acad Sci U S A. 2008;105:15791–15796. doi: 10.1073/pnas.0804399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Roche KW. Posttranslational regulation of AMPA receptor trafficking and function. Curr Opin Neurobiol. 2012;22:470–479. doi: 10.1016/j.conb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78:615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- Markwardt ML, Kremers GJ, Kraft CA, Ray K, Cranfill PJ, Wilson KA, Day RN, Wachter RM, Davidson MW, Rizzo MA. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden KC, Shemesh A, Bayer KU, Carroll RC. Selective translocation of Ca2+/calmodulin protein kinase IIalpha (CaMKIIalpha) to inhibitory synapses. Proc Natl Acad Sci U S A. 2010;107:20559–20564. doi: 10.1073/pnas.1010346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill MA, Chen Y, Strack S, Hell JW. Activity-driven postsynaptic translocation of CaMKII. Trends Pharmacol Sci. 2005;26:645–653. doi: 10.1016/j.tips.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82 doi: 10.1103/PhysRevE.82.041914. 041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc Natl Acad Sci U S A. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Bosch M, Hayashi Y. The roles of CaMKII and F-actin in the structural plasticity of dendritic spines: a potential molecular identity of a synaptic tag? Physiology (Bethesda) 2009;24:357–366. doi: 10.1152/physiol.00029.2009. [DOI] [PubMed] [Google Scholar]
- Opazo P, Labrecque S, Tigaret CM, Frouin A, Wiseman PW, De Koninck P, Choquet D. CaMKII triggers the diffusional trapping of surface AMPARs through phosphorylation of stargazin. Neuron. 2010;67:239–252. doi: 10.1016/j.neuron.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Otmakhov N, Tao-Cheng JH, Carpenter S, Asrican B, Dosemeci A, Reese TS, Lisman J. Persistent accumulation of calcium/calmodulin-dependent protein kinase II in dendritic spines after induction of NMDA receptor-dependent chemical long-term potentiation. J Neurosci. 2004;24:9324–9331. doi: 10.1523/JNEUROSCI.2350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, Srivastava DP. Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 2008;18:405–413. doi: 10.1016/j.tcb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Petersen JD, Chen X, Vinade L, Dosemeci A, Lisman JE, Reese TS. Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J Neurosci. 2003;23:11270–11278. doi: 10.1523/JNEUROSCI.23-35-11270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncer JC, Esteban JA, Malinow R. Multiple mechanisms for the potentiation of AMPA receptor-mediated transmission by alpha-Ca2+/calmodulin-dependent protein kinase II. J Neurosci. 2002;22:4406–4411. doi: 10.1523/JNEUROSCI.22-11-04406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo RL, Okuno H, Spooner PA, Frenguelli BG, Bito H, Morris RG. Synaptic tagging and capture: differential role of distinct calcium/calmodulin kinases in protein synthesis-dependent long-term potentiation. J Neurosci. 2010;30:4981–4989. doi: 10.1523/JNEUROSCI.3140-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M, Domanov Y, Sandrin F, Izeddin I, Bassereau P, Triller A. Lateral diffusion on tubular membranes: quantification of measurements bias. PLoS One. 2011;6:e25731. doi: 10.1371/journal.pone.0025731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Bass MA, Jiao Y, MacMillan LB, Carmody LC, Bartlett RK, Colbran RJ. Multivalent interactions of calcium/calmodulin-dependent protein kinase II with the postsynaptic density proteins NR2B, densin-180, and alpha-actinin-2. J Biol Chem. 2005;280:35329–35336. doi: 10.1074/jbc.M502191200. [DOI] [PubMed] [Google Scholar]
- Sanabria H, Digman MA, Gratton E, Waxham MN. Spatial diffusivity and availability of intracellular calmodulin. Biophys J. 2008;95:6002–6015. doi: 10.1529/biophysj.108.138974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savin T, Doyle PS. Static and dynamic errors in particle tracking microrheology. Biophys J. 2005;88:623–638. doi: 10.1529/biophysj.104.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Sharma K, Fong DK, Craig AM. Postsynaptic protein mobility in dendritic spines: long-term regulation by synaptic NMDA receptor activation. Mol Cell Neurosci. 2006;31:702–712. doi: 10.1016/j.mcn.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Shen K, Meyer T. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science. 1999;284:162–166. doi: 10.1126/science.284.5411.162. [DOI] [PubMed] [Google Scholar]
- Shen K, Teruel MN, Subramanian K, Meyer T. CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron. 1998;21:593–606. doi: 10.1016/S0896-6273(00)80569-3. [DOI] [PubMed] [Google Scholar]
- Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- Shim SH, Xia C, Zhong G, Babcock HP, Vaughan JC, Huang B, Wang X, Xu C, Bi GQ, Zhuang X. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc Natl Acad Sci U S A. 2012;109:13978–13983. doi: 10.1073/pnas.1201882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DT, Sheng M, Lee SH. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci. 2012;15:1655–1666. doi: 10.1038/nn.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Steiner P, Higley MJ, Xu W, Czervionke BL, Malenka RC, Sabatini BL. Destabilization of the postsynaptic density by PSD-95 serine 73 phosphorylation inhibits spine growth and synaptic plasticity. Neuron. 2008;60:788–802. doi: 10.1016/j.neuron.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack S, Colbran RJ. Autophosphorylation-dependent targeting of calcium/calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem. 1998;273:20689–20692. doi: 10.1074/jbc.273.33.20689. [DOI] [PubMed] [Google Scholar]
- Sumioka A, Yan D, Tomita S. TARP phosphorylation regulates synaptic AMPA receptors through lipid bilayers. Neuron. 2010;66:755–767. doi: 10.1016/j.neuron.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KA, Suzuki KG, Shirai YM, Shibutani ST, Miyahara MS, Tsuboi H, Yahara M, Yoshimura A, Mayor S, Fujiwara TK, Kusumi A. Membrane molecules mobile even after chemical fixation. Nat Methods. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- Walikonis RS, Oguni A, Khorosheva EM, Jeng CJ, Asuncion FJ, Kennedy MB. Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin. J Neurosci. 2001;21:423–433. doi: 10.1523/JNEUROSCI.21-02-00423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YP, Holbro N, Oertner TG. Optical induction of plasticity at single synapses reveals input-specific accumulation of alphaCaMKII. Proc Natl Acad Sci U S A. 2008;105:12039–12044. doi: 10.1073/pnas.0802940105. [DOI] [PMC free article] [PubMed] [Google Scholar]