Torres-Padilla and colleagues investigate the chromatin-based mechanisms behind the transition from totipotency to pluripotency in the developing mouse embryo. Tracking histone dynamics by FRAP in vivo reveals that core histone mobility decreases as development proceeds, defining different cellular states (totipotency, pluripotency, and differentiation). Strikingly, totipotent cells in vitro display the same high chromatin mobility as totipotent cells in the embryo. The data suggest that changes in chromatin dynamics underlie the transitions in cellular plasticity and that higher chromatin mobility is at the nuclear foundations of totipotency.
Keywords: pluripotency, chromatin dynamics, reprogramming, cell fate, totipotent cells
Abstract
The fusion of the gametes upon fertilization results in the formation of a totipotent cell. Embryonic chromatin is expected to be able to support a large degree of plasticity. However, whether this plasticity relies on a particular conformation of the embryonic chromatin is unknown. Moreover, whether chromatin plasticity is functionally linked to cellular potency has not been addressed. Here, we adapted fluorescence recovery after photobleaching (FRAP) in the developing mouse embryo and show that mobility of the core histones H2A, H3.1, and H3.2 is unusually high in two-cell stage embryos and decreases as development proceeds. The transition toward pluripotency is accompanied by a decrease in histone mobility, and, upon lineage allocation, pluripotent cells retain higher mobility than the differentiated trophectoderm. Importantly, totipotent two-cell-like embryonic stem cells also display high core histone mobility, implying that reprogramming toward totipotency entails changes in chromatin mobility. Our data suggest that changes in chromatin dynamics underlie the transitions in cellular plasticity and that higher chromatin mobility is at the nuclear foundations of totipotency.
Embryonic cells are characterized by a large degree of plasticity, which is the ability to generate different cell types upon differentiation and is necessary to start a full developmental program. After fertilization, the mouse embryo has the transient capacity to generate both embryonic and extraembryonic cell types, a feature that is referred to as totipotency (Tarkowski 1959; Ishiuchi and Torres-Padilla 2013). This is in contrast to pluripotent cells, which contribute to all three germ layers of the embryo proper, but not to extraembryonic lineages, and therefore have a more restricted potential than totipotent cells. In mice, only the zygote and two-cell stage blastomeres are, strictly speaking, totipotent, since they have the ability to develop into a full organism without the need of carrier cells (Tarkowski 1959; Tarkowski and Wroblewska 1967; Kelly et al. 1978; Ishiuchi and Torres-Padilla 2013). As development progresses, pluripotent cells form in the inner cell mass (ICM) of the blastocyst, accompanied by the activation of pluripotency-associated transcription factors like Nanog and Pou5f1/Oct4 (Nichols et al. 1998; Chambers et al. 2003). The first differentiated embryonic tissue, the trophectoderm (TE), appears morphologically distinguishable and surrounds the ICM in the blastocyst. Thus, during the early stages of development, the mouse embryo undergoes dramatic changes in cellular plasticity.
Upon fertilization, embryonic chromatin undergoes intense chromatin remodeling. Indeed, this epigenetic reprogramming of the gametes is thought to be essential to gain totipotency (Sado and Ferguson-Smith 2005; Surani et al. 2007; Hemberger et al. 2009). However, the precise conformation of embryonic chromatin and the way it is remodeled to sustain totipotency and subsequent pluripotency remain largely unknown. In particular, whether and which changes in chromatin dynamics and organization underlie the transitions in cellular plasticity have not been established. It is generally assumed that a more plastic chromatin is present in pluripotent cells. Although this has been analyzed to some extent in pluripotent stem (embryonic stem [ES]) cells in culture (Meshorer et al. 2006; Melcer et al. 2012), it has not been addressed in vivo, and the molecular and epigenetic features of totipotent cells are scarce. Moreover, whether chromatin plasticity is functionally linked to cellular potency and fate has not been addressed experimentally.
Results and Discussion
To address whether chromatin plasticity parallels cellular potency in vivo, we first examined chromatin mobility. For this, we set up fluorescence recovery after photobleaching (FRAP) of chromatin proteins in embryos. Although FRAP is an approach routinely used in cultured cells, it has not yet been used to track chromatin dynamics in the developing mammalian embryo. We therefore first established conditions for FRAP of chromatin proteins that are compatible with normal embryonic development (Supplemental Fig. S1) We injected mRNA for GFP-tagged histones into zygotes at the fertilization cone stage before pronuclei formation based on previously titrated histone mRNA concentrations (Santenard et al. 2010). We then cultured these embryos and performed FRAP on individual two-cell or eight-cell stage nuclei (Fig. 1A). To ensure that the GFP signal that we observed derives from histones in chromatin, we first verified incorporation of exogenously expressed histones by analyzing mitotic chromosomes, which revealed a strong GFP signal on chromatin (Supplemental Fig. S2). Second, we also imaged interphase nuclei after treatment with Triton X-100, which releases nonbound chromatin proteins. Triton pre-extraction did not detectably alter the GFP signal in the nucleoplasm. Together, this suggests that under the experimental conditions that we established, GFP-tagged histones are efficiently incorporated into embryonic chromatin (Supplemental Fig. S2). We initially analyzed chromatin mobility of histone H2A, carefully avoiding the nucleoli precursors. A representative nucleus of an eight-cell stage embryo during FRAP acquisition for H2A and single typical FRAP curves of two-cell and eight-cell stage nuclei are shown in Figure 1, B and C, respectively. H2A-GFP showed a striking, reproducible high mobility in two-cell stage embryos, with relatively fast recovery kinetics and an ∼29% mobile fraction (Fig. 1D,E; Supplemental Table S1). Interestingly, H2A-GFP mobility was significantly reduced at the eight-cell stage compared with the two-cell stage (P < 0.0001) (Fig. 1D,E; Supplemental Tables S3), suggesting that chromatin dynamics decrease as development proceeds. Importantly, the mobility of H2A-GFP and its changes during development were independent of the amount of mRNA injected or the timing of microinjection (Supplemental Fig. S4).
Figure 1.
FRAP analysis of chromatin components in the developing mouse embryo reveals a decrease in chromatin mobility of H2A-GFP. (A) Experimental setup for FRAP in embryos. Zygotes were collected and microinjected with in vitro transcribed mRNA and cultured until the indicated developmental stages, when they were subjected to FRAP. After imaging acquisition, embryos were cultured until the blastocyst stage, and their development was scored. (B) A representative nucleus of an eight-cell stage embryo expressing H2A-GFP during a FRAP experiment is shown. The bleached region is represented by a rectangle. Bar, 10 μm. (C) Representative single FRAP curves of H2A-GFP at the two-cell (red) and eight-cell (blue) stages. The bleach time point is indicated by an arrow. Recovery of H2A-GFP is significantly faster at the two-cell stage compared with the eight-cell stage. (D) Recovery curves of H2A-GFP at the two-cell (red) and eight-cell (blue) stages. Recovery was quantified in the bleached area over a 60-sec period, and the curves were normalized to zero to account for differences in bleach depth between experiments. Individual points are mean ± SEM, and mean values were fit into an exponential curve. (E) Estimated mobile fractions (±SEM) of H2A-GFP in two-cell and eight-cell stage embryos.
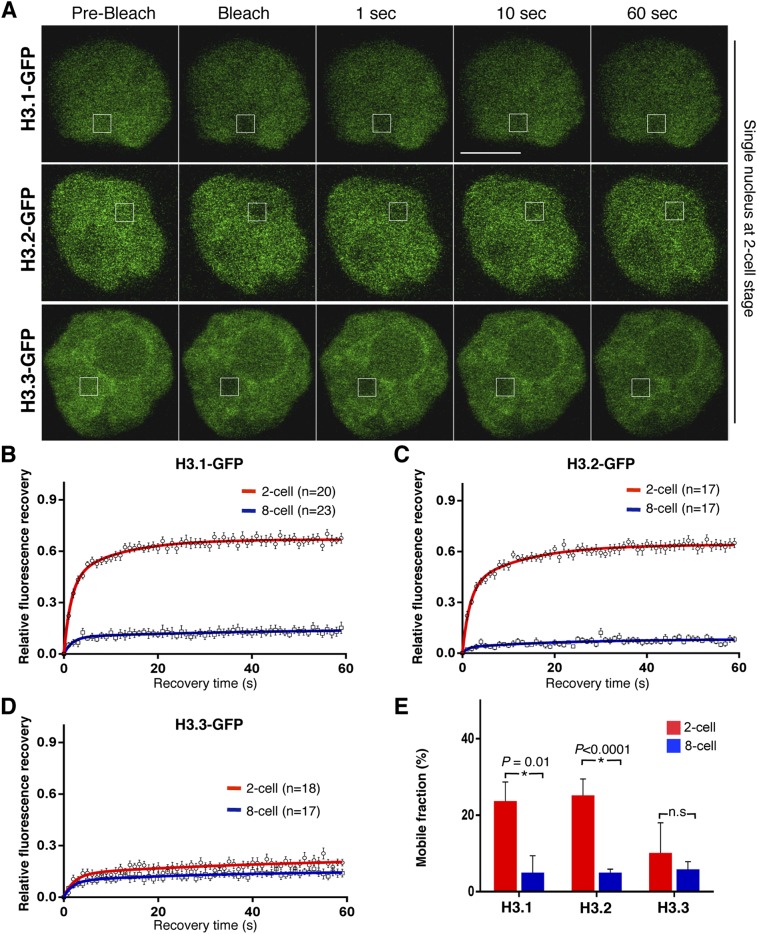
Upon chromatin remodeling, H2A/H2B dimers are known to be released prior to H3 and H4 tetramers (Groth et al. 2007; Xu et al. 2010; Winkler et al. 2012). To address whether the above observations are specific for H2A or reflect a general property of embryonic chromatin, we expanded our analysis to histone H3. We used the same experimental design as above and verified chromatin incorporation of the three H3 variants H3.1, H3.2, and H3.3 fused to GFP (Supplemental Figs. S2, S3; Santenard et al. 2010). Similarly to H2A-GFP, GFP-tagged H3.1 and H3.2 displayed a remarkable high mobility in two-cell stage embryos (Fig. 2A–C), with a mobile fraction of 24% ± 5% (n = 20) and 25% ± 5% (n = 17), respectively, which was independent of the amount of mRNA injected (Supplemental Fig. S4). Importantly, the mobility of both H3.1-GFP and H3.2-GFP decreased significantly as development proceeded to the eight-cell stage, reaching a mobile fraction of ∼5% for both histones (P = 0.01 for H3.1-GFP and P < 0.0001 for H3.2-GFP) (Fig. 2B,C,E; Supplemental Tables S1, S3, S4). Thus, globally, canonical core histones display a remarkably high mobility at the beginning of embryogenesis, as measured by FRAP, which diminishes as development proceeds, suggesting that chromatin mobility might be linked to cellular potency. Next, we analyzed the replacement histone variant H3.3. In contrast to H3.1 and H3.2, H3.3-GFP showed much lower mobility in two-cell stage embryos, with an estimated mobile fraction of 10% ± 7.8% (n = 18), which did not change significantly in eight-cell embryos (6% ± 2%; n = 17; P = 0.59) (Fig. 2D,E; Supplemental Table S3), suggesting that some chromatin components may not change their mobility between the two-cell and eight-cell stages.
Figure 2.
Mobility of core histones H3.1-GFP and H3.2-GFP decreases between the two-cell and the eight-cell stages, but H3.3-GFP mobility remains unchanged. (A) Nuclei of two-cell stage embryos expressing either H3.1-GFP (top), H3.2-GFP (middle), or H3.3-GFP (bottom) at the indicated time points during representative FRAP experiments. The bleached region is indicated by a rectangle. (B) FRAP curves for H3.1-GFP in two-cell and eight-cell stage embryos reveal higher mobility of H3.1-GFP at the earlier developmental stage. (C) H3.2-GFP FRAP curves at the two-cell and eight-cell stages. Two-cell stage embryos show an unusual, high mobility behavior of H3.2-GFP, which is dramatically decreased with developmental progression. (D) FRAP curves for H3.3-GFP at the two-cell and eight-cell stages show no significant change in H3.3 dynamics between the two stages. (E) Calculated mobile fractions (±SEM) of H3 variants at the two-cell and eight-cell stages. (B–D) Values represent mean ± SEM of multiple embryos, where n indicates the number of nuclei analyzed. FRAP was performed in only one nucleus per embryo. The mean values were fit into an exponential curve. P-values were calculated using unpaired t-test between each two groups.
To investigate whether the decreased mobility of canonical core histones at the eight-cell stage is regulated through the action of histone modifiers, we performed FRAP for H2A-GFP and H3.1-GFP in the presence of chemical inhibitors. Incubation of eight-cell stage embryos with TSA, a global HDAC inhibitor known to affect H1e mobility in ES cells (Melcer et al. 2012), did not seem to alter the mobility of H2A-GFP or H3.1-GFP compared with control embryos (P = 0.92 and P = 0.67, respectively) (Supplemental Fig. S5; Supplemental Table S5). We also asked whether decreasing global H3K9me2 levels through the action of BIX-01294, a compound reported to inhibit the G9a methyltransferase (Kubicek et al. 2007), would impact histone mobility in eight-cell stage embryos. Culturing eight-cell stage embryos with BIX-01294 led to a reduction in global H3K9me2 levels. While H3.1-GFP mobility remained virtually unchanged upon BIX-01294 treatment, H2A-GFP displayed a slightly higher mobility in the presence of BIX-01294 (Supplemental Fig. S5). However, this difference was not statistically significant under the experimental conditions used (Supplemental Table S5). Although not statistically significant, the trend change in H2A-GFP dynamics in the presence of BIX-01294 may suggest that global changes in H3K9 methylation might potentially be involved in the changes in histone mobility from the two-cell to the eight-cell stage. Indeed, it is known that global H3K9me3 levels increase from the eight-cell stage onward, when embryonic constitutive heterochromatin state is replaced by the canonical Suv39h-mediated state (Puschendorf et al. 2008). The sharp down-regulation of the H3K9me2/me3 demethylase Kdm4b at the eight-cell stage is also in line with this suggestion (Burton et al. 2013). Since BIX-01294 is reported to inhibit specifically G9a (Kubicek et al. 2007), it will be important to manipulate additional H3K9me pathways to address this in full.
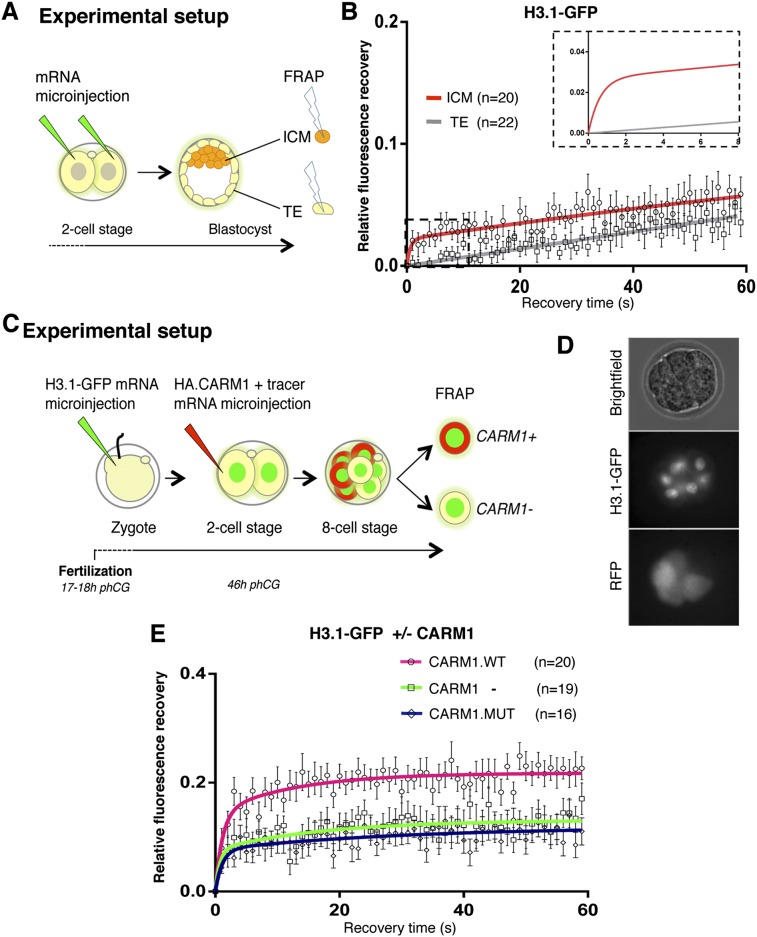
To further explore the hypothesis that chromatin dynamics might be linked to cellular potency, we next investigated chromatin mobility in the two lineages of the early blastocyst, the ICM and the TE, which are characterized by different degrees of cellular potency. We devised an experimental setup to perform FRAP in individual nuclei of ICM and TE cells (Fig. 3A). FRAP analysis revealed that H3.1-GFP displays higher mobility in pluripotent cells of the ICM compared with the differentiated TE counterpart (Fig. 3B). Remarkably, the recovery kinetics between the ICM and TE differed drastically, by an order of magnitude higher (Fig. 3B, inset; Supplemental Table S2), and the H3.1-GFP mobile fraction was approximately twofold higher in ICM cells compared with the TE (Fig. 3B), suggesting that pluripotent cells retain high chromatin mobility, while TE cells do not.
Figure 3.
Pluripotent cells retain high chromatin mobility upon lineage allocation. (A) The two blastomeres of a two-cell stage embryo were microinjected with equal amounts of H3.1-GFP mRNA, and embryos were developed to the blastocyst stage. At embryonic day 3.5 (E3.5), a single nucleus of the ICM and in the TE was subjected to FRAP as in Figure 1. (B) Mean FRAP values for H3.1-GFP in the ICM (red line) and TE (gray line) over 60-sec period of recovery immediately after bleaching. The smaller graph in the top right corner represents a zoom of the dashed rectangle, which includes the first 8 sec post-bleach, in which data points are omitted for clarity. (C) Schematic representation of FRAP experiments to address chromatin mobility during lineage allocation. Zygotes were microinjected with H3.1-GFP mRNA as in Figure 1 and cultured until the late two-cell stage, when one blastomere was microinjected with HA.CARM1 mRNA and mRFP mRNA as tracer. Microinjection of HA.CARM1 mRNA at this stage allocates the progeny of the injected cell to the ICM (Torres-Padilla et al. 2007). (D) Representative eight-cell stage embryo after double microinjection. (Middle image) While all nuclei are H3.1-GFP-positive, only four blastomeres are RFP-positive (and HA.CARM1-positive). (E) FRAP curves for H3.1-GFP in HA.CARM1 wild-type-positive (pink line), HA.CARM1-negative (green line), and CARM1 catalytic death-positive (dark blue line) blastomeres at the eight-cell stage. In each embryo, one RFP-positive and one RFP-negative cell were analyzed by FRAP. Allocation of ICM fate through CARM1 expression increases H3.1-GFP mobility at the eight-cell stage. Under the same experimental conditions, the CARM1 catalytic mutant does not alter H3.1-GFP mobility.
The difference in H3.1-GFP mobility between ICM versus TE that we observed and the fact that ICM cells display higher mobility further prompted us to ask whether chromatin mobility is functionally linked to lineage allocation. To directly address this possibility, we sought to determine chromatin mobility in embryonic cells immediately before the segregation of these two lineages, which first occurs upon the formation of outer and inner cells during the division of the eight-cell to the 16-cell stage (Kelly et al. 1978; Johnson and Ziomek 1981). In the ideal experimental setup, tracking chromatin mobility in eight-cell stage blastomeres that are destined to become ICM should be performed. However, because the cells in the mammalian embryo are not predetermined but are subject to regulative development (Rossant and Tam 2004), performing this is technically not feasible without perturbing development. We therefore took advantage of earlier findings showing that expression of PRMT4/CARM1 in a late two-cell stage blastomere drives the progeny of this cell to the ICM (Torres-Padilla et al. 2007). We microinjected H3.1-GFP mRNA in zygotes as before and performed a second microinjection of Carm1 mRNA together with RFP mRNA as a lineage tracer (Fig. 3C). We then conducted FRAP in CARM1-positive and CARM1-negative cells at the eight-cell stage, which were distinguishable by the presence of the RFP tracer (Fig. 3C,D). Notably, while the CARM1-negative cells displayed the H3.1-GFP mobility of eight-cell blastomeres described above, CARM1-expressing cells showed a significantly higher H3.1-GFP mobility and a mobile fraction of approximately twofold (14% ± 3% compared with 7.7% ± 2% for the fast fraction and 7.3% ± 2.9% compared with 5.4% ± 1.7% for the slower fraction; P < 0.01) (Fig. 3E; Supplemental Table S6). Importantly, cells expressing a CARM1 catalytic mutant that is unable to direct cells toward the ICM (Torres-Padilla et al. 2007) displayed an H3.1-GFP mobility similar to noninjected cells (P = 0.97) (Fig. 3E; Supplemental Table S6), suggesting that cells destined to become pluripotent ICM retain higher histone mobility than future TE cells.
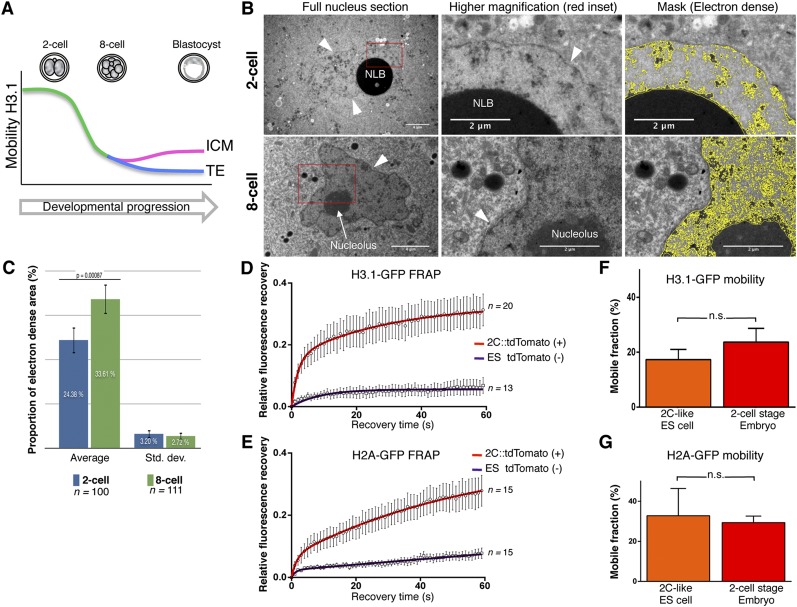
The above results together suggest that, in totipotent cells in the early embryo, the core histones H3.1-GFP, H3.2-GFP, and H2A-GFP display a very high mobility and that their mobility decreases as development proceeds, with pluripotent cells maintaining higher histone mobility than the TE upon lineage allocation (Fig. 4A). Given the unusually high mobility that we observed in two-cell stage nuclei, we next wondered whether any particular conformational or organizational feature of embryonic chromatin could be the basis for this. We thus established conditions to analyze embryonic chromatin ultrastructurally using transmission electron microscopy (TEM). Analysis of two-cell stage nuclei with TEM revealed a rather dispersed nucleoplasm largely devoid of the typical heterochromatic, electron-dense regions found in differentiated cells (Supplemental Fig. S6; Davies 1967), with a prominent nucleolar-like body (NLB) (Fig. 4B). Instead, only few electron-dense foci were visible throughout the nucleus, with no obvious enrichment of electron-dense heterochromatin in the vicinity of the nuclear membrane, a known feature of somatic cells (Cremer and Cremer 2001; Towbin et al. 2013), in agreement with reported electron spectroscopic imaging (Ahmed et al. 2010). Eight-cell stage chromatin appeared slightly more compacted than in two-cell nuclei, with larger electron-dense areas and stronger accumulation of compacted chromatin regions around the nuclear membrane and the nucleolus (Fig. 4B). Indeed, quantification of the electron-dense area across >100 TEM sections for each stage revealed a significant increase in the proportion of the electron-dense area between the two-cell and the eight-cell stage (P = 0.0008) (Fig. 4C), suggesting progressive compaction of a significant part of the chromatin between these two stages. Furthermore, the global increase in the proportion of electron-dense regions correlates with the decrease in histone mobility from the two-cell to the eight-cell stage that we observed. Thus, early embryonic chromatin displays an “atypical” loose chromatin ultrastructure, in line with the high chromatin protein mobility that we report.
Figure 4.
Totipotent cells display unusually high chromatin mobility and loose chromatin ultrastructure. (A) Schematic summary of H3.1-GFP mobility throughout development. (B) Electron micrographs of two-cell (top) and eight-cell (bottom) stage nuclei. Nucleolar-like bodies (NLBs) and nucleoli are indicated; arrowheads point to the nuclear membrane. The higher magnification corresponds to the red inset. The right column shows the mask to segment electron-dense regions for quantification (see Supplemental Fig. S8 for details). Note that the NLBs are very electron-dense, presumably due to the presence of negatively charged proteins such as nucleoplasmin (Inoue and Aoki 2010). (C) Quantification of the relative electron-dense area in two-cell and eight-cell stage nuclei reveals an increase of chromatin compaction. The average and SD of the indicated number of sections analyzed are shown. (D,E) Mean FRAP values of H3.1-GFP (D) and H2A-GFP (E) in tdTomato-positive two-cell-like (2C) ES cells (red curve) compared with tdTomato-negative ES cells (purple curve) grown in LIF+2i medium. The number of cells analyzed is shown at the side of each curve. ES cells with 2C properties are characterized by dramatically higher H3.1 mobility compared with non-2C, pluripotent ES cells within the same population. (F,G) Mobile fractions (±SEM) of H3.1-GFP (F) and H2A-GFP (G) in 2C ES cells and two-cell stage embryos. While the variability is higher in the in vitro totipotent cells (2C-like), the high mobility of H3.1-GFP is comparable between in vitro 2C cells and in vivo in two-cell embryos. (n.s.) Non-significant (unpaired t-test).
Interestingly, the eight-cell stage changes in histone mobility correlate with global changes in chromatin organization and in the developmental program. Namely, major changes in heterochromatin organization, including formation of chomocenters (Probst et al. 2007), the global repression of retrotransposons (Peaston et al. 2004; Fadloun et al. 2013), and a significant increase in the electron-dense regions in the nuclei (this study), take place by the eight-cell stage. Also, these changes in histone mobility coincide with the time when individual cells develop polarity (Johnson and Ziomek 1981), which we previously suggested to co-occur with major changes in the expression of chromatin modifiers at the eight-cell stage (Burton et al. 2013).
Totipotent “two-cell-like” (2C) cells have recently been shown to emerge stochastically in ES cell cultures in vitro and can be identified by the activation of a specific endogenous retroelement of the ERVL family, MuERVL (Macfarlan et al. 2012). We reasoned that if high chromatin mobility is indeed an inherent property of totipotent cells, 2C ES cells should display equally high chromatin mobility as two-cell stage embryos. To address this, we generated ES cells stably expressing tdTomato under the control of MuERVL regulatory sequences, as previously described (Macfarlan et al. 2012). We confirmed that tdTomato expression occurs in a rather low percentage of the ES cell population (<0.5%) and that these ES cells are devoid of detectable levels of OCT4 protein (Supplemental Fig. S7; Macfarlan et al. 2012; data not shown). We then transfected plasmids harboring the same H3.1-GFP and H2A-GFP cDNAs as above and performed FRAP in 2C ES cells as well as in “normal” ES cells within the same colonies (tdTomato-positive and tdTomato-negative, respectively). Remarkably, 2C ES cells displayed a much higher mobility for both H3.1-GFP and H2A-GFP in tdTomato-positive compared with tdTomato-negative ES cells (Fig. 4D,E). The average mobile fraction of both proteins, albeit more variable, was similar to that in two-cell stage blastomeres (Fig. 4F,G). Thus, totipotent cells in vivo and in vitro are characterized by a very mobile chromatin configuration, suggesting that induction of totipotency entails changes in chromatin mobility.
Our results suggest that, globally, embryonic chromatin is extremely dynamic, more than in ES cells, which, although pluripotent, have a more restricted developmental potential than the totipotent cells in the early embryo. A loose chromatin conformation in the early embryo, in terms of both its dynamics and ultrastructure, might be permissive for the large-scale remodeling underlying epigenetic reprogramming after fertilization. While all core histones assayed were highly mobile, the replacement histone variant H3.3-GFP was the least mobile of the histones analyzed. Because H3.3 has been shown to be the major H3 variant deposited after fertilization (Torres-Padilla et al. 2006; Santenard et al. 2010; Akiyama et a. 2011), this initial incorporation of H3.3 may serve as a “placeholder” mechanism prior to the subsequent incorporation of the replication-dependent histones H3.1 and H3.2. It is possible that the high dynamics observed for H3.2 and H3.1 might be the result of this global wave of incorporation of H3.1/H3.2 and/or the transition toward a more “mature chromatin” where H3.3 is no longer the major histone variant. This is in agreement with the known dynamics of incorporation of H3 variants after fertilization as well as with the proposed model of H3.3 as a “placeholder” (Akiyama et al. 2011; Dunleavy et al. 2011) and as protective nucleosome gap filling (Ray-Gallet et al. 2011).
Our data suggest that increased chromatin mobility in vitro and in vivo is a key feature of totipotent cells and distinguishes them from pluripotent cells. Whether altering chromatin mobility is sufficient to induce totipotency and modulate cellular plasticity is an attractive possibility that remains to be addressed.
Materials and methods
Embryo collection and microinjection
Embryos were collected from F1 (C57BL/6 × CBA/H) crosses on superovulation. Human H3.1 (Santenard et al. 2010), H3.2, H3.3 (Santenard et al. 2010), and H2A (HIST1H2AK) cDNAs were subcloned into pRN3P plasmid, and corresponding mRNAs were transcribed in vitro, as described previously (Santenard et al. 2010). All fusions were cloned with EGFP in the C terminus, and all plasmids have identical 3′ and 5′ untranslated regions (UTRs). For the FRAP analysis in blastocysts, the levels of GFP in the blastocyst were too noisy when microinjection was performed in the zygote. Thus, to overcome this limitation, both blastomeres of two-cell stage embryos, collected at 45 h after administration of human chorionic gonadotropin (hphCG), were injected with identical concentrations of mRNA. We previously determined that recovery rates are independent of protein levels (see Supplemental Fig. S4). We verified incorporation of histones into chromatin (see the Supplemental Material).
FRAP microscopy
FRAP was performed using a Leica SP2 confocal microscope and at 37°C using a 63.0 × 1.4 oil objective. Embryos were placed in drops of M2 medium on a glass-bottomed dish. A rectangular region of interest of 2.28 μm2 was chosen randomly within a nucleus, avoiding the NLBs, and was subjected to FRAP. Ten prebleach frames were acquired followed by two bleach pulses without acquisition. Recovery of fluorescence was followed during 60 sec, with 1 frame per second. The raw data were processed with Fiji software (ImageJ). All analysis was done on background-subtracted values using EasyFRAP (see the Supplemental Material for a thorough description). The obtained curves were normalized using the full-scale normalization method so that the first post-bleach frame was set to 0. Normalized curves were then subjected to curve fitting.
Curve fitting and statistical analysis
Experimentally obtained and normalized recovery curves were fit using Prism6 software (GraphPad Software). A two-phase exponential association equation, Y = Ymax1 × [1 − e(− K1 × X)] + Ymax2 × [1 − e(− K2 × X)], was used to obtain mobile fractions and reaction rates, as this has been previously described to be appropriate for nuclear proteins (Phair and Misteli 2000). Unpaired t-tests were used for comparing two groups. Throughout the study, Ymax1 values are used for mobile fraction estimation, as they reflect the steady-state protein pool, unless otherwise stated.
Electron microscopy
Embryos at the two-cell (n = 3) and eight-cell (n = 3) stages were collected after natural matings of B6CBAF1/J mice, fixed, and contrasted with osmium tetroxide and 1% uranyl acetate for 1 h. Samples were observed with a transmission electron microscope using an Orius 1000 CCD camera. Quantification of electron-dense regions was performed with a dedicated analysis pipeline implemented in ImageJ and MatLab (Supplemental Fig. S8).
Acknowledgments
We thank members of the M.E.T.-P. laboratory for discussions, and P. Kessler and M. Koch of the IGBMC-ICS imaging facility. M.E.T.-P. acknowledges funding from EpiGeneSys, ERC-Stg “NuclearPotency,” and FP7 Marie-Curie Actions ITN Nucleosome4D. E.M. acknowledges funding from ERC-Stg “ExprES” (281781), Nucleosome4D ITN, and the Israel Ministry of Science. A.B. was a Nucleosome4D and Association pour la Recherche contre le Cancer fellow. T.I. holds a post-doctoral HFSP fellowship, and A.E. holds a fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche. E.V.S.R.R. was a Nucleosome4D ITN fellow and a Lady Davis Fellow.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.238881.114.
References
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE 5: e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, Suzuki O, Matsuda J, Aoki F 2011. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet 7: e1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Muller J, Tu S, Padilla-Longoria P, Guccione E, Torres-Padilla ME 2013. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep 5: 687–701 [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2: 292–301 [DOI] [PubMed] [Google Scholar]
- Davies HG 1967. Fine structure of heterochromatin in certain cell nuclei. Nature 214: 208–210 [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2: 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol 20: 332–338 [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G 2007. Chromatin challenges during DNA replication and repair. Cell 128: 721–733 [DOI] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W 2009. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol 10: 526–537 [DOI] [PubMed] [Google Scholar]
- Inoue A, Aoki F 2010. Role of the nucleoplasmin 2 C-terminal domain in the formation of nucleolus-like bodies in mouse oocytes. FASEB J 24: 485–494 [DOI] [PubMed] [Google Scholar]
- Ishiuchi T, Torres-Padilla ME 2013. Towards an understanding of the regulatory mechanisms of totipotency. Curr Opin Genet Dev 23: 512–518 [DOI] [PubMed] [Google Scholar]
- Johnson MH, Ziomek CA 1981. The foundation of two distinct cell lineages within the mouse morula. Cell 24: 71–80 [DOI] [PubMed] [Google Scholar]
- Kelly SJ, Mulnard JG, Graham CF 1978. Cell division and cell allocation in early mouse development. J Embryol Exp Morphol 48: 37–51 [PubMed] [Google Scholar]
- Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25: 473–481 [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, Skoultchi A, Stewart CL, Bustin M, Meshorer E 2012. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat Commun 3: 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606 [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Probst AV, Santos F, Reik W, Almouzni G, Dean W 2007. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma 116: 403–415 [DOI] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. 2008. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 40: 411–420 [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 44: 928–941 [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PP 2004. Emerging asymmetry and embryonic patterning in early mouse development. Dev Cell 7: 155–164 [DOI] [PubMed] [Google Scholar]
- Sado T, Ferguson-Smith AC 2005. Imprinted X inactivation and reprogramming in the preimplantation mouse embryo. Hum Mol Genet 14: R59–R64 [DOI] [PubMed] [Google Scholar]
- Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME 2010. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 12: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P 2007. Genetic and epigenetic regulators of pluripotency. Cell 128: 747–762 [DOI] [PubMed] [Google Scholar]
- Tarkowski AK 1959. Experiments on the development of isolated blastomers of mouse eggs. Nature 184: 1286–1287 [DOI] [PubMed] [Google Scholar]
- Tarkowski AK, Wroblewska J 1967. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol 18: 155–180 [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M 2006. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol 50: 455–461 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445: 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Sandoval A, Gasser SM 2013. Mechanisms of heterochromatin subnuclear localization. Trends Biochem Sci 38: 356–363 [DOI] [PubMed] [Google Scholar]
- Winkler DD, Zhou H, Dar MA, Zhang Z, Luger K 2012. Yeast CAF-1 assembles histone (H3–H4)2 tetramers prior to DNA deposition. Nucleic Acids Res 40: 10139–10149 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B 2010. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98 [DOI] [PubMed] [Google Scholar]