The spliceosome machinery is composed of multimeric protein complexes that generate a diverse repertoire of mRNA. Here, Adler et al. discover that PRPF6, a member of the tri-snRNP spliceosome complex, drives cancer proliferation. Inhibition of PRPF6 and other tri-snRNP complex proteins selectively abrogated growth in cancer cells with high tri-snRNP levels. Reducing PRPF6 altered the splicing of a discrete number of genes, including an oncogenic isoform of the ZAK kinase. This study identifies an essential role for PRPF6 in cancer via splicing of distinct growth-related gene products.
Keywords: RNAi, PRPF6, tri-snRNP, spliceosome, colon cancer
Abstract
The spliceosome machinery is composed of multimeric protein complexes that generate a diverse repertoire of mRNA through coordinated splicing of heteronuclear RNAs. While somatic mutations in spliceosome components have been discovered in several cancer types, the molecular bases and consequences of spliceosome aberrations in cancer are poorly understood. Here we report for the first time that PRPF6, a member of the tri-snRNP (small ribonucleoprotein) spliceosome complex, drives cancer proliferation by preferential splicing of genes associated with growth regulation. Inhibition of PRPF6 and other tri-snRNP complex proteins, but not other snRNP spliceosome complexes, selectively abrogated growth in cancer cells with high tri-snRNP levels. High-resolution transcriptome analyses revealed that reduced PRPF6 alters the constitutive and alternative splicing of a discrete number of genes, including an oncogenic isoform of the ZAK kinase. These findings implicate an essential role for PRPF6 in cancer via splicing of distinct growth-related gene products.
Colorectal carcinoma (CRC) is one of the most prevalent and fatal types of cancers, accounting for >600,000 deaths worldwide annually (Haggar and Boushey 2009). Genetic and genomic analyses of colorectal tumors have uncovered a number of key somatic and germline mutations that drive tumorigenesis at a molecular level and can be linked to well-defined disease stages of tumor progression (Fearon and Vogelstein 1990; Fearon 1995; Sillars-Hardebol et al. 2010). Colorectal tumors can be divided into three main genetic subtypes based on these initiating molecular alterations: (1) chromosomal instability (CIN), (2) CpG island methylator phenotype (CIMP), and (3) microsatellite instability (MSI). CIN is the most frequent alteration found in colon cancer (85%) and arises from the accumulation of genetic alterations (Rajagopalan et al. 2003). CIN tumors initiate with mutations in the tumor suppressor protein APC or β-catenin and lead to constitutive activation of the Wnt/β-catenin pathway (Bienz and Clevers 2000). Additional mutations in KRAS and p53 are acquired in later stages of tumor development, leading to genomic instability and chromosomal copy number alterations (CNA) (Baker et al. 1989; Fearon and Vogelstein 1990).
Chromosomal alterations in colon cancer are nonrandom events and are most often characterized by recurrent broad gains in chromosomal regions 7, 8q, 13, and 20q (Martin et al. 2007; Firestein et al. 2008). The clinical relevance of these alterations is highlighted by their association to metastatic progression and poor prognosis in CRC patients (Sheffer et al. 2009; Loo et al. 2013). Structural and functional interrogations of these regions have described putative oncogenes at chromosomes 7 (MET), 8q (MYC), 13 (CDK8 and CDX2), and 20q (AURKA and TPX2) that may play a role in colon cancer cell proliferation (Firestein et al. 2008; Dulak et al. 2012; Salari et al. 2012; Sillars-Hardebol et al. 2012; Bardelli et al. 2013). However, given the broad nature of chromosomal gain in colonic tumors that frequently span the entire chromosomal arm, we hypothesized that additional genes within these amplicons are necessary for tumor growth.
Spliceosome-coordinated RNA splicing is an essential cellular process that can generate an immensely diverse repertoire of RNA (Matlin et al. 2005; Wahl et al. 2009). Components of the spliceosome machinery have been shown to be dysregulated in human disease. One example is the tri-snRNP (small ribonucleoprotein) complex, which consists of U4/U5/U6 proteins and is known to cause an autosomal dominant form of retinitis pigmentosa (adRP) when mutated (Sharp 1988; Tanackovic et al. 2011; Will and Lührmann 2011). More recently, recurrent somatic mutations have been identified in a number of spliceosome components in cancers (Ebert and Bernard 2011; Wang et al. 2011; Yoshida et al. 2011; Graubert et al. 2012; Hahn and Scott 2012; Imielinski et al. 2012; Quesada et al. 2012), raising speculation that the spliceosome may play a role in tumor growth. Nevertheless, the molecular bases and consequences of spliceosome dysregulation in cancer development are not yet known.
In this study, we used an integrative genomic approach to systematically identify oncogenic drivers of colon cancer proliferation. We report that the coordinate amplification or overexpression of PRPF6 and other components of the tri-snRNP spliceosome complex drive proliferation in colorectal cancer. We further show that impaired PRPF6 activity leads to splicing alterations in a discrete number of genes. We characterize one of these isoform variants and its oncogenic properties.
Results
Integrative analysis of colon cancer identified PRPF6 as a regulator of colon cancer proliferation
We conducted an integrative genomic analysis of human and mouse colon tumors to identify novel genes with key roles in colon cancer development. We first identified 1257 genes and microRNAs (miRNA) that were both differentially expressed and localized to regions of copy number gain (CNG) using 505 colon tumors and 194 normal colonic tissue samples. A loss-of-function RNAi screen was then performed in 14 colon cancer cell lines that were annotated by expression, copy number, and mutational profiling (Fig. 1A). In parallel, we also performed gene expression profiling of three independent murine colon cancer models ([1] ApcMin, [2] ApcMin; KrasG12D; Vil-Cre, and [3] ApcLoxp/+; CDX2-Cre), comparing the intestinal tumors with normal mucosa. A primary screen using pooled siRNAs identified 184 genes, including the known oncogenes KRAS, MYC, and CTNNB1, for which knockdown correlated with the expression, copy number, or mutation status of the given gene (Supplemental Fig. 1; Supplemental Table 1). Subsequent secondary screening of these 184 genes using four independent siRNAs per gene identified 17 genes that exhibited a consistent phenotype with two or more scoring siRNAs (Supplemental Table 2). We prioritized these genes based on (1) the correlation between expression and CNG in colon tumors, (2) the degree of genomic amplification, (3) the correlation between protein expression and RNA expression in cancer cell lines, and (4) the conserved expression up-regulation in the three aforementioned genetically engineered murine colon cancer models. Of these genes, PRPF6, a spliceosome component that undergoes recurrent CNG in 40% of colon cancers, met all filtering thresholds and was further pursued (Supplemental Tables 3, 4).
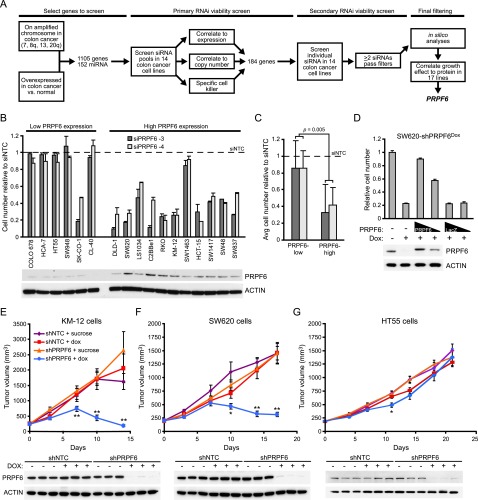
Figure 1.
Integrative genomic analysis identifies PRPF6 as a driver of colon cancer growth in vitro and in vivo. (A) Schematic of RNAi viability screens and filtering methods. (B) Relative cell number (compared with siNTC; mean ± SD) is shown for two independent PRPF6 siRNAs 6 d after siRNA transfection. Cell lines are separated by high or low PRPF6 protein expression levels as determined by immunoblot analysis. (C) The average relative cell number for each siRNA in each group of cells lines. P-value is a Student’s t-test. (D) The bar graph shows cell number relative to nontreated control in SW620 cells with stable integration of a doxycycline (Dox)-inducible PRPF6 shRNA (SW620-shPRPF6Dox). Doxycycline-induced PRPF6 knockdown is indicated. Adenoviruses expressing either shRNA-resistant PRPF6 or a LacZ control were infected 2 d after doxycycline-induced PRPF6 knockdown at both a low and high multiplicity of infection. Cell number was measured 7 d after doxycycline induction. Immunoblot shows PRPF6 expression under the indicated conditions. (E–G) Xenograft tumor volume measurements over time (n = 10–15 mice per group) in KM-12 cells (E), SW620 cells (F), or HT55 cells (G). Mean ± SEM is shown. Immunoblot of PRPF6 protein levels in xenograft tumors were at day 7. (*) P-value < 0.05; (**) P-value < 0.001 for shPRPF6 + Dox as compared with shNTC + Dox groups.
We first examined the correlation between PRPF6 expression, amplification, and dependence in colon cancer. We found PRPF6 to be overexpressed in a subset of primary and metastatic colon cancers (Supplemental Fig. 2A). PRPF6 overexpression in colon cancer was confirmed by immunohistochemistry (IHC) and positively correlated to PRPF6 CNG (Supplemental Fig. 2B–E). Consistent with this correlation in colon tumors, colon cancer cell lines with PRPF6 CNG also exhibited higher expression of the PRPF6 protein (Fig. 1B; Supplemental Fig. 2F). Mechanisms other than CNG must contribute to increased PRPF6 expression, as PRPF6 protein was also elevated in four cell lines with disomy at the PRPF6 locus (Supplemental Fig. 2F). In nearly all cell lines tested, loss of PRPF6 specifically inhibited the growth of colon cancer cell lines with high levels of PRPF6 protein (Fig. 1B,C). Equivalent protein knockdown was achieved in both high- and low-PRPF6-expressing cell lines (Supplemental Fig. 3A,B). To validate the specificity of these effects, we generated and tested multiple doxycycline-inducible lentiviral PRPF6 shRNAs. We found that only those shRNAs that reduced PRPF6 protein inhibited cell growth (Supplemental Fig. 3C). Importantly, restoration of PRPF6 by adenovirus-mediated expression efficiently rescued the cell growth defect in a PRPF6-high-expressing cell line (Fig. 1D). To characterize the effect of acute loss of PRPF6 on tumor growth in vivo, we used an inducible shRNA system to deplete PRPF6 in implanted tumors (Adler et al. 2012). Consistent with cell growth defects seen in vitro after PRPF6 loss, doxycycline-induced acute knockdown of PRPF6 in fully formed xenografts led to a significant shrinkage only in tumor models that show evidence of high PRPF6 expression (Fig. 1E–G). Together, these data identify PRPF6 as an important regulator of growth in colon cancer.
RNAi-mediated depletion of tri-snRNP components leads to selective growth defects in PRPF6-high colon cancer cells
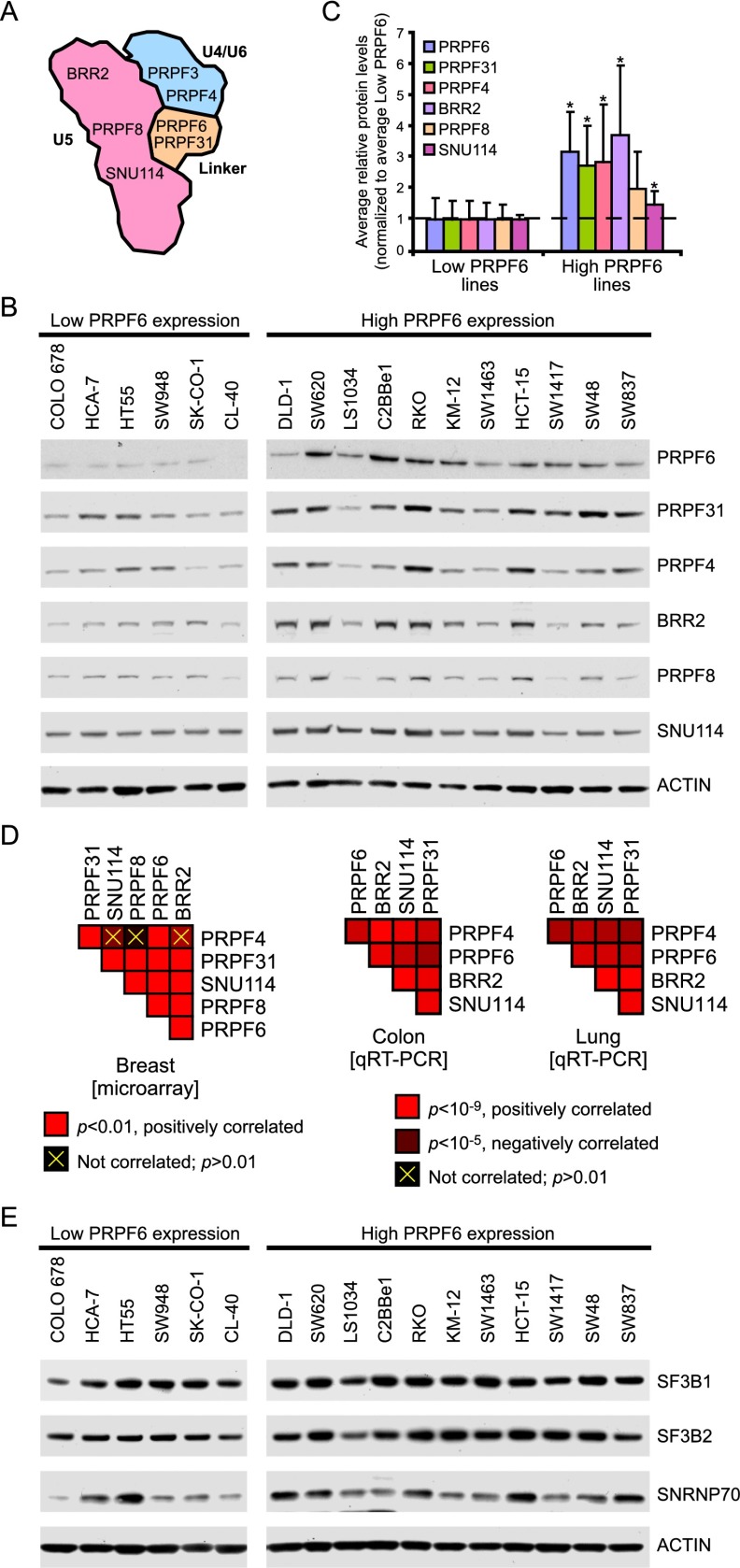
Intron splicing occurs through a series of coordinated steps mediated by multimeric snRNP complexes (U1, U2, U4, U5, and U6) collectively termed the major spliceosome (Wahl et al. 2009). PRPF6 is thought to act as a molecular bridge linking the U5 and U4/U6 proteins to form the tri-snRNP complex (Fig. 2A; Makarov et al. 2000). To determine whether other tri-snRNP components are also implicated in colon cancer growth, we characterized the expression patterns of multiple tri-snRNP proteins in cancer. We found that tri-snRNP protein expression was coordinately increased in colon cancer cell lines that exhibit high levels of PRPF6 (Fig. 2B,C). Similarly, gene expression and copy number analysis by either microarray or quantitative PCR (qPCR) in different tumor types (colon, lung, and breast) showed that multiple tri-snRNP components were significantly coexpressed or coamplified (Fig. 2D; data not shown). No consistent expression differences between PRPF6-high- and PRPF6-low-expressing cancers were seen for non-tri-snRNP spliceosome components, suggesting that coexpression is specific to the tri-snRNP complex components (Fig. 2E). To examine whether PRPF6 loss directly affects tri-snRNP expression, we depleted PRPF6 and examined individual tri-snRNP components before cell growth defects were observed (3 d after PRPF6 knockdown). We found that PRPF6 knockdown led to reduced expression of other tri-snRNP proteins as well as their mislocalization to Cajal bodies (Supplemental Fig. 4). The Cajal body phenotype has been previously observed in other cell lines (Schaffert et al. 2004) and is consistent with tri-snRNP disruption.
Figure 2.
The tri-snRNP complex is coordinately overexpressed in colon cancer. (A) A schematic of the tri-snRNP complex with select components indicated. Adapted by permission from Macmillan Publishers Ltd.: Nature Structural Biology (Häcker et al. 2008), © 2008. (B) Immunoblot of several tri-snRNP complex components in low- and high-PRPF6-expressing cell lines. The PRPF6 blot from Figure 1B that was used to classify each cell line as having high or low PRPF6 expression is shown here for reference. (C) The level of each protein from the immunoblot shown in B was quantified relative to ACTIN, and the mean ± SD protein level for each group of cell lines is provided. The values were normalized to the average expression of the low-PRPF6-expressing cell lines. (*) P < 0.05, Student’s t-test. (D) Gene expression data from microarray (left) or qRT–PCR (right) for the indicated tri-snRNP complex genes were correlated to each other in the indicated tumor types using Pearson correlation across all tumor samples. Red boxes indicate that the two genes are positively correlated according to the P-value scales provided. Correlations that are not significant are crossed out in yellow (P > 0.01). P-values are one-tailed t-tests. (E) Immunoblot of spliceosome components in low- and high-PRPF6-expressing cell lines.
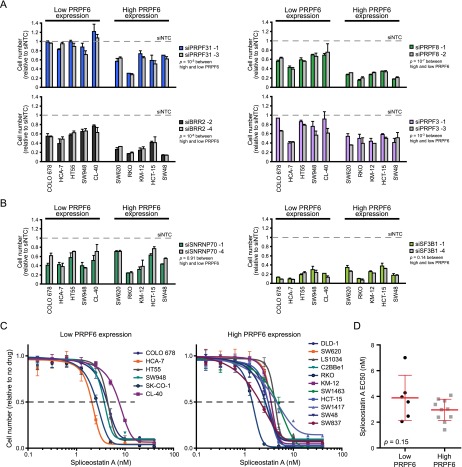
We then characterized the phenotypic effect of inhibiting tri-snRNP complex proteins in PRPF6-high versus PRPF6-low cell lines. Knockdown of the tri-snRNP protein PRPF31, PRPF8, BRR2, or PRPF3 in colon cancer cells led to reduced growth specifically in cell lines that expressed high levels of PRPF6 (Fig. 3A). Next, we tested the effect of PRPF6 modulation in human primary colon cells. We note that while the growth effect was not as robust as seen in PRPF6-high cell lines, PRPF6 inhibition reduced cell growth by ∼40% in primary colonic cells (Supplemental Fig. 3D). To assess whether PRPF6-high cell lines are generally more sensitive to impairment of the splicing machinery, we inhibited components of the U1 (SNRNP70) and U2 (SF3B1) spliceosome by RNAi-mediated depletion. We found that both PRPF6-high and PRPF6-low cell lines were equally sensitive to SNRNP70 and SF3B1 inhibition (Fig. 3B). Efficient knockdown was confirmed for the RNAi conditions tested (Supplemental Fig. 3E). Knockdown of PLK1, a ubiquitously expressed cell cycle kinase, led to reduced growth in PRPF6-high and PRPF6-low cell lines, indicating that both groups of cell lines are equally sensitive to a more generic proliferation inhibitor (Supplemental Fig. 3F). Similar results were obtained with a larger panel of cell lines using spliceostatin A, a small molecule inhibitor of SF3B1 (Fig. 3C,D; Kaida et al. 2007). These observations highlight a selective requirement for components of the tri-snRNP complex in regulating growth of cancers in which they are amplified or overexpressed.
Figure 3.
The tri-snRNP complex is required for proliferation in colon cancer. (A,B) Relative cell number (compared with siNTC; mean ± SD) is shown for two independent siRNAs 6 d after siRNA transfection in the indicated cell lines. P-values were calculated between each group of cell lines; Student’s t-test. (C) Spliceostatin A was added in twofold dilutions at the indicated final concentration to low- or high-PRPF6-expressing cell lines. The cell number (relative to no drug addition) 5 d after drug addition is provided. The data were fitted to a sigmoidal dose response curve, and the EC50 values were determined. (D) Mean ± SD EC50 values are shown for each group of cell lines from C. P-value is a Student’s t-test.
Haploinsufficiency of PRPF genes, leading to impaired tri-snRNP activity, can be generally tolerated at the organismal level (Yin et al. 2011); however, complete ablation results in embryonic lethality (Graziotto et al. 2008; Bujakowska et al. 2009). To further characterize the role of PRPF6 in cancer cells, we used the CRISPR system to generate and characterize knockouts of PRPF6 and other tri-snRNP components (Cong et al. 2013; Mali et al. 2013). To do so, we stably expressed the CAS9 nuclease in three PRPF6-high (RKO, SW620, and SW48) and three PRPF6-low (HT55, COLO 678, and HCA-7) cell lines. Two independent single guide RNAs (sgRNAs) targeting PRPF6 and two other tri-snRNP proteins, BRR2 and PRPF8, were introduced, and cells were monitored for cell viability. We found that individual deletion of all three tri-snRNP genes led to consistent cell viability defects in both PRPF6-high and PRPF6-low cell lines. Immunoblot analysis confirmed efficient reduction in protein expression in PRPF6-high and PRPF6-low cell lines for all sgRNAs tested (Supplemental Fig. 5A–C). To generate and characterize cells that are null for tri-snRNP components, we transduced HT55-CAS9 cells with PRPF6 or BRR2 sgRNAs and isolated single-cell clones after selection for sgRNA expression. PCR analysis of genomic DNA from 18 PRPF6 sgRNA and 23 BRR2 sgRNA clones revealed wild-type or heterozygous alleles but no homozygous deletions in the targeted PRPF6 and BRR2 loci, respectively (Supplemental Fig. 5D,E). Immunoblot analysis showed PRPF6 and BRR2 expression in all clones, albeit at lower levels in heterozygous cells. PCR and immunoblot analyses of representative wild-type and heterozygous clones are shown in Supplemental Figure 5, D and E. These results suggest that while complete depletion of tri-snRNP components is systematically deleterious, as would be expected for essential spliceosome components, impaired activity of PRPF6 and other tri-snRNP proteins (vis a vis RNAi) can be tolerated in some cancer cell lines but not others.
PRPF6 is essential for splicing of a small subset of genes in colon cancer
Recent studies have described mutations in spliceosomal components and suggest an emerging role for splicing regulation in human disease (Ebert and Bernard 2011; Wang et al. 2011; Yoshida et al. 2011; Graubert et al. 2012 ; Hahn and Scott 2012; Imielinski et al. 2012; Quesada et al. 2012). A mutation in PRPF6 (PRPF6R729W) in its C-terminal tetraco-peptide repeat (TPR) domain has been reported to impair tri-snRNP assembly and function and lead to a heritable form of RP (Tanackovic et al. 2011). To assess whether PRPF6-mediated growth in cancer requires its splicing activity, we tested the ability of PRPF6 mutants that either harbor the PRPF6R729W RP mutation or lack the TPR domain to restore growth after PRPF6 depletion. We found that both mutants were unable to restore cell growth (Fig. 4A), implying an important role for PRPF6-mediated splicing in cancer growth.
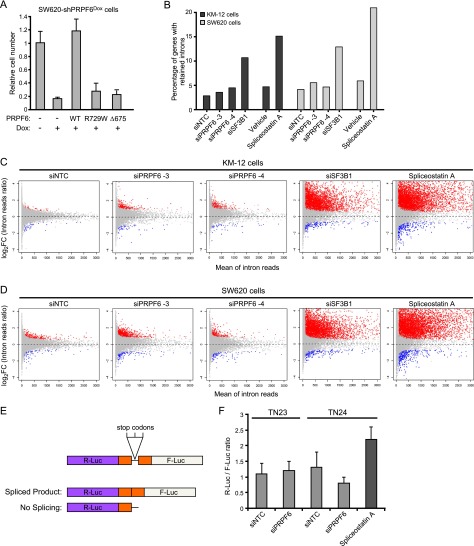
Figure 4.
PRPF6 inhibition leads to intron retention in a small subset of genes. (A) The bar graph depicts cell number relative to nontreated control in SW620-shPRPF6Dox cells. Doxycycline-induced PRPF6 knockdown is indicated. Adenoviruses expressing wild-type or different mutant forms of shRNA-resistant PRPF6 are indicated. (B) The bar graph shows percentage of intron reads over total uniquely mapped reads based on RNA-seq data. Each bar represents the average percentage of two or three replicates. (C,D) The scatter plot shows the log2 fold change of normalized intron reads as a function of average intron read count for all genes in KM-12 cells (C) and SW620 cells (D). The normalized intron reads ratio is treatment/control (three replicates per group; control is no treatment). The mean intron reads is the average of all intron reads within the annotated intron regions for a given gene across all samples in the comparison (the three control samples and the three treatment samples). The genes with significant intron retention are shown in red, while genes with significant decreased intron reads are shown in blue. (E) Schematic representation of splicing dual-reporter assay that mimics endogenous splicing by insertion of recombinant gene fragments flanked by an intronic region with stop codons. (F) The bar graph depicts splicing activity as measured by the Renilla (R-Luc) to firefly (F-Luc) luciferase ratio 3 d after transfection of the indicated siRNAs along with two splicing reporters, TN23 and TN24. Spliceostatin A was incubated with siNTC-treated cells for 8 h and was used as a control. Representative results are shown for one of three independent experiments (mean ± SD).
Alterations to the splicing machinery in cancer have recently been shown to lead to broad changes in constitutive splicing and intronic retention events (Yoshida et al. 2011). To further investigate PRPF6-mediated splicing in cancer, we used RNA sequencing (RNA-seq) and exon microarrays to characterize the PRPF6-regulated spliceome in colon cancer. To do this, we depleted PRPF6 and collected RNA shortly after initiation of knockdown. We confirmed that equivalent levels of PRPF6 knockdown were achieved in all cell lines and that the independent siRNAs used exhibited high correlation (Supplemental Fig. 6). We found that PRPF6 inhibition led to intronic retention in a relatively small subset of genes (Fig. 4B–D). In contrast, U2 spliceosome inhibition by either spliceostatin A or SF3B1 depletion led to global changes in intron retention of thousands of genes in the human transcriptome. Consistent with these observations, spliceostatin A-mediated SF3B1 inhibition, but not PRPF6 depletion, affected splicing in a generic splicing reporter assay (Fig. 4E,F; Nasim and Eperon 2006). These data indicate that the consequences of inhibiting the splicing machinery can be either broad or more gene-specific depending on the spliceosome component that is targeted.
Alternative splicing of an oncogenic form of the ZAK kinase is regulated by PRPF6
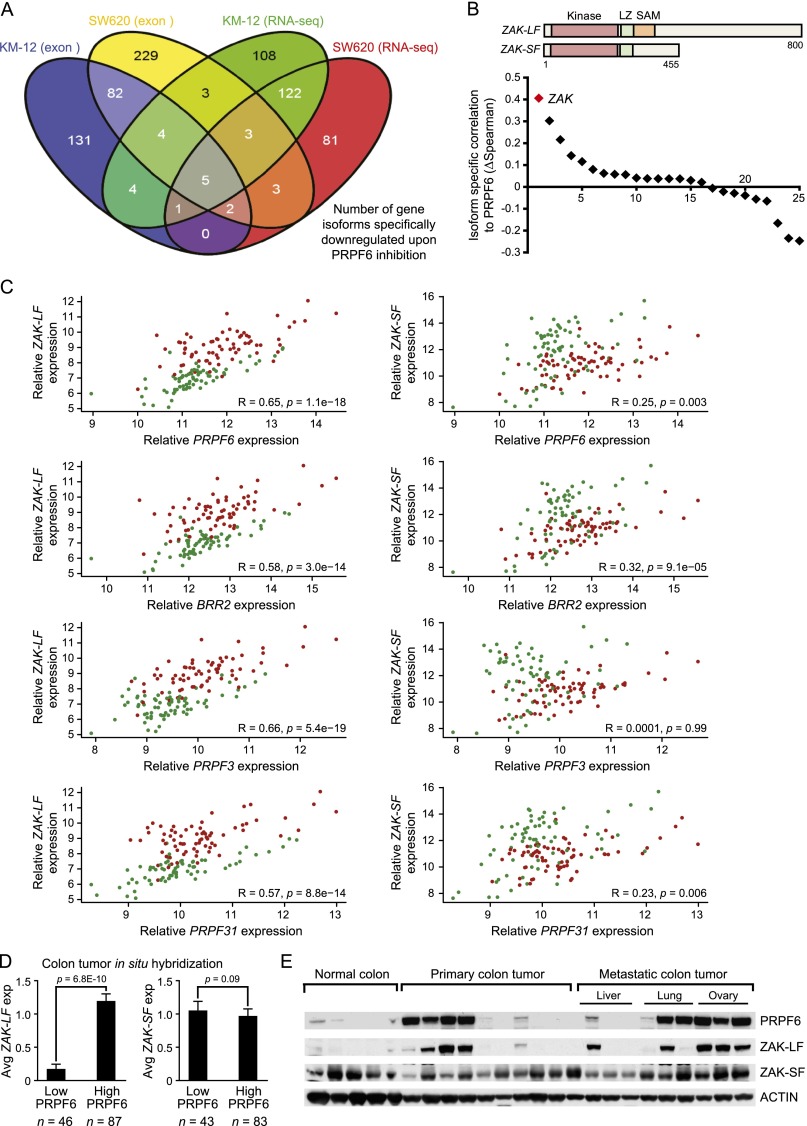
Alternative RNA splicing is known to generate oncogenic variants with cancer-specific functions (David and Manley 2010). To characterize alternative splicing events that are sensitive to PRPF6 loss, we analyzed gene transcript variant changes by both RNA-seq and exon microarray in two PRPF6-high cell lines: KM-12 and SW620. After compiling the two data sets, we found 25 gene isoforms that were specifically down-regulated after PRPF6 inhibition (Fig. 5A; Supplemental Table 5). We used the Homer algorithm (Heinz et al. 2010) to search for sequence motifs in the 25 genes displaying intron retention upon PRPF6 inhibition but did not find any significant enrichment. To identify genes with potential biological relevance, we performed Spearman correlation analysis in a RNA-seq data set of colon cancer and normal tissue samples (Seshagiri et al. 2012), comparing PRPF6 levels with the isoform-specific expression of each transcript (Supplemental Table 6). Candidate genes were then ranked based on the difference of Spearman correlation coefficient (ΔSpearman) between the perturbed and nonperturbed isoform per given gene. From this analysis, we identified the long form of the ZAK kinase (ZAK-LF, NM_016653) as correlating highly with PRPF6 expression as compared with the ZAK short form (ZAK-SF, NM_133646) (Fig. 5B). We found that ZAK-LF RNA levels specifically correlated with not only PRPF6 but also expression of multiple tri-snRNP complex components when compared with ZAK-SF (Fig. 5C). To examine ZAK isoform expression more directly, we performed in situ hybridization (ISH) analysis on colonic tissues using ZAK isoform-specific probes. We found that the ZAK-LF was specifically up-regulated in colon cancer, while ZAK-SF was expressed in both normal colon and colon cancer samples (Supplemental Fig. 7). Next, we correlated ZAK mRNA expression with PRPF6 protein expression using a tissue microarray of 133 colon cancer specimens that were probed for ZAK mRNA (by ISH) and PRPF6 expression (by IHC). Consistent with our RNA-seq analysis, the ZAK-LF isoform, but not ZAK-SF, was highly correlated to PRPF6 expression (Fig. 5D). These findings were confirmed at the protein level, where ZAK-LF and PRPF6 were similarly expressed in normal colon tissue and colon cancer samples (Fig. 5E). These results illustrate the differential ZAK isoform usage in normal colon and colon cancer and suggest a relationship between ZAK-LF mRNA expression and PRPF6 levels.
Figure 5.
PRPF6 regulates splicing of the ZAK protein kinase. (A) Gene isoform usage changes are analyzed after PRPF6 loss. A four-way Venn diagram illustrates the number of genes that have significantly reduced isoform expression upon PRPF6 depletion. Criteria for gene selection are reduced isoform usage in exon microarray (adjusted P-value <0.05) and RNA-seq data (adjusted P-value ≤0.005 and mean read counts >50) using two independent siRNAs targeting PRPF6 in at least one of two cell lines tested (KM-12 and SW620). (B) The distribution plot ranks the degree of correlation between PRPF6 expression and the 25 gene isoforms down-regulated after PRPF6 loss (ΔSpearman = [Spearmandownregulated isoform − Spearmanunchanged isoform]). A schematic of the domains of the different isoforms of the top-scoring gene (ZAK) is shown. (LZ) Leucine zipper domain; (SAM) sterile α motif. (C) The scatter plot shows expression correlation of tri-snRNP genes PRPF6, BRR2, PRPF3, and PRPF31 to long (ZAK-LF; NM_016653) and short (ZAK-SF; NM_133646) transcript variants of the ZAK gene. Each dot represents one sample. Green and red circles denote normal colon and colon cancer, respectively. Spearman’s rank correlation coefficients (R-values) and associated P-values are shown in each plot. (D) The bar graph shows the correlation between PRPF6 protein (IHC) and ZAK mRNA isoform expression (ISH) in human colon cancer samples. PRPF6-high and PRPF6-low cutoffs are defined as tumors in which ≥30% or <30% of the cancer cells express PRPF6 protein, respectively. ZAK mRNA expression is scored on a qualitative scale. (0) No expression; (1) weak; (2) moderate; (3) strong staining. Sample size for each cohort is indicated below the bar graph. Mean ± SD is shown, and P-values are indicated (Student’s t-test). (E) Immunoblot showing PRPF6 and ZAK isoform expression in the indicated human colon specimens.
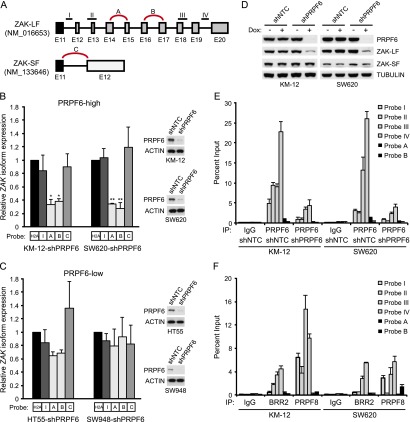
We next assessed whether PRPF6 regulates ZAK alternative splicing. To examine ZAK isoform usage in cell lines, we used TaqMan probes specific to the ZAK pre-mRNA and the two spliced ZAK isoforms ZAK-LF and ZAK-SF (Fig. 6A). We then tested the effect of acute PRPF6 knockdown on ZAK expression in both PRPF6-high (SW620 and KM-12) and PRPF6-low (HT55 and SW948) cell lines stably expressing a doxycycline-inducible PRPF6 shRNA. Depletion of PRPF6 led to a significant reduction in ZAK-LF levels in PRPF6-high cell lines (Fig. 6B,C). Importantly, both ZAK pre-mRNA and ZAK-SF mRNA levels were not significantly changed in any of the four cell lines, strongly suggesting that these effects are occurring at the post-transcriptional level. Immunoblot analysis confirmed that efficient PRPF6 knockdown was achieved in all four cell lines (Fig. 6B,C). Consistent with these findings, we found that PRPF6 depletion in PRPF6-high colon cancer cell lines specifically reduced ZAK-LF protein levels but not the ZAK-SF isoform (Fig. 6D). Based on these findings, we then tested whether PRPF6 directly associates with ZAK pre-mRNA. To examine PRPF6 binding to ZAK mRNA, RNA immunoprecipitation (RNA-IP) experiments were carried out in KM-12 and SW620 cells. We found strong enrichment of ZAK pre-mRNA in PRPF6 but not IgG immunoprecipitates (as detected with primers I–IV) (Fig. 6E). Importantly, the level of unspliced ZAK mRNA bound to PRPF6 was significantly reduced after PRPF6 knockdown (Fig. 6E). Consistent findings were also found using antibodies against the tri-snRNP components BRR2 and PRPF8 (Fig. 6F). Together, these data strongly suggest that PRPF6 and other tri-snRNP components regulate ZAK alternative splicing in a subset of cancer cells.
Figure 6.
PRPF6 regulates ZAK-LF alternative splicing and associates with unspliced ZAK mRNA. (A) The schematic cartoon shows the TaqMan probe locations that were used to detect ZAK spliced (A–C) and unspliced (I–IV) mRNA species ([E] exon number). (B,C) qRT–PCR analysis of spliced ZAK isoforms and unspliced ZAK pre-mRNA after PRPF6 knockdown. RNA was isolated from the indicated cell lines 4 d after exposure to doxycycline to induce PRPF6 knockdown. Changes in ZAK mRNA species were quantitated by TaqMan qRT–PCR from doxycycline-treated or untreated cells and normalized to an intronless control gene, H2A. Primers were used as indicated in A. The bar graph shows mean ± SD. (*) P < 0.05; (**) P < 0.001 compared with ZAK pre-mRNA expression (probe I), Student’s t-test. (D) Immunoblot showing ZAK-LF and ZAK-SF expression changes 4 or 5 d after doxycycline-induced PRPF6 knockdown in SW620 and KM-12 cell lines. (E) RNA-IP analysis of PRPF6 binding to ZAK mRNA. RNA samples were purified from whole cellular lysates (input) or PRPF6 and IgG control immunoprecipitates in the indicated cell lines. ZAK mRNA was detected using TaqMan qRT–PCR with the indicated primers. The bar graphs depict the relative binding compared with input levels. Error bars are SDs from triplicate measurements. (F) RNA-IP analysis of BRR2 and PRPF8 binding to ZAK mRNA. Experiments were performed as in E.
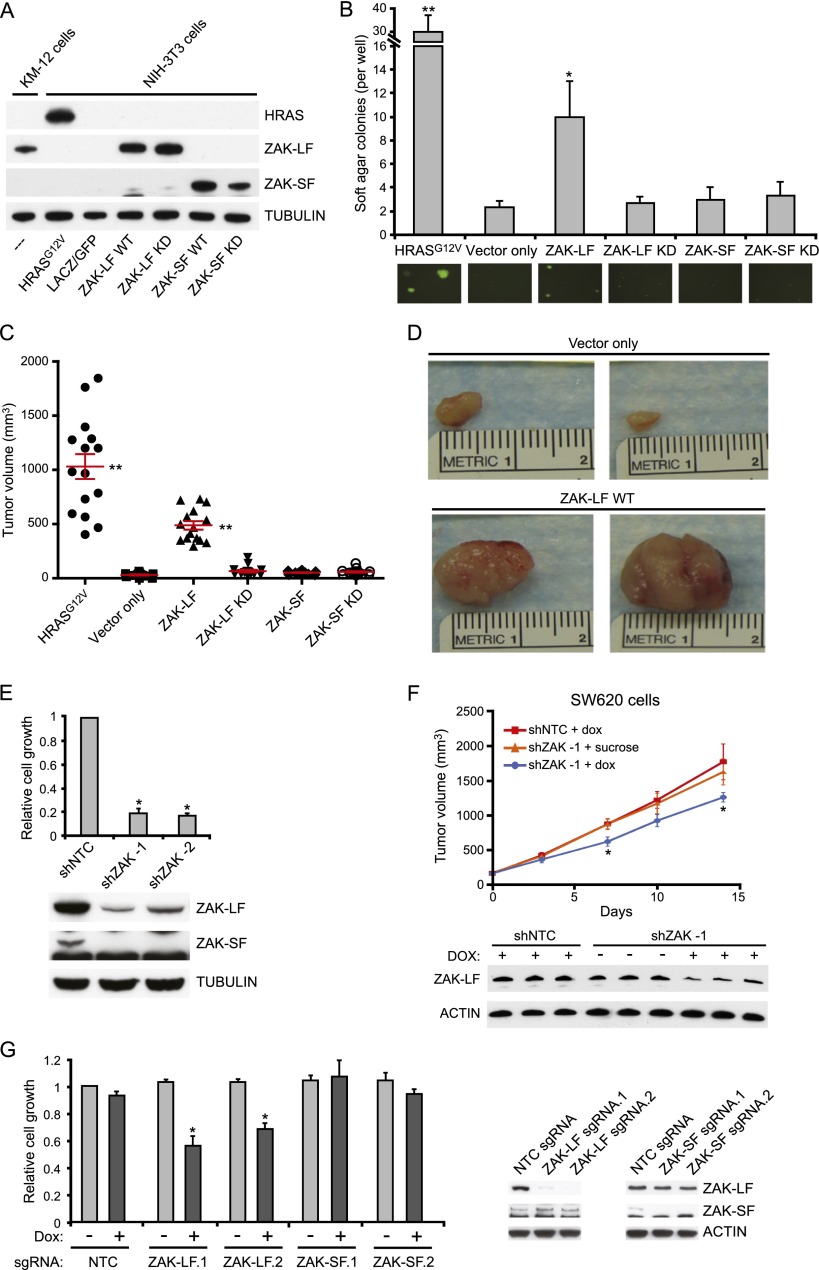
ZAK is a stress-activated protein kinase shown to activate the JNK/SAPK signaling pathway (Liu et al. 2000). Prior studies have suggested distinct functions for ZAK isoforms, with ZAK-LF being implicated in histone phosphorylation (Choi et al. 2005) and stress response (Gotoh et al. 2001). To address whether the ZAK isoforms have oncogenic activity, we overexpressed either wild-type or kinase-dead versions of ZAK-LF and ZAK-SF in immortalized murine fibroblasts (NIH-3T3) (Fig. 7A). ZAK-LF expression induced anchorage-independent colony growth and xenograft tumor formation in immunodeficient mice, whereas ZAK-SF or kinase-dead forms of either ZAK isoform failed to transform the cells (Fig. 7B–D). We then expressed the different ZAK isoforms in colonic cell lines that are resistant to PRPF6 knockdown (SW948 and SW1463) and assessed their growth rate or oncogenic activity. While SW948 and SW1463 proliferation was not affected by ZAK-LF expression, we detected a significant increase in the number of colonies in a focus formation assay in the SW948 cell line (Supplemental Fig. 8A,B). We also tested whether exogenous ZAK expression can compensate for PRPF6 knockdown effects. We found that ectopic expression of ZAK-LF or ZAK-SF did not restore cell growth after PRPF6 knockdown, implying that additional critical genes besides ZAK are necessary for PRPF6-mediated growth effects (Supplemental Fig. 8C).
Figure 7.
The PRPF6-regulated ZAK protein isoform is oncogenic. (A) Immunoblot showing expression levels of wild-type (WT) and kinase-dead (KD) ZAK variants in NIH-3T3 immortalized murine fibroblasts that were transduced with the indicated constructs. Expression of ZAK in untransfected KM-12 colon cancer cells is shown for reference. (B) The bar graph shows anchorage-independent growth in NIH-3T3 cells infected with the indicated lentiviral expression vectors. Representative photomicrographs show discrete colony formation in ZAK-LF (WT)- and HRAS-expressing NIH-3T3 cells (transduced cells express GFP). Experiments were performed in triplicate (mean ± SD is shown). (*) P < 0.01 compared with vector only; (**) P < 0.005 compared with vector only, Student’s t-test. (C) The graph shows the tumor volume of transduced NIH-3T3 cells grown as subcutaneous tumors in immunodeficient mice. Tumor size was measured 14 d after xenografting. Each point represents a single tumor measurement (n = 15 animals per group; mean ± SD is shown). (**) P < 0.0001 compared with vector only. (D) Representative photographic images of tumors are shown at day 14 after injection of NIH-3T3 cells transduced with either ZAK-LF wild-type or negative control vector. (E) The bar graph shows relative cell number (compared with shNTC; mean ± SD) for two independent shRNAs 7 d after shRNA infection in the KM-12 cell line. (*) P < 0.001 compared with shNTC, Student’s t-test. The immunoblot shows expression of ZAK-LF and ZAK-SF 4 d after shRNA infection. (F) Xenograft tumor volume measurements over time (n = 10–15 mice per group) in SW620 cells. Mean ± SEM is shown. (*) P < 0.05 compared with the shNTC + Dox control group; Student’s t-test. Immunoblot of ZAK-LF protein levels in xenografted tumors 7 d after knockdown induction is shown. (G) The bar graph shows relative cell number (compared with NTC sgRNA, no doxycycline; mean ± SD) for KM-12 cells in the absence or presence of doxycycline-inducible CAS9 expression. (*) P < 0.01 compared with the respective ZAK-LF sgRNA, no doxycycline, Student’s t-test. Immunoblot shows expression of ZAK-LF and ZAK-SF 7 d after induction of CAS9 and sgRNA expression.
While ZAK is not sufficient to compensate for PRPF6 inhibition, we explored whether it was necessary for colon cancer growth. We assessed the consequences of ZAK depletion in colon cancer cells marked by high PRPF6 levels (KM-12 and SW620). shRNAs that reduce both ZAK isoforms led to reduced growth in vitro in the PRPF6-dependent colon cancer cell line KM-12 (Fig. 7E). To assess the effect of ZAK RNAi-mediated inhibition in vivo, we induced acute knockdown of ZAK in fully formed SW620 xenografts (using the same vector system in Fig. 1E–G). We found that inhibition of ZAK expression by 50% was associated with a 39% reduction of tumor growth (based on area under the curve [AUC] measurement) as compared with the shNTC + Dox group in xenografted animals (Fig. 7F). To perform isoform-specific ZAK depletion, we used the CRISPR system to target genomic deletions in exons specific to either the ZAK-LF or ZAK-SF isoform; of note, multiple isoform-specific shRNAs to ZAK were tested but did not yield robust knockdown (data not shown). Two independent sgRNAs targeting ZAK-LF or ZAK-SF were introduced into an engineered KM-12 cell line that carries a doxycycline-inducible CAS9 gene. In the presence of CAS9 expression, we found that KM-12 cells were uniquely sensitive to loss of ZAK-LF but not ZAK-SF expression (Fig. 7G). Specific depletion of ZAK-LF and ZAK-SF by the respective sgRNAs was confirmed by immunoblot analysis. Together, these data show that a PRPF6-dependent ZAK isoform is required for cancer growth and has specific kinase-dependent oncogenic activity.
Discussion
In this study, we used an integrative genomic approach to comprehensively interrogate amplicon-specific genetic drivers of colon cancer. We identified a subset of cancers that are specifically dependent on increased expression of PRPF6 and other members of the tri-snRNP spliceosome complex. We show that PRPF6 inhibition alters the splicing of a discrete number of gene products that are important for tumor growth. Collectively, our results have important implications for understanding how regulation of splicing impinges on the growth of tumors and identifiy possible targets for future cancer therapeutics.
In recent years, components of the spliceosome machinery have been shown to be dysregulated in cancer. Overexpression of Sm domain spliceosome proteins has been found in a number of malignancies and defines a subset of cancers with aggressive features (Quidville et al. 2013). A number of studies have identified frequent and recurrent somatic mutations in spliceosome machinery components such as SF3B1, ZRSR2, U2AF1, and SRSF2 in multiple cancer types. Of particular interest is SF3B1, which is frequently mutated in myelodysplastic syndrome, chronic lymphocytic leukemia (Quesada et al. 2012), and uveal melanoma (Harbour et al. 2013; Martin et al. 2013). SF3B1 is a drug target for the small molecule spliceostatin A, generating hope that inhibiting SF3B1 may have therapeutic benefits. We found that, unlike PRPF6 inhibition, which leads to specific splicing alterations, loss of SF3B1 function by either siRNA knockdown or small molecule inhibition leads to global inhibition of splicing, with broad toxic effects to cells. While it remains to be determined whether SF3B1 mutant cancers display greater sensitivity to spliceostatin A, our findings suggest that targeting SF3B1 and other U2 complex components may offer a limited therapeutic window.
PRPF6 and other tri-snRNP components are essential for cell viability and vertebrate development (Urushiyama et al. 1997; Graziotto et al. 2008; Bujakowska et al. 2009). Our work similarly shows that complete ablation of tri-snRNP genes is not tolerated in both PRPF6-high and PRPF6-low cancer cells. Our finding that some cancer cells (i.e., PRPF6-amplified/overexpressing cell lines) are more acutely sensitive to PRPF6 inhibition (vis a vis RNAi knockdown) is therefore particularly interesting and suggests that impaired tri-snRNP activity is better tolerated by a subset of cancers with low tri-snRNP levels or activity. This suggestion is supported by the tissue-specific defects seen in several genetic syndromes that are characterized by tri-snRNP insufficiency but are compatible with life. adRP, spinal muscular atrophy (SMA), and mandibulofacial dysostosis with microcephaly syndrome (MFDM) are heritable genetic syndromes that stem from heterozygous mutations or deletions in the SMN gene (SMA) or discrete tri-snRNP factors (adRP and MFDM) and consequently lead to haploinsufficiency and impaired tri-snRNP function (Boulisfane et al. 2011; Tanackovic et al. 2011; Lines et al. 2012). Although these mutated splicing factors are ubiquitously expressed, defects are seen in specific cell types or tissues consistent with our data that some cells are more sensitive to perturbations in the tri-snRNP complex. Furthermore, consistent with our own spliceome analysis in PRPF6-depleted cancer cells, these patients have been found to harbor discrete splicing alterations in a relatively small number of genes (Tanackovic et al. 2011). Taken together, these data strongly suggest that while complete loss of PRPF6 is not compatible with viability, some cells and tissues are more sensitive to a state of reduced activity. Given the recent interest in spliceosome inhibitors for cancer therapy (Bonnal et al. 2012), our finding will have important implications for future drug development efforts in this field.
A number of studies have revealed that the expression of alternatively spliced gene isoforms can drive malignant phenotypes (Christofk et al. 2008; Poulikakos et al. 2011). Our study shows that increased PRPF6 expression is required for alternative splicing of ZAK-LF. Moreover, we show that the PRPF6-regulated ZAK transcript variant has kinase-dependent oncogenic properties. Previous studies have suggested that the long and short ZAK isoforms have distinct functions, with ZAK-LF implicated in histone phosphorylation and actin cytoskeletal organization (Gotoh et al. 2001). While we found that kinase activity is required for ZAK-mediated oncogenic transformation, the catalytic domain is retained in both ZAK-SF and ZAK-LF, suggesting that the difference in ZAK activity may be imparted through allosteric modulation of its kinase domain or interaction with different protein partners. Interestingly, ZAK has been shown to activate several cancer-related signaling pathways such as NF-κB, Wnt/β-catenin, and AP1 (Liu et al. 2000; Firestein et al. 2008). Because we found that ZAK was not sufficient to rescue the growth defects following PRPF6 inhibition, the effects of PRPF6 loss on cell growth likely involves a number of genes. We note that from our analysis of 25 PRPF6-regulated alternatively spliced isoforms, we found several genes with previously described roles in promoting cancer. This includes CAPRIN1 in osteosarcoma growth and metastasis (Sabile et al. 2013), RAB11FIP1/RCP in breast cancer growth (Zhang et al. 2009), and JAG2 in breast cancer metastasis (Xing et al. 2011). Future studies will be needed to further delineate the role of specific ZAK isoforms and other candidate PRPF6-regulated oncogenes in cancer signaling and tumorigenesis.
Together, these data suggest that PRPF6, an essential component of the tri-snRNP complex, regulates the splicing of discrete gene products, some of which are implicated in driving cancer phenotypes such as proliferation and oncogenic transformation. We propose that specific components of the spliceosome machinery that are dysregulated and necessary for cancer growth when overexpressed, such as the tri-snRNP complex, may be amenable to therapeutic inhibition.
Materials and methods
Cell line culturing
All cancer cell lines were grown in DMEM (high glucose), 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (Invitrogen). Primary human colon cells were obtained from Applied Biological Materials and grown in Prigrow III medium, 5% FBS, and 1% penicillin/streptomycin on collagen-coated plates.
RNAi viability screening
Identification of candidate targets to screen
Genes that were both amplified and overexpressed in colon cancer were picked to screen for viability in a panel of colon cancer cell lines. The most commonly amplified chromosomal arms in colon cancer are chromosomes 7, 8q, 13, and 20q (Martin et al. 2007; Firestein et al. 2008). These same regions were also found to be commonly amplified in colon cancer cell lines that were used for the viability screen (Martin et al. 2007). Starting with all genes on the indicated chromosomal regions, the genes that were overexpressed in human colon cancer as compared with normal colon were identified using the following microarray data sets with the indicated statistical cutoff: (1) 86 normal and 227 tumor/metastases (GeneLogic data on Affymetrix Human Genome U133A/B array; P < 0.01, Student’s t-test), (2) 79 normal and 154 tumor/metastases (GeneLogic data on Affymetrix Human Genome U133 Plus array; P < 0.01, Student’s t-test), (3) five normal and 100 tumor (Gene Expression Omnibus GSE5206 [Kaiser et al. 2007]; P < 0.05, Student’s t-test), and (4) 24 normal and 24 tumor (Gene Expression Omnibus GSE10950 [Jiang et al. 2008]; P < 0.05, Student’s t-test).
Genes that were expressed significantly higher in tumor versus normal in one or more of the four expression data sets were selected for the primary RNAi viability screen. The miRNA genes screened were those found within chromosome 7, 8q, 13, or 20q; no expression data were used to filter the miRNA genes. The final number of genes/miRNAs analyzed was 1257: chromosome 7 = 531 genes, chromosome 8q = 264 genes, chromosome 13 = 215 genes, and chromosome 20q = 247 genes.
Primary RNAi viability screen
The cell lines used for the RNAi screen are listed in Supplemental Figure 1 and Supplemental Table 1. Each cell line underwent extensive optimization to determine the best conditions to carry out siRNA transfection in a 384-well format. First, cells were plated at different densities, and their growth was observed in real time for 6 d using the IncuCyte live-cell imaging system (Essen Bioscience). The cell density that led to confluency at day 6 was selected. Next, negative (nontarget control; NTC) and positive (TOX and siPLK1) controls were transfected into each cell line using multiple lipids and different lipid amounts. The conditions that led to the strongest growth effects from addition of TOX and siPLK1, but had few toxic effects from the negative control, were selected.
For genes, Dharmacon siGENOME SMARTpool siRNAs were used at a 100 nM final concentration. For miRNAs, Dharmacon miRIDIAN miRNA Mimic or miRIDIAN miRNA hairpin inhibitors were used at a 100 nM final concentration. siRNA/miRNAs (referred to as siRNAs hereafter) were reverse-transfected in duplicate in Aurora Biotechnologies white 384-IQ SQ/EB 384-well assay plates. PBS was added to all wells of the evaporation barrier. Individual siRNAs were arrayed in 384-well deep well plates (Greiner), and then a master mix of lipid and Opti-MEM (Invitrogen) was added to each well and then arrayed to individual 384-well assay plates using a Mini-Oasis liquid-handling robot (Dynamic Devices). Next, >30 min after mixing lipid and siRNA, cells were added to each well using a Finstruments Multidrop dispenser (MTX Lab Systems). Cells were grown for 6 d, and the final cell number was measured with CellTiter-Glo (Promega).
Duplicate readings of the raw values for all cell lines were very similar (range of R2 values after comparing replicates: 0.73–0.95, average is 0.86). For each plate, all gene values were first normalized to the plate average: GeneNorm = GeneX/plate average. To normalize the different levels of growth effects seen across multiple cell lines, the cell number readings were then converted to Z-scores. The following calculation was completed for each cell line: (GeneNorm − mean All GeneNorm)/stdev All GeneNorm. Each of the duplicate readings was normalized independently and then averaged. The normalized, averaged data for all targets in the primary RNAi screen in 14 cell lines are in Supplemental Table 1.
For each gene, we obtained gene expression and copy number data, and for each miRNA, we obtained miRNA expression data (Supplemental Table 1). For subsequent analyses, we focused only on genes and not miRNAs for secondary screening. We used three different methods to filter the gene targets that would be used for a secondary RNAi screen. The results for each filtering method described below are in Supplemental Table 1; the final list of 184 genes that passed one or more filters described below is provided in Supplemental Table 2.
Correlation to gene expression. We identified genes where the cell growth defect was negatively correlated with its relative gene expression level across all 14 cell lines; i.e., if the gene is expressed higher, it should have a stronger growth defect when inhibited (negative Z-score). We considered only genes that had a growth defect in at least two cell lines (Z-score less than −0.5 in two or more cell lines). Next, a Pearson correlation was calculated between the Z-score representing cell number and relative log2 gene expression levels. Genes with a Pearson correlation less than −0.36 were selected for the secondary RNAi screen (P < 0.1, one-tailed t-test).
Correlation to copy number. We identified genes in which the cell growth defect was negatively correlated with its copy number across all 14 cell lines; i.e., if the gene has a higher copy number, it should have a stronger growth defect when inhibited (negative Z-score). We considered only genes that had a growth defect in at least two cell lines (Z-score less than −0.5 in two or more cell lines). Next, a Pearson correlation was calculated between the Z-score representing cell number and copy number levels. Genes with a Pearson correlation less than −0.36 were selected for the secondary RNAi screen (P < 0.1, one-tailed t-test).
Specific killer. We identified genes in which, although there was no correlation between cell growth and expression or copy number, they were strongly inhibitory for cell growth in many (but not all) cell lines when knocked down. We considered only genes that did not have strong growth defects in all cell lines (Z-score >0 in two or more cell lines). Next, the Z-scores across all cell lines were averaged. Genes with an average Z-score less than −0.68 were selected for the secondary RNAi screen.
Secondary RNAi viability screen
The same cell lines used for the primary RNAi screen were used in the secondary RNAi screen using the same transfection conditions. The individual Dharmacon siGENOME oligos that made up the siRNA pools were individually arrayed and used at a 25 nM final concentration. The siRNA reverse transfection and cell growth assay was completed exactly as described for the primary RNAi screen. Note that two of the 14 cell lines were removed from further analysis due to technical issues: For LoVo cells, the negative control had a strong cell growth defect across all assay plates; for COLO 741 cells, none of the individual siRNA oligos had an effect on cell growth.
Because this list of genes was biased toward identifying the targets that had cell growth effects across multiple cell lines, we decided against a Z-score analysis, since Z-scores are preferred for data sets in which only a small percentage of targets are expected to have an effect. Instead, all values were normalized to the average of the NTCs on each plate (average siNTC was set to 1), and then duplicate values for each cell line were averaged. The normalized, averaged data for all targets in the secondary RNAi screen for 12 cell lines are in Supplemental Table 2.
To filter the secondary gene targets, we used the same three methods described above for the primary RNAi screen with minor modification described below. The 17 genes that passed these filters are provided in Supplemental Table 3.
Additional analysis of top 17 RNAi targets
Several criteria were analyzed for the top 17 targets identified from the secondary RNAi screen (Supplemental Table 3). First, antibodies were identified that could detect each endogenous protein by immunoblotting, and then a protein was considered to be correlated to its RNA expression if at least half of the cell lines showed concordance of their protein and RNA expression levels. Antibody specificity was confirmed by detecting the respective overexpressed protein in 293 cells (data not shown). Second, it was determined whether any of these genes were significantly overexpressed in multiple mouse colon cancer models: in the ApcLoxp/+; CDX2-Cre mouse model of colon cancer (Hinoi et al. 2007) compared with normal colon tissue (P < 10−6, Student’s t-test) or in ApcMin or ApcMin; KrasG12D; Vil-Cre small intestine tumors compared with normal tissue (P < 0.05, Student’s t-test). Only genes expressed at least 1.4-fold higher in the cancer tissues compared with normal were considered significantly up-regulated. Third, 122 primary colon cancers were analyzed to determine whether the expression of each gene was directly correlated with its copy number (P < 10−4, one-sided t-test; The Cancer Genome Atlas Data Portal, http://tcga-portal.nci.nih.gov). Last, the same 122 tumors were analyzed for high-level amplification of each gene: More than 20% patients with three or more gene copies was considered high-level amplification.
siRNA, shRNA, and sgRNA experiments
For RNAi of individual genes for cell growth or immunoblot analysis, individual siRNAs were reverse-transfected in 96-well format using Dharmafect 4 transfection reagent. For RNAi of individual genes for total RNA harvesting, individual siRNAs were reverse-transfected in six-well format using Dharmafect 4 transfection reagent.
For in vitro shRNA experiments, pLKO vectors containing the respective shRNA sequences were obtained from Sigma. Lentivirus production and transduction of cells were carried out as previously described (Adler et al. 2012). For in vivo shRNA experiments, a doxycycline-inducible shRNA against PRPF6 was constructed using the lentivirus pHush-shRNA system (Gray et al. 2007) as described (Adler et al. 2012). For rescue experiments, site-directed mutagenesis was used to generate PRPF6 mutants that were resistant to shRNA-mediated knockdown and contained the R729W mutation (PRPF6R729W) or encoded a premature stop codon at amino acid 675 (PRPF6Δ675). PRPF6 constructs were inserted into the pAd/CMV/V5-DEST adenoviral vector through gateway recombineering and expressed per the manufacturer’s instructions (Invitrogen). Adenoviral particles were titered using the Adeno-X rapid titering kit (Clontech) on 293 cells.
For CAS9/sgRNA experiments, the CAS9 gene was cloned into the pInducer20 lentiviral vector (Meerbrey et al. 2011) or pLenti6.3 (Invitrogen) and stably integrated into cell lines through lentiviral transduction and neomycin or blasticidin selection, respectively. sgRNA sequences were identified using the CRISPR design program (http://crispr.mit.edu) and cloned into the pLKO lentiviral plasmid as KpnI/EcoRI fragments. A sgRNA targeting the firefly luciferase gene was used as a negative control. sgRNAs were introduced into cell lines by lentiviral transduction and puromycin selection.
Spliceostatin A treatment
Spliceostatin A is a general inhibitor of splicing that acts by binding to the SF3B complex of the spliceosome (Kaida et al. 2007; Lo et al. 2007). We kindly received an aliquot of spliceostatin A from Minoru Yoshida (RIKEN). To determine the effect of spliceostatin A treatment on colon cancer growth, colon cancer cell lines were first plated in 96-well format. Spliceostatin A was provided at 100 μg/mL solution in methanol (∼190 μM). The drug was diluted in PBS and added to cells in twofold dilutions ranging from 40 nM to 0.16 nM final concentration in triplicate wells. Five days later, the final cell number was measured with CellTiter-Glo (Promega) and normalized to cells without drug addition (the small amount of methanol added to the cells did not affect cell growth) (data not shown). The half-maximal effective concentration (EC50) for each cell line was determined by fitting the data to a sigmoidal dose response curve using GraphPad Prism. For RNA-seq analysis and splicing reporter assays, cells were incubated for 8 h with spliceostatin A at a final concentration of 100 nM.
Antibodies
The following antibodies were used: ACTIN (clone C4, MP Biomedicals), PRPF6 (Bethyl Laboratories), PRPF31 (clone 8E1, Abnova), PRPF4 (Abnova), BRR2 (Sigma), PRPF8 (ProteinTech), SNU114 (Novus), PRPF3 (Sigma), SF3B1 (Bethyl Laboratories), SF3B2 (Bethyl Laboratories), SNRNP70 (Sigma), Alexa Fluor 488 donkey anti-rabbit IgG (Invitrogen), and Alexa Fluor 568 donkey anti-mouse IgG (Invitrogen).
Histology, IHC, and ISH analyses
Hematoxylin and eosin and IHC analyses were performed by a board-certified pathologist (R. Firestein). IHC was performed on 4-µm-thick FFPE tissue sections mounted on glass slides. All IHC steps were carried out on the Ventana Discovery XT autostainer (Ventana Medical Systems). Pretreatment was done with Cell Conditioner 1 with the standard time. PRPF6 primary antibody was used at 2 μg/mL. Slides were incubated with primary antibody for 60 min at 37°C. Ventana Rabbit OmniMap was used as the detection system. Ventana DAB and Hematoxylin II were used for chromogenic detection and counterstain. For each sample, the H-score was determined by the following equation: (3 × percentage of strongly staining nuclei) + (2 × percentage of moderately staining nuclei) + (percentage of weakly staining nuclei) (the scale is 0–300). Immunofluorescence was carried out as described (Adler et al. 2012).
Nonisotopic ISH was performed on 4-µm FFPE sections using QuantiGene ViewRNA ISH tissue assay (Affymetrix/Panomics) following the manufacturer’s protocol on a Tecan platform equipped to carry out nonisotopic ISH. Gene-specific probe sets for detection of human ZAK isoform 1 mRNA (VA1-15607) and ZAK isoform 2 mRNA (VA1-15608)—target regions 1190–2158 and 1626–2774, respectively in GenBank accessions NM_016653 and NM_133646—were used on tissue samples. A probe set to Bacillus subtilis dihydropicolinate reductase (dapB) (VF1-11712)—target region 1363–2044 in GenBank accession L38424—was used as a negative control. Horseradish peroxidase (HRP)-conjugated label probe was used, followed by TSA (tyramide signal amplification) to increase sensitivity (Perkin Elmer). Briefly, TSA Plus DIG stock solution (digoxigenin) was diluted 1:50 in 1× Plus Amplification Diluent, applied to sections, and incubated for 10 min at room temperature. This was followed by incubation with anti-DIG-AP (Roche) diluted 1:500 in TNB blocking buffer with 4% lamb serum (Gibco) for 30 min at room temperature. Vulcan Fast Red substrate (Biocare) was used for chromogenic detection.
Focus formation, soft agar colony formation, and xenograft studies
Focus formation, soft agar, and xenograft tumor growth studies were carried out as described (Firestein et al. 2008; Adler et al. 2012). For the xenograft studies, cells stably expressing doxycycline-inducible shRNAs were injected subcutaneously into the flank area of mice. All sucrose and doxycycline treatments were delivered via water bottles. Kinase-dead forms of ZAK were introduced by site-directed mutagenesis and confirmed by sequencing as previously described (Gotoh et al. 2001).
Human tissue samples
Normal human colon, colon tumors, and metastatic colon tumors were obtained from Asterand, Integrated Laboratory Services, Cooperative Human Tissue Network, or ProteoGenex. Each tumor was verified to contain a high percentage of tumor cells by a board-certified pathologist (R. Firestein).
Gene expression and copy number analysis
For qRT–PCR experiments, total RNA was isolated with the RNeasy minikit (Qiagen). Reverse transcription followed by qPCR was performed with the TaqMan one-step RT–PCR master mix (Applied Biosystems). Samples were normalized to expression of GUSB or the H2A gene.
For quantitative copy number analysis, total DNA was isolated with the DNeasy blood and tissue kit (Qiagen). qPCR for PRPF6 copy number was performed with the TaqMan genotyping master mix using TaqMan copy number probes (Applied Biosystems). For colon cell lines and tumors, samples were normalized to copy number at the TERT locus, which is rarely amplified or deleted in colon cancer (Tumorscape, http://www.broadinstitute.org/tumorscape); lung tumor samples were normalized to copy number at the RPL15 locus, which has a stable copy number in lung cancer.
RNA-IP and TaqMan PCR
RNA-IPs were performed using the EZ-Magna RIP immunoprecipitation kit according to the manufacturer’s protocol (Millipore). Briefly, cells were harvested 4 d after induction with doxycycline or no treatment control and subjected to immunoprecipitation with PRPF6, BRR2, PRPF8, or rabbit IgG antibody. RNA was isolated from total cell extract (input) as well as the immunoprecipitate reactions. qRT–PCR was performed using TaqMan probes (Hs00370456_m1, Hs00928997_m1, and Hs00928996_m1, Applied Biosystems) targeting specific ZAK isoforms.
Microarray hybridization
For cell lines used in the primary RNAi screen, total RNA was extracted from duplicate samples using the mirVana miRNA isolation kit (Ambion). Genomic DNA was harvested from cells using the DNeasy blood and tissue kit with RNase treatment (Qiagen). RNA and DNA were quantified using UV-spec Nanodrop (Thermo Scientific) and then profiled on a 2100 Bioanalyzer (Agilent). RNA samples were labeled and hybridized to human gene expression 4x44K or miRNA expression 8x15K (release 12.0) arrays according to the manufacturer’s protocol (Agilent). For gene expression, Cy5-labeled test samples were hybridized against Cy3-labeled universal human reference RNA (Stratagene). DNA samples were hybridized to human 1M CGH arrays (Agilent). Following hybridization, the arrays were washed, dried, and scanned on Agilent’s scanner. Agilent’s Feature Extraction software was used to analyze array images.
For ApcMin and ApcMin; KrasG12D; Vil-Cre samples, RNA was isolated from pooled small intestinal tumors and matched normal murine intestine. For ApcLoxp/+; CDX2-Cre samples, colon tumor and normal colon tissue of 265- to 360-d-old mice were flash-frozen upon removal. Total RNA and DNA were extracted using the AllPrep DNA/RNA minikit (Qiagen). RNA and DNA samples were labeled and hybridized to mouse gene expression 4x44K and mouse 1M CGH arrays, respectively, according to the manufacturer’s instructions (Agilent). Following array hybridization, washing, and scanning, analysis of array images was done using Agilent’s Feature Extraction software.
For siPRPF6 samples, total RNA was harvested in triplicate using the RNeasy minikit with on-column DNase digestion (Qiagen) 3 d after siRNA transfection. RNA was labeled and hybridized to G3 human exon 2x400K arrays as described in the manufacturer’s hybridization kit (Agilent). All samples were labeled with Cy5 and hybridized against Cy3-labeled universal human reference (Stratagene). Following hybridization, the arrays were washed, scanned, and analyzed with Agilent’s Feature Extraction software. Gene expression data are available at Gene Expression Omnibus (accession no. GSE56846).
RNA-seq and analysis
Total RNA from the siPRPF6 samples used for the exon microarray analysis was subjected to oligo (dT) capture and enrichment, and the resulting mRNA fraction was used to construct complementary DNA libraries. Transcriptome sequencing (RNA-seq) was performed on the Illumina HiSeq platform using the standard paired-end protocol. Each group had two or three biological replicates. In total, 28 million to 125 million 75- to 100-base-pair (bp) reads were generated per sample. The RNA-seq reads were then mapped to the human genome (NCBI build 37) by using GSNAP (Wu and Nacu 2010), allowing a maximum of three mismatches per 75-bp sequencing end, where 65%–83% of reads were uniquely mapped to the human reference genome. To quantify the gene expression, isoform-specific expression, and intron retention level, the number of reads mapping to exons, introns, and specific isoforms of each RefSeq gene was summarized, and the mapped reads for each gene were normalized by the sequencing depth of each sample. The differential gene expression and intron retention analyses were performed with DESeq (Anders and Huber 2010), which included normalizing the sequence reads by the effective library size following an inherent protocol in DESeq. Isoform-specific expression was analyzed with DEXseq (Anders et al. 2012). Both of these programs are based on a negative binomial distribution model. Spearman’s rank correlations between expression values of ZAK isoforms and tri-snRNP components were calculated. RNA-seq data are publically available (European Genome–phenome Archive [EGA], accession no. EGAS0000100076).
Exon microarray transcriptome data analysis
Analysis of exon microarray data was performed with GeneSpring GX 12 (Agilent). GeneSpring gene level analysis was used to identify differentially expressed genes between siNTC and each individual siPRPF6 for each cell line. Genes were considered differentially expressed if the expression of each gene in siPRPF6 samples was >1.3-fold increased or decreased compared with siNTC in each cell line and significantly different between siNTC and the siPRPF6 samples (P < 0.05, Student’s t-test with Benjamini-Hochberg multiple testing correction). The significance of gene overlap between the cell lines was determined with a hypergeometric distribution.
For the heat map of gene expression changes, the gene expression data were first exported from GeneSpring. Before exporting, the data were log2-transformed and normalized to the median expression of the respective siNTC replicates for each cell line. Only genes that were significantly changing in both KM-12 and SW620 cells are shown.
To determine whether the two independent PRPF6 siRNAs had similar changes, Pearson correlations were carried out genome-wide on the exported gene expression data. Every pairwise combination of each siRNA (siNTC, siPRPF6-3, and siPRPF6-4) was compared. For each pair-wide analysis, each microarray replicate was compared with each other (three replicates for each siRNA equals nine correlations for each siRNA pairwise comparison). The nine Pearson correlations values were averaged. Positive correlations indicate that the siRNAs are targeting similar genes, while correlations near 0 indicate a lack of correlation between siRNAs.
GeneSpring exon level analysis was used to identify alternatively spliced genes between siNTC and each individual siPRPF6 for each cell line. The splicing index, which is the log2 fold change of individual exon expression between samples that are being compared, was first used to select genes that have one or more exons that are differentially expressed between siNTC and siPRPF6 samples (splicing index >1 or less than −1). Next, t-tests were used to identify the changes that were significantly different (P < 0.05, Student’s t-test with Benjamini-Hochberg multiple testing correction).
Competing interest statement
All authors with the exception of E.R.F. were employed by Genentech, Inc., during the time the study was done.
Acknowledgments
We thank Jens Reeder, Michael Lawrence, Melanie Huntley, William Forrest, and Hanbin Liu for computational assistance and valuable discussion, and Minoru Yoshida for the spliceostatin A aliquot. A.S.A., J.-P.S., and R.F. designed the RNAi screen; A.S.A., M.L.M., G.Q., E.T., B.H., and J.-P.S. performed the RNAi screen; A.S.A. conducted follow-up analysis of top RNAi screen hits; A.S.A., V.S.C., M.L.M., S.H., S.Y., M.S., and R.F. contributed to the analysis of tri-snRNA and/or ZAK; E.C., S.Y., and E.M.B. performed the xenograft experiments; M.Y. performed and analyzed ISH studies of ZAK expression; A.S.A., Z.M., S.S., J.L., Z.Z., G.P., M.J., E.R.F., and Z.J. performed the microarray and RNA-seq analysis; and A.S.A. and R.F. wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.237206.113.
References
- Adler AS, McCleland ML, Truong T, Lau S, Modrusan Z, Soukup TM, Roose-Girma M, Blackwood EM, Firestein R 2012. CDK8 maintains tumor dedifferentiation and embryonic stem cell pluripotency. Cancer Res 72: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W 2012. Detecting differential usage of exons from RNA-seq data. Genome Res 22: 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. 1989. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244: 217–221 [DOI] [PubMed] [Google Scholar]
- Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D, et al. 2013. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 3: 658–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M, Clevers H 2000. Linking colorectal cancer to Wnt signaling. Cell 103: 311–320 [DOI] [PubMed] [Google Scholar]
- Bonnal S, Vigevani L, Valcárcel J 2012. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 11: 847–859 [DOI] [PubMed] [Google Scholar]
- Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonné R 2011. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet 20: 641–648 [DOI] [PubMed] [Google Scholar]
- Bujakowska K, Maubaret C, Chakarova CF, Tanimoto N, Beck SC, Fahl E, Humphries MM, Kenna PF, Makarov E, Makarova O, et al. 2009. Study of gene-targeted mouse models of splicing factor gene Prpf31 implicated in human autosomal dominant retinitis pigmentosa (RP). Invest Ophthalmol Vis Sci 50: 5927–5933 [DOI] [PubMed] [Google Scholar]
- Choi HS, Choi BY, Cho Y-Y, Zhu F, Bode AM, Dong Z 2005. Phosphorylation of Ser28 in histone H3 mediated by mixed lineage kinase-like mitogen-activated protein triple kinase α. J Biol Chem 280: 13545–13553 [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24: 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Schumacher SE, van Lieshout J, Imamura Y, Fox C, Shim B, Ramos AH, Saksena G, Baca SC, Baselga J, et al. 2012. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res 72: 4383–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert B, Bernard OA 2011. Mutations in RNA splicing machinery in human cancers. N Engl J Med 365: 2534–2535 [DOI] [PubMed] [Google Scholar]
- Fearon ER 1995. Molecular genetics of colorectal cancer. Ann N Y Acad Sci 768: 101–110 [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767 [DOI] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. 2008. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 455: 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh I, Adachi M, Nishida E 2001. Identification and characterization of a novel MAP kinase kinase kinase, MLTK. J Biol Chem 276: 4276–4286 [DOI] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, et al. 2012. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 44: 53–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DC, Hoeflich KP, Peng L, Gu Z, Gogineni A, Murray LJ, Eby M, Kljavin N, Seshagiri S, Cole MJ, et al. 2007. pHUSH: a single vector system for conditional gene expression. BMC Biotechnol 7: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziotto JJ, Inglehearn CF, Pack MA, Pierce EA 2008. Decreased levels of the RNA splicing factor Prpf3 in mice and zebrafish do not cause photoreceptor degeneration. Invest Ophthalmol Vis Sci 49: 3830–3838 [DOI] [PubMed] [Google Scholar]
- Häcker I, Sander B, Golas MM, Wolf E, Karagöz E, Kastner B, Stark H, Fabrizio P, Lührmann R 2008. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol 15: 1206–1212 [DOI] [PubMed] [Google Scholar]
- Haggar FA, Boushey RP 2009. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 22: 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CN, Scott HS 2012. Spliceosome mutations in hematopoietic malignancies. Nat Genet 44: 9–10 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Roberson EDO, Anbunathan H, Onken MD, Worley LA, Bowcock AM 2013. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet 45: 133–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, Fearon ER 2007. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res 67: 9721–9730 [DOI] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. 2012. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Tan J, Li J, Kivimäe S, Yang X, Zhuang L, Lee PL, Chan MTW, Stanton LW, Liu ET, et al. 2008. DACT3 is an epigenetic regulator of Wnt/β-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 13: 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583 [DOI] [PubMed] [Google Scholar]
- Kaiser S, Park Y-K, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et al. 2007. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol 8: R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines MA, Huang L, Schwartzentruber J, Douglas SL, Lynch DC, Beaulieu C, Guion-Almeida ML, Zechi-Ceide RM, Gener B, Gillessen-Kaesbach G, et al. 2012. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet 90: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TC, Huang CJ, Chu YC, Wei CC, Chou CC, Chou MY, Chou CK, Yang JJ 2000. Cloning and expression of ZAK, a mixed lineage kinase-like protein containing a leucine-zipper and a sterile-α motif. Biochem Biophys Res Commun 274: 811–816 [DOI] [PubMed] [Google Scholar]
- Lo C-W, Kaida D, Nishimura S, Matsuyama A, Yashiroda Y, Taoka H, Ishigami K, Watanabe H, Nakajima H, Tani T, et al. 2007. Inhibition of splicing and nuclear retention of pre-mRNA by spliceostatin A in fission yeast. Biochem Biophys Res Commun 364: 573–577 [DOI] [PubMed] [Google Scholar]
- Loo LW, Tiirikainen M, Cheng I, Lum-Jones A, Seifried A, Church JM, Gryfe R, Weisenberger DJ, Lindor NM, Gallinger S, et al. 2013. Integrated analysis of genome-wide copy number alterations and gene expression in microsatellite stable, CpG island methylator phenotype-negative colon cancer. Genes Chromosomes Cancer 52: 450–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Achsel T, Lührmann R 2000. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J Mol Biol 298: 567–575 [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ES, Tonon G, Sinha R, Xiao Y, Feng B, Kimmelman AC, Protopopov A, Ivanova E, Brennan C, Montgomery K, et al. 2007. Common and distinct genomic events in sporadic colorectal cancer and diverse cancer types. Cancer Res 67: 10736–10743 [DOI] [PubMed] [Google Scholar]
- Martin M, Maßhöfer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, et al. 2013. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 45: 933–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CWJ 2005. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398 [DOI] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci 108: 3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim MT, Eperon IC 2006. A double-reporter splicing assay for determining splicing efficiency in mammalian cells. Nat Protoc 1: 1022–1028 [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480: 387–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al. 2012. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 44: 47–52 [DOI] [PubMed] [Google Scholar]
- Quidville V, Alsafadi S, Goubar A, Commo F, Scott V, Pioche-Durieu C, Girault I, Baconnais S, Le Cam E, Lazar V, et al. 2013. Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Cancer Res 73: 2247–2258 [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C 2003. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer 3: 695–701 [DOI] [PubMed] [Google Scholar]
- Sabile AA, Arlt MJE, Muff R, Husmann K, Hess D, Bertz J, Langsam B, Aemisegger C, Ziegler U, Born W, et al. 2013. Caprin-1, a novel Cyr61-interacting protein, promotes osteosarcoma tumor growth and lung metastasis in mice. Biochim Biophys Acta 1832: 1173–1182 [DOI] [PubMed] [Google Scholar]
- Salari K, Spulak ME, Cuff J, Forster AD, Giacomini CP, Huang S, Ko ME, Lin AY, van de Rijn M, Pollack JR 2012. CDX2 is an amplified lineage-survival oncogene in colorectal cancer. Proc Natl Acad Sci 109: E3196–E3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Lührmann R 2004. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J 23: 3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. 2012. Recurrent R-spondin fusions in colon cancer. Nature 488: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA 1988. RNA splicing and genes. JAMA 260: 3035–3041 [PubMed] [Google Scholar]
- Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA, Domany E 2009. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci 106: 7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillars-Hardebol AH, Carvalho B, de Wit M, Postma C, Delis-van Diemen PM, Mongera S, Ylstra B, van de Wiel MA, Meijer GA, Fijneman RJA 2010. Identification of key genes for carcinogenic pathways associated with colorectal adenoma-to-carcinoma progression. Tumour Biol 31: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillars-Hardebol AH, Carvalho B, Tijssen M, Beliën JAM, de Wit M, Delis-van Diemen PM, Pontén F, van de Wiel MA, Fijneman RJA, Meijer GA 2012. TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut 61: 1568–1575 [DOI] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Ayuso C, Harper S, Berson EL, Rivolta C 2011. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am J Hum Genet 88: 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushiyama S, Tani T, Ohshima Y 1997. The prp1+ gene required for pre-mRNA splicing in Schizosaccharomyces pombe encodes a protein that contains TPR motifs and is similar to Prp6p of budding yeast. Genetics 147: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. 2011. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 365: 2497–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing F, Okuda H, Watabe M, Kobayashi A, Pai SK, Liu W, Pandey PR, Fukuda K, Hirota S, Sugai T, et al. 2011. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene 30: 4075–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Brocher J, Fischer U, Winkler C 2011. Mutant Prpf31 causes pre-mRNA splicing defects and rod photoreceptor cell degeneration in a zebrafish model for retinitis pigmentosa. Mol Neurodegener 6: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69 [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY, et al. 2009. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest 119: 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]