The resolution of recombination intermediates containing Holliday junctions (HJs) is critical for genome maintenance. Three pathways for HJ processing involve the BTR complex, the SLX–MUS complex, and GEN1. It has been suggested that SLX–MUS and GEN1 provide backup resolution pathways during mitosis. West and colleagues now show that cells lacking SLX–MUS and GEN1 exhibit impaired replication fork progression and chromosome instability. Furthermore, the effects of SLX4, MUS81, and GEN1 depletion extend throughout the cell cycle, revealing vital and expansive functions for these factors in genome integrity.
Keywords: DNA repair, chromosome segregation, structure-selective endonuclease, Holliday junction, Bloom’s syndrome, Fanconi anemia
Abstract
The resolution of recombination intermediates containing Holliday junctions (HJs) is critical for genome maintenance and proper chromosome segregation. Three pathways for HJ processing exist in human cells and involve the following enzymes/complexes: BLM–TopoIIIα–RMI1–RMI2 (BTR complex), SLX1–SLX4–MUS81–EME1 (SLX–MUS complex), and GEN1. Cycling cells preferentially use the BTR complex for the removal of double HJs in S phase, with SLX–MUS and GEN1 acting at temporally distinct phases of the cell cycle. Cells lacking SLX–MUS and GEN1 exhibit chromosome missegregation, micronucleus formation, and elevated levels of 53BP1-positive G1 nuclear bodies, suggesting that defects in chromosome segregation lead to the transmission of extensive DNA damage to daughter cells. In addition, however, we found that the effects of SLX4, MUS81, and GEN1 depletion extend beyond mitosis, since genome instability is observed throughout all phases of the cell cycle. This is exemplified in the form of impaired replication fork movement and S-phase progression, endogenous checkpoint activation, chromosome segmentation, and multinucleation. In contrast to SLX4, SLX1, the nuclease subunit of the SLX1–SLX4 structure-selective nuclease, plays no role in the replication-related phenotypes associated with SLX4/MUS81 and GEN1 depletion. These observations demonstrate that the SLX1–SLX4 nuclease and the SLX4 scaffold play divergent roles in the maintenance of genome integrity in human cells.
DNA joint molecules arise during the recombinational repair of DNA double-strand breaks (DSBs) and form a physical connection between sister chromatids or homologous chromosomes. In mitotic cells, the primary mechanism for their removal involves the BLM–TopoisomeraseIIIα–RMI1–RMI2 (BTR) complex, which gives rise to noncrossover products (Wu and Hickson 2003). This constitutes an important mechanism of crossover avoidance, which otherwise can lead to loss of heterozygosity and cancer predisposition.
Inactivation of the BLM helicase leads to the genome instability disorder known as Bloom’s syndrome (BS). Cells derived from BS patients display an increased frequency of sister chromatid exchanges (SCEs) and genome instability (Chaganti et al. 1974). These SCEs arise through the actions of three structure-selective endonucleases, SLX1–SLX4, MUS81–EME1, and GEN1, which resolve recombination intermediates containing Holliday junctions (HJs) to form both crossovers and noncrossovers (Wechsler et al. 2011; Castor et al. 2013; Garner et al. 2013; Wyatt et al. 2013). SLX1–SLX4 and MUS81–EME1 interact to form an SLX1–SLX4–MUS81–EME1 complex (SLX–MUS) that cleaves HJs by a nick and counternick mechanism (Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009; Castor et al. 2013; Wyatt et al. 2013). These interactions occur at G2/M phase of the cell cycle, when the SLX–MUS complex forms in response to CDK-mediated phosphorylation of EME1 (Wyatt et al. 2013). In addition to SLX–MUS, the HJ resolvase GEN1 provides an independent and genetically distinct resolution pathway and cleaves HJs by introducing two coordinated and symmetrically related nicks across the junction (Ip et al. 2008; Rass et al. 2010; Garner et al. 2013; Wyatt et al. 2013). The GEN1 protein is predominantly cytoplasmic and is thought to act later in the cell cycle than SLX–MUS, when breakdown of the nuclear envelope enables it to gain access to chromatin (Matos et al. 2011).
BS cells depleted for one or more HJ resolvase exhibit reduced SCE formation and high levels of mortality (Wechsler et al. 2011; Wyatt et al. 2013). Interestingly, depletion of SLX4 + GEN1 or MUS81 + GEN1 from BS cells also leads to the formation of elongated and segmented chromosomes (Wechsler et al. 2011). Similar chromosome abnormalities were observed in SLX4-deficient cells depleted for BLM (Garner et al. 2013). It has been suggested that this segmentation phenotype might result from abnormal chromosome condensation caused by the presence of persistent sister chromatid entanglements that accumulate in cells lacking both BLM and HJ resolution activities (Wechsler et al. 2011). Consistent with this proposal, segmented/elongated chromosomes have not been observed in cells with a functional BTR pathway.
Although it is thought that SLX–MUS and GEN1 process persistent HJs that have escaped BTR-mediated dissolution, normal cells depleted for SLX–MUS and GEN1 exhibit defects in chromosome segregation exemplified by the appearance of DAPI-positive chromosome bridges and lagging chromosomes (Wyatt et al. 2013). The activities of SLX–MUS and GEN1 are also required for the repair of DNA interstrand cross-links (ICLs), and cells lacking these proteins display increased sensitivity to cross-linking agents such as cisplatin or mitomycin C (MMC) and show reduced damage-induced SCE formation (Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009; Garner et al. 2013; Wyatt et al. 2013).
Mutations in SLX4 have been reported in patients with Fanconi anemia (FA), a recessive genetic disorder marked by the presence of genome instability, bone marrow failure, cancer predisposition, and hypersensitivity to DNA-cross-linking agents (Stoepker et al. 2011; Kim et al. 2013). Cells derived from these patients exhibit gross mitotic defects upon depletion of BLM or GEN1, as indicated by the presence of micronuclei, nuclear bridges, and irregular or catastrophic nuclei (Garner et al. 2013). They also commonly undergo mitotic failure, resulting in the formation of binucleate cells that fail to divide. Additionally, metaphase chromosomes from GEN1-depleted SLX4-null cells exhibit paired acentric chromosome fragments (Garner et al. 2013). These mitotic defects contribute to the high levels of mortality observed in BLM- or GEN1-depleted SLX4-null cells. However, whether the deficiency of these enzymes also leads to genome instability in other phases of the cell cycle is presently unknown.
In addition to its interaction with MUS81–EME1, SLX4 provides a scaffold for the binding of several other DNA repair proteins, including XPF–ERCC1, the mismatch repair proteins MSH2–MSH3, the SNM1B/Apollo 5′ exonuclease, and the shelterin proteins TRF2–RAP1 (Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009; Salewsky et al. 2012). MUS81 and ERCC1 are important for common fragile site breakage (or “expression”) during G2/M phase, a process that aids faithful chromosome disjunction by severing intertwined DNA strands at sites of ongoing/incomplete replication (Naim et al. 2013; Ying et al. 2013). Consistent with this, MUS81- and ERCC1-depleted cells exhibit high frequencies of segregation defects and 53BP1-positive G1 nuclear bodies (NBs), in particular when subjected to exogenous replication stress. Moreover, micronuclei and 53BP1 NBs in MUS81-depleted cells are frequently associated with fragile site DNA sequences (Ying et al. 2013).
The MUS81 nuclease has also been implicated in replication fork restart in response to treatment with replication inhibitors such as hydroxyurea (HU) and aphidicolin (APH), although increased fork stalling in the absence of exogenous replication stress was not observed (Hanada et al. 2007; Shimura et al. 2008; Naim et al. 2013; Ying et al. 2013). SLX4-depleted cells are sensitive to replisome-blocking agents such as camptothecin (Munoz et al. 2009; Svendsen et al. 2009; Kim et al. 2013), but it is presently unclear whether the SLX1–SLX4 nuclease plays a role in replication fork maintenance, as studies investigating the sensitivity of SLX1-depleted human cells to replisome-blocking agents have produced conflicting results (Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009; Kim et al. 2013). It is also unknown whether the SLX4 scaffold coordinates the activities of its interacting nucleases at replication forks.
Our present understanding of the functions of GEN1 relate almost exclusively to its role in the resolution of HJs at mitosis (especially in the absence of BTR and/or SLX–MUS HJ processing pathways), and it has been suggested that GEN1 simply provides a backup resolution pathway that is required for the avoidance of mitotic defects (Wechsler et al. 2011; Garner et al. 2013; Wyatt et al. 2013). However, in the work described here, we show that SLX4, MUS81, and GEN1 depletion not only impedes the timely and faithful completion of mitosis in undamaged or damaged BLM-proficient human cells but also results in increased genome instability throughout the cell cycle. We found that SLX4-, MUS81-, and GEN1-depleted cells exhibit impaired replication fork movement/S-phase progression, endogenous checkpoint activation, chromosome instability, and multinucleation. The functions of SLX4 and MUS81 are epistatic in protecting against these phenotypes but synthetic with the depletion of GEN1. Furthermore, we found that SLX1 is dispensable for these functions, providing evidence for divergent roles for the SLX1–SLX4 nuclease and the SLX4 scaffold in the maintenance of genome integrity in human cells.
Results
Anaphase bridge and lagging chromosome formation in resolvase-depleted cells
Cells depleted of SLX–MUS and GEN1 exhibit high frequencies of anaphase bridges and lagging chromosomes, particularly after treatment with DNA-damaging agents (Garner et al. 2013; Wyatt et al. 2013). To characterize these abnormalities in detail, HeLa cells were depleted for SLX1–SLX4 using siRNA against SLX4, treated with the DNA-damaging agent cisplatin, and visualized by immunofluorescence (Fig. 1). As shown previously, SLX4 depletion leads to destabilization of the SLX1–SLX4 complex and a reduction in the levels of both SLX1 and SLX4 (Supplemental Fig. 1A). In contrast, SLX1 depletion does not affect the stability of SLX4 (Munoz et al. 2009; Castor et al. 2013; Wyatt et al. 2013).
Figure 1.
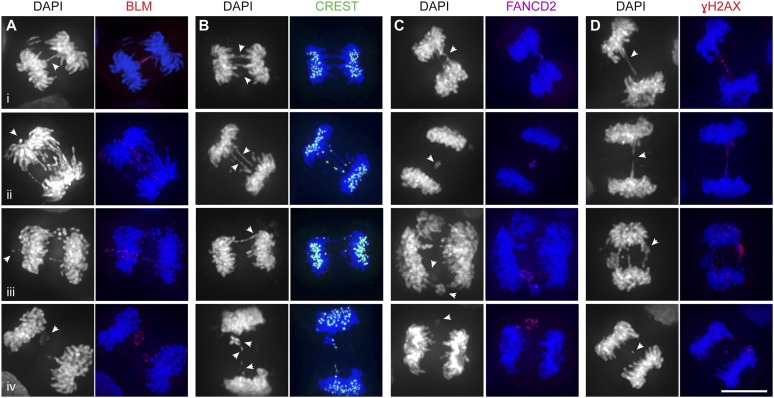
DAPI bridges and lagging chromosomes in SLX4-depleted cells contain BLM, centromeric DNA, γH2AX, and FANCD2. (A) Visualization of BLM (red) on DAPI bridges and lagging chromosomes in cisplatin treated SLX4-depleted HeLa cells. (Panel i) BLM staining coats a DAPI-positive bridge. (Panels ii,iii) BLM-positive bridges link lagging chromosomes to segregating DNA. (Panel iv) BLM coats a mass of lagging chromosomes. (B) Analysis of centromeric DNA (CREST) (green) on DAPI bridges and lagging chromosomes. (Panel i) Bridges and lagging chromosomes lacking centromeric DNA. (Panels ii,iii) Anaphase bridges associated with lagging centromeres at one or both ends of the bridge. (Panel iv) CREST foci on individual/pairs of lagging chromosomes. (C) Analysis of anaphase bridges and laggards for FANCD2 (purple). (Panel i) An anaphase bridge with a FANCD2 focus at the central connection point of the bridge. (Panels ii–iv) Lagging chromosomes containing FANCD2 foci or coating. (D) Appearance of γH2AX (red) on DAPI-positive bridges and lagging chromosomes. (Panels i,ii) γH2AX coats DAPI bridges. (Panels iii,iv) γH2AX coating or foci on lagging chromosomes. Bars, 10 μm. Arrows indicate points of interest.
When we examined 450 anaphase bridges and lagging chromosomes from SLX4-depleted cells, we found that ∼90% of the DAPI-stained DNA bridges also stained positively for BLM (Figs. 1A [panel i], 2A), indicating that the BTR dissolution pathway continues to attempt to resolve joint molecules that persist until mitosis. Consistent with this, BLM-deficient BS cells exhibit a higher frequency of DAPI-positive bridges relative to BLM-complemented isogenic cell lines (Supplemental Fig. 1B; Chan et al. 2007). Moreover, depletion of SLX4 from BS cells resulted in a further increase in DAPI bridges, indicating that BTR and SLX–MUS constitute alternative pathways for joint molecule processing/anaphase bridge resolution in cycling cells (Supplemental Fig. 1B).
Figure 2.
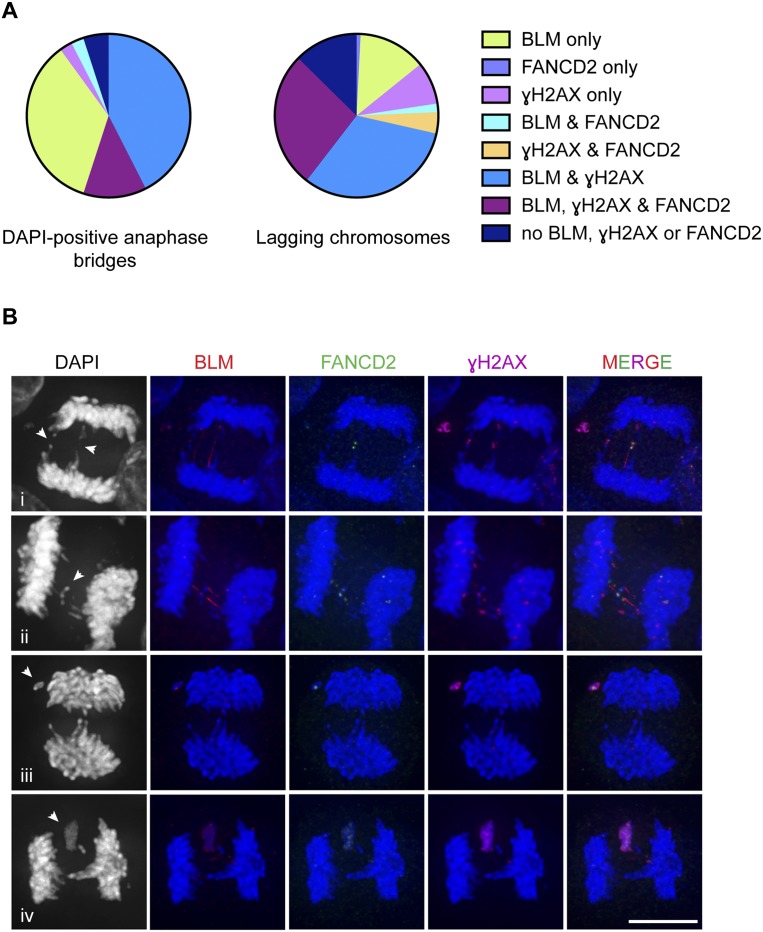
The presence of BLM, γH2AX, and, to a lesser extent, FANCD2 on anaphase bridges and lagging chromosomes in SLX4-depleted cells. (A) Proportion of DAPI-positive anaphase bridges and lagging chromosomes containing BLM, FANCD2, and/or γH2AX. More than 150 DAPI bridges and lagging chromosomes were analyzed. (B) BLM (red), FANCD2 (green), and γH2AX (purple) staining on the indicated gapped DAPI-staining bridges (panel i), lagging chromosomes and fragments (panels i–iii), and masses of lagging DNA (panel iv). All quantifications and representative images were obtained from cisplatin-treated, SLX4-depleted HeLa cells. Bar, 10 μm. Arrows indicate points of interest.
BLM foci were also present on the segregating DNA masses in SLX4-depleted cells (Supplemental Fig. 1C). A significant proportion of these foci colocalized with γH2AX, a marker for DNA DSBs (data not shown), indicating that they mark sites of damage that arise from either the aberrant processing of recombination intermediates or the persistence of unusual DNA structures or breaks. Similarly, BLM was found on extensive bridge-like structures that connected lagging chromosomes to one another as well as to one or both masses of segregating daughter DNA (Figs. 1A [panels ii,iii], 2A). Occasionally, BLM staining was found to coat masses of lagging DNA (Fig. 1A, panel iv).
All of the key observations made in cells depleted for SLX4 were also seen in cells depleted for SLX1, MUS81, or GEN1 (data not shown), indicating that these findings are representative of a general defect in the resolution of recombination intermediates. Due to the phenotypic similarities observed after resolvase depletion, the following section focuses primarily on cells depleted for SLX4.
BLM-stained DAPI bridges link centromeric DNA
BLM-positive anaphase bridges are commonly classified into (1) centromeric bridges that contain unresolved fully replicated DNA catenanes or (2) FANCD2/FANCI-associated bridges, which localize to chromosome arms (at loci representing common fragile sites) and mostly contain DNA replication intermediates (Chan et al. 2007, 2009; Naim and Rosselli 2009; Chan and Hickson 2011). We therefore examined whether the DAPI-positive bridges observed in resolvase-depleted cells were primarily centromeric or FANCD2-associated.
Using an anti-centromere antibody (CREST), we found that ∼90% of the DAPI bridges contained centromeric DNA at one or both ends of the bridge. In most cases, these foci lagged behind the main cluster of centromeres present within the two segregating daughter DNA masses (Fig. 1B, panels ii,iii). Consistent with the bridges being predominantly centromeric, FANCD2 was detected in only a small subset of DAPI-positive bridges (∼15%). When present, FANCD2 localized to either the base or the center of the bridge (Fig. 1C, panel i). The presence of FANCD2 and centromeric DNA on DAPI bridges was mutually exclusive, and only in very rare instances did we find FANCD2 at the central connection point of centromeric DAPI bridges (data not shown).
In contrast to the anaphase bridges, 34% of lagging chromosomes contained centromeric DNA (Fig. 1B, panel iv). Conversely, FANCD2 was more abundant on laggards than on anaphase bridges (Fig. 1C, panels ii–iv). The distinct differences observed with DAPI-positive bridges and lagging chromosomes indicate that these structures may contain different types of DNA intermediates.
DAPI bridges and laggards in resolvase-depleted cells contain broken DNA
Our finding that the majority of lagging chromosomes lack centromeres excludes the possibility that merotelic kinetochore attachments contribute to their formation. However, since resolvase-deficient cells exhibit elevated levels of genome instability (Fekairi et al. 2009; Garner et al. 2013), we reasoned that the lagging chromosomes might represent broken DNA fragments that are left behind as mitotic spindle forces pull segregating daughter nuclei toward opposing spindle poles. This hypothesis was supported by observations indicating that >70% of laggards showed γH2AX foci/coating (Fig. 1D, panels iii,iv).
Interestingly, γH2AX was found on 58% of the DAPI-positive bridges in SLX4-depleted anaphase cells, often localizing to visible gaps at the base or in the middle of a bridge (Supplemental Fig. 1D) and, in some cases, extending from these gaps to coat almost the entire bridge (Fig. 1D, panels i,ii). This was clearly the case with late anaphase/telophase cells (Supplemental Fig. 1E) and could be a consequence of the breakage of persistent bridges by mitotic spindle forces.
BLM and γH2AX often localized to the same anaphase bridges (Fig. 2A), whereas FANCD2 was less abundant and was only present on bridges that also contained BLM and/or γH2AX (Fig. 2B, panel i). On the lagging chromosomes, most FANCD2 foci (>90%) localized with γH2AX, consistent with them representing sites where DSBs are present (Fig. 2A,B). The extensive γH2AX staining observed in these cells indicates that DNA damage occurs and/or persists in the absence of normal levels of resolution activities. Moreover, the requirement for BLM at anaphase indicates that it is particularly important in SLX4-depleted cells.
Elevated levels of micronuclei and multinucleation in SLX–MUS- and GEN1-depleted cells
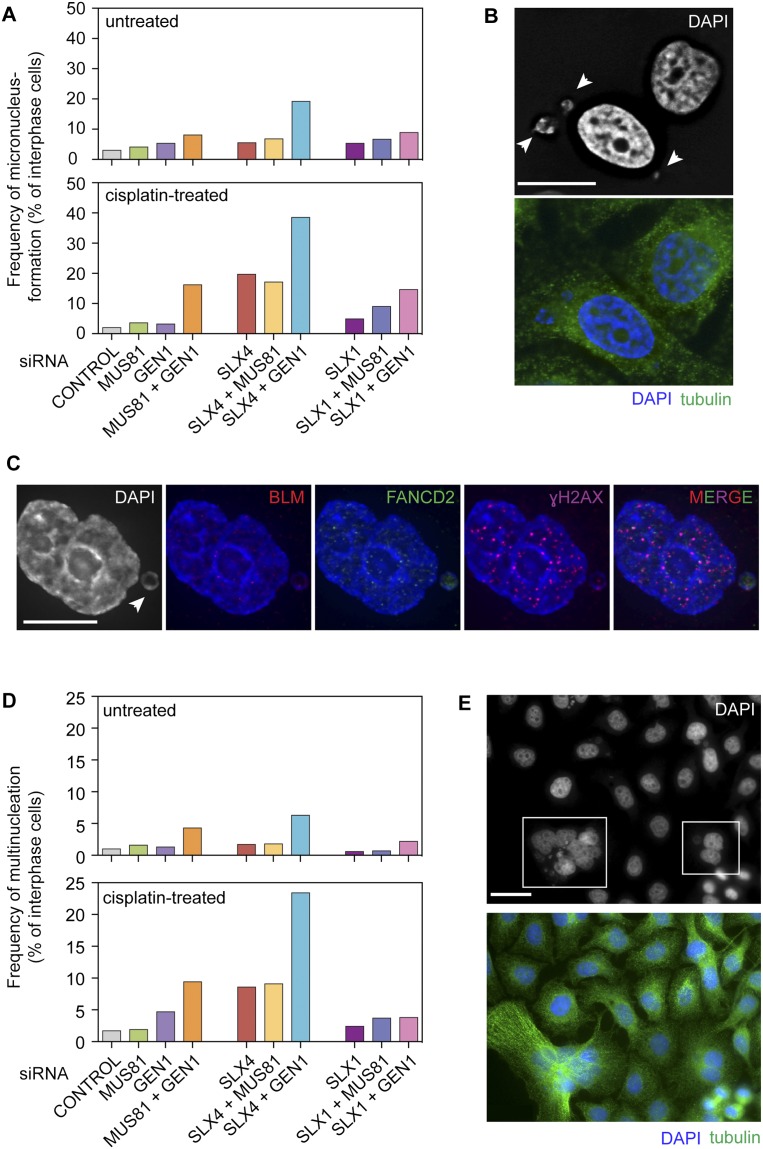
The presence of anaphase bridges and lagging chromosomes leads to micronucleus formation upon cytokinesis (Fenech and Neville 1992; Hoffelder et al. 2004; Utani et al. 2010). We therefore analyzed the effect of resolvase depletion on the formation of micronuclei in both undamaged and damaged cells. Consistent with the levels of bridges and laggards observed in undamaged resolvase-depleted cells (Wyatt et al. 2013), cells lacking SLX1, SLX4, MUS81, or GEN1 showed only minor increases in micronucleus formation (Fig. 3A, top panel; Supplemental Table S1). Cells lacking both SLX4 and GEN1, however, exhibited a significant increase in their formation (Supplemental Table S1). Moreover, pairwise depletion of SLX1 + GEN1, SLX4 + GEN1, and MUS81 + GEN1 followed by cisplatin treatment led to high levels of micronucleus formation (Fig. 3A, bottom panel; Supplemental Table S1). Further increases in the levels of micronuclei were not observed upon depleting MUS81 with either SLX1 or SLX4, consistent with observations showing that SLX–MUS and GEN1 constitute two distinct pathways of resolution that are important for ensuring faithful chromosome segregation (Castor et al. 2013; Garner et al. 2013; Wyatt et al. 2013). Interestingly, micronuclei levels in SLX4-depleted cells were greater relative to SLX1- and/or MUS81-depleted cells. This is consistent with SLX4-depleted cells showing a higher incidence of DAPI-positive bridges and laggards relative to SLX1/MUS81 depletions (Wyatt et al. 2013) and likely reflects additional SLX4 scaffold functions mediated through one of its other interacting partners. As with DAPI bridges and laggards, we also observed γH2AX, BLM, and FANCD2 on micronuclei, suggesting that these too contain damaged DNA (Fig. 3C).
Figure 3.
Micronucleus formation and multinucleation in SLX1-, SLX4-, MUS81-, and/or GEN1-depleted cells. (A) Micronuclei were quantified in untreated and cisplatin-treated HeLa cells depleted for the indicated proteins (n = 6992 interphase cells). (B) Representative image of micronuclei (arrowheads) found in SLX4-depleted HeLa cells following cisplatin treatment. Bar, 16 μm. (C) Visualization of BLM (red), FANCD2 (green), and γH2AX (purple) in a micronucleus (arrowhead) in a SLX4-depleted and cisplatin-treated HeLa cell. Bar, 10 μm. (D) Quantification of the frequencies of multinucleate cells following treatment with the indicated siRNAs with or without cisplatin (n = 7403 interphase cells). (E) Examples of multinucleation (boxed) in damaged HeLa cells depleted for SLX4. Bar, 32 μm.
SLX4 + GEN1- and MUS81 + GEN1-depleted cells additionally exhibited significant levels of multinucleation, relative to cells treated with any of the single siRNAs (Fig. 3D, top panel; Supplemental Table S1). Multinucleate cells were characterized by the presence of more than one (up to eight) nuclear masses of varying sizes accompanied by an expanded cytoplasm, as revealed by α-tubulin staining. Representative examples observed in SLX4-depleted cells are shown in Figure 3E. The proportion of multinucleate cells increased further upon exposure to cisplatin such that 10% and >20% of MUS81 + GEN1- and SLX4 + GEN1-depleted cells were multinucleate, respectively (Fig. 3D, bottom panel; Supplemental Table S1). Depletion of MUS81 + SLX4 led to no additional effects on the levels of multinucleation.
In contrast to cisplatin-treated cells in which SLX4 + GEN1 were depleted, loss of SLX1 + GEN1 did not result in significant levels of multinucleation (Fig. 3D; Supplemental Table S1). This was not due to inefficient depletion of SLX1 (Supplemental Fig. 1A), since SCE levels in these cells were comparable with those treated with SLX4 siRNA (Supplemental Fig. 2). Indeed, both SLX1- and SLX4-depleted cells exhibited significantly lower levels of SCEs than control cells. These data indicate that multinucleation does not result from the depletion of the SLX1–SLX4 nuclease but is instead associated with the deficiency of SLX4’s scaffold functions.
Elevated levels of 53BP1-positive G1 NBs
DNA lesions, such as DSBs, can be transmitted to daughter cells following mitosis, where they are sequestered into 53BP1 NBs (Harrigan et al. 2011; Lukas et al. 2011). Since a large proportion of anaphase/telophase cells depleted for HJ resolvases showed γH2AX foci, we next analyzed the levels of 53BP1-positive G1 NBs to determine whether at least some of this damage was transmitted to the daughter cells.
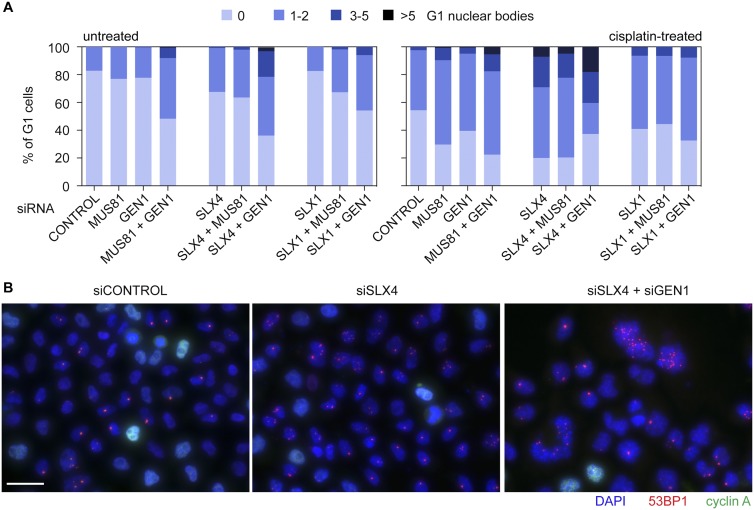
We found that 53BP1-positive NBs were more common in SLX1-, SLX4-, MUS81-, and/or GEN1-depleted cells, relative to cells treated with control siRNA, even in the absence of exogenous damage (Fig. 4A). The highest levels were observed in cells depleted for both SLX–MUS and GEN1 pathway proteins. This was further exacerbated by the introduction of exogenous damage, with a subset of SLX4 + GEN1- and MUS81 + GEN1-depleted G1 cells in particular showing more than five 53BP1 NBs. Indeed, we observed that SLX4 + GEN1 depletions led to significantly more 53BP1 NBs than the corresponding SLX1 + GEN1 depletions (Fig. 4A). Representative images of the SLX4 and SLX4 + GEN1 depletions are shown in Figure 4B. Collectively, these data show that the DNA lesions present in the resolvase-depleted anaphase cells remain unrepaired and are transmitted to the daughters.
Figure 4.
Resolvase-depleted cells exhibit elevated levels of 53BP1-positive G1 NBs. (A) Quantification of the proportion of G1 cells containing zero, one to two, three to five, and more than five 53BP1-positive NBs following treatment with the indicated siRNAs and with or without cisplatin treatment (n = 8894 G1 cells). (B) Representative images of 53BP1-positive NBs (red) in control, SLX4, or SLX4 + GEN1 siRNA-treated, damaged cyclin A-negative HeLa cells. Bar, 32 μm.
In normal cells, 53BP1 foci are rarely observed in the S or G2 phases of the cell cycle unless the cells are exposed to DNA-damaging agents such as ionizing radiation (Lukas et al. 2011). However, we found that undamaged cyclin A-positive cells, characteristic of those in S/G2 phase, exhibited increased levels of 53BP1 foci after depletion of SLX–MUS and GEN1 (Supplemental Fig. 3). This was particularly prominent in the SLX4 + GEN1 and MUS81 + GEN1 depletions, as 33%–37% of the cyclin A-stained cells also contained 53BP1 foci. The appearance of these foci indicates that depletion of SLX–MUS and GEN1 leads to increased genome instability not only in mitosis but also during S/G2 phases of the cell cycle. However, whether they represent damage transmitted from the preceding cell cycle or that acquired during interphase in the absence of SLX–MUS and GEN1 is presently unclear.
Severe chromosome abnormalities in SLX4-, MUS81-, and GEN1-depleted cells
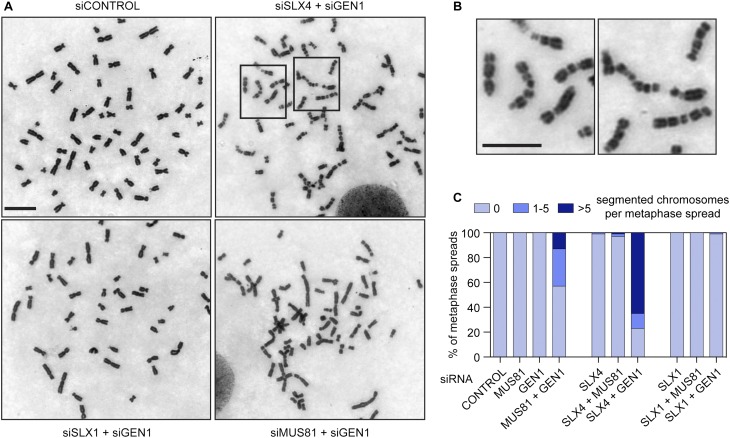
BS cells depleted for SLX4 + GEN1 or MUS81 + GEN1 exhibit severe chromosomal abnormalities, such as the appearance of highly elongated and segmented chromosomes (Wechsler et al. 2011). Similar observations have been made in SLX4-null cells, derived from individuals with FA, after depletion of BLM (Garner et al. 2013). To determine whether loss of BLM function is directly related to the segmentation phenotype, we analyzed their formation in BLM-proficient cells following resolvase depletion. We found that depletion of SLX4 + GEN1 or MUS81 + GEN1 from cisplatin-treated HeLa cells led to the formation of segmented chromosomes (Fig. 5A,B), providing the first demonstration that this phenotype occurs in cells proficient for BLM. Chromosome segmentation was not observed in SLX1, SLX4, MUS81, or GEN1 single knockdowns or when SLX–MUS pathway proteins were depleted together (Fig. 5C).
Figure 5.
Appearance of segmented chromosomes in cisplatin-treated cells depleted for SLX4 + GEN1 and MUS81 + GEN1. (A) Representative images of Giemsa-stained metaphase spreads prepared from HeLa cells treated with control, SLX4 + GEN1, SLX1 + GEN1, or MUS81 + GEN1 siRNA and exposed to cisplatin. Bar, 10 μm. (B) The two boxed regions in A showing segmented chromosomes are presented at higher magnification. Bar, 7 μm. (C) Quantification of segmented chromosomes per metaphase spread in cisplatin-treated, resolvase-depleted cells (n = 1000 metaphase spreads).
Consistent with our earlier observations with the multinucleation phenotype, treatment with SLX1 + GEN1 siRNAs did not lead to the formation of elongated/segmented chromosomes, in contrast to the results obtained with SLX4 + GEN1 depletion (Fig. 5A,C). These results indicate that cells lacking the nuclease activity of SLX1–SLX4 are phenotypically distinct from those depleted for SLX4’s scaffold function. Similar observations were made in undamaged BS cells treated with SLX1 + GEN1 siRNAs, which exhibited significantly lower levels of chromosome segmentation compared with SLX4 + GEN1-depleted cells (Supplemental Fig. 4A,B). Furthermore, upon exposure to exogenous damage, SLX4-depleted BS cells, but not those depleted for SLX1, displayed severe chromosome segmentation (Supplemental Fig. 4C,D), highlighting the importance of the SLX4 scaffold, but not the SLX1–SLX4 nuclease, for maintaining normal chromosome architecture in the absence of BLM function.
Loss of SLX4, MUS81, and GEN1 triggers a checkpoint response
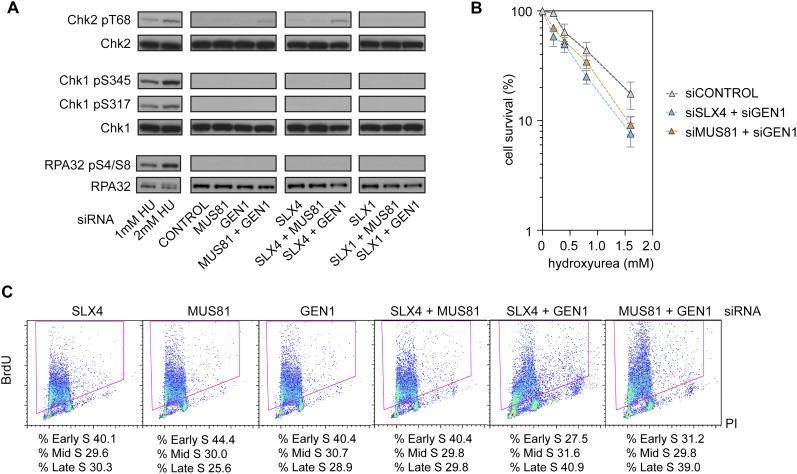
BS cells, which display elevated levels of genomic instability, exhibit an ATM/CHK2-mediated DSB checkpoint response (Rao et al. 2007). We therefore determined whether elevated levels of genomic instability associated with SLX–MUS and GEN1 depletion also lead to activation of the same checkpoint. Analysis of the levels of phospho-Thr68 of CHK2 revealed that CHK2 was activated by depletion of SLX4 + GEN1 and, to a lesser extent, by MUS81 + GEN1 (Fig. 6A). Marginal levels of CHK2 activation were observed after SLX4 or SLX4 + MUS81 depletion. Interestingly, CHK2 activation was not observed in SLX1-, SLX1 + MUS81-, or SLX1 + GEN1-depleted cells, indicating that a deficiency of the SLX4 scaffold, rather than the SLX1–SLX4 nuclease, is responsible for triggering the checkpoint. Loss of SLX–MUS and GEN1 pathway proteins, however, failed to activate CHK1, exemplified by the absence of Ser317 or Ser345 phosphorylation (Fig. 6A). Activation of CHK2, rather than CHK1, indicates that the checkpoint response in SLX4/MUS81- and GEN1-depleted cells is predominantly triggered by DSBs rather than single-stranded breaks. This was further corroborated by the absence of significant RPA32 phosphorylation (S4/S8) in SLX–MUS- and GEN1-depleted cells (Fig. 6A).
Figure 6.
Checkpoint activation and DNA damage sensitivity of SLX4-, MUS81-, and GEN1-depleted cells. (A) Analysis of phosphorylated CHK1, CHK2, and RPA32 in whole-cell extracts prepared from undamaged HeLa cells treated with the indicated siRNAs. Controls: Cells were treated with the indicated concentrations of HU for 24 h. (B) HU sensitivity of the indicated siRNA-treated cells. Error bars indicate ±SEM. (C) Quantification of early, mid-, and late S-phase populations (boxed) in asynchronous cultures of BrdU-labeled cells treated with the indicated siRNAs.
SLX4, MUS81, and GEN1 are required for replication fork progression
Cells depleted for SLX4 + GEN1 or MUS81 + GEN1, but not SLX1, SLX4, MUS81, GEN1, SLX1 + MUS81, SLX4 + MUS81, or SLX1 + GEN1, were found to be sensitive to HU (Fig. 6B; Supplemental Fig. 5A). We therefore determined whether SLX4 + GEN1 or MUS81 + GEN1 depletion affected the proportion of cells in S phase. Previously, it was shown that increased fractions of SLX4 + GEN1- and MUS81 + GEN1-depleted cells possess a “4n” (or G2/M) DNA content (Wyatt et al. 2013). However, using phospho-histone H3 labeling, we found that these cells had similar mitotic fractions compared with those treated with either control siRNA or SLX4, MUS81, or GEN1 siRNA (Supplemental Fig. 5B). Using a flow cytometric assay for bromodeoxyuridine (BrdU) labeling, we found that SLX4 + GEN1 or MUS81 + GEN1 depletion instead led to the enrichment of cells in late S phase (Fig. 6C), in which the DNA content is similar to that of cells in G2 or M phase. Elevated populations of late S-phase cells were not observed in the single depletions or other pairwise combinations.
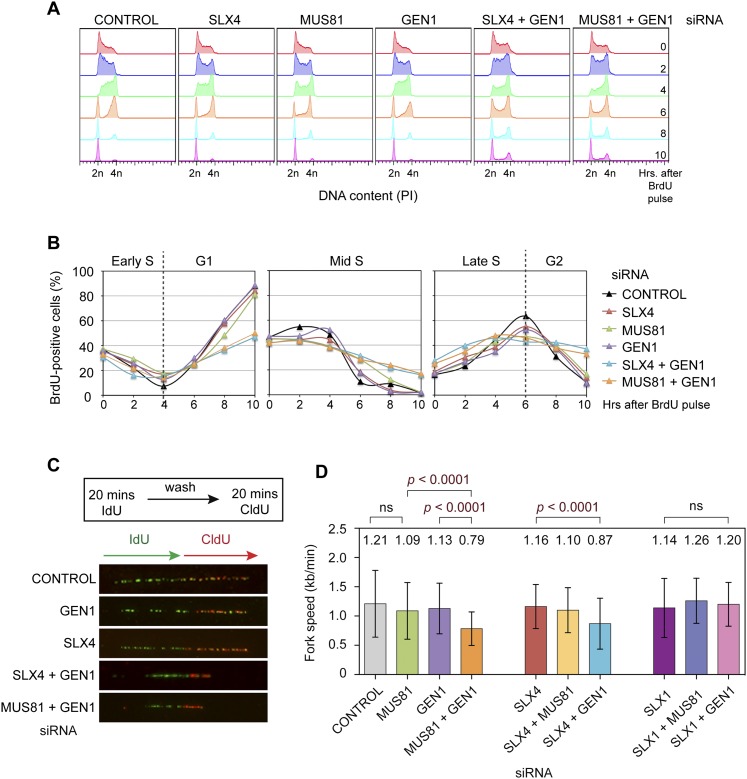
Enriched S-phase populations can stem from cells that slow replication or arrest during S phase, leading us to determine whether SLX4 + GEN1- or MUS81 + GEN1-depleted cells exhibited a delay in S-phase progression. To do this, we pulsed asynchronous populations of siRNA-treated cells with BrdU for 30–45 min (to selectively label S-phase cells) and then followed the progression of labeled cells over the next 10 h using flow cytometry. Significant delays in S-phase progression were observed in both SLX4 + GEN1- and MUS81 + GEN1-depleted cells relative to those treated with either control or SLX4, MUS81, or GEN1 siRNA (Fig. 7A,B). Indeed, after 10 h, when 81%–89% of the single siRNA-treated cells had completed cell division and progressed to a new cell cycle (G1), only 47%–50% of SLX4 + GEN1- or MUS81 + GEN1-depleted cells had progressed normally, and the rest of the cells remained in S or G2 phase (Fig. 7B). Delays in cell cycle progression were further exacerbated by the presence of exogenous damage, with significant proportions of SLX4 + GEN1- or MUS81 + GEN1-depleted cells exhibiting 8n DNA contents (Supplemental Fig. 6A). In contrast to SLX4 + GEN1-depleted cells, those lacking SLX1 + GEN1 displayed normal cell cycle progression kinetics (Supplemental Fig. 6B,C), consistent with our observations showing that they were insensitive to HU treatment and exhibited normal DNA content distributions (Supplemental Fig. 5A,B).
Figure 7.
Impaired replication fork progression in SLX4-, MUS81-, and GEN1-depleted cells. (A) Cell cycle progression of BrdU-positive (S-phase) cells following treatment with the indicated siRNAs. (B) Kinetics of cell cycle progression, quantified as in A. (C) Representative images of IdU-labeled (green) and CldU-labeled (red) combed DNA fibers prepared from HeLa cells treated with the indicated siRNAs. (D) Replication fork velocity (kilobases per minute) in undamaged siRNA-treated HeLa cells. Data represent the mean fork velocity after each siRNA treatment (values are indicated at the top of the graph). P-values were determined using two-tailed Mann-Whitney tests. (ns) Not significant. Error bars indicate ±SD.
Endogenous checkpoint activation, sensitivity to replication inhibitors, and slow S-phase progression are indicative of defects in DNA replication. We therefore examined replication dynamics in SLX4-, MUS81-, and/or GEN1-depleted cells using molecular combing, in which replicating DNA was visualized by consecutively pulsing cells with iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU). Since the stretching factor of combed DNA fibers is constant, the lengths of IdU and CldU tracts provide reliable estimates of fork velocity. We found that depletion of SLX4 + GEN1 or MUS81 + GEN1 reduced replication fork velocity by 28%–35% relative to control cells (Fig. 7C,D). We did not observe any significant reductions in fork speed in SLX1-, SLX4-, MUS81-, GEN1-, SLX4 + MUS81-, SLX1 + MUS81-, or SLX1 + GEN1-depleted cells. These data bolster the concept that the SLX1–SLX4 nuclease is dispensable for replication fork maintenance and thus for efficient S-phase progression. Depletion of the SLX4 scaffold or MUS81 in combination with GEN1, however, leads to defects in fork progression. Given that MUS81 is known to play a role in replication fork restart following exposure to HU (Hanada et al. 2007; Ying et al. 2013), these results implicate the SLX4 scaffold in facilitating MUS81 actions at stalled forks.
Discussion
In mitotic human cells, HJs that arise during recombinational repair are preferentially dissolved by the BTR pathway. The HJ resolvases have largely been viewed as backups for this pathway, assisting in the removal of persistent HJs in G2/M phase, especially in the absence of BTR-mediated dissolution (as in BS) or when the damage load on the cells is high (e.g., after exposure to genotoxic agents such as cisplatin) (Wechsler et al. 2011; Garner et al. 2013; Wyatt et al. 2013). Here, we show that depletion of SLX4, MUS81, and GEN1 from undamaged BLM-proficient cells leads to genome instability not only in mitosis but also throughout the cell cycle.
Importantly, we found that SLX4/MUS81 and GEN1 are important for normal DNA replication and S-phase progression in human cells. Indeed, depletion of SLX4/MUS81 and GEN1 led to reduced replication fork speed and delayed S-phase progression, resulting in skewed DNA content distributions with enriched late S-phase populations. Significant replication fork impediments were observed only when both the SLX4/MUS81 and GEN1 pathways were depleted, consistent with functional redundancies between these proteins. The observed reduced replication fork velocity may result from a defect in replication fork recovery, as observed previously in MUS81-deficient human and mouse cells after HU or APH treatment (Hanada et al. 2007; Ying et al. 2013). Consistent with this proposal, we observed the activation of CHK2 in SLX4/MUS81- and GEN1-depleted cells, most likely caused by replication fork collapse and DSB formation.
SLX4/MUS81- and GEN1-depleted, cisplatin-treated cells exhibit gross chromosomal abnormalities characterized by the appearance of highly segmented chromosomes. Similar phenotypes have been observed in BLM-depleted SLX4-null cells and SLX4/MUS81- and GEN1-depleted BS cells (Wechsler et al. 2011; Garner et al. 2013). It has been suggested that segmentation might result from abnormal chromosome condensation in cells lacking HJ dissolution/resolution functions. Consistent with this, the chromosome indentations found within intact chromosomes are free of the condensin SMC2 and can be suppressed by expression of the cryptic bacterial resolvase RusA (Wechsler et al. 2011; Garner et al. 2013).
How might the presence of DNA intermediates that are normally processed by BTR, SLX4/MUS81, and/or GEN1 impede chromosome condensation? One possibility is that delays in replication timing/progression promote defects in chromosome condensation. For example, human and mouse cells exposed to ionizing radiation exhibit delayed replication timing and impaired chromosome condensation (Breger et al. 2004). Similarly, mutations in Drosophila replication genes such as orc2 and orc5 have been linked with abnormal chromosome condensation (Pflumm and Botchan 2001). These observations indicate that a normal density of active replication origins as well as the timely completion of S phase are important for proper chromosome condensation. It is therefore possible that the condensation defects seen in SLX4/MUS81- and GEN1-depleted cells are also causally linked to the impaired replication dynamics observed in these cells. In accord with this, SLX1 + GEN1-depleted cells, which show normal S-phase progression, are largely devoid of chromosome segmentation.
We found that depletion of SLX4/MUS81 and GEN1 led to significant levels of multinucleation. Hyperploidy has been reported in BLM- and GEN1-depleted SLX4-null human cells (Garner et al. 2013), Drosophila mus312 (SLX4) and mus309 (BLM) double mutants (Andersen et al. 2011), and Saccharomyces cerevisiae sgs1 (BLM) and mms4 (EME1) double mutants (Matos et al. 2013). Although the precise cause for multinucleation remains unclear, it is possible that significant replication delays and DNA repair defects in SLX4/MUS81- and GEN1-depleted cells lead to the inhibition of cytokinesis and promote multinucleation. Indeed, delayed replication timing is associated with hyperploidy in irradiated mouse primary cells and with multinucleation in the absence of Orc6 in Drosophila (Breger et al. 2004). This might also explain why SLX1 + GEN1-depleted cells, which display normal replication kinetics, do not display multinucleation.
Although a role for MUS81 in replication fork maintenance is well established, GEN1 has not previously been linked with DNA replication. However, S. cerevisiae mutants lacking both Yen1 and Dna2, an essential replication gene, are synthetic-lethal (Budd et al. 2005), and yen1 mus81 double mutants are exquisitely sensitive to DNA-damaging agents that perturb replisome progression (Blanco et al. 2010). Importantly, we found that SLX1-depleted cells displayed normal replication fork velocity and S-phase progression. Moreover, SLX1-, SLX1 + MUS81-, or SLX1 + GEN1-depleted cells did not exhibit CHK2 activation, multinucleation, or chromosome segmentation. The absence of replication defects even in the absence of both SLX1 and GEN1 indicates that the SLX1–SLX4 nuclease does not contribute to the striking SLX4 + GEN1 replication-related phenotypes. Similar observations have been reported in S. cerevisiae, where Slx4, but not Slx1, is important for replication fork recovery following DNA damage (Roberts et al. 2006; Flott et al. 2007). Since SLX4 and MUS81 were epistatic in ensuring normal replication in our study, it is tempting to speculate that SLX4 is important for targeting MUS81 to replication forks during S phase and/or stimulating its activity on these structures. Consistent with this, SLX4 stimulates the cleavage activity of MUS81 on model replication fork structures (Munoz et al. 2009). Partially nonoverlapping functions for SLX1 and MUS81 within the context of the SLX4 scaffold also explains why SLX1 + MUS81-depleted cells show reduced survival relative to the respective single knockdowns (Wyatt et al. 2013).
Although the SLX1–SLX4 nuclease appears to be dispensable for replication progression, it is nonetheless important for damage-induced SCE formation and faithful chromosome segregation (Wyatt et al. 2013). Accordingly, SLX1–SLX4-, MUS81-, and/or GEN1-depleted anaphase cells show elevated levels of DAPI-positive chromosome bridges and lagging chromosomes, particularly in the presence of exogenous damage (Wyatt et al. 2013). We found that the BLM helicase is ubiquitous on these structures. Since most DAPI-positive bridges in resolution-deficient cells are centromeric (and FANCD2-negative), it is likely that BLM dissolves persistent, fully replicated DNA catenanes that are present within these structures. In this respect, the BTR pathway in turn serves as a backup for defects in HJ resolution during anaphase.
The anaphase bridges and laggards found in resolvase-depleted cells are also frequently positive for the DSB marker γH2AX, with BLM and γH2AX staining often apparent on the same structures. The recruitment of BLM to γH2AX-positive bridges and laggards presumably indicates its involvement in DSB repair in the absence of the SLX–MUS and GEN1 proteins. In addition to containing γH2AX, many laggards lack centromeric DNA, suggesting that they represent broken DNA fragments that fail to attach to the mitotic spindle apparatus. Increased genome instability in SLX4-, MUS81-, and GEN1-depleted S/G2 cells may thus contribute to the mitotic defects observed in these cells. This, to some extent, also explains why laggards are more common in SLX4/MUS81- and GEN1-depleted undamaged cells relative to those lacking SLX1 and GEN1 (Wyatt et al. 2013).
Finally, we found that depletion of SLX–MUS and GEN1 pathway proteins is associated with increased levels of 53BP1-positive G1 NBs, indicating that at least some of the damage observed during anaphase remains unrepaired and is transmitted to the daughter cells. The hallmark features of genome instability associated with their deficiency are thus explicit throughout the cell cycle in the form of elevated levels of 53BP1-positive NBs in G1, impaired replication fork progression during S phase, and chromosomal instability and missegregation during M phase.
Materials and methods
Cell lines and cell culture
HeLa Kyoto cells, BLM-deficient GM08505 fibroblasts, and BLM-complemented (GM08505-derived) PSNF5 fibroblasts were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All cultures were grown at 37°C in a humidified atmosphere containing 10% CO2.
siRNA transfections and Western blotting
siRNA-mediated protein depletions were carried out as described (Wyatt et al. 2013). The efficiency of protein depletion was monitored by Western blotting 48 and/or 72 h after the first transfection. Cells were lysed on ice in buffer containing 20 mM Tris-HCl (pH 7.5 or 8.0), 150 mM NaCl, 2 mM EDTA, 0.1% (v/v) Triton X-100, 10% (v/v) glycerol, and a cocktail of complete, EDTA-free protease inhibitors (Roche). Aliquots (20–50 μg) of whole-cell extracts were subjected to SDS-PAGE, and the proteins were transferred onto nitrocellulose membranes for Western blotting. The primary antibodies used were sheep anti-SLX1 and SLX4 (a gift from J. Rouse), rabbit anti-GEN1 (Wechsler et al. 2011), mouse anti-MUS81 (MTA30 2G10/3, Santa Cruz Biotechnology), mouse anti-XPF (ab3299, Abcam), mouse anti-CHK2 (05-649, Millipore), mouse anti-CHK1 (C9358, Sigma), rabbit anti-phosphoCHK2 (Thr68) (2661, Cell Signaling Technology), rabbit anti-phosphoCHK1 (Ser345) (2348, Cell Signaling Technology), rabbit anti-phosphoCHK1 (Ser317) (2344S, Cell Signaling Technology), mouse anti-RPA32 (ab2175, Abcam), and rabbit anti-phosphoRPA32 (S4/S8) (A300-245A, Bethyl Laboratories).
Drug treatment
Unless otherwise indicated, cells were treated with 2 μM cisplatin 48–60 h after the first transfection. After 1 h, the cells were washed with PBS and then maintained in fresh medium.
Immunofluorescence
Cells were harvested 36 h after the first transfection and replated on coverslips in six-well cell culture plates. After 12 h, when required, the cells were treated with 2 μM cisplatin for 1 h, washed in PBS, and released into fresh medium for 60 h. The cells were then washed three times in PBS and fixed in 4% formaldehyde for 10 min. After washing twice more with PBS, the cells were permeabilized with 0.2% Triton X-100 for 5 min and washed again. Cells were then blocked in 2% BSA and 20 mM glycine (in PBS) for 15 min, followed by 2% BSA for another 15 min. The primary antibodies used were mouse anti-γH2AX (05-636, Millipore), goat anti-BLM (C-18, Santa Cruz Biotechnology), rabbit anti-FANCD2 (NB100-182, Novus Biologicals), human anti-centromere (CREST) (HCT-0100, Immunovision), rat anti-tubulin (MCA 78G, AbD Serotec), rabbit anti-53BP1 (ab36823, Abcam), and mouse anti-cyclin A (ab16726, Abcam). Alexa 488-, Alexa 555-, or Alexa 647-conjugated donkey anti-rabbit, donkey anti-mouse, donkey anti-rat, donkey anti-goat, and goat anti-human secondary antibodies were purchased from Molecular Probes (Invitrogen). All antibodies were diluted in 2% BSA in PBS, and antibody incubations were carried out for 1 h at either room temperature (primary antibodies) or 37°C (secondary antibodies). Cells were washed three times in PBS after the primary and secondary antibody incubations. Finally, coverslips were mounted on glass slides using ProLong gold anti-fade reagent with DAPI (Life Technologies). The slides were viewed using a Zeiss Axio Imager M2 microscope with an EC Plan-Neofluar 40×/1.3 objective. Images were captured using a Hamamatsu ORCA-R2 (C10600-10B) camera with Volocity 5.5 software (PerkinElmer).
Metaphase spreads
HeLa Kyoto cells were treated for 1 h with 0.2 μg/mL colcemid 48 h after exposure to cisplatin. Both undamaged and damaged GM08505 cells were treated with colcemid 72 h after the first transfection (where indicated, GM08505 cells were treated with 1 μM cisplatin for 24 h prior to colcemid treatment). The cells were then harvested, swollen in 75 mM KCl at 37°C for 15–30 min, fixed with 3:1 methanol:acetic acid, and spread on glass slides. After air-drying, the slides were stained with 7% Giemsa for 10 min. After mounting, the slides were viewed, and images were captured as described above using a Plan-Apochromat 63×/1.4 objective.
SCE assays
The assay was adapted from a published protocol (Bayani and Squire 2005). Briefly, cells were incubated in medium containing 110 μM BrdU for 48 h, followed by 0.2 μg/mL colcemid for 1 h. Cells were harvested, fixed, and spread on glass slides as described above. After air-drying, the slides were incubated in 0.1 mg/mL Hoechst-33258 for 30 min, washed with PBS, and exposed to UV light for 10 min. The slides were then soaked in 2× SSC for 1 h and stained with 7% Giemsa for 10 min. Slides were viewed, and images were captured as described above.
Clonogenic cell survival assay
Cells were harvested 48 h after the first transfection and replated in 10-cm tissue culture plates. After 12 h, the cells were treated with 0, 0.2, 0.4, 0.8, or 1.6 mM HU for 24 h. The cells were then washed three times with PBS and maintained in fresh, drug-free medium for 2 wk. Colonies were stained for 2–10 min in 40 mg/mL crystal violet solution containing 20% ethanol. Survival (percentage) following exposure to HU was calculated against an undamaged sample treated with the same siRNA.
Flow cytometry
Analyses of DNA content distribution and estimation of proportions of mitotic and S-phase cells were performed in parallel with immunofluorescence and DNA-combing experiments. Cells were harvested 36 h after the first transfection and replated on 10-cm cell culture plates for another 72 h. To determine the proportions of S-phase populations following siRNA treatments, cells were incubated in medium containing 10 μM BrdU for 30 min prior to harvest. For analyses of cell cycle progression, cells were replated 36 h after the first transfection. After 12 h, when required, cells were treated with 2 μM cisplatin for 1 h, washed in PBS, and released into fresh medium for 60 h. Next, cells were pulsed with 10 μM BrdU for 30–45 min and released into fresh medium for 0, 2, 4, 6, 8, or 10 h. Following harvest, cells were washed three times with PBS and fixed in ice-cold ethanol for at least 30 min at 4°C.
For DNA analysis, fixed cells were washed twice in PBS, treated with 50 μL of 100 μg/mL RNase, and stained with 50 μg/mL propidium iodide (PI) (Sigma). Samples were run on an LSRFortessa (Becton Dickinson) using 561-nm excitation for PI, with fluorescence being detected using a 610/10-nm filter. Cell doublets and debris were excluded from analysis, and at least 10,000 events were acquired per sample. DNA histogram analysis was performed using the Watson pragmatic algorithm with FlowJo software (Watson et al. 1987).
For BrdU staining, cells were washed twice in PBS, treated with 2N HCl for 20 min, and then washed three times in PBS with 0.1% Tween-20. Cells were treated with mouse anti-BrdU antibody (Becton Dickinson) for 30 min at room temperature, washed twice, and stained with rabbit anti-mouse FITC (Dako) for 30 min. Following two more washes, the cells were stained with PI as above. Samples were run on an LSR Fortessa using 561-nm excitation for PI and 488-nm excitation for FITC. Fluorescence was collected using a 610/10-nm bandpass filter and a 530/30-nm bandpass filter, respectively. Debris and doublets were excluded, and at least 10,000 BrdU-positive events were acquired. Data were analyzed in FlowJo with a gate set on BrdU-positive events, and PI histogram data of these events were obtained using the Watson pragmatic algorithm.
For phospho-histone H3 analysis, samples were washed twice and treated with mouse anti-phospho-histone H3 antibody (Ser10) (Cell Signaling Technology) for 60 min at room temperature. Cells were washed twice and stained with rabbit anti-mouse FITC (Dako) for 30 min. Following two more washes, the cells were stained with PI as above. Samples were run on an LSR Fortessa using 561-nm excitation for PI and 488-nm excitation for FITC. Fluorescence was collected using a 610/10-nm bandpass filter and a 530/30-nm bandpass filter, respectively. Debris and doublets were excluded, and at least 10,000 events were acquired. Data were analyzed in FlowJo.
DNA combing
DNA combing was performed as described (Michalet et al. 1997). Briefly, cells were sequentially pulsed with medium containing 20 μM IdU and 200 μM CldU for 20 min prior to harvest. DNA fibers were extracted in agarose plugs and stretched on salianized coverslips using the molecular combing system (Genomic Vision). CldU and IdU were detected using rat (ab6326, Abcam) and mouse FITC-conjugated (347583, Becton Dickinson) anti-BrdU antibodies, respectively. Alexa 594- or Alexa 488-conjugated goat anti-rat, chicken anti-goat, rabbit anti-mouse, and donkey anti-rabbit secondary antibodies were purchased from Molecular Probes (Invitrogen). The coverslips were mounted on glass slides using VectaShield mounting medium. Slides were viewed, and images were captured using an EC Plan-Neofluar 40×/1.3 objective. Images were processed as described (Michalet et al. 1997).
Acknowledgments
We thank Jean-Baptiste Vannier for help with the DNA-combing assays, John Rouse for providing SLX antibodies, and members of our laboratory for discussions. This work was supported by Cancer Research UK, the European Research Council, the Louis-Jeantet Foundation, the Swiss Bridge Foundation, and the Breast Cancer Campaign.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.238303.114.
References
- Andersen SL, Kuo HK, Savukoski D, Brodsky MH, Sekelsky J 2011. Three structure-selective endonucleases are essential in the ansence of BLM helicase in Drosophila. PLoS Genet 7: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayani J, Squire JA 2005. Sister chromatid exchange. Curr Protoc Cell Biol 25: 22.7.1–22.7.4 [DOI] [PubMed] [Google Scholar]
- Blanco MG, Matos J, Rass U, Ip SCY, West SC 2010. Functional overlap between the structure-specific nucleases Yen1 and Mus81–Mms4 for DNA damage repair in S. cerevisiae. DNA Repair 9: 394–402 [DOI] [PubMed] [Google Scholar]
- Breger KS, Smith L, Turker MS, Thayer MJ 2004. Ionizing radiation induces frequent translocations with delayed replication and condensation. Cancer Res 64: 8231–8238 [DOI] [PubMed] [Google Scholar]
- Budd ME, Tong AHY, Polaczek P, Boone C, Campbell JL 2005. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet 1: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castor D, Nair N, Déclais AC, Lachaud C, Toth R, Macartney TJ, Lilley DMJ, Arthur JS, Rouse J 2013. Cooperative control of Holliday junction resolution and DNA repair by the SLX1 and MUS81–EME1 nucleases. Mol Cell 52: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J 1974. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci 71: 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Hickson ID 2011. New insights into the formation and resolution of ultra-fine anaphase bridges. Semin Cell Dev Biol 22: 906–912 [DOI] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 26: 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying SM, Hickson ID 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, Russell P, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M, Neville S 1992. Conversion of excision-repairable DNA lesions to micronuclei within one cell cycle in human lymphocytes. Environ Mol Mutagen 19: 27–36 [DOI] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27: 6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E, Kim Y, Lach FP, Kottemann MC, Smogorzewska A 2013. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 5: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R 2007. The structure-specific endonuclease MUS81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP 2011. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 193: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM, Saunders WS 2004. Resolution of anaphase bridges in cancer cells. Chromosoma 112: 389–397 [DOI] [PubMed] [Google Scholar]
- Ip SCY, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC 2008. Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361 [DOI] [PubMed] [Google Scholar]
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A 2013. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood 121: 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13: 243–253 [DOI] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen SL, Skehel JM, West SC 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, West SC 2013. Cell cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Rep. 4: 76–86 [DOI] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A 1997. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science 277: 1518–1523 [DOI] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35: 116–127 [DOI] [PubMed] [Google Scholar]
- Naim V, Rosselli F 2009. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 11: 761–768 [DOI] [PubMed] [Google Scholar]
- Naim V, Wilhelm T, Debatisse M, Rosselli F 2013. ERCC1 and MUS81–EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol 15: 1008–1015 [DOI] [PubMed] [Google Scholar]
- Pflumm MF, Botchan MR 2001. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development 128: 1697–1707 [DOI] [PubMed] [Google Scholar]
- Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao ZH, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y 2007. Endogenous γ-H2AX–ATM–Chk2 checkpoint activation in Bloom’s syndrome helicase deficient cells is related to DNA replication arrested forks. Mol Cancer Res 5: 713–724 [DOI] [PubMed] [Google Scholar]
- Rass U, Compton SA, Matos J, Singleton MR, Ip SCY, Blanco MG, Griffith JD, West SC 2010. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev 24: 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW 2006. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell 17: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salewsky B, Schmiester M, Schindler D, Digweed M, Demuth I 2012. The nuclease hSNM1B/Apollo is linked to the Fanconi anemia pathway via its interaction with FANCP/SLX4. Hum Mol Genet 21: 4948–4956 [DOI] [PubMed] [Google Scholar]
- Shimura T, Torres MJ, Martin MM, Rao VA, Pommier Y, Katsura M, Miyagawa K, Aladjerm MI 2008. Bloom’s syndrome helicase and MUS81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. J Mol Biol 375: 1152–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AWM, et al. 2011. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet 43: 138–141 [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utani K, Kohno Y, Okamoto A, Shimizu N 2010. Emergence of micronuclei and their effects on the fate of cells under replication stress. PLoS ONE 5: e10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JV, Chambers SH, Smith PJ 1987. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry 8: 1–8 [DOI] [PubMed] [Google Scholar]
- Wechsler T, Newman S, West SC 2011. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Wyatt HDM, Sarbajna S, Matos J, West SC 2013. Coordinated actions of SLX1–SLX4 and MUS81–EME1 for Holliday junction resolution in human cells. Mol Cell 52: 234–247 [DOI] [PubMed] [Google Scholar]
- Ying SM, Minocherhomji S, Chan KL, Palmai-Pallag T, Chu WK, Wass T, Mankouri HW, Liu Y, Hickson ID 2013. MUS81 promotes common fragile site expression. Nat Cell Biol 15: 1001–1007 [DOI] [PubMed] [Google Scholar]