Abstract
Aqueous humor (AH) is one of the body fluids in the eye, which is known to be related with various ocular diseases, but the complete RNAs characteristic of the AH in patients is not yet known. The aim of this study was, with a microarray analysis, to reveal the disease-related extracellular miRNAs profiles in individual patients AH. 100 μl of AH was collected by anterior chamber paracentesis from 10 glaucoma, 5 cataract, and 5 epiretinal membrane patients. The extracted total RNAs were shorter than 200 nt, and their amount was 5.27 ± 0.41 ng in average. Among 530.5 ± 44.6 miRNA types detected in each sample with a microarray detectable 2019 types of matured miRNAs, 172 miRNAs were detected in all 10 glaucoma or control patients. From the glaucoma group, 11 significantly up-regulated and 18 significantly down-regulated miRNAs (P < 0.05 for both) were found to have areas under the curve better than 0.74 in a receiver operating characteristic analysis. They also formed a cluster composed only of glaucoma patients in a hierarchal cluster analysis. AH had a possibility of becoming a source of miRNA that can serve as a biomarker and a therapeutic target.
Increasing the quality of care expected by patients requires the development of improved diagnostic, prognostic and therapeutic monitoring tools for disease. Biomarkers of disease, including extracellular microRNAs (miRNAs) in bodily fluids, may play a role in achieving this purpose, as well as elucidating pathogenic mechanisms. MiRNAs have recently attracted attention for the prospect of earlier diagnosis and more precise application of existing therapies1.
MiRNAs are small, non-coding RNA molecules, consisting of 19–22 nucleotides. They have been intensely studied since the discovery two decades ago of the role of the lin-4 gene in regulating protein abundance2,3 with the general goals of understanding their role in translational repression and messenger RNA cleavage in various biological processes4,5,6, and elucidating pathologies in these processes6,7,8,9. After the recent discovery of circulating extracellular miRNAs in bodily fluids (e.g. plasma and serum), new searches for disease-related extracellular miRNAs have started in various fields4,10,11,12,13.
There are three sources of extracellular miRNAs in the eye: tears, the vitreous humor, and the aqueous humor (AH)11,14,15. Studies of ocular fluids and other cellular tissues that used a proteomic approach have identified several disease-related proteins16,17,18,19 including multiple Alzheimer's disease related peptides in the AH of eyes with glaucoma20,21. In comparison, research into ocular extracellular miRNAs is only at an early stage11,14,15 and still has great potential to uncover novel, accurate and valuable biomarkers and therapeutic targets.
Glaucoma is one of the leading causes of blindness worldwide22,23,24. The axons of retinal ganglion cells, which extend to the brain through the optic nerve, are irreversibly damaged in glaucoma, but with early detection and proper medical treatment, it is possible to slow the progress of this damage. The present study focused on finding AH miRNAs related to glaucoma. The AH is the most frequently sampled ocular fluid in examinations and clinical studies of ocular disease. It is secreted by the ciliary epithelium, which supplies nutrients and removes metabolic waste from the avascular tissues of the eye such as the lens and cornea. The AH is excreted through the angle of the anterior chamber, which consists of the trabecular meshwork, uvea, and sclera. Pressure regulation and dysregulation in this system is associated with glaucoma25. Luna et al reported that miR-29b has a potential role in glaucoma with an in vitro assay of the trabecular meshwork cells26,27. MiRNAs in the AH may have a key role in pathological conditions related to glaucoma, but the entire miRNA profile of the glaucomatous AH has not yet been determined.
The discovery of novel miRNAs has been accelerating with advances in analytical technologies28, and the total number of known human mature miRNAs has reached 2019 (miRBase Version 19, released on Aug. 1, 2012). The possibility of finding new and more useful biomarkers thus continues to rise. In this study, we used a microarray system able to detect all 2019 known mature miRNAs to analyze individual AH samples from patients with glaucoma, cataracts, and epiretinal membrane (ERM), with the aim of revealing specific diseases, individual expression characteristics, and the number of detectible miRNA types in the AH.
Results
Total RNA extraction from AH of ocular patients
Table 1 and supplementary Table S1 shows patient characteristics, quantity of RNA yielded, and the number of miRNAs detected by the microarray system. There was no significant difference in age between the 10 glaucoma patients (72.8 ± 3.0 years old) and the 10 control patients (cataract and epiretinal membrane, 71.1 ± 2.6 years old), who participated in this study (Supplementary Figure. S2a online). Intra ocular pressure of glaucoma group was significantly higher than control group (Supplementary Figure. S2b online). We were able to obtain high quality purified total RNA with the bioanalyzer. There was a significant quantity of RNA shorter than 200 nt, and no ribosomal RNA longer than 200 nt (specifically, 18S and 28S rRNA were absent) (Figure 1a). From a 100 μl sample of AH, we obtained 5.27 ± 0.54 ng of purified total RNA from the glaucoma patients and 6.19 ± 0.62 ng from the 10 control patients (the average for both groups was 5.73 ± 0.41 ng). There was no significant difference in the quantity of purified total RNA obtained from the two groups (Figure 1b).
Table 1. Patient characteristics, summary of RNA yield and number of miRNAs detected by microarray. POAG : primary open angle glaucoma PEX : pseudoexfoliation glaucoma, PACG : primary angle closure glaucoma.
| Patient no. | Age (years) | Sex | Preoperative diagnosis | Note | Amount of total RNA (ng) | Number of detected genes |
|---|---|---|---|---|---|---|
| Cont. 1 | 71 | M | Cataract | Hypertension | 6.54 | 539 |
| Cont. 2 | 77 | M | Cataract | Dislipidemia, diabetic melllitus | 4.59 | 821 |
| Cont. 3 | 66 | F | Cataract | Hypertension, dislipidemia | 1.43 | 626 |
| Cont. 4 | 66 | F | Cataract | 7.33 | 518 | |
| Cont. 5 | 69 | F | Cataract | Hypertension, dislipidemia, | 6.64 | 432 |
| Cont. 6 | 69 | F | Epiretinal Membrane | Hypertension | 5.81 | 490 |
| Cont. 7 | 84 | F | Epiretinal Membrane | Hypertension, dislipidemia, diabetic melllitus | 8.47 | 298 |
| Cont. 8 | 80 | M | Epiretinal Membrane | Hypertension | 7.24 | 465 |
| Cont. 9 | 74 | F | Epiretinal Membrane | Hypertension | 7.24 | 395 |
| Cont. 10 | 55 | F | Epiretinal Membrane | Dislipidemia | 6.62 | 1026 |
| Gla. 1 | 83 | F | Glaucoma (POAG), Cataract | Hypertension, diabetic melllitus | 6.06 | 602 |
| Gla. 2 | 64 | M | Glaucoma (PACG), Cataract | 5.56 | 801 | |
| Gla. 3 | 87 | M | Glaucoma (POAG), Cataract | Hypertension, dislipidemia | 6.24 | 583 |
| Gla. 4 | 75 | F | Glaucoma (POAG) | Hypertension, dislipidemia | 6.64 | 455 |
| Gla. 5 | 58 | F | Glaucoma (PEX) | 7.21 | 519 | |
| Gla. 6 | 70 | M | Glaucoma (PEX) | 3.40 | 345 | |
| Gla. 7 | 63 | M | Glaucoma (POAG) | Hypertension | 2.75 | 186 |
| Gla. 8 | 70 | M | Glaucoma (POAG) | Hypertension | 7.42 | 740 |
| Gla. 9 | 82 | M | Glaucoma (POAG) | Diabetic melllitus | 3.48 | 344 |
| Gla. 10 | 76 | F | Glaucoma (POAG) | 3.96 | 425 |
Figure 1. (a) Total RNA validation, screens capture of Agilent 2100 Bioanalyzer electropherograms of representative human AH samples (total RNA), (b) comparison of total RNA quantity in glaucoma and control patients.
Microarray analysis of miRNAs in AH
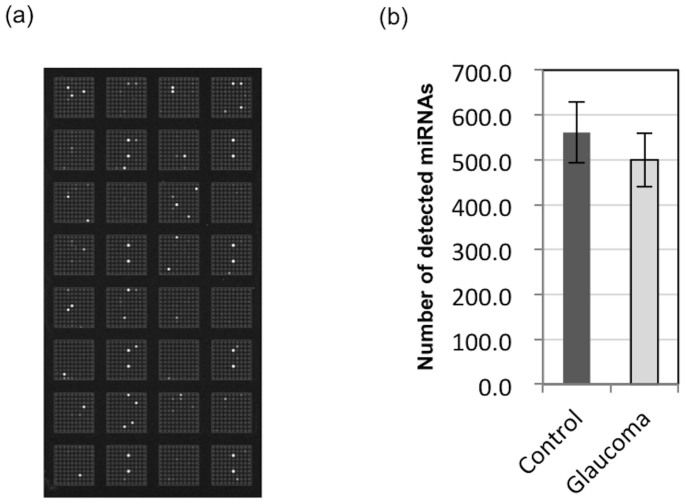
Even with the small amount of purified total RNA and without any amplification steps, bright spots were present on the microarray (Figure 2a). An average of 530.5 ± 44.6 miRNA types were detected in the 20 samples, with no correlation to the quantity of purified total RNA (R2 = 0.0197, Table 1, Supplementary Fig. S2 online), and no significant difference in the number of miRNAs detected in the glaucoma patients (500.0 ± 59.7) or control patients (561.0 ± 68.2) (Figure 2b).
Figure 2. (a) MiRNA microarray analysis, representative scanned image of a microarray capable of detecting 2019 miRNAs, and (b) comparison of detected miRNAs number in glaucoma and control patients.
Statistical analysis of microarray data of AH miRNAs
All of the values observed were normalized to the expression value of hsa-miR- 3940-5p, which was detected in all the samples (p = 0.985, ratio of 1.005, Supplementary Table S2 online). 57 miRNAs showed a statistically significant difference (p < 0.05, q < 0.2) in expression levels in the control patients and glaucoma patients (Supplementary Figure S2 online). In this group, hsa-let-7b-3p (Supplementary Table S3 online), has-miR-4507, has-miR-3620-5p, has-miR-1587, and has-miR-4484 were most significantly different (p < 0.01).
The numbers of miRNA types detected in the all of 10 glaucoma patients and the all of 10 control patients were 158 and 135, respectively (Figure 3a). 121 of these miRNAs (group A) were detected in all 20 patients. 14 miRNAs (group B) were detected in all glaucoma patients, but not detected in any control patients. 37 miRNAs (group C) were detected in all control patients, but not detected in any glaucoma patients. Among the 121 miRNAs of group A, 18 miRNAs showed a statistically significant difference (<0.05) in expression level in the control patients and glaucoma patients, as shown in Fig. 3b and supplementary Table S4 online. Among these 18 miRNAs, 8 were up-regulated and 10 were down-regulated in the glaucoma patients. Among the 14 miRNAs of group B, 3 miRNAs were up-regulated in the glaucoma patients, a significant difference (<0.05) (Figure 3c, and Supplementary Table S5 online). Among the 37 miRNAs of group C, 8 miRNAs were down-regulated in the glaucoma patients, a significant difference (<0.05) dFigure 3c, and Supplementary Table S6 online). A receiver operating characteristic (ROC) analysis of the 11 up-regulated and 18 down-regulated miRNAs showed that they all had an area under the curve (AUC) greater than 0.74 (Figure 3b, c, and d). A hierarchal cluster analysis with these 29 miRNA markers showed a cluster composed only of glaucoma patients was formed (Figure 4).
Figure 3.
(a) Venn diagram of the miRNAs detected in all control patients and all glaucoma patients. We detected 121 miRNAs in all 20 patients (group A), 14 miRNAs in all glaucoma patients, but not in all control patients (group B), and 37 miRNAs in all control patients, but not in all glaucoma patients (group C). (b) List of 18 miRNAs with a significant difference in glaucoma and control patients, screened from group A. (c) List of 3 miRNAs up-regulated in glaucoma patients and exhibiting a significant difference in the glaucoma and control patients, screened from group B. (d) List of 8 miRNAs down-regulated in glaucoma patients and exhibiting a significant difference in glaucoma and control patients, screened from group C.
Figure 4. Cluster analysis of selected 29 miRNA markers for glaucoma.
Targets and pathway analysis of glaucoma specific AH miRNAs
DIANA-microT analysis was performed on 29 miRNA markers and hsa-let-7b-3p. Sixty-two and 266 sets of up- and down- regulated miRNAs/targets were predicted to have miTG scores > 0.97000 (Supplementary Table S7 and S8 online). Ten and 9 molecules, respectively, were targeted by more than one up- or down-regulated miRNA (Table 2). The top 2 molecular networks predicted by the IPA analysis were (1) RNA post-transcriptional modification, developmental disorders, hereditary disorders, and (2) tissue development, neurological diseases, and auditory diseases (Table 3). The top 2 predicted canonical pathways were (1) protein kinase A signaling and (2) calcium signaling (Table 4).
Table 2. Molecules predicted by DIANA-microT (v3.0) to be targeted by more than one of the miRNAs that were significantly up- or down-regulated in the glaucoma patients. AUC: Area under the curve.
| Average | ||||||
|---|---|---|---|---|---|---|
| Name of miRNA | Control | Glaucoma | p-value (t-test) | Fold change | Difference ((Glaucoma)-(Control)) | AUC |
| hsa-miR-4507 | 0.095 | 0.069 | 0.005 | 0.728 | −0.026 | 0.85 |
| hsa-miR-3620-5p | 0.211 | 0.131 | 0.006 | 0.622 | −0.080 | 0.88 |
| hsa-miR-4484 | 0.071 | 0.117 | 0.008 | 1.639 | 0.045 | 0.84 |
| hsa-miR-5001-5p | 0.230 | 0.166 | 0.011 | 0.718 | −0.065 | 0.82 |
| hsa-miR-6132 | 0.514 | 0.365 | 0.011 | 0.710 | −0.149 | 0.84 |
| hsa-miR-6515-3p | 0.053 | 0.075 | 0.011 | 1.413 | 0.022 | 0.78 |
| hsa-miR-4467 | 0.972 | 0.768 | 0.013 | 0.790 | −0.204 | 0.82 |
| hsa-miR-3663-3p | 0.135 | 0.193 | 0.015 | 1.432 | 0.058 | 0.86 |
| hsa-miR-187-5p | 0.089 | 0.047 | 0.020 | 0.524 | −0.042 | 0.80 |
| hsa-miR-4433-3p | 0.044 | 0.064 | 0.022 | 1.474 | 0.021 | 0.80 |
| hsa-miR-6717-5p | 0.207 | 0.395 | 0.027 | 1.909 | 0.188 | 0.88 |
| hsa-miR-6722-3p | 0.213 | 0.110 | 0.027 | 0.517 | −0.103 | 0.78 |
| hsa-miR-4725-3p | 0.049 | 0.078 | 0.029 | 1.588 | 0.029 | 0.76 |
| hsa-miR-1202 | 0.044 | 0.061 | 0.032 | 1.388 | 0.017 | 0.84 |
| hsa-miR-3197 | 0.258 | 0.375 | 0.045 | 1.454 | 0.117 | 0.78 |
| hsa-miR-4749-5p | 0.159 | 0.126 | 0.046 | 0.794 | −0.033 | 0.75 |
| hsa-miR-1260b | 0.683 | 0.450 | 0.046 | 0.659 | −0.233 | 0.85 |
| hsa-miR-4634 | 0.185 | 0.136 | 0.050 | 0.738 | −0.048 | 0.75 |
Table 3. Top 5 molecular networks predicted by an IPA analysis of miRNA targets.
| Score | Focus Molecules | Top Diseases and Functions |
|---|---|---|
| 48 | 27 | RNA Post-Transcriptional Modification, Developmental Disorder, Hereditary Disorder |
| 37 | 22 | Tissue Development, Neurological Disease, Auditory Disease |
| 33 | 20 | Connective Tissue Disorders, Dermatological Diseases and Conditions, Gastrointestinal Disease |
| 32 | 22 | Organ Morphology, Renal and Urological System Development and Function, Cellular Assembly and Organization |
| 30 | 19 | Cellular Assembly and Organization, DNA Replication, Recombination, and Repair, Cell Morphology |
Table 4. Top 5 canonical pathways predicted by an IPA analysis of miRNA targets.
| Ingenuity Canonical Pathways | -log(p-value) | Ratio | Molecules |
|---|---|---|---|
| Protein Kinase A Signaling | 1.60E+00 | 2.46E-02 | PDE2A,NFAT5,FLNA,DUSP1,GNG2,CHP1,ELK1,PTPN12,CDC25A,PDE1C |
| Calcium Signaling | 1.54E+00 | 2.76E-02 | NFAT5,CHRFAM7A,CHP1,CHRNA7,TPM4,TRPC3 |
| Relaxin Signaling | 1.50E+00 | 3.05E-02 | PDE2A,GNAO1,GNG2,ELK1,PDE1C |
| Clathrin-mediated Endocytosis Signaling | 1.49E+00 | 3.03E-02 | SNX9,RAB5A,USP9X,CHP1,NUMB,MYO1E |
| Role of CHK Proteins in Cell Cycle Checkpoint Control | 1.47E+00 | 5.08E-02 | PPM1J,CDKN1A,CDC25A |
Discussion
The AH is an attractive source of novel miRNA biomarkers of ocular diseases, because of its accessibility, specificity, independence from other organs and novelty. Even though many researchers have reported various risk factors and biomarkers for ocular diseases epidemiologically and genetically29,30,31,32,33,34,35, studies of AH RNA are rare. The primary reason is that the AH is not a cellular sample, and the amount of total RNA in the AH is so small that it is undetectable with the usual spectophotometrical procedures. Even against such a challenging background, Dunmire JJ et al. reported a preliminary result suggesting that miRNAs exist in the AH in 201314, which raised interest in further study of AH RNA profiles and their use as ocular disease biomarkers.
In the present study, we evaluated the concentration of total RNA in the AH (14–85 ng/ml) for the first time, and found that it is similar to human breast milk (9.7–228.2 ng/ml) as reported by Kosaka N. et al.10, but less than in other body fluids (e.g. amniotic fluid, tears, seminal fluids, and so on) as reported by Jessica AW11. Additionally, the size distribution of RNA in the AH (<200 nt, Fig. 1a) is shorter than in breast milk (<300 nt). The high miRNA/small RNA purity in the AH may compensate for miRNAs detection and relevancy as a source of biomarkers.
One of the advancements of our study was the larger number of tested miRNAs than in any other study of miRNAs in other body fluids (e.g. 264 targets with a PCR array for an AH mixture from 5 cataract patients14, 723 targets with a micro array for breast milk10, and 706 targets with a PCR array for 12 body fluids11). Detected miRNAs in an individual AH sample reached five hundred on average, which was approx. 5 times more than that were detected in a mixture of 5 AH samples. Furthermore, our study is the first to reveal that individual miRNA profiles vary for every patient, and that the number of miRNA(s) detected in common was limited to 121 (26%, in average) in 20 patients.
This is the first report to identify a number of miRNAs in the AH, and compare them between glaucoma and control patients (cataract, and ERM) (Figure 3b, c, and d). We also found a higher AUC for these miRNAs in the glaucoma patients. In addition, Hsa-let-7b-3p was revealed as having the best odds ratio for glaucoma (7 glaucoma patients and 0 control patients) in this study (Supplementary Table S2 online). Furthermore, combining these markers has the potential to increase the sensitivity of glaucoma diagnosis (Figure 4).
We also found that miRNA profiles had the potential to reveal the stage of pathology, which is not possible with a genome analysis. For example, Let-7b miRNA, which had the best odds ratio for glaucoma in this study, has been reported to be an age-related marker of cataracts when expressed in the lens epithelium36, as well as a down-regulation marker of tumorigenesis and retinoblastomas37. Additionally, bioinformatics predicted which molecular targets (Table 2, Supplementary Table S6 online), molecular networks (Table 3), and canonical pathways (Table 4) were related to the detected miRNA biomarker candidates. Several predicted target molecules such as ENFB3, ESRRG, and BCL2 family (Table 2, Supplementary Table S7 online) have been already reported as glaucoma-related-molecules in clinical study and/or animal model38,39,40. The predicted data obtained in this study, may lead to useful findings on the pathology of glaucoma and may reveal novel therapeutic targets.
Glaucoma can be categorized into primary open angle glaucoma (POAG), normal tension glaucoma (NTG), primary angle closure glaucoma (PACG), and other subtypes, and their pathological conditions are extremely varied. In addition, medication for patients has a potential to influence in AH miRNA profiles. In a future study, we expect to find miRNA biomarkers that can diagnose these glaucoma subtypes, pathological conditions, and pharmacological effects. We also expect that discovery of new miRNAs will be dramatically accelerated by the development of new analyzer machines, such as next-generation sequencers28,41,42,43. Exhaustive analysis of miRNAs is expected to be carried out periodically to update our knowledge of biomarkers and therapeutic targets.
In summery, we found short RNA in the AH of our patients, and profiling with a microarray system identified several candidate miRNAs related to glaucoma. Additionally, more than a hundred miRNAs were found in most individual AH miRNA profiles that had potential as biomarkers and therapeutic targets for various ocular and other diseases.
Methods
AH sampling
This research (University Hospital Medical Information Network; UMIN Study ID N.: UMIN000011121) was conducted in accordance with the Declaration of Helsinki and Tohoku University Medical School's Institutional Review Board approved the protocol (2012-1-546). Informed consent was obtained from patients undergoing glaucoma, cataract, and epiretinal membrane surgery at Tohoku University Hospital, Sendai, Miyagi, Japan. AH was collected from patients who met the inclusion criterion of having had no history of cancer, asthma, and ocular diseases other than glaucoma, cataract, and epiretinal membrane.
The enrolment and grouping criteria were listed as follows: (1) POAG was diagnosed as glaucomatous optic neuropathy on fundoscopic examination, characteristic visual field defects on the Humphrey Field Analyzer according to the Anderson-Pattela classification confirmed in at least two visual field examinations, open anterior-chamber angles on gonioscopy, and elevated intraocular pressure (intraocular pressure (IOP) > 21 mm Hg by the Goldmann applanation tonometer); (2) The diagnosis criteria for NTG were identical to those for POAG, except that the IOP never exceeded 21 mm Hg; (3) PACG was defined as glaucomatous optic neuropathy, above characteristic visual field defect, and an occludible angle; (4) Pseudoexfoliation glaucoma (PEX) was defined in this study as glaucomatous optic neuropathy with the same characteristic visual field defects as the previously mentioned forms of glaucoma, an open anterior chamber angle in a gonioscopic examination, and the finding of characteristic exfoliative material on the anterior lens surface and/or in the iris in a slit-lamp examination, in one or both eyes. The presence of abnormal visual field defects meant that the results of a glaucoma hemifield test were outside normal limits and that a cluster of three or more nonedge points were present, all depressed on the pattern deviation plot at P < 0.05 (with one depressed at P < 0.01), as well as that the corrected pattern SD was significant at P < 0.05. An occludible angle was classified when the posterior trabecular meshwork could not be seen over an angle of 180 degrees or more without indentation.
Cataract and macular disorders such ERM are the most common non-exudative retinal diseases used as control groups in clinical ophthalmological studies, including those of glaucoma44. The eyes with ERM included in this study were exclusively eyes with idiopathic ERM, and did not have any other macular abnormalities. Eyes with secondary ERM (e.g., attributable to diabetic retinopathy, venous occlusion, retinal detachment, uveitis, or trauma) were thus excluded.
AH was collected from patients who met the inclusion criterion of having had no history of cancer, asthma, and ocular diseases other than glaucoma, cataract, and epiretinal membrane.
AH sampling and RNA isolation
Approximately 100 μl of AH was collected from each patient by anterior chamber paracentesis, using a 30-gauge needle inserted through the peripheral cornea at the beginning of the procedure. The needle did not contact any iris or lens tissue during the sample collection. AH was transferred to 1.5 ml siliconized tubes and immediately placed on ice. Samples were subsequently centrifuged for 10 min at 300 g for removing cellular components, and its aqueous phase was gently collected. The AH was mixed with 700 μl QIAzol Lysis reagent and stored at −80°C until further processing. Purification of miRNA from thawed AH/QIAzol samples were performed with miRNeasy Mini Kit (Qiagen, Valencia, CA). And fraction of small RNAs (<200 nucleotides) was conformed by Agilent 2100 bioanalyzer (Agilent teqnology) with a RNA 6000 Pico kit.
miRNA array preparation and analysis
MiRNA in purified RNA was measured using the Toray Industries miRNA analysis system, in which AH miRNA samples were hybridized to 3D-Gene human miRNA ver. 1.60 chips containing 2019 miRNAs (Toray Industries, Inc., Tokyo, Japan). MiRNA gene expression data were scaled by global normalization, and differential expression was analyzed45. Two-tailed, Welch's t-test was used to detect significant associations. P-values were adjusted for multiple testing based on the false discovery rate with the Bioconductor package q-value46.
Bioinformatical analysis
After a determination was made of which miRNAs had significantly different levels of expression in the control subjects and glaucoma patients, the molecular targets of these miRNAs were predicted with DIANA-microT v3.0 (http://diana.cslab.ece.ntua.gr/microT/)47. Pathway and global functional analyses were performed with IPA software48,49. The dataset uploaded to the IPA application was the list of target molecules with miTG scores > 0.97000 from the DIANA-microT prediction. Values of plus one and minus one were assigned to the molecules predicted to be targeted by down- and up- regulated miRNAs, respectively.
Author Contributions
Y.T. designed the research. H.K. and T.N. sampled the aqueous humor from the patients. Y.T. and J.S. analyzed the aqueous humor samples with the microarray system. Y.T., S.T., T.K., M.Y., K.M.N. and T.I. analyzed the data thus obtained. Y.T. prepared the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank the patients and medical staff from Tohoku university Hospital for their kind cooperation in this study. This study was supported by Leave a Nest - TORAY Industries award (Y.T.), Grants-in-Aid from the Ministry of Education, Science and Technology of Japan (24659756 for T.N., and 40625513 and 26670263 for Y.T.), and donative funds from Senju Pharmaceutical Co., Ltd and NIDEK Co., Ltd.
References
- Weiland M., Gao X. H., Zhou L. & Mi Q. S. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 9, 850–859 (2012). [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L. & Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75, 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I. & Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75, 855–862 (1993). [DOI] [PubMed] [Google Scholar]
- Ouellet D. L., Perron M. P., Gobeil L. A., Plante P. & Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J Biomed Biotech. 2006, 69616 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N. & Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 9, 102–114 (2008). [DOI] [PubMed] [Google Scholar]
- Belevych A. E. et al. MicroRNA-1 and -133 increase arrhythmogenesis in heart failure by dissociating phosphatase activity from RyR2 complex. PLoS One. 6, e28324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. MicroRNA expression profiles classify human cancers. Nature. 435, 834–883 (2005). [DOI] [PubMed] [Google Scholar]
- Baker M. RNA interference: MicroRNAs as biomarkers. Nature. 464, 1227 (2010). [DOI] [PubMed] [Google Scholar]
- Ju J. miRNAs as biomarkers in colorectal cancer diagnosis and prognosis. Bioanalysis. 2, 901–906 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Izumi H., Sekine K. & Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 1, 7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. A. et al. The microRNA spectrum in 12 body fluids. Clin Chem. 56, 1733–1741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubakov D. et al. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int J Legal Med. 124, 217–226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez M. A. et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 8, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunmire J. J., Lagouros E., Bouhenni R. A., Jones M. & Edward D. P. MicroRNA in aqueous humor from patients with cataract. Exp Eye Res 108, 68–71 (2013). [DOI] [PubMed] [Google Scholar]
- Ragusa M. et al. MicroRNAs in vitreus humor from patients with ocular diseases. Mol Vis 19, 430–440 (2013). [PMC free article] [PubMed] [Google Scholar]
- Richardson M. R. et al. Alterations in the aqueous humor proteome in patients with Fuchs endothelial corneal dystrophy. Mol Vis. 16, 2376–2383 (2010). [PMC free article] [PubMed] [Google Scholar]
- Kunikata H. et al. Intraocular concentrations of cytokines and chemokines in rhegmatogenous retinal detachment and the effect of intravitreal triamcinolone acetonide. Am J Ophthalmol. 155, 1028–1037 (2013). [DOI] [PubMed] [Google Scholar]
- Duan X. et al. Proteomic analysis of aqueous humor from patients with myopia. Mol Vis. 14, 370–377 (2008). [PMC free article] [PubMed] [Google Scholar]
- Bouhenni R. A. et al. Identification of differentially expressed proteins in the aqueous humor of primary congenital glaucoma. Exp Eye Res. 92, 67–75 (2011). [DOI] [PubMed] [Google Scholar]
- Inoue T., Kawaji T. & Tanihara H. Elevated levels of multiple biomarkers of Alzheimer's disease in the aqueous humor of eyes with open-angle glaucoma. Invest Ophthalmol Vis Sci. 54, 5353–5358 (2013). [DOI] [PubMed] [Google Scholar]
- Janciauskiene S., Westin K., Grip O. & Krakau T. Detection of Alzheimer peptides and chemokines in the aqueous humor. Eur J Ophthalmol. 21, 104–111 (2011). [DOI] [PubMed] [Google Scholar]
- Quigley H. A. Number of people with glaucoma worldwide. Br J Ophthalmol. 80, 389–393 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley H. A. & Broman A. T. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 90, 262–267 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnikoff S. et al. Global data on visual impairment in the year 2002. Bull World Health Organ. 82, 844–851 (2004). [PMC free article] [PubMed] [Google Scholar]
- Coca-Prados M. & Escribano J. New perspectives in aqueous humor secretion and in glaucoma: the ciliary body as a multifunctional neuroendocrine gland. Prog Retin Eye Res. 26, 239–262 (2007). [DOI] [PubMed] [Google Scholar]
- Luna C., Li G., Qiu J., Epstein D. L. & Gonzalez P. Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol Vis. 28, 2488–2497 (2009). [PMC free article] [PubMed] [Google Scholar]
- Luna C., Li G., Qiu J., Epstein D. L. & Gonzalez P. Cross-talk between miR-29 and transforming growth factor-betas in trabecular meshwork cells. Invest Ophthalmol Vis Sci. 52, 3567–3572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H. & Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 4, e22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana E. N. et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 20, 649–658 (2011). [DOI] [PubMed] [Google Scholar]
- Jaimes M., Rivera-Parra D., Miranda-Duarte A., Valdés G. & Zenteno J. C. Prevalence of high-risk alleles in the LOXL1 gene and its association with pseudoexfoliation syndrome and exfoliation glaucoma in a Latin American population. Ophthalmic Genet. 33, 12–17 (2012). [DOI] [PubMed] [Google Scholar]
- Burdon K. P. et al. Glaucoma risk alleles at CDKN2B-AS1 are associated with lower intraocular pressure, normal-tension glaucoma, and advanced glaucoma. Ophthalmol. 119, 1539–1545 (2012). [DOI] [PubMed] [Google Scholar]
- Carbone M. A. et al. Genes of the unfolded protein response pathway harbor risk alleles for primary open angle glaucoma. PLoS One. 6, e20649 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primus S., Harris A., Siesky B. A. & Guidoboni G. Diabetes: a risk factor for glaucoma? Br J Ophthalmol. 95, 1621–1622 (2011). [DOI] [PubMed] [Google Scholar]
- Marcus M. W., de Vries M. M., Junoy Montolio F. G. & Jansonius N. M. Myopia as a risk factor for open-angle glaucoma: a systematic review and meta-analysis. Ophthalmol. 118,1989–1994 (2011). [DOI] [PubMed] [Google Scholar]
- Marcus D. M. et al. Sleep disorders: a risk factor for normal-tension glaucoma? J Glaucoma. 10, 177–183 (2001). [DOI] [PubMed] [Google Scholar]
- Peng C. H. et al. MicroRNAs and cataracts: correlation among let-7 expression, age and the severity of lens opacity. Br J Ophthalmol. 96, 747–751 (2012). [DOI] [PubMed] [Google Scholar]
- Mu G. et al. Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let-7 down-regulation in retinoblastomas. Hum Pathol. 41, 493–502 (2010). [DOI] [PubMed] [Google Scholar]
- Libby R. T. et al. Susceptibility to neurodegeneration in a glaucoma is modified by Bax gene dosage. PloS Genet. 1, 17–26 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajaranant T. S. & Pasquale L. R. Estrogen deficiency accelerates aging of the optic nerve. Menopause. 19, 942–947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C. T., Tran T. & Sretavan D. Axonal/glial upregulation of EphB/ephrin-B signaling in mouse experimental ocular hypertension. Invest Ophthalmol Vis Sci. 51, 991–1001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M., Kawaji H., Grandjean V., Kiani J. & Rassoulzadegan M. Novel small noncoding RNAs in mouse spermatozoa, zygotes and early embryos. PLoS One. 7, e44542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H. et al. The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Genome Biol. 10, R40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin J. et al. FANTOM4 EdgeExpressDB: an integrated database of promoters, genes, microRNAs, expression dynamics and regulatory interactions. Genome Biol. 10, R39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkanen R. A. et al. Vitreous amino acid concentrations in patients with glaucoma undergoing vitrectomy. Arch Ophthalmol. 121, 183–188 (2003). [DOI] [PubMed] [Google Scholar]
- Ohno M. et al. The flavonoid apigenin improves glucose tolerance through inhibition of microRNA maturation in miRNA103 transgenic mice. Sci Rep. 3, 2553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- John D. S., Robert T. et al. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M. et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 10, 295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas Y. et al. Liver transcriptome profile in pigs with extreme phenotypes of intramuscular fatty acid composition. BMC Genomics. 13, 547 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat-Vidal C. et al. Identification of temporal and region-specific myocardial gene expression patterns in response to infarction in swine. PLoS One 8, e54785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information