Abstract
Cutaneous melanoma is an aggressive malignant tumor of melanocytes which accounts for 80% of skin cancer-related deaths. A number of driver mutations have been identified in melanoma, with the mutually exclusive BRAF V600E and NRAS Q61A mutations together accounting for roughly 70% of mutations. Simultaneous BRAF V600E and NRAS Q61A mutations in melanoma are rare, with evidence suggesting that up to 2.9% (2/69) of primary cutaneous melanomas carry both mutations. Here we describe a 42-year-old man with concurrent BRAF E586K and NRAS Q81K driver mutations. To our knowledge, this is the first description of these driver mutations occurring simultaneously in primary cutaneous melanoma.
Key Words: Cutaneous melanoma, Concurrent BRAF E586K and NRAS Q81K mutations, Skin cancer
Introduction
Primary cutaneous melanoma is an aggressive malignancy of melanocytes which accounts for only 4% of dermatologic malignancies, but is responsible for 80% of skin cancer-related deaths and, with metastatic disease, has a 5-year survival rate of only 14% [1]. The risk factors for melanoma are immunosuppression, ultraviolent radiation exposure, a family history of melanoma, and multiple benign or atypical nevi [2]. Histologically, melanomas are characterized by different growth patterns, such as superficial spreading, nodular, acral lentiginous, and lentigo maligna melanomas [3]. Many different melanoma driver mutations have been identified, with approximately 50–60% being mutated BRAF and 15–20% mutated NRAS Interestingly, the V600E and Q16R mutations account for roughly 90% of BRAF and NRAS mutations, respectively [4, 5]. Less common driver mutations include other BRAF and NRAS mutations as well as mutations in other genes such as c-Kit (2–6%), CTNNB1 (2–3%), GNA11 (2%), GNAQ (1%), and MEK1 (2–6%) [4, 5, 6]. In general, melanomas with concurrent driver mutations are rare; Goel et al. [6] examined 69 cases of primary cutaneous melanoma and found that only 2.9% (2/69) carried concurrent BRAF V600E and NRAS Q16R mutations.
The activating BRAF E586K mutation, causing constitutive enzyme heterodimerization and activation, is rare in melanomas [7, 8]. Similarly, the NRAS Q81K mutation accounts for a small percentage of melanoma NRAS driver mutations [5]. Here we describe the first reported case of concurrent BRAF E586K and NRAS Q16R driver mutations in the presence of primary cutaneous melanoma.
Case Report
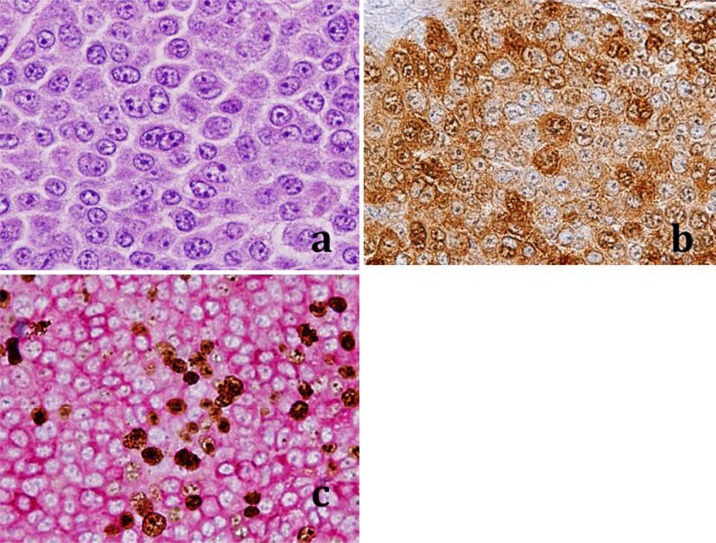
A 42-year-old male Caucasian with a history of chronic sun exposure presented with a mole on his left shoulder that had recently become red, itchy, and inflamed. The lesion was excised and HE sections were prepared. Histologic examination revealed sheets of pleomorphic epithelial cells and a histology consistent with cutaneous melanoma (fig. 1a). Immunohistochemical (IHC) S100 staining showed diffuse membranous and cytoplasmic staining in most malignant cells (fig. 1b). Dual Mart-1 and Ki-67 IHC staining showed strong membranous Mart-1 staining (red stain) and strong Ki-67 nuclear staining (brown stain). Dual S100 and Mart-1 IHC positivity is typical of melanomas (fig. 1b, c) [2]. The nuclear Ki-67 positivity showed a brisk mitotic index of approximately 20% (fig. 1c). IHC staining was performed at Reliapath (Opelousas, La., USA) according to regulations of the College of American Pathologists.
Fig. 1.
Representative HE and IHC staining results of the cutaneous melanoma removed from the patient's back. a High-power staining of the melanoma showing epithelioid pleomorphic malignant cells. b IHC staining showing S100 positivity typical of melanomas. c IHC staining showing dual Mart-1 (membranous) and Ki-67 (nuclear) positivity, also consistent with melanomas.
Several weeks later, a PET/CT scan of the patient revealed an abnormal fluorodeoxyglucose uptake by soft tissue nodules in the anterolateral chest wall which were suggestive of metastatic disease. Additionally, the resection of a jejunal mass and 4 local lymph nodes showed a metastatic melanoma. Next-generation sequencing was done on the metastatic jejunal melanoma to analyze the tumor for possible druggable driver mutations. A sequencing analysis of the BRAF exons 11 and 15 and the NRAS exons 2 and 3 revealed concurrent BRAF E586K and NRAS Q16R driver mutations. The analysis of the c-Kit exons 9, 11, 13, 17, and 18 revealed no mutations.
Discussion
NRAS mutations in melanoma commonly compromise the ability of the enzyme to hydrolyze guanosine triphosphate, allowing the enzyme to be constitutively active and activate downstream signaling proteins such as RAS and BRAF [9]. RAS and BRAF activities contribute to the malignant proliferation by inducing the activities of proteins including mTOR, cyclin D1, and NF-κB [9]. Additionally, RAS also activates the phosphatidylinositol 3-kinase/PTEN/Akt pathway, leading to apoptosis inhibition and malignant cell survival [9]. Concurrent NRAS and BRAF activating driver mutations are not expected to be common events, as they lie on the same molecular signal transduction pathway, and once one activating driver mutation occurs there would be little selective pressure for a second mutation to occur [6, 9].
To our knowledge, concurrent BRAF E586K and NRAS Q81K driver mutations have not been previously identified in melanomas. The clinical treatment of this rare combination of concurrent driver mutations in this case may be difficult, as it is unknown whether the BRAF E586K mutation responds to BRAF inhibitors. Additionally, there is in vitro evidence that NRAS mutations may confer resistance to BRAF inhibitors [5, 10].
References
- 1.Cancer Facts and Figures. Atlanta American Cancer Society; 2003. [Google Scholar]
- 2.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 3.Smoller BR. Histologic criteria for diagnosing primary cutaneous malignant melanoma. Mod Pathol. 2006;19(suppl 2):S34–S40. doi: 10.1038/modpathol.3800508. [DOI] [PubMed] [Google Scholar]
- 4.My Cancer Genome: molecular profiling of melanoma. http://www.mycancergenome.org/content/disease/melanoma
- 5.Kunz M. Oncogenes in melanoma: an update. Eur J Cell Biol. 2014;93:1–10. doi: 10.1016/j.ejcb.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 7.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R, Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 8.Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan RJ, Flaherty K. MAP kinase signaling and inhibition in melanoma. Oncogene. 2013;32:2373–2379. doi: 10.1038/onc.2012.345. [DOI] [PubMed] [Google Scholar]
- 10.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]