Abstract
Physical inactivity contributes to cardiovascular disease, type II diabetes, obesity, and some types of cancer. While the literature is clear that there is genetic regulation of physical activity with existing gene knockout data suggesting that skeletal muscle mechanisms contribute to the regulation of activity, actual differences in end-protein expression between high- and low-active mice have not been investigated. This study used two-dimensional differential gel electrophoresis coupled with mass spectrometry to evaluate the proteomic differences between high-active (C57L/J) and low-active (C3H/HeJ) mice in the soleus and extensor digitorum longus (EDL). Furthermore, vivo-morpholinos were used to transiently knockdown candidate proteins to confirm their involvement in physical activity regulation. Proteins with higher expression patterns generally fell into the calcium-regulating and Krebs (TCA) cycle pathways in the high-active mice (e.g., annexin A6, P = 0.0031; calsequestrin 1; P = 0.000025), while the overexpressed proteins in the low-active mice generally fell into cytoskeletal structure- and electron transport chain-related pathways (e.g., ATPase, P = 0.031; NADH dehydrogenase, P = 0.027). Transient knockdown of annexin A6 and calsequestrin 1 protein of high-active mice with vivo-morpholinos resulted in decreased physical activity levels (P = 0.001). These data suggest that high- and low-active mice have unique protein expression patterns and that each pattern contributes to the peripheral capability to be either high- or low-active, suggesting that different specific mechanisms regulate activity leading to the high- or low-activity status of the animal.
Keywords: mouse wheel running, annexin A6, calsequestrin 1, 2D-DIGE, vivo-morpholinos
regular physical activity prevents cardiovascular disease, obesity, and type II diabetes (33). When objectively measured, ∼3.5% of adults meet the recommended daily physical activity guidelines (38). Data have shown that physical activity levels are influenced by genetic mechanisms (14, 20, 29, 30, 33, 35, 51); however, the identity of the genetic mechanisms involved remains unclear despite efforts to determine genomic variance and transcripts associated with activity levels. Genetic variance and transcript data only illustrate a portion of the potential regulatory mechanisms for voluntary physical activity, and neither avenue of investigation accurately predicts the final protein products that are associated with the phenotype (6, 10, 13).
Two-dimensional differential gel electrophoresis (2D-DIGE) is an accurate and reliable method to identify global proteomic differences (8, 11, 54). While 2D-DIGE paired with mass spectroscopy provides protein identification, the results are correlative with end phenotype. To determine causal mechanisms 2D-DIGE results must be confirmed with an in vivo method to knockdown identified proteins to determine the proteins' role in the phenotype of interest. Vivo-morpholinos are a novel tool developed by Morcos and colleagues (39) that allows for transient knockdown of a targeted protein. Our lab has evaluated vivo-morpholino effectiveness in the physical activity model in terms of delivery to brain and skeletal muscle tissue and washout time courses of vivo-morpholinos, as well as appropriate vivo-morpholino dosages and controls (9). Our results show that vivo-morpholinos are an ideal tool to be paired with 2D-DIGE and mass spectroscopy to identify mechanisms associated with the physical activity phenotype.
Kelly et al. (23) has hypothesized that the regulation of physical activity arising through genetic control mechanisms has both central (brain) and peripheral (skeletal muscle) components. Indeed, several studies have shown that alteration of skeletal muscle gene expression [e.g., Glut4(50); IL-15Rα (42)] without alteration in central brain gene expression can markedly change voluntary physical activity. Thus the purpose of this study was to identify proteomic differences between high- and low-active mice in slow- and fast-twitch skeletal muscle, followed by causal determination of the role of strong candidate proteins in regulating physical activity.
METHODS
This study was conducted in two experiments; both experiments were submitted and approved by the IACUC at Texas A&M University.
Experiment 1: proteome determination (Fig. 1).
Fig. 1.
General study timeline for the 2 experiments. Experiment 1 was the global proteome experiment whereas experiment 2 was the transient-silencing experiment to confirm calsequestrin 1 (Casq1) and annexin A6 (Anxa6) roles in physical activity.
Twelve 8-wk-old, low-active C3H/HeJ mice (6 male and 6 female) and twelve 8-wk-old high-active C57L/J mice (6 male and 6 female) were obtained from The Jackson Laboratory (Bar Harbor, ME). Previous extensive work from our lab has shown that the low C3H/HeJ mice have a lower physical activity level (1.2 ± 1.7 km run per day) compared with the high C57L/J (10.7 ± 2.7 km run per day) mice (26–30, 34, 35). Our standard operating procedure has been to individually house mice at 9 wk of age with running wheels equipped with computers (Sigma Sport, St. Charles, IL) to measure average daily distance run (km/day), duration of activity (min/day), and average speed of activity (m/min) (34). However, due to our concern that exposure to the running wheel would alter protein expression in the skeletal muscle (4), the mice, after 1 wk of acclimation, were individually housed with locked running wheels for 1 wk. Locked running wheels were used to simulate the environment of previous physical activity studies from this lab (34) while preventing wheel running. All animals were housed in an AAALAC-certified vivarium maintained at 18–21°C and 20–40% humidity with 12:12-h light-dark cycles that initiated at 6:00 AM. Food (Harland Teklad 8604 Rodent Diet, Madison, WI) and water were provided ad libitum. Body masses (to the nearest 0.1 g) were collected once per week throughout the study and body composition was determined prior to euthanasia using a GE Lunar Piximus dual X-ray absorptiometer (GE Healthcare Waukesha, WI).
At 10 wk of age, mice were anesthetized using vaporized isoflurane followed by cervical dislocation. The soleus (peripheral slow-twitch skeletal muscle) and extensor digitorum longus (EDL, peripheral fast-twitch skeletal muscle) (2) were removed and flash frozen for later analysis. Subsequently, for protein extraction, tissues were placed in Tris/CHAPS lysis buffer and homogenized. Protein concentration was determined by Bradford assay.
Two-dimensional differential in-gel electrophoresis (2-D DIGE) and protein identification were initiated using techniques previously published (21, 22). In brief, protein was precipitated with chloroform/methanol and dissolved in DIGE labeling buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, pH 8.5). Samples were fluorescently labeled by combining either 50 μg of EDL protein or 45 μg of soleus protein with 200 pmol CyDye DIGE Fluors (GE Healthcare). One sample was labeled with Cy3 while the other was labeled with Cy5 to allow for preferential labeling. A pooled sample containing equal amounts of each sample was labeled with Cy2. The labeling reactions were quenched with 10 mM lysine. The samples were randomly mixed so that one Cy3- and one Cy5-labeled sample were loaded on a single gel, along with the Cy2-labeled pooled sample, which was used as an internal standard and allowed for each resolved protein to be semiquantitatively assessed relative to the standard within and between all gels, thereby minimizing gel-to-gel variation. Unlabeled protein samples were added to the labeled proteins (800 μg total protein for EDL and 328.5 μg total protein for soleus) and used to rehydrate the IPG strips (either 24 cm, pH 4–7, Immobiline DryStrip for EDL or 13 cm, pH 4–7, Immobiline DryStrip for soleus, GE Healthcare) by passive diffusion. Isoelectric focusing was performed on an IPGPhor (GE Healthcare) with a program of 500 V for 1 h followed by 1,000 V for 1 h followed by a linear gradient to 8,000 V until ∼60,000 V·h for EDL or 40,000 V·h for soleus was achieved. The focused strips were equilibrated in two steps: 15 min in SDS equilibration buffer I (6 M urea, 2% SDS, 30% glycerol, 50 mM Tris, pH 8.8, 0.01% bromophenol blue, and 10 mg/ml DTT) followed by 15 min with equilibration buffer II in which the DTT was replaced by 25 mg/ml iodoacetamide. The equilibrated IPG strips were placed directly on top of polymerized 12% SDS gels and covered with low-melt agarose. Gels were run in cooled tanks at 1 W per gel until the dye front was at the bottom of the gel.
Gel images were obtained using a Typhoon Trio, Variable Mode Imager (GE Healthcare). DeCyder software (GE Healthcare, version 6.5) was used to detect spots, subtract background, and to normalize spots against the pooled standard. Further, DeCyder was used to match spots between gels and determine significant changes in protein expression (P < 0.05) and the average ratio. Average ratio was derived from the normalized spot volume standardized against the intragel standard, thus providing a measure of the magnitude of expression differences between identified proteins. Spot detection and matching were verified visually. Gels to be used for spot picking were fixed in 10% methanol and 7.5% acetic acid overnight and poststained with Deep Purple Total Protein Stain (GE Healthcare) and imaged on the Typhoon. The poststained spots were matched to the CyDye gel images using DeCyder software. Picking and digestion were performed using Ettan robotic components (GE Healthcare).
Spots that showed significant (P < 0.05) differences in abundance between low-active (C3H/HeJ) and high-active (C57L/J) mice were robotically picked, washed, and digested (GE Healthcare, Ettan system) with recombinant porcine trypsin (Promega, Madison, WI) as described elsewhere (40). Extracted tryptic peptides were concentrated by SpeedVac and analyzed by nanoelectrospray ionization/ion-trap mass spectrometry (LC/MS/MS with an LTQ XL linear; ThermoFinnigan, San Jose, CA). Subsequently, peptides from the MS were identified using the MASCOT search engine. The MASCOT program (v2.2) searched the mouse genome using the following parameters for protein identification: 1) one missed cleavage by trypsin; 2) monoisotopic peptide masses; 3) peptide mass tolerance of 1.2 Da; and 4) fragment mass tolerance of 0.8 Da. Further, oxidation of methionine (variable modification) and carbamidomethylation (fixed modification) of cysteine were taken into consideration by MASCOT in the protein identification. Peptides were matched to proteins at a minimum of two peptides. Protein identifications were verified by Scaffold (Proteome Software, Portland, OR). Proteins were then categorized based on their function using String 9.0 (48).
Based on the 2D-DIGE results, two highly significantly expressed (P < 0.001) proteins [annexin A6 (Anxa6) and calsequestrin 1 (Casq1), see results] were validated using standard Western blot techniques. Briefly, proteins from three randomly chosen samples of each strain were separated by SDS-PAGE, and then transferred to a nitrocellulose membrane with transfer confirmed by Ponceau S stain. Membranes were incubated overnight in 1:1,000 ratio of primary antibody recognizing Anxa6 (Biorbyt, San Francisco CA) or Casq1 (GeneTex, Irvine, CA) and blocking buffer (5% nonfat dried milk, 0.5% Tween 20). Membranes were then incubated in the secondary horseradish peroxidase antibody (Cell Signaling Technology, Beverly, MA). Chemiluminescence was imaged and all analysis was performed with a Flurochem analyzer (Derbyshire, UK). Additionally GAPDH (GeneTex) was analyzed as a loading control. GAPDH was used as a loading control as opposed to actin or tubulin because actin and tubulin structures were expressed differentially between the low C3H/HeJ and high C57L/J mice. There was no difference in GAPDH expression between the high- and low-active mice. Western blots were analyzed using the individual protein band's optical density that allowed for a semiquantitative estimate of protein level.
Experiment 2: role of candidate proteins in physical activity (Fig. 1).
The 2D-DIGE results showed that Anxa6 and Casq1 were highly significantly overexpressed (P < 0.001) in high-active mice. Given that both proteins are involved in calcium homeostasis of skeletal muscle (5, 44), Anxa6 and Casq1 were determined to be strong candidates for the peripheral regulation of physical activity and selected for further evaluation using transient gene silencing vivo-morpholinos. Forty-two high-active C57L/J male mice were obtained from The Jackson Laboratory. Male mice were chosen since there was no difference in protein expression between males and females (see 2D-DIGE results) and to prevent any effects of the estrous cycle of female mice on physical activity response (12). The mice were randomly divided into three treatment groups: 1) control mice (n = 15) that were treated with physiological saline; 2) Anxa6-targeted mice (n = 15); and 3) Casq1-targeted mice (n = 12). Previous results from our lab have shown that physiological saline is an appropriate control compared with vivo-morpholino scramble in terms of physical activity response and expression of targeted protein (9).
The vivo-morpholino study was a partial mirror of the proteomic study because the goal was to keep the mice of both studies at a similar age. We worked to keep the mice at the same age between the two experiments because physical activity level of mice is altered with aging (51). At 8 wk of age, mice were individually housed on running wheels with ad libitum access to food and water for 3 days to establish baseline wheel running. Physical activity was measured by using a computer (Sigma Sport BC500) that monitored revolutions of a solid-surface running wheel mounted in the mouse's cage. As noted earlier, this system has been extensively validated in past studies (e.g., 34). Daily distance (km/day) and duration of activity (min/day) were measured and averaged for each treatment week (baseline, treatment, recovery). Following baseline wheel running the running wheel was locked for 4 days to mirror the settings of the 2D-DIGE study (described previously) and prevent the expression of proteins because of a training effect. At the end of the first week (3 days running, 4 days locked wheel), three mice were euthanized as described above, and the soleus and EDL were extracted to determine baseline levels of Anxa6 and Casq1 expression using Western blotting techniques described earlier. During the subsequent treatment week, mice were administered either vivo-morpholinos targeting Anxa6 or Casq1 (11 mg/kg, GeneTools, Philomath, OR) or equal volume physiological saline (∼56 μl) via tail-vein injection on treatment days 1, 3, 5, and 7. After the vivo-morpholino treatment week, mice were allowed to recover for 1 wk (days 8–15). Throughout both the treatment and recovery weeks, the wheels were unlocked and activity was measured by distance run per day (9).
To establish the protein expression pattern during the treatment (days 1–7) and recovery weeks (days 8–15) a cohort (n = 3) of mice were euthanized at intervals throughout the protocol. Control mice soleus and EDL were analyzed on days 2, 4, 8, and 15, the Anxa6 vivo-morpholino group's tissue was analyzed on days 2, 3, 4, and 15, and the Casq1 group's tissue was analyzed on days 2, 3, and 15. Initially, tissues from the vivo-morpholino groups were to be analyzed on a similar pattern, as was the control group; however, due to vivo-morpholino production issues, several mice had to be excluded from analysis. Protein knockdown was measured by Western blot as described above. Additionally, to provide a comparison to the low-active (C3H/HeJ) mice, Anxa6 and Casq1 expression in the soleus from the vivo-morpholino-treated animals (treatment day 3) and low C3H/HeJ mice were analyzed using Western blots.
Statistics.
Mouse physical characteristics were analyzed with an ANOVA to compare strain (C57L/J and C3H/HeJ), sex (male and female), weight, and percent body fat. An alpha level of 0.05 was set a priori and if a significant main effect was found, a Tukey's HSD post hoc test was used. 2D-DIGE results were analyzed using ANOVA, with significantly differentially expressed protein spot determination denoted with an a priori alpha value of 0.05, with subsequent protein identification (mass spectroscopy identification) using an a priori alpha value of 0.05. The conformational Western blot optical densities (experiment 1) were analyzed using a Student's t-test (GraphPad Software, La Jolla, CA) to compare protein levels between high-active and low-active mice with an alpha level of 0.05 set a priori. For the Western blots used to analyze vivo-morpholino results (experiment 2), a 2 × 2 ANOVA was used to compare day of treatment and treatment group with an alpha level of 0.05 set a priori and a Tukey's HSD post hoc test if main effects were significant. Further comparisons of wheel running with vivo-morpholino treatment were analyzed using a 3 × 3 ANOVA with week (baseline, treatment, and recovery) and treatment group (control, Anxa6 vivo-morpholino, and Casq1 vivo-morpholino) as the main effects. An alpha level of 0.05 was set a priori. If a main effect was found significant, a Tukey's HSD post hoc test was used.
RESULTS
Experiment 1: proteome determination.
The physical characteristics of the mice showed that the male mice in both strains were significantly (P = 0.001) heavier than the females (C3H/HeJ males 21.25 ± 1.60 g; C3H/HeJ females 18.91 ± 1.06 g; C57L/J males 22.21 ± 1.16 g; C57L/J females 17.21 ± 0.92 g), yet there was no difference (P = 0.0501) in the percent body fat (C3H/HeJ males 16.54 ± 3.75%; C3H/HeJ females 14.64 ± 1.64%; C57L/J males 13.23 ± 1.92%; C57L/J females 12.62 ± 1.75%) of the mice between or within strains.
There were 42 differentially expressed proteins identified in the soleus (Table 1) and eight differentially expressed proteins in the EDL (Table 2). Of the differentially expressed soleus proteins, the coding genes for 11 are located in known physical activity quantitative trait loci (QTL; Table 1) while the coding genes for three EDL proteins are located in QTL for physical activity (Table 2). In general, when examining protein differences based on their function, the low-active C3H/HeJ mice exhibited the overexpression of proteins associated with the electron transport chain and cytoskeletal structure while the high-active C57L/J animals overexpressed proteins associated with the Krebs (TCA) cycle and calcium regulation (Table 3).
Table 1.
Soleus differentially expressed proteins between high- and low-active mice
| Protein | Strain Expressed In | P | Avg Ratio | No. of Identified Peptides | % Sequence Coverage | QTL |
|---|---|---|---|---|---|---|
| Calsequestrin 1 | C57L/J | 0.000025 | 1.78 | 2 | 5 | N/A |
| Radixin | C3H/HeJ | 0.00028 | 1.28 | 2 | 3 | N/A |
| Annexin A6 | C57L/J | 0.00031 | 1.53 | 15 | 26 | Mini Muscle (14) |
| Phosphoenolpyruvate carboxykinase | C3H/HeJ | 0.00042 | 1.5 | 6 | 15 | N/A |
| Peroxiredoxin-6 | C3H/HeJ | 0.00079 | 1.46 | 8 | 32 | N/A |
| [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1 | C57L/J | 0.0022 | 1.37 | 2 | 4 | N/A |
| Transferrin | C3H/HeJ | 0.0026 | 1.44 | 12 | 17 | N/A |
| Lumican | C57L/J | 0.004 | 1.49 | 3 | 9 | N/A |
| Synaptic vesicle membrane protein VAT-1 homolog | C57L/J | 0.0046 | 1.27 | 3 | 10 | N/A |
| Serine protease inhibitor A3K | C57L/J | 0.005 | 1.91 | 6 | 15 | N/A |
| Epoxide hydrolase 2 | C57L/J | 0.0051 | 1.21 | 8 | 13 | Duration (29) |
| Thioredoxin reductase 1 | C57L/J | 0.0051 | 1.21 | 3 | 5 | Duration (29) |
| Myosin, light polypeptide 3 | C57L/J | 0.0062 | 1.9 | 3 | 14 | N/A |
| Apolipoprotein A-I | C57L/J | 0.0062 | 1.9 | 3 | 12 | N/A |
| Atp5b | C3H/HeJ | 0.007 | 1.38 | 12 | 31 | Speed (29) |
| Tubulin beta-3 | C3H/HeJ | 0.007 | 1.38 | 2 | 12 | Duration (29) |
| Vimentin | C3H/HeJ | 0.007 | 1.38 | 2 | 5 | N/A |
| Hemopexin | C3H/HeJ | 0.018 | 1.34 | 2 | 4 | Distance/Duration (23) |
| Phosphomannomutase 2 | C57L/J | 0.018 | 1.31 | 4 | 19 | N/A |
| Hypothetical protein Hoch_2831 | C57L/J | 0.023 | 2.09 | 2 | 1 | N/A |
| Annexin A4 | C57L/J | 0.023 | 1.16 | 8 | 27 | N/A |
| Pyruvate dehydrogenase E1 component subunit beta | C57L/J | 0.023 | 1.16 | 6 | 18 | N/A |
| Alpha-2-HS-glycoprotein | C3H/HeJ | 0.026 | 1.26 | 2 | 24 | N/A |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 | C3H/HeJ | 0.027 | 1.31 | 4 | 17 | Average Speed (23) |
| Cofilin2 | C3H/HeJ | 0.027 | 1.31 | 2 | 12 | N/A |
| Vinculin | C3H/HeJ | 0.028 | 1.23 | 3 | 3 | Duration (29) |
| Tripartite motif-containing protein 72 | C57L/J | 0.029 | 1.27 | 7 | 16 | N/A |
| Sarcalumenin | C57L/J | 0.029 | 1.27 | 7 | 15 | N/A |
| Pyruvate dehydrogenase protein X component | C57L/J | 0.029 | 1.27 | 3 | 7 | N/A |
| FK506 binding protein 4 | C57L/J | 0.029 | 1.27 | 2 | 5 | N/A |
| Alpha-1 protease inhibitor 2 | C3H/HeJ | 0.031 | 1.28 | 4 | 21 | N/A |
| Alpha-1-antitrypsin 1-4 | C3H/HeJ | 0.031 | 1.28 | 4 | 21 | N/A |
| Serpina1c protein | C3H/HeJ | 0.031 | 1.28 | 7 | 16 | N/A |
| Alanyl-tRNA synthase | C3H/HeJ | 0.031 | 1.15 | 8 | 8 | N/A |
| ATPase | C3H/HeJ | 0.031 | 1.15 | 7 | 8 | N/A |
| Aldehyde dehydrogenase | C3H/HeJ | 0.031 | 1.14 | 5 | 10 | N/A |
| 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 1 | C3H/HeJ | 0.031 | 1.14 | 2 | 4 | N/A |
| 2-Oxoglutarate dehydrogenase | C3H/HeJ | 0.035 | 1.16 | 5 | 6 | Mini-muscle (14) |
| Annexin A5 | C3H/HeJ | 0.036 | 1.25 | 9 | 29 | N/A |
| 14-3-3 Protein gamma subtype | C57L/J | 0.036 | 1.2 | 5 | 16 | N/A |
| Ubiquinone biosynthesis protein COQ9 | C57L/J | 0.042 | 1.24 | 4 | 16 | Distance (23) |
| Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase | C57L/J | 0.22 | 1.24 | 5 | 11 | Distance (23) |
Protein, name of the identified protein; strain expressed in, the mouse strain in which the identified protein was expressed; P, significance level of differential protein expression as obtained from DeCyder; Avg ratio was derived from the normalized spot volume standardized against the intra-gel standard provided by Decyder-software analysis, thus providing a measure of the magnitude of expression differences between identified proteins; No. of Identified Peptides, the number of peptides identified from the picked gel spot; % Sequence Coverage, the percentage of the number of amino acids identified in comparison to the total number of amino acids in the protein sequence, thus allowing for a confidence interval of 95% that the peptide sequence corresponds to the identified protein; QTL indicates if the identified protein is in a Quantitative Trait Loci for a coding gene associated with voluntary physical activity along with the literature reference for that QTL.
Table 2.
EDL differentially expressed proteins between high and low-active mice
| Protein | Strain Expressed In | P | Avg Ratio | No. of Identified Peptides | % Sequence Coverage | QTL |
|---|---|---|---|---|---|---|
| V-type proton ATPase catalytic subunit A | C57L/J | 0.0016 | 3.72 | 2 | 4 | N/A |
| Annexin A6 | C57L/J | 0.0016 | 2.11 | 15 | 24 | Mini Muscle (14) |
| Sdha | C3H/HeJ | 0.0016 | 1.43 | 6 | 10 | N/A |
| Electron transfer flavoprotein-ubiquinone oxidoreductase | C3H/HeJ | 0.0016 | 1.43 | 3 | 6 | N/A |
| Alpha-actinin-2 | C3H/HeJ | 0.0044 | 1.82 | 2 | 2 | Speed (29) |
| Apolipoprotein A-I | C57L/J | 0.0046 | 2.2 | 2 | 7 | N/A |
| T-complex protein 1 subunit zeta | C3H/HeJ | 0.035 | 1.28 | 4 | 7 | N/A |
| Alpha-tubulin 8 | C3H/HeJ | 0.05 | 1.45 | 3 | 6 | Speed (29) |
See Table 1 for description of column headings.
Table 3.
Protein expression profiles based on function of the protein
| Protein | Strain Overexpressed In | Tissue Expressed In |
|---|---|---|
| Electron transport chain | ||
| Atp5b protein | C3H/HeJ | Soleus |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 | C3H/HeJ | Soleus |
| ATPase | C3H/HeJ | Soleus |
| Electron transfer flavoprotein ubiquinone oxidoreductase | C3H/HeJ | Soleus |
| TCA cycle | ||
| Pyruvate dehydrogenase protein X component | C57L/J | Soleus |
| Pyruvate dehydrogenase E1 component subunit beta | C57L/J | Soleus |
| Pyruvate dehydrogenase [acetyl-transferring]-phosphatase 1 | C57L/J | Soleus |
| Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | C57L/J | Soleus |
| 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 1 | C3H/HeJ | Soleus |
| Phosphomannomutase 2 | C57L/J | Soleus |
| Sdha protein | C3H/HeJ | EDL |
| Oxygen transport | ||
| Transferrin | C3H/HeJ | Soleus |
| Oxidative stress response | ||
| Hemopexin | C3H/HeJ | Soleus |
| Thioredoxin reducatse 1 | C57L/J | Soleus |
| Peroxiredoxin-6 | C3H/HeJ | Soleus |
| Ubiquinone biosynthesis protein COQ9 | C57L/J | Soleus |
| Protein regulation | ||
| Serine protease inhibitor A3K | C57L/J | Soleus |
| alanyl-tRNA Synthase | C3H/HeJ | Soleus |
| FK506 binding protein 4 | C57L/J | Soleus |
| Alpha-1 protease inhibitor 2 | C3H/HeJ | Soleus |
| Alpha-1-antitrypsin 1-4 precursor | C3H/HeJ | Soleus |
| Calcium regulation | ||
| Annexin A6 | C57L/J | Soleus/EDL |
| Calsequestrin 1 | C57L/J | Soleus |
| Sarcalumenin | C57L/J | Soleus |
| Annexin A4 | C57L/J | Soleus |
| Structural proteins | ||
| Radixin | C3H/HeJ | Soleus |
| Lumican | C57L/J | Soleus |
| Tripartite motif-containing protein 72 | C57L/J | Soleus |
| Vinculin | C3H/HeJ | Soleus |
| Vimentin | C3H/HeJ | Soleus |
| Tubulin beta-3 | C3H/HeJ | Soleus |
| Coflin-2-like | C3H/HeJ | Soleus |
| Alpha-actinin-2 | C3H/HeJ | EDL |
| T-complex protein 1 subunit zeta | C3H/HeJ | EDL |
| Alpha tubulin 8 | C3H/HeJ | EDL |
| Other | ||
| Epoxide hydrolase 2 | C57L/J | Soleus |
| Aldehyde dehydrogenase | C3H/HeJ | Soleus |
| Hoch 2831 | C57L/J | Soleus |
| Apolipoprotein A-I | C57L/J | Soleus/EDL |
| Myosin, light polypeptide 3 | C57L/J | Soleus |
| Phosphoenolpyruvate carboxykinase | C3H/HeJ | Soleus |
| Serpina1c protein | C3H/HeJ | Soleus |
| Synaptic vesicle membrane protein VAT-1 | C57L/J | Soleus |
| 14-3-3 Protein gamma subtype | C57L/J | Soleus |
| V-type proton ATPase catalytic subunit A | C57L/J | Soleus |
| Annexin A5 | C3H/HeJ | Soleus |
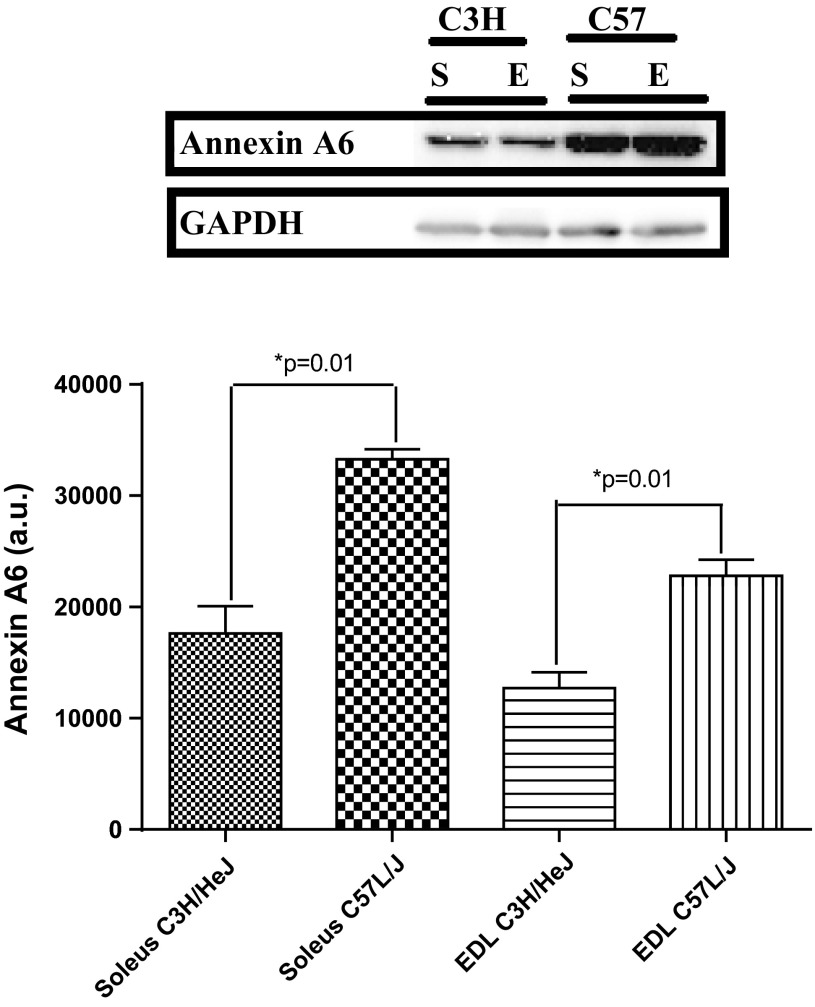
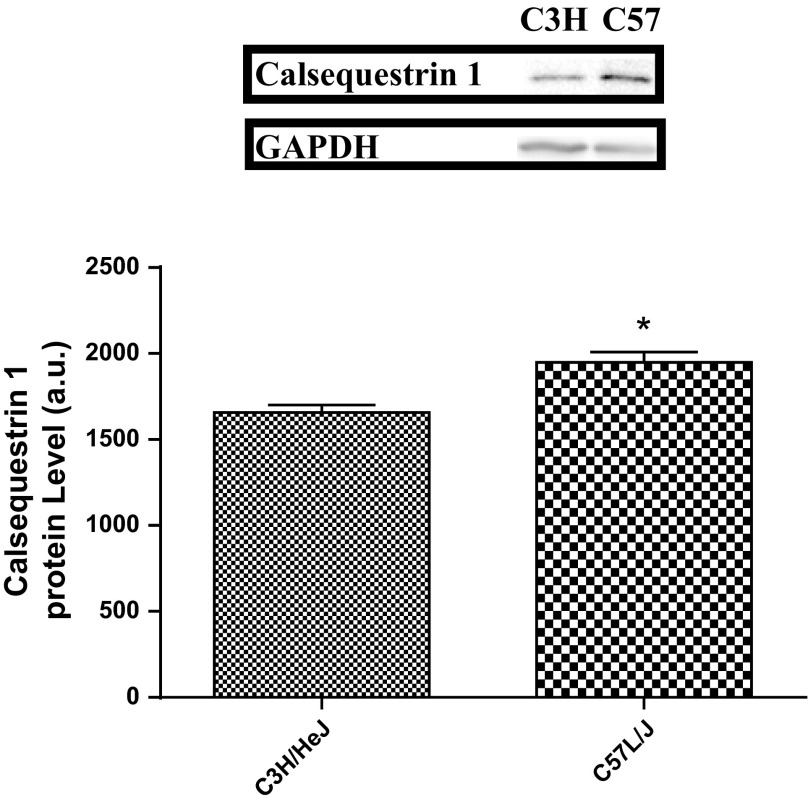
Two proteins, annexin A6 (Anxa6, P = 0.00031) and calsequestrin 1 (Casq1, P = 0.000025), were differentially expressed between the high- and low-active mice (Figs. 2 and 3). Anxa6 was overexpressed in the soleus and EDL of high-active (C57L/J) mice, while Casq1 was overexpressed in the soleus of high-active (C57L/J) mice. Anxa6 and Casq1 were categorized as strong potential candidates for the peripheral regulation of physical activity due to their role in skeletal muscle calcium regulation.
Fig. 2.
Confirmatory Western blots for the overexpression of annexin A6 in high-active (C57L/J) mice compared with low-active (C3H/HeJ) mice in soleus and extensor digitorum longus (EDL) tissue. “E” represents a blot image for the EDL and “S” represents the blot image for the soleus. GAPDH was used as a loading control. The optical density of the blot is represented as mean ± SD along with a representative blot. The high-active animals significantly (*P = 0.01) overexpressed annexin A6 in the soleus and EDL.
Fig. 3.
Confirmatory Western blots for the overexpression of calsequestrin 1 in high-active (C57L/J) mice compared with low-active (C3H/HeJ) mice in soleus tissue. GAPDH was used as a loading control. The optical density of the blot is represented as mean ± SD along with a representative blot. The high-active animals significantly (*P = 0.0024) overexpressed calsequestrin 1 in the soleus.
Experiment 2: role of candidate proteins in physical activity.
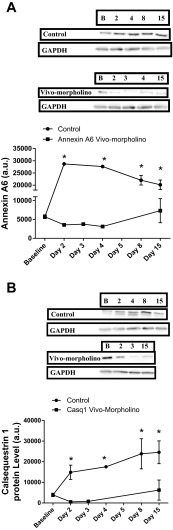
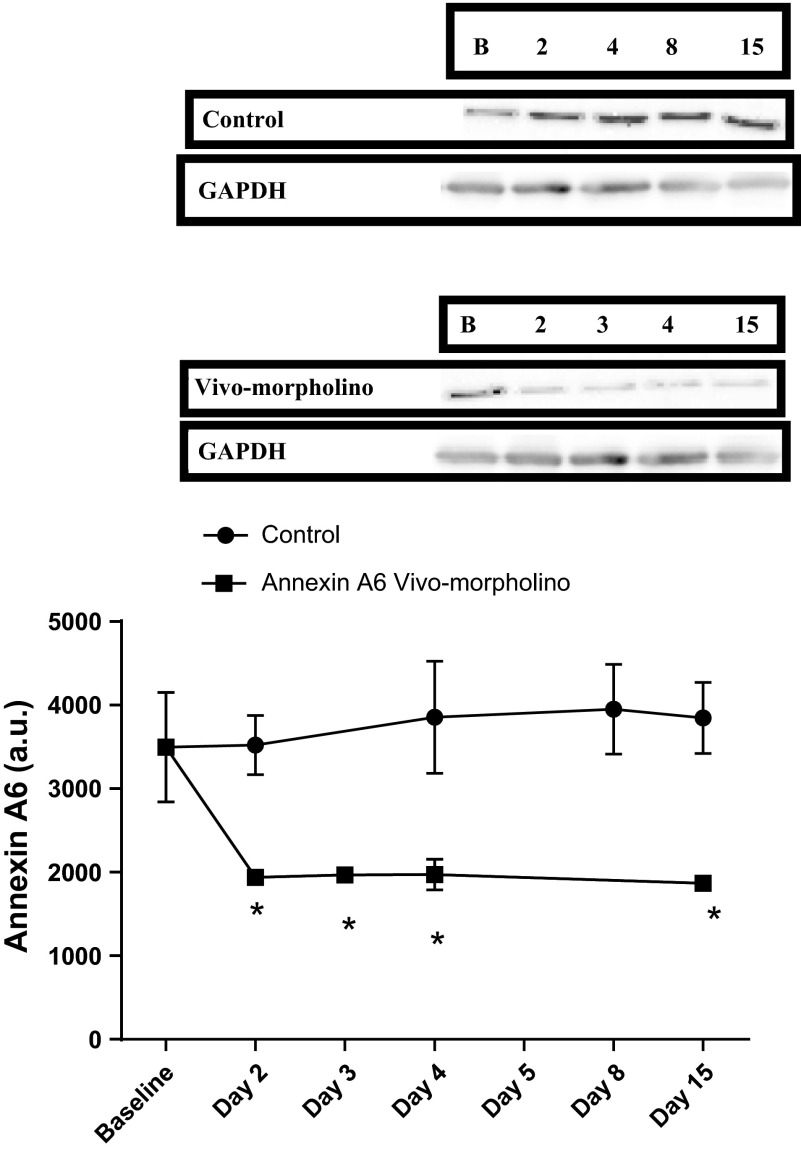
Application of vivo-morpholinos successfully transiently knocked down the expression of both Anxa6 (P = 0.0048, Fig. 4A) and Casq1 (P = 0.0152, Fig. 4B) in the soleus during the treatment week compared with baseline. Interestingly the control animals overexpressed both Anxa6 and Casq1 compared with baseline values during the treatment and recovery weeks. In the EDL, Anxa6 was significantly (P = 0.0019) knocked down from baseline (Fig. 5) and remained knockdown throughout the study.
Fig. 4.
A: Western blot data and a representative Western blot image for annexin A6 expression in the soleus for control and annexin A6 vivo-morpholino-treated animals. The optical density of the blot is represented as mean ± SD. There was a significant (*P = 0.0048) decrease from baseline in the annexin A6 protein expression in the vivo-morpholino group and an increase in annexin A6 protein expression in the control group from baseline. B: Western blot data and a representative Western blot image for calsequestrin 1 expression in the soleus for control and calsequestrin 1 vivo-morpholino-treated animals. The optical density of the blot is represented as mean ± SD. There was a significant (*P = 0.0152) decrease from baseline in the calsequestrin 1 protein expression in the vivo-morpholino group and an increase in calsequestrin 1 protein expression in the control group from baseline. GAPDH is included as a loading control.
Fig. 5.
Western blot data and a representative Western blot image for annexin 6 expression in the EDL for control and annexin A6 vivo-morpholino-treated animals. The optical density of the blot is represented as mean ± SD. There was a significant (*P = 0.0019) decrease from baseline in the annexin A6 protein expression in the vivo-morpholino group. GAPDH is included as a loading control.
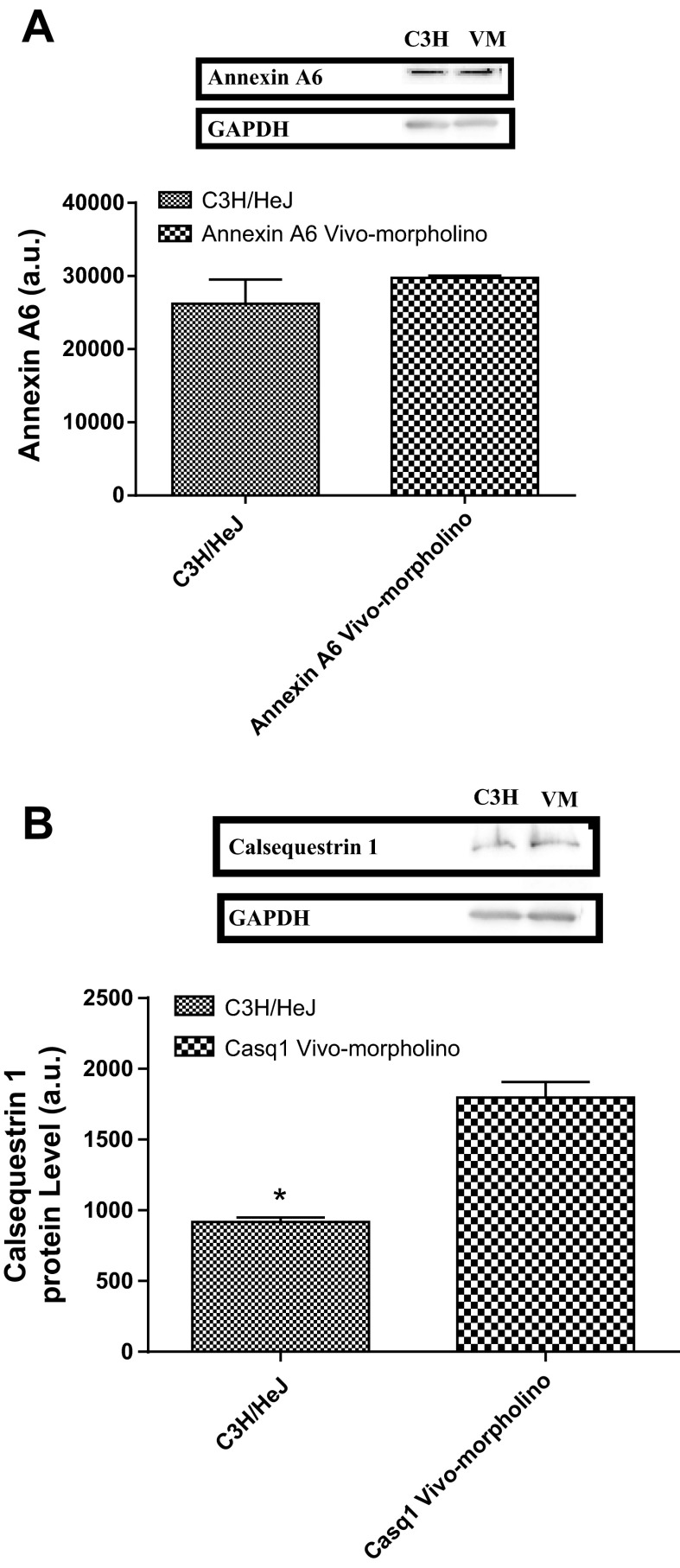
When comparing soleus samples from day 3 of vivo-morpholino treatment to C3H/HeJ (low-active mice) samples there was no difference in Anxa6 protein expression (Fig. 6A; P = 0.14), i.e., treatment with vivo-morpholinos titrated the high-active C57L/J animals' Anxa6 protein expression to the level of the low-active C3H/HeJ animals. However, while vivo-morpholino treatment did decrease Casq1 levels in the high-active C57L/J animals (80% knockdown from baseline), the low-active C3H/HeJ animals had a lower expression of Casq1 (Fig. 6B, P = 0.002).
Fig. 6.
A: Western blot data and a representative Western blot image for annexin A6 expression in the soleus of C3H/HeJ mice and annexin A6 vivo-morpholino-treated animals on day 3 of treatment. The optical density of the blot is represented as mean ± SD. There was no difference in annexin A6 protein expression (P = 0.14). B: Western blot data and a representative Western blot image for calsequestrin 1 expression in the soleus of C3H/HeJ mice and calsequestrin 1 vivo-morpholino-treated animals on day 3 of treatment. C3H/HeJ mice had significantly (*P = 0.002) less calsequestrin 1 than vivo-morpholino-treated animals. GAPDH is included as a loading control.
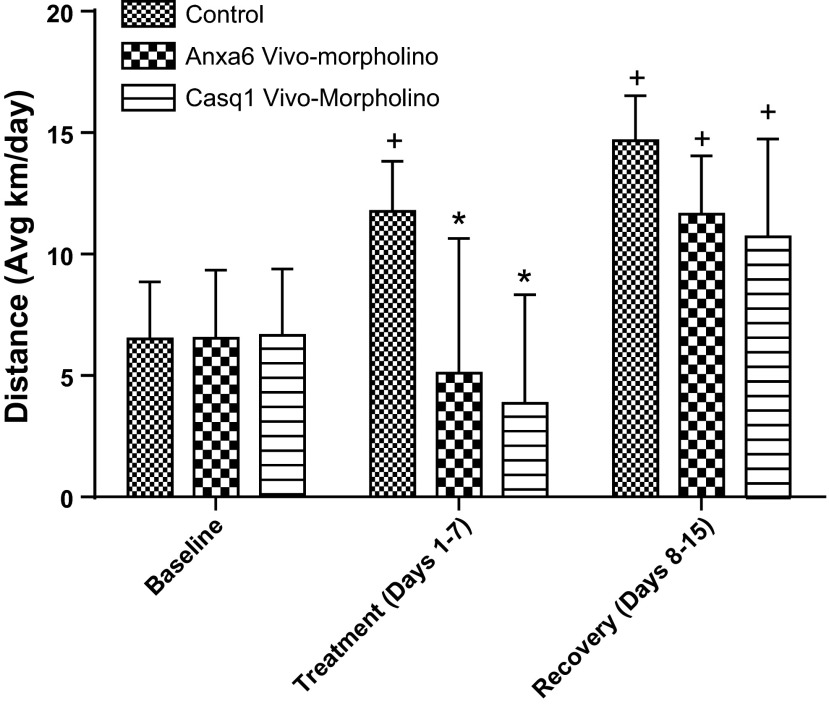
The average physical activity response during the treatment week when Anxa6 and Casq1 in the vivo-morpholino treatment group were knocked down was significantly less than the control group (P = 0.001, Fig. 7). At the end of the recovery week, activity was similar between all groups. Interestingly the control group during the treatment week and all groups during the recovery week ran significantly (P = 0.01) further than the corresponding groups during the baseline week.
Fig. 7.
Physical activity responses (average km/day) of C57L/J mice receiving vivo-morpholino treatment knocking down annexin A6 or calsequestrin 1. There was a significant decrease (*P = 0.001) in activity for the annexin A6 and calsequestrin 1 vivo-morpholino-treated animals during the treatment week. Additionally there was a significant (+P = 0.01) increase in activity in the control group during the treatment week and all groups in the recovery week compared with baseline.
DISCUSSION
The aim of this study was to utilize proteomic and gene-silencing techniques to identify skeletal muscle proteins that may be involved in the regulation of physical activity utilizing a high-active vs. low-active mouse model. We observed different protein signature patterns in the high C57L/J and low C3H/HeJ active mice, with the differentially expressed proteins falling into specific physiological function groups. The high-active C57L/J mice overexpressed proteins associated with the Krebs (TCA) cycle and proteins involved in calcium regulation while the low-active C3H/HeJ mice overexpressed proteins in the electron transport chain and cytoskeletal structure that influence cell motility and force transduction. Further, when we transiently silenced two of the most differentially expressed proteins found in the high-active mice (Anxa6 and Casq1), we observed significant decreases in physical activity suggesting that these proteins are involved in regulating physical activity levels.
While it has been shown that there is a significant genetic influence on physical activity regulation (33), the site of regulation and mechanisms regulating physical activity are still ambiguous. Kelly (24) has suggested that both peripheral and central mechanisms may be involved in activity regulation; indeed, several studies have considered central brain mechanisms focused around dopaminergic (27) and endocannabinoid mechanisms (7). These central mechanisms have been suggested to provide the “drive” to be active. However, other studies have suggested that without the “capability” to be active, an organism will not exhibit high levels of physical activity. Indeed, Tsao et al. (50) showed that a transgenic overexpression of Glut4 in skeletal muscle induced a fourfold increase in daily activity, and Pistilli et al. (42) showed that knocking out IL-15Rα in skeletal muscle also increased wheel running activity. These data suggest that skeletal muscle mechanisms can markedly influence the amount of physical activity completed irrespective of central drive. Thus our study focused on skeletal muscle proteomic differences between high and low physically active mice to provide foundational targets for further mechanistic studies as well as potential interventions to influence physical activity level.
High-active animals.
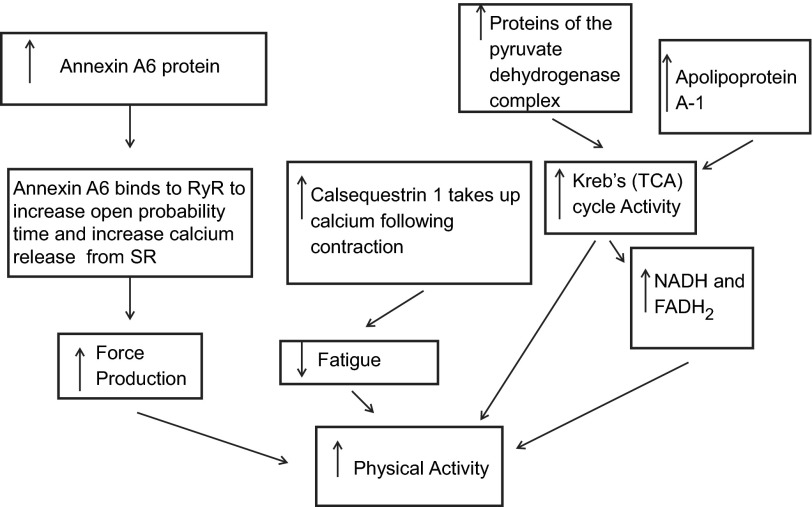
The high-active C57L/J mice overexpressed multiple calcium-regulatory proteins, including Casq1 and Anxa6. From these results, we would suggest that the high-active C57L/J mice have an increased capability to be active because their skeletal muscles can more efficiently release and take up calcium due to the relative overexpression of these calcium-regulatory proteins. Casq1 is a calcium-binding protein located in the sarcoplasmic reticulum (SR) (44) while Anxa6 influences calcium release from the SR (45) by increasing the open probability time of the calcium channel (3, 5). Further, the Anxa6 gene is located in a previously identified QTL for the “mini muscle” phenotype, which is a phenotype associated with the size of the triceps surae of mice selectively bred for high physical activity (14). Thus the overexpression of both Casq1 and Anxa6, in conjunction with their biological roles, could form a potential mechanism by which high-active mice exhibit decreased fatigability of skeletal muscle and an increase in force production resulting in an increased capability to do physical activity (Fig. 8).
Fig. 8.
Proposed schema of how peripheral skeletal muscle protein mechanisms could increase physical activity in high-active animals. 1) As calcium levels rise during EC coupling, annexin A6 binds to the RyR increasing the open probability time, thereby releasing more calcium. 2) Calcium binds to troponin C allowing for actin-myosin cross-bridge formation. The higher concentration of calcium results in high force production and fatigability. 3) Energy in the form of ATP is generated by the Krebs (TCA) cycle and electron transport chain (ETC). A higher amount of proteins in the pyruvate dehydrogenase complex leads to high acetyl CoA levels, which results in more turns of the Krebs (TCA) cycle. 4) This leads to more NADH and FADH2, which yields more ATP from the ETC chain. 5) Following contraction there is elevated calcium in the myoplasm, which causes fatigue. 6) Increased levels of calsequestrin 1 bind free calcium and store it in the sarcoplasmic reticulum, thereby reducing fatigability. The combination of increased force production, increased substrate availability by the Krebs (TCA) cycle, and decreased fatigue leads to increased physical activity.
It has been shown that there is an inverse relationship between skeletal muscle force production and skeletal muscle fatigue resistance (19) due in large part to calcium concentration during excitation coupling (19, 36). A higher level of Anxa6 in the high-active C57L/J animals would result in a higher concentration of calcium released from the SR, which would cause greater force production (47). However, this higher myoplasmic calcium concentration following contraction would actually increase the fatigability of skeletal muscle (19, 47). Offsetting this potential increase in fatigability with Anxa6, Casq1's calcium-binding properties in the SR (44) would lead to an increased binding of calcium following the Anxa6-induced increased release of calcium from the SR. Thus Casq1 would act to decrease myoplasmic calcium levels and thereby reduce fatigability, which could conceptually increase duration of activity, while an increased force production in both slow- and fast-twitch fibers would increase speed of activity. Supporting this hypothesis is previous work from our lab that has shown that the high-active C57L/J run significantly farther (892%), longer (464%) and faster (228%) than the low-active C3H/HeJ on a daily basis (34).
As a means to confirm the hypothesis that Anxa6 and Casq1 influence physical activity level, we used transient gene silencing to knockdown both proteins in high active C57L/J mice which caused a significant reduction in wheel running (Fig. 7). Further, to confirm that Anxa6 and Casq1 was not knocked out in skeletal muscle, we compared Anxa6 and Casq1 vivo-morpholino-treated animals to the low-active C3H/HeJ animals. We observed that with knockdown, Anxa6 levels were similar between treated high-active C57L/J animals and low-active C3H/HeJ animals (Fig. 6A). Casq1, while 80% reduced from the control state, was still higher in the treated animals vs. the low-active C3H/HeJ animals (Fig. 6B). Thus, while protein knockdown is associated with decreased activity (1.2 ± 0.1 km run on day 3) in the high-active C57L/J mice these distances were still higher than typical low-active C3H/HeJ male mice (0.6 ± 1.1 km) (34).
Interestingly during the treatment week, the control animals exhibited a significant increase in physical activity compared with baseline (Fig. 7). This increased activity corresponded to an increase in Anxa6 (Fig. 4A) and Casq1 (Fig. 4B) in the soleus. Yang et al. (55) hypothesized that when a running wheel is returned after it has been removed—which was the case in our study—there is an increase in physical activity level, due to an increased action of the reward system of the brain (55). We would suggest that in addition to Yang's postulated change in central drive, the increase in physical activity after reintroduction of the running wheel could also be a result of the overexpression of Anxa6 and Casq1 in the control animals' solei.
In addition to calcium regulation, the overexpression of proteins associated with the Krebs (TCA) cycle could positively affect maintenance of high physical activity. The most significant proteins overexpressed in the Krebs (TCA) cycle were three proteins in the pyruvate dehydrogenase complex [i.e., pyruvate dehydrogenase protein X component, pyruvate dehydrogenase (acetyl-transferring)-phosphatase 1, and pyruvate dehydrogenase E1 component subunit beta]. The overexpression of the subunits of pyruvate dehydrogenase could conceptually lead to an increased capability to convert pyruvate to acetyl-CoA, with a potential subsequent increase in Krebs (TCA) cycle activity and an increased NADH and FADH2 availability (15). This process would increase available ATP to meet the demands of an increased physical activity level. Further, an overexpression of the key Krebs (TCA) regulator dihydrolipoyllysine-residue succinyltransferase component of the 2-oxoglutarate dehydrogenase complex (52) influences the physical activity response by regulating the production of succinyl-CoA. An overexpression of the dihydrolipoyllysine-residue succinyltransferase component would increase the capability to produce both succinyl-CoA as well as increase levels of both NADH and FADH2 (52), thereby increasing the capability for high physical activity. Furthermore, the coding gene for dihydrolipoyllysine-residue succinyltransferase component is located in an identified QTL associated with both wheel running distance (34) and the mini muscle (14). Additionally, apolipoprotein A-1 (Apo1) was overexpressed in the soleus and EDL of high-active mice and is associated with cholesterol and fat storage, which would facilitate the use of fat as an additional substrate source for the Krebs (TCA) cycle (56). This suggestion is supported by Meek et al. (37) who showed that mice bred for high activity for over 60 generations only increased their activity level when fed a high-fat diet. Thus an overexpression of Apo1 and proteins associated with the Krebs (TCA) cycle would further contribute to the high wheel running activity of the C57L/J mice.
Low-active animals.
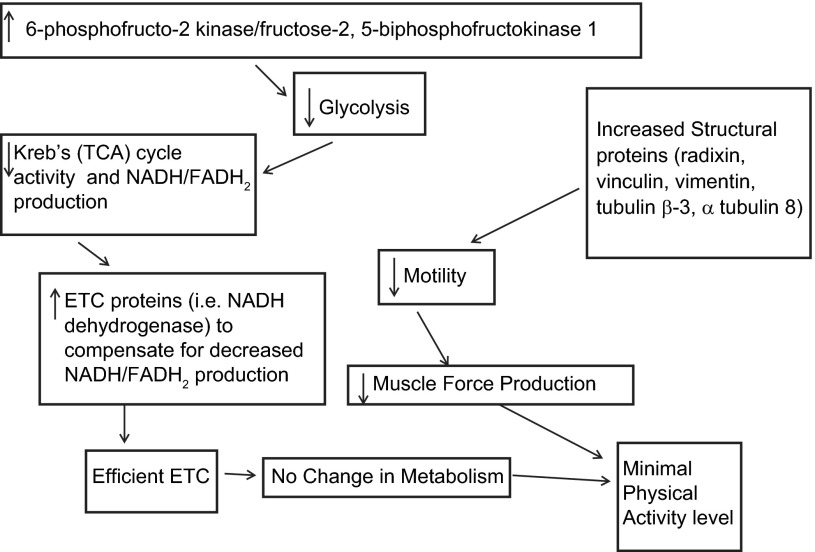
While we observed several protein expression patterns that would support maintenance of high activity, there were also protein expression patterns in the low-active animals that could conceptually inhibit activity (Fig. 9). The low-active animals overexpressed the enzyme 6-phosphofructo-2 kinase/fructose-2,6-bisphosphatase 1, which decreases the activity of the primary regulator of glycolysis, phosphofructokinase 1 (17), which would, in turn, lead to a higher level of degradation of 2,6-bisphosphate with a subsequent decrease in glycolysis and increased gluconeogenesis (17). The decrease in glycolysis would decrease the generation of pyruvate and acetyl-CoA, thereby leading to a decrease in electron donors for the electron transport chain (ETC) and a potential decrease in available energy needed to support activity. Interestingly, we also observed that the C3H/HeJ mice overexpressed several proteins associated with the ETC [e.g., electron transfer flavoprotein ubiquinone oxidoreductase, ATPase, and NADH dehydrogenase (ubiquinone)]. The overexpression of these proteins would generally lead to a more efficient generation of ATP via the electron transport chain (53). We would speculate that in the low-active animals a more efficient generation of ATP through the ETC could serve as a compensatory mechanism for the inhibited glycolytic production of electron donors as well as the relative underexpression of proteins in the Krebs (TCA) cycle compared with the high-active animals.
Fig. 9.
Proposed schema of how peripheral skeletal muscle protein mechanisms might inhibit physical activity in low-active animals. 1) An increase in 6-phosphofructo-2 kinase/fructose-2,5-bisphosphofructokinase 1 decreases glycolysis and Krebs (TCA) cycle activity. 2) With a decrease in Krebs (TCA) cycle activity, there is a decrease in NADH and FADH2 production, which in turn decreases ATP production and physical activity. 3) However, with an increase in the ETC proteins (NADH dehydrogenase) there is an increase in the efficiency of the ETC thereby compensating for the decrease in Krebs (TCA) cycle activity. 4) The increase in structural proteins, radixin, vinculin, vimentin, tubulin beta-3, and alpha tubulin 8 decreases mobility of actin and thereby reduces force production and physical activity.
In addition to the metabolic proteins, the low-active C3H/HeJ animals overexpressed proteins associated with cytoskeletal structure, specifically radixin, lumican, tubulin beta 3, coflin 2, alpha actinin 2, and tubulin 8. These proteins bind actin and microtubules to influence cell membrane/cytoskeletal organization (16, 18, 31, 49). The overexpression of these proteins suggests that there is increased rigidity of the muscle cells resulting in an overall decreased motility of the low-active C3H/HeJ skeletal muscle fibers compared with the high-active C57L/J mice (41). Peng et al. (41) suggested that the decreased motility of the cell can decrease force transduction and skeletal muscle force production. Thus the overexpression of these cytoskeletal structural proteins, as well as the observed overexpression of the specific glycolytic and ETC proteins, could theoretically hinder the capability for wheel running and physical activity. These proteins are attractive targets for further vivo-morpholino studies investigating whether the knockdown of these proteins would increase the activity of the low-active animals.
It should be noted that intravenous injection of vivo-morpholinos could cause knockdown of the targeted proteins systemically. However, it has been shown by Morcos et al. (39) that vivo-morpholino delivery to the brain, vasculature, and heart is limited, excluding these organs from probable gene silencing. Calsequestrin 1 is expressed in skeletal muscle, blood, and platelets, and therefore based on the findings of Morcos et al. (39) it can be hypothesized that knockdown of calsequestrin 1 only occurred in skeletal muscle. Further, annexin A6 is found in kidney, liver, lung, heart, brain, and skin, as well as multiple blood cells (mononuclear, neutrophil, beta-lymphocyte, and platelets) (1), and thus there is potential that Anxa6 was transiently silenced in kidney, liver, and lung, in addition to skeletal muscle. Given our results, we cannot conclusively say that just the skeletal muscle knockdown of Casq1 and Anxa6 was solely responsible for the decreased physical activity. Therefore, future studies are needed to tease out the skeletal muscle contribution vs. any other potential peripherally involved organ.
A potential limitation of this study is that the differential proteomic expression between activity levels simply arose because we used two differing inbred strains of mice and that these proteome expression differences are not related to physical activity regulation. While this scenario is a possibility, it is worth noting that the proteins identified are primarily in pathways that influence muscle contraction and therefore, at least conceptually, could affect physical activity capacity (25). As an attempt to verify that these protein differences were not merely a result of genetic background, but arose from the differences in the control of physical activity, we used vivo-morpholinos to transiently silence the primary candidate proteins we found. The silencing resulted in knockdown levels similar to the low-active animals as well as reducing the high-active animals' activity levels to those resembling the low-active mice. These results, while not providing absolute elimination of a possibility that our protein results were due solely to background genomic differences, are an indication that Casq1 and Anxa6 do influence physical activity level and thus are candidate proteins for the peripheral control of physical activity. Certainly, the next step in understanding how skeletal muscle properties may affect physical activity is to replicate the vivo-morpholino knockdown of these candidate proteins and subject the skeletal muscle to various functional analyses. For example, several papers (32, 43, 46) have specifically investigated calcium flux, mitochondria efficiency, and ATP turnover in rats selectively bred for exercise endurance, and these measurements have contributed significantly to the characterization of that model.
In conclusion, our results support the hypothesis that peripheral, skeletal muscle factors participate in the regulation of voluntary physical activity and these factors arise from multiple protein pathways. Specifically, there appears to be a different mechanism regulating high vs. low activity. High-active animals overexpressed proteins associated with the Krebs (TCA) cycle and calcium regulation, suggesting a decreased fatigability, increased force production, and increased capability of providing substrates for the electron transport chain. We confirmed the role of two calcium-handling proteins in regulating activity by transiently knocking down the proteins in healthy animals with a subsequent reduction in physical activity. Conversely, low-active animals overexpressed structural proteins that could decrease muscle fiber motility, as well as overexpressing proteins that would decrease glycolysis with a potential compensation through an increased efficiency of the electron transport chain.
GRANTS
This project was funded by National Institutes of Health Grant AR-050085 and start-up funds from the Department of Health and Kinesiology at Texas A&M University.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: D.P.F., L.J.D., and J.T.L. conception and design of research; D.P.F., L.J.D., E.E.S., and H.L.V. performed experiments; D.P.F. analyzed data; D.P.F. and J.T.L. interpreted results of experiments; D.P.F. prepared figures; D.P.F. drafted manuscript; D.P.F., L.J.D., E.E.S., H.L.V., and J.T.L. edited and revised manuscript; D.P.F., L.J.D., E.E.S., H.L.V., and J.T.L. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank S. Marek, C. Irwin, and A. Jimenez of the Biology of Physical Activity lab at Texas A&M University for help in animal handling and tissue collection over the course of the study.
REFERENCES
- 1.Babiychuk EB, Draeger A. Annexins in cell membrane dynamics. Ca(2+)-regulated association of lipid microdomains. J Cell Biol 150: 1113–1124, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barclay CJ, Constable JK, Gibbs CL. Energetics of fast- and slow-twitch muscles of the mouse. J Physiol 472: 61–80, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrientos G, Hidalgo C. Annexin VI is attached to transverse-tubule membranes in isolated skeletal muscle triads. J Membrane Biol 188: 163–173, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Czarkowska-Paczek B, Zendzian-Piotrowska M, Bartlomiejczyk I, Przybylski J, Gorski J. The effect of acute and prolonged endurance exercise on transforming growth factor-beta1 generation in rat skeletal and heart muscle. J Physiol Pharmacol 60: 157–162, 2009 [PubMed] [Google Scholar]
- 5.Diaz-Munoz M, Hamilton SL, Kaetzel MA, Hazarika P, Dedman JR. Modulation of Ca2+ release channel activity from sarcoplasmic reticulum by annexin VI (67-kDa calcimedin). J Biol Chem 265: 15894–15899, 1990 [PubMed] [Google Scholar]
- 6.DiPetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet 21: 683–692, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Dubreucq S, Durand A, Matias I, Bénard G, Richard E, Soria-Gomez E, Glangetas C, Groc L, Wadleigh A, Massa F, Bartsch D, Marsicano G, Georges F, Chaouloff F. Ventral tegmental area cannabinoid type-1 receptors control voluntary exercise performance. Biol Psychiatry 73: 895–903, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Egan B, Dowling P, O'Connor PL, Henry M, Meleady P, Zierath JR, O'Gorman DJ. 2-D DIGE analysis of the mitochondrial proteome from human skeletal muscle reveals time course-dependent remodelling in response to 14 consecutive days of endurance exercise training. Proteomics 11: 1413–1428, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Ferguson D, Schmitt E, Lightfoot J. Vivo-morpholinos induced transient knockdown of physical activity related proteins. PLoS One. 2013. April 22; 8(4): e61472. 10.1371/journal.pone.0061472 Print 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nature Rev Genet 6: 271–286, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Gelfi C, Vigano A, Ripamonti M, Pontoglio A, Begum S, Pellegrino MA, Grassi B, Bottinelli R, Wait R, Cerretelli P. The human muscle proteome in aging. J Proteome Res 5: 1344–1353, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Gorzek JF, Hendrickson KC, Forstner JP, Rixen JL, Moran AL, Lowe DA. Estradiol and tamoxifen reverse ovariectomy-induced physical inactivity in mice. Med Sci Sports Exerc 39: 248–256, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartmann J, Garland T, Jr, Hannon RM, Kelly SA, Munoz G, Pomp D. Fine mapping of “mini-muscle,” a recessive mutation causing reduced hindlimb muscle mass in mice. J Hered 99: 679–687, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horecker BL. The biochemistry of sugars. Int Z Vitam Ernahrungsforsch Beih 15: 1–21, 1976 [PubMed] [Google Scholar]
- 16.Howard J. The movement of kinesin along microtubules. Annu Rev Physiol 58: 703–729, 1996 [DOI] [PubMed] [Google Scholar]
- 17.Hue L, Rider MH. Role of fructose 2,6-bisphosphate in the control of glycolysis in mammalian tissues. Biochem J 245: 313–324, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivetic A, Ridley AJ. Ezrin/radixin/moesin proteins and Rho GTPase signalling in leucocytes. Immunology 112: 165–176, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James RS, Walter I, Seebacher F. Variation in expression of calcium-handling proteins is associated with inter-individual differences in mechanical performance of rat (Rattus norvegicus) skeletal muscle. J Exp Biol 214: 3542–3548, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Jonas I, Vaanholt LM, Doornbos M, Garland T, Jr, Scheurink AJ, Nyakas C, van Dijk G. Effects of selective breeding for increased wheel-running behavior on circadian timing of substrate oxidation and ingestive behavior. Physiol Behav 99: 549–554, 2010 [DOI] [PubMed] [Google Scholar]
- 21.Karp NA, Lilley KS. Maximising sensitivity for detecting changes in protein expression: experimental design using minimal CyDyes. Proteomics 5: 3105–3115, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Karp NA, Lilley KS. Design and analysis issues in quantitative proteomics studies. Proteomics 7, Suppl 1: 42–50, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Kelly SA, Nehrenberg DL, Hua K, Garland T, Jr, Pomp D. Functional genomic architecture of predisposition to voluntary exercise in mice: expression QTL in the brain. Genetics 191: 643–654, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly SA, Nehrenberg DL, Peirce JL, Hua K, Steffy BM, Wiltshire T, Pardo-Manuel de Villena F, Garland T, Jr, Pomp D. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol Genomics 42: 190–200, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinnunen S, Manttari S. Specific effects of endurance and sprint training on protein expression of calsequestrin and SERCA in mouse skeletal muscle. J Muscle Res Cell Motil 33: 123–130, 2012 [DOI] [PubMed] [Google Scholar]
- 26.Knab AM, Bowen RS, Hamilton AT, Gulledge AA, Lightfoot JT. Altered dopaminergic profiles: implications for the regulation of voluntary physical activity. Behav Brain Res 204: 147–152, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knab AM, Bowen RS, Hamilton AT, Lightfoot JT. Pharmacological manipulation of the dopaminergic system affects wheel-running activity in differentially active mice. J Biol Regul Homeostat Agents 26: 119–129, 2012 [PMC free article] [PubMed] [Google Scholar]
- 28.Knab AM, Bowen RS, Moore-Harrison T, Hamilton AT, Turner MJ, Lightfoot JT. Repeatability of exercise behaviors in mice. Physiol Behav 98: 433–440, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leamy LJ, Pomp D, Lightfoot JT. An epistatic genetic basis for physical activity traits in mice. J Hered 99: 639–646, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leamy LJ, Pomp D, Lightfoot JT. A search for quantitative trait loci controlling within-individual variation of physical activity traits in mice. BMC Genet 11: 83, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lek M, Quinlan KG, North KN. The evolution of skeletal muscle performance: gene duplication and divergence of human sarcomeric alpha-actinins. Bioessays 32: 17–25, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Lessard SJ, Rivas DA, Stephenson EJ, Yaspelkis BB, 3rd, Koch LG, Britton SL, Hawley JA. Exercise training reverses impaired skeletal muscle metabolism induced by artificial selection for low aerobic capacity. Am J Physiol Regul Integr Comp Physiol 300: R175–R182, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lightfoot JT. Current understanding of the genetic basis for physical activity. J Nutr 141: 526–530, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lightfoot JT, Leamy L, Pomp D, Turner MJ, Fodor AA, Knab A, Bowen RS, Ferguson D, Moore-Harrison T, Hamilton A. Strain screen and haplotype association mapping of wheel running in inbred mouse strains. J Appl Physiol 109: 623–634, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lightfoot JT, Turner MJ, Daves M, Vordermark A, Kleeberger SR. Genetic influence on daily wheel running activity level. Physiol Genomics 19: 270–276, 2004 [DOI] [PubMed] [Google Scholar]
- 36.MacIntosh BR, Holash RJ, Renaud JM. Skeletal muscle fatigue—regulation of excitation-contraction coupling to avoid metabolic catastrophe. J Cell Sci 125: 2105–2114, 2012 [DOI] [PubMed] [Google Scholar]
- 37.Meek TH, Eisenmann JC, Garland T., Jr Western diet increases wheel running in mice selectively bred for high voluntary wheel running. Int J Obesity 34: 960–969, 2010 [DOI] [PubMed] [Google Scholar]
- 38.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA 291: 1238–1245, 2004 [DOI] [PubMed] [Google Scholar]
- 39.Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques 45: 613–614, 616, 618 passim, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Nabity MB, Lees GE, Dangott LJ, Cianciolo R, Suchodolski JS, Steiner JM. Proteomic analysis of urine from male dogs during early stages of tubulointerstitial injury in a canine model of progressive glomerular disease. Vet Clin Pathol 40: 222–236, 2011 [DOI] [PubMed] [Google Scholar]
- 41.Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol 287: 191–231, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pistilli EE, Siu PM, Alway SE. Interleukin-15 responses to aging and unloading-induced skeletal muscle atrophy. Am J Physiol Cell Physiol 292: C1298–C1304, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Rivas DA, Lessard SJ, Saito M, Friedhuber AM, Koch LG, Britton SL, Yaspelkis BB, 3rd, Hawley JA. Low intrinsic running capacity is associated with reduced skeletal muscle substrate oxidation and lower mitochondrial content in white skeletal muscle. Am J Physiol Regul Integr Comp Physiol 300: R835–R843, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi AE, Dirksen RT. Sarcoplasmic reticulum: the dynamic calcium governor of muscle. Muscle Nerve 33: 715–731, 2006 [DOI] [PubMed] [Google Scholar]
- 45.Song G, Harding SE, Duchen MR, Tunwell R, O'Gara P, Hawkins TE, Moss SE. Altered mechanical properties and intracellular calcium signaling in cardiomyocytes from annexin 6 null-mutant mice. FASEB J 16: 622–624, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Stephenson EJ, Stepto NK, Koch LG, Britton SL, Hawley JA. Divergent skeletal muscle respiratory capacities in rats artificially selected for high and low running ability: a role for Nor1? J Appl Physiol 113: 1403–1412, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summermatter S, Thurnheer R, Santos G, Mosca B, Baum O, Treves S, Hoppeler H, Zorzato F, Handschin C. Remodeling of calcium handling in skeletal muscle through PGC-1alpha: impact on force, fatigability, and fiber type. Am J Physiol Cell Physiol 302: C88–C99, 2012 [DOI] [PubMed] [Google Scholar]
- 48.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–D568, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tischfield MA, Engle EC. Distinct alpha- and beta-tubulin isotypes are required for the positioning, differentiation and survival of neurons: new support for the “multi-tubulin” hypothesis. Biosci Rep 30: 319–330, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsao TS, Li J, Chang KS, Stenbit AE, Galuska D, Anderson JE, Zierath JR, McCarter RJ, Charron MJ. Metabolic adaptations in skeletal muscle overexpressing GLUT4: effects on muscle and physical activity. FASEB J 15: 958–969, 2001 [DOI] [PubMed] [Google Scholar]
- 51.Turner MJ, Kleeberger SR, Lightfoot JT. Influence of genetic background on daily running-wheel activity differs with aging. Physiol Genomics 22: 76–85, 2005 [DOI] [PubMed] [Google Scholar]
- 52.Tylicki A, Bunik VI, Strumilo S. [2-Oxoglutarate dehydrogenase complex and its multipoint control]. Postepy Biochem 57: 304–313, 2011 [PubMed] [Google Scholar]
- 53.Yagi T, Seo BB, Di Bernardo S, Nakamaru-Ogiso E, Kao MC, Matsuno-Yagi A. NADH dehydrogenases: from basic science to biomedicine. J Bioenerg Biomembr 33: 233–242, 2001 [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi W, Fujimoto E, Higuchi M, Tabata I. A DIGE proteomic analysis for high-intensity exercise-trained rat skeletal muscle. J Biochem 148: 327–333, 2010 [DOI] [PubMed] [Google Scholar]
- 55.Yang HS, Vitaterna MH, Laposky AD, Shimomura K, Turek FW. Genetic analysis of daily physical activity using a mouse chromosome substitution strain. Physiol Genomics 39: 47–55, 2009 [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Wang Y, Li J, Yu J, Pu J, Li L, Zhang H, Zhang S, Peng G, Yang F, Liu P. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. J Proteome Res 10: 4757–4768, 2011 [DOI] [PubMed] [Google Scholar]