Summary
Tet enzymes (Tet1/2/3) convert 5-methylcytosine (5mC) to 5-hydroxy-methylcytosine (5hmC) and are dynamically expressed during development. While loss of individual Tet enzymes or combined deficiency of Tet1/2 allows for embryogenesis, the effect of complete loss of Tet activity and 5hmC marks in development is not established. We have generated Tet1/2/3 triple knockout (TKO) mouse embryonic stem cells (ESCs) and examined their developmental potential. Combined deficiency of all three Tets depleted 5hmC and impaired ESC differentiation as seen in poorly differentiated TKO embryoid bodies (EBs) and teratomas. Consistent with impaired differentiation, TKO-ESCs contributed poorly to chimeric embryos, a defect rescued by Tet1 re-expression, and could not support embryonic development. Global gene expression and methylome analyses of TKO-EBs revealed promoter hypermethylation and deregulation of genes implicated in embryonic development and differentiation. These findings suggest a requirement for Tet- and 5hmC-mediated DNA demethylation in proper regulation of gene expression during ESC differentiation and development.
Keywords: Tet1, Tet2, Tet3, 5-hydroxymethylcytosine, differentiation, DNA methylation
Introduction
DNA methylation is a prominent epigenetic modification in the eukaryotic genome and dynamic changes in the DNA methylation landscape are essential for normal regulation of genes during development (Smith et al., 2012; Gifford et al., 2013; Xie et al., 2013). While it is well defined how the establishment and maintenance of DNA methylation mediated by DNA methyltransferases is achieved, the mechanisms for removal of this modification are poorly understood (Wu and Zhang, 2010; 2014). The Ten eleven translocation (Tet) family of enzymes (Tet1/2/3) has been implicated in DNA demethylation (Tahiliani et al., 2009; Ito et al., 2010). These enzymes contain a C-terminal catalytic domain that has dioxygenase activity and converts 5-methylcytosine (5mC) sequentially to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (He et al., 2011; Ito et al., 2011). Given that DNMT1 recognizes 5hmC poorly, it is believed that this modification promotes passive DNA demethylation (Wu and Zhang, 2010). However, it has also been proposed that these modified bases function as intermediates in the process of active DNA demethylation where they are enzymatically recognized and removed from the genome by components of the DNA repair machinery. Since 5hmC has also been found as a stable base in the genome that reaches significant abundance in several cell types, it may also serve as a distinct epigenetic mark and not just as a DNA demethylation intermediate (Wu and Zhang, 2010; 2014). Genomic studies have identified 5hmC as highly abundant in CpG-rich promoters and gene bodies in mouse ESCs (Pastor et al., 2011; Williams et al., 2011) and in enhancer elements in human embryonic stem cells (Stroud et al., 2011), where it may have critical regulatory roles in orchestrating gene expression.
Tet enzymes and 5hmC are present in various embryonic and adult cell types including the zygote (Wossidlo et al., 2011), embryonic stem cells (Ito et al., 2010; Koh et al., 2011), primordial germ cells (Hajkova et al., 2010) and neurons (Kriaucionis and Heintz, 2009). Despite their roles in epigenetic regulation of the genome, notably DNA demethylation, and their presence in various stages of embryonic and adult development, the physiological relevance of these enzymes has not been well established. The recent generation of Tet1, Tet2 and Tet3 single mutant (Dawlaty et al., 2011; Gu et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011) and Tet1/2 double knockout mice and ESCs (Dawlaty et al., 2013), has shed light on the roles of these proteins in pluripotency as well as embryonic and adult development. Both Tet1 and Tet2 are expressed in mouse ES cells and depletion of either of these proteins reduces 5hmC levels but does not affect pluripotency (Dawlaty et al., 2011; Koh et al., 2011). Tet1 knockout ES cells remain pluripotent and Tet1 knockout mice are viable and fertile although displaying reduced body and litter sizes, suggesting a subtle role for Tet1 in embryonic development and gametogenesis (Dawlaty et al., 2011). Like Tet1, Tet2 is also dispensable for embryonic development and adult mice are viable and fertile. However, Tet2 deficiency promotes chronic myelomonocytic leukemia (CMML) formation in mice and humans (Li et al., 2011; Moran-Crusio et al., 2011). In contrast to Tet1 and Tet2, Tet3 is not expressed in ES cells and is only induced upon differentiation, consistent with its presence in various differentiated cell types (Dawlaty et al., 2013). In addition, Tet3 is expressed in the oocytes and zygote where it participates in hydroxylating the paternal pronucleus and promotes DNA demethylation (Gu et al., 2011; Wossidlo et al., 2011). Conditional deletion of Tet3 in the oocyte leads to delayed demethylation of the paternal genome and increased developmental failure suggesting an essential role for Tet3 during pre-implantation development. Tet3 homozygous mutant mice can develop to term but die at birth (Gu et al., 2011) suggesting that loss of Tet3 alone does not block differentiation and embryonic development and is possibly compensated for, at least in part, by Tet1 and Tet2.
Recently, we have shown that combined loss of Tet1 and Tet2 depletes 5hmC levels in ESCs and germ cells but is compatible with embryonic development and allows development of viable and overtly healthy adult mice (Dawlaty et al., 2013). However, a large fraction of double mutant embryos exhibited mid gestation defects, perinatal lethality and compromised imprinting and had reduced 5hmC and increased 5mC levels. This prompted us to investigate whether Tet3 compensates for loss of Tet1 and Tet2 during development and whether DNA hydroxymethylation is critical for proper development. To address this, we have generated Tet1/2/3 triple knockout (TKO) mouse ES cells and examined their differentiation and developmental potential. Our results identify Tet-mediated epigenetic regulation of developmental genes during differentiation of ES cells and establish a critical role of these enzymes in differentiation and development.
Results
Generation and characterization of Tet triple knockout (TKO) mouse embryonic stem cells
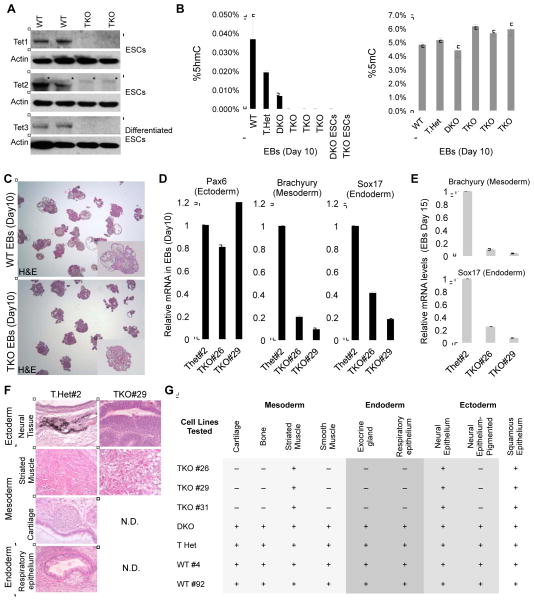
To study the effect of combined deficiency of Tet1, Tet2 and Tet3 on ES cell pluripotency and differentiation potential, we generated Tet TKO ESCs by intercrossing Tet1/2/3 triple heterozygote (THet) mice and deriving THet and Tet1−/−|Tet2−/−|Tet3+/− ES cell lines. The latter cell lines were subsequently targeted to delete the wild type Tet3 allele and obtain Tet TKO ESCs. Genotyping by Southern Blot confirmed the loss of the wild type allele of all three Tet genes (Figure S1A). All TKO ESC lines maintained normal ES cell morphology, expressed the pluripotency markers Oct4 and Nanog and could form embryoid bodies (EBs) (Figure S1B). Further experiments confirmed depletion of all three Tet transcripts (Figure S1C) and proteins (Figure 1A) in ESCs, and in differentiated cell types such as EBs or retinoic acid treated ES cells. To investigate how loss of Tet enzymes affects global 5hmC levels, we applied mass spectrometry to measure levels of 5mC and 5hmC in genomic DNA isolated from TKO, THet, DKO and wild type EBs (Figure 1B). While THet and DKO EBs had ~50% and ~80% reduction in 5hmC levels, respectively, TKO EBs were completely depleted of 5hmC, suggesting that Tet1, Tet2 and Tet3 collaborate in establishing and maintaining 5hmC marks in the genome. Concomitant with loss of 5hmC, TKO EBs had a subtle increase in global 5mC levels, suggesting that depletion of 5hmC levels leads to increased global hypermethylation during ESC differentiation, as observed previously (Dawlaty et al., 2013).
Figure 1. Loss of 5hmC and restricted differentiation potential of Tet TKO ES cells in embryoid body and teratoma formation assays.
(A) Confirmation of loss of Tet enzymes in Tet TKO ESCs and differentiated cells by Western blot.
(B) Quantification of 5hmC and 5mC in ESCs and EBs of the indicated genotypes by mass spectrometry.
(C) Sections of paraffin-embedded EBs derived from ES cell lines of indicated genotypes stained with Hematoxylin and Eosin (H&E).
(D &E) RT-qPCR for markers of embryonic germ layers in day 10 EBs (D) and day 15 EBs (E) of the indicated genotypes. Data are normalized to Gapdh.
(F) Representative images of each embryonic germ layer from hematoxylin and eosin (H&E)-stained sections of teratomas derived from ESCs of the indicated genotypes. N.D.= none detected.
(G) Summary table of the various germ layer tissues detected in teratomas of indicated genotypes. For all panels, error bars indicate standard deviation.
(See also Figure S1)
Combined loss of all three Tet enzymes compromises differentiation in embryoid body formation and teratoma assays
To assess how combined loss of all three Tet proteins affect differentiation of ESCs, we differentiated TKO and WT or THet control ESCs to EBs. While both, TKO and control ESCs, formed EBs, histologic examination of TKO EBs revealed poorly differentiated tissues with substantially fewer differentiated structures compared to control EBs (Figure 1C). Moreover, TKO EBs expressed reduced levels of mesodermal and endodermal markers (Figure 1D). The expression levels of these markers also remained low in late-stage day 15 TKO EBs (Figure 1E) suggesting that the poor differentiation of TKO ESCs is not due to a delay in differentiation and rather likely due to restricted developmental potency of ESCs. Consistent with these observations, teratomas derived from WT, THet and DKO ESCs contained multiple tissue types of all three germ layers whereas TKO teratomas lacked endodermal and selected mesodermal structures and did not contain more advanced ectodermal structures such as pigmented neural epithelium (Figure 1F&G). These findings suggest that Tet enzymes are critical for proper differentiation of ES cells.
TKO ES cells contribute poorly to chimeric embryos and cannot support development
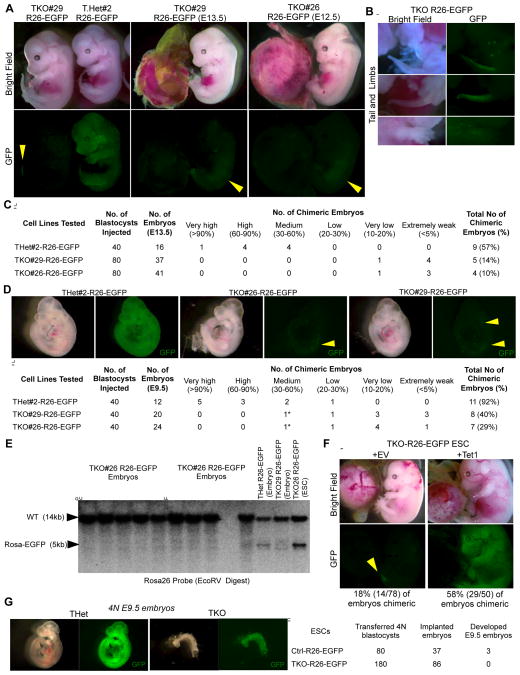
To more stringently assess the differentiation potential of TKO ESCs during embryogenesis we injected Rosa26-EGFP targeted TKO and THet control ESCs, which ubiquitously express EGFP (Figure S2A&B), into blastocysts. After transplantation into foster mothers embryos were dissected at E13.5 revealing widespread contribution of THet-R26-EGFP cells to embryos with nearly 60% being chimeric and with most having high and medium GFP signals. In contrast, two independent TKO ESCs clones (TKO#26-R26-EGFP and TKO#29-R26-EGFP) exhibited very poor contribution to developing embryos (Figure 2A) with only ~15% being chimeric and displaying an extremely low GFP signal, mostly in tail, appendages and dorso-lateral areas (Figure 2A–C). We also inspected chimeric embryos at E9.5 for TKO ESC contribution and observed, similarly to more advanced embryos, a significantly lower incidence of chimeric embryos (~35%) as compared to control THet ESCs (92%) with the majority of TKO chimeric embryos displaying very poor contribution (Figure 2D). The few embryos that had slightly higher contributions were growth retarded or morphologically defective.
Figure 2. Limited contribution of Tet TKO ES cells to developing embryos in a chimera assay.
A&B) Bright field and fluorescence images of E13.5 chimeric embryos generated by injecting Rosa26-EGFP targeted ESCs of the indicated genotypes into WT blastocysts. The very poor and weaker GFP signal in TKO ESC chimeras are highlighted in the images in B.
C) Summary table for all cell lines tested in the chimera assay.
D) Top: Bright field and fluorescence images of E9.5 chimeric embryos generated by injecting Rosa26-EGFP targeted ESCs of the indicated genotypes into WT blastocysts. Bottom: Summary table for all cell lines tested in this chimera assay. Asterisk indicates growth retarded and defective embryos.
E) Southern blot confirming the negligible to no detection of the TKO-Rosa26-EGFP allele in DNA extracted from E13.5 TKO chimeric embryos.
F) E14.5 chimeric embryos generated by injecting Rosa26-EGFP targeted ESCs of indicated genotypes into WT blastocysts.
G) Left: Bright field and fluorescence images of E9.5 4N embryos generated by injecting Rosa26-EGFP targeted ESCs of the indicated genotypes into 4N WT blastocysts. Right: Summary table for all cell lines tested in the tetraploid complementation assay.
For all panels, arrowheads point to poor GFP signal.
(See also Figure S2)
To exclude the possibility that lack of contribution was due to silencing of the GFP transgene in the 5hmC-deficient cells, we assayed for the presence of the donor cells by Southern blot analysis on whole embryo DNA using a Rosa26 locus probe. As shown in Figure 2E, no Rosa-EGFP targeted DNA was detected confirming the lack or at most very low presence of TKO cells in chimeric embryos. Consistent with these observations, examination of selected organs from postnatal mice derived from embryos injected with TKO-R26-EGFP cells revealed no detectable incorporation of EGFP positive cells. (Figure S2C).
Both independent clones of TKO ESCs tested in this assay exhibited a similar compromised differentiation potential and poor contribution to chimeras strongly suggesting that the observed phenotypes are due to the engineered Tet enzyme mutations rather than any unlinked mutations. To further support this, we over expressed Tet1 in TKO-R26-EGFP ESCs and found that ectopic expression of Tet1 in TKO ESCs rescued their differentiation defects and restored contribution to chimeras. Fifty eight percent of embryos injected with TKO-R26EGFP+Tet1 were chimeric with majority of the chimeric embryos having medium to high degree of contribution and comparable to the chimeric embryos derived from THet-R26-EGFP ESCs. In contrast, only 18% of embryos injected with TKO-R26EGFP cells transduced with an empty vector were chimeric with all embryos having a very poor level of contribution (Figure 2F).
To assess whether the poor contribution of TKO ESCs to a developing embryo is due to reduced proliferation of these cells, we quantified the growth rate of TKO ESCs, which was indistinguishable from that of control THet ESCs (Figure S2D). This suggests that TKO ESCs are not out competed by host ICM cells due to a proliferation defect. We also tested the ability of TKO ESCs to support embryonic development in a tetraploid complementation assay where the absolute differentiation potential of TKO ESCs and their ability to develop into an embryo can be assessed in the absence of competition of wild type host cells. When analyzed at E9.5, the TKO 4N-injected blastocysts displayed only rudimentary structures and failed to support development of an embryo proper in contrast to the three normally developed THet embryos (Figure 2G). These findings further support the notion that Tet deficiency compromises the differentiation potential of ESCs and restricts their developmental potential during embryogenesis.
Deficiency of Tet enzymes leads to aberrant promoter hypermethylation and deregulation of developmental genes during differentiation
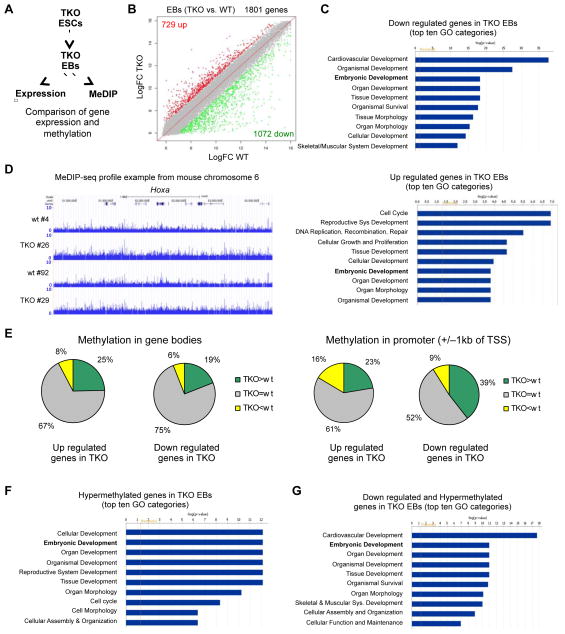
To determine the effects of loss of Tet enzymes and 5hmC depletion on DNA methylation and gene expression during differentiation, we differentiated TKO and WT ESCs to EBs and subjected them to gene expression profiling and methylation analyses (Figure 3A). We analyzed mRNA levels from WT and TKO EBs by microarray (Figure 3B) and found that the majority (1072/1801) of deregulated genes in TKO EBs were down regulated in contrast to 729 up regulated genes. Gene ontology analysis revealed that both up and down regulated genes in TKO EBs were implicated in various developmental processes including embryonic development and differentiation (Figure 3C). Nevertheless, the specific enrichment for developmental categories was much more significant (lower p values) in the set of down regulated genes than in the set of up regulated genes. The latter set was associated with a broader variety of biological functions, which included development, but also cell cycle, replication, DNA-maintenance and proliferation.
Figure 3. Aberrant promoter hypermethylation and deregulation of developmental genes in Tet TKO embryoid bodies.
A) Schematic of the MeDIP and gene expression analyses to identify hypermethylated and down regulated genes during differentiation of TKO ES cells.
B) Scatter plots showing differentially expressed genes in red (up regulated) or in green (down regulated) across a panel of TKO EBs when compared to WT EBs.
C) Gene ontology analysis of differentially expressed genes in TKO EBs.
D) MeDIP-seq profile of a 3 Mb region from mouse chromosome 6 with the Hoxa cluster in the center in two independent TKO and WT EB clones as an example for the general hypermethylation observed in TKO EBs. Enrichments are indicated as normalized read counts.
E) Analysis of deregulated genes in TKO EBs correlating their expression to the methylation status of their promoters or gene bodies. The pie charts show the percentage of deregulated genes in TKO EBs that show hypermethylation compared to WT (green), hypomethylation compared to WT (yellow) or do not change (grey), in the gene body or the promoter (+/− 1000 bp from the TSS).
F) Gene ontology analysis of all genes with hypermethylated promoter regions (normalized average read counts > 4 in the region +/− 1000 bp from the TSS) in TKO EBs.
G) Gene ontology analysis of all genes down regulated in TKO EBs that also have differentially hypermethylated promoters (TKO vs. WT).
(See also Table S1)
Since loss of Tet enzymes promotes global hypermethylation in TKO EBs (Figure 1B), we analyzed the methylome of TKO EBs by 5-methylcytosine DNA immunoprecipitation using specific antibodies against 5mC, followed by massive parallel sequencing (MeDIP-seq). Consistent with the mass spectrometry analysis (Figure 1B) we found a significant increase in total 5mC reads across all chromosomes in TKO EBs compared to WT EBs. Also, baseline 5mC levels were substantially increased across the genome, as exemplified by a 3 Mb region from mouse chromosome 6 containing the Hoxa locus (Figure 3D). To further characterize the TKO methylation landscape, we analyzed the position-wise coverage of 5mC peaks in gene bodies and promoters (+/− 1kb of TSS) of deregulated genes in TKO EBs (Figure 3E). We found a weak correlation between increased gene body methylation and higher gene expression and a strong enrichment for promoter hypermethylation among down regulated genes. Thirty nine percent of genes with reduced expression in TKO EBs had higher promoter methylation levels than WT EBs. Gene ontology analyses revealed that the majority of genes with hypermethylated promoter regions in TKO EBs were implicated in developmental processes including embryonic development and cellular differentiation (Figure 3F). Similarly, down regulated genes with hypermethylated promoters were enriched for developmental categories (Figure 3G).
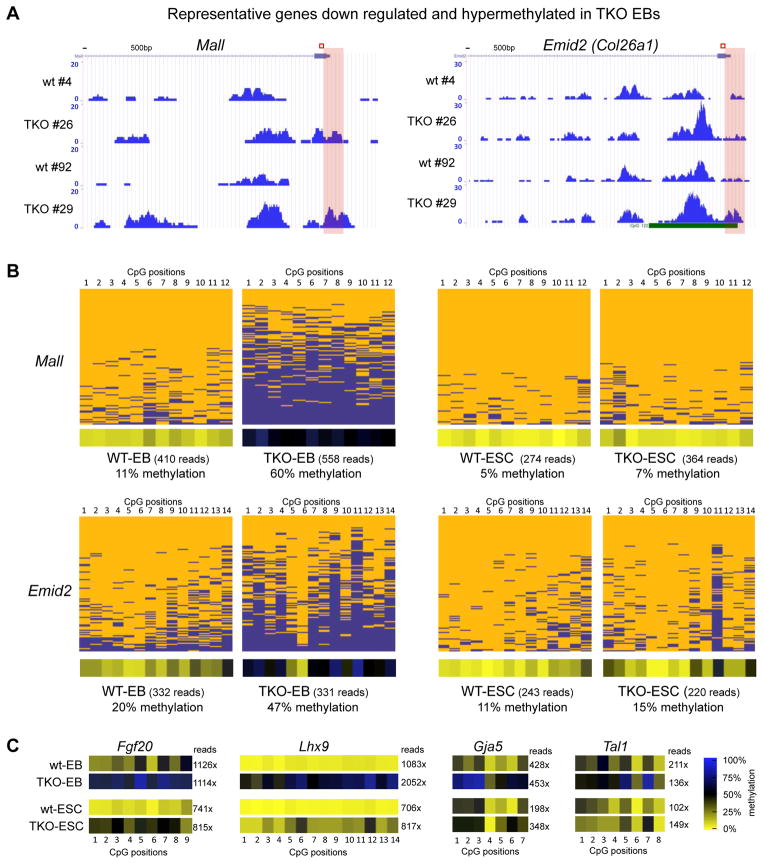
To validate the hypermethylation of deregulated developmental regulators during differentiation of TKO EBs, we applied locus specific 454 bisulfite sequencing to analyze promoter methylation patterns of representative hypermethylated genes from our MeDIP analysis (Figure 4A). Analysis of 200–300 bp promoter regions of the downregulated and hypermethylated genes Emid2, Mall, Gja5 and Tal1 in DNA from TKO EBs and ESCs confirmed robust hypermethylation of these regions (2 to 5 fold more than controls) during differentiation to EBs but not in ESCs (Figure 4B and C, Figure S3). Two other genes Lhx9 and Fgf20, which are hypermethylated but not expressed in EBs, exhibited increased hypermethylation in both EBs and ESCs (Figure 4C and Figure S3). Our findings indicate that the promoter regions of many developmental genes become hypermethylated in Tet-deficient EBs. A subset of these differentiation specific genes is active in WT EBs with their promoter hypermethylated in TKO EBs, which correlates with reduced expression and likely causes the observed developmental phenotypes.
Figure 4. Gene specific bisulfite sequencing for validation of hypermethylation of deregulated developmental genes in Tet TKO embryoid bodies.
A) MeDIP-seq profiles of representative developmental genes in TKO EBs. Enrichments are indicated as normalized read counts. Red boxes indicate the genomic region analyzed by gene specific 454 bisulfite sequencing in B and C.
B) Validation of MeDIP-data by gene specific 454 bisulfite sequencing. Results for a 200–300 bp region (see red box in A) at the promoters of indicated genes are shown as heatmaps in which each row represents one sequence read and each column an individual CpG site within the analyzed region. Individual blue boxes indicate methylated and yellow boxes indicate unmethylated CpG dinucleotides. Panels below heatmaps show the average methylation of each interrogated CpG for the analyzed DNA fragment. For color bar see C. Sequencing coverage (reads) are indicated.
C) Heat maps for four additional promoter regions of the genes analyzed. Panels show the average methylation of each interrogated CpG according to the color bar shown on the right. Numbers indicate the sequencing coverage.
(See also Figure S3)
Discussion
Defining the biological significance of Tet proteins and 5hmC marks during development has been a primary focus of recent investigations. Here, we have established that the loss of all three Tet enzymes impairs differentiation of ES cells. This phenotype is less severe than that observed with Dnmt1 mutant ESCs where the loss of methyltransferase activity and severely decreased 5mC levels blocks differentiation and contribution to embryoid bodies in chimera assays (Lei et al., 1996; Panning and Jaenisch, 1996). In contrast to Dnmt1 mutant cells, Tet TKO ESCs can form most germ layers in teratoma assays and contribute, albeit very poorly, to a developing embryo in chimera assays. This indicates that Tet activity and 5hmC modifications are essential for proper differentiation during development. Expression profiles and genome wide methylation maps of WT and TKO EBs show that the majority of deregulated genes in TKO EBs are down regulated, show promoter hypermethylation and include developmental regulators that are repressed by DNA methylation in ESCs (Gifford et al., 2013; Xie et al., 2013). A subset of these affected genes are also potential targets of DNA methylation during early embryonic development where transition from blastocyst to epiblast stage of a developing embryo is associated with modulation of DNA methylation levels for proper orchestration of gene expression (Smith et al., 2012). These observations suggest that Tet enzymes confer their biologically critical effects during ESC differentiation by regulating promoter methylation levels of a subset of developmental regulators and lineage commitment genes and thus enabling their activation by differentiation induced and lineage specific demethylation.
Tet3 is not expressed in ESCs therefore the epigenetic landscape of TKO ESCs is likely to be similar to that of Tet1/2 DKO ESCs. Our previous work as well as recent methylation analysis of Tet1/2 DKO ESCs (Hackett et al., 2013) have confirmed increased methylation in these cells. Postnatal survival of some Tet1/2 double mutant mice (Dawlaty et al., 2013) supports the notion that Tet3 activity partially rescues Tet1/2 deficiency. However, as we find here, complete loss of Tet activity impairs proper promoter demethylation and leads to silencing of target genes, resulting in reduced developmental potential of ESCs.
The experiments described here address a long-standing question in the field regarding the requirements of Tet proteins and the 5hmC mark in development. Our findings complement previous studies on the role of DNA methyltransferases in development (Li et al., 1992; Okano et al., 1999; Sakaue et al., 2010) and propose that both DNA methyltransferases and Tet dioxygenases work in concert to establish and maintain the appropriate pattern of DNA methylation during ESCs differentiation and development. Recent work has shown differential genomic localization of Tet1 and Tet2 in ESCs (Huang et al., 2014). It will be interesting to further investigate the genomic occupancy of Tet enzymes upon differentiation to establish whether Tet1, Tet2 and Tet3 bind and activate similar or different classes of target genes and can substitute for one another. In addition, further studies are needed to define whether demethylation-independent functions of 5hmC as a stable epigenetic mark are also implicated in epigenetic regulation of development. This should also entail examining how loss of 5hmC impairs localization of its readers or binding partners. As this study primarily underpins the requirements of Tet enzymes in differentiation of embryonic stem cells during embryogenesis, future work using conditional deletion systems will be critical in defining the exact role of Tet enzymes in regulating the homoeostasis of many adult tissues. This will help to further define physiological implications of Tet-mediated epigenetic mechanisms of gene regulation in development and diseases including cancer, where Tet enzymes are mutated or down regulated and 5hmC levels are significantly reduced.
Experimental Procedures
Derivation and culture of TKO ESCs
Tet1 and Tet2 knockout alleles have been described previously (Dawlaty et al., 2013). The Tet3 null allele was generated by deleting exon4 of Tet3 in V6.5 ESCs (Dawlaty and Jaenisch unpublished). Tet1/2/3 triple heterozygote and Tet1−/−|Tet2−/−|Tet3+/− mouse embryonic stem cells were derived by intercrossing Tet1/2/3 triple heterozygote mice. Due to the very low expected Mendelian frequency of TKO embryos from this cross, we did not derive TKO ES cells from blastocysts but subjected Tet1−/−|Tet2−/−|Tet3+/− ESCs to one round of gene targeting to delete the Tet3 wild type allele and generate TKO ESCs. All ES cell lines were expanded on feeders using regular ESCs media containing Leukemia Inhibitory Factor (LIF), and were genotyped by PCR and Southern blot using the protocols and probes previously outlined (Dawlaty et al., 2013). For the labeling of ESCs with EGFP, an EGFP-pgk-puro cassette was targeted to the Rosa26 locus and positive clones were screened by Southern blot.
Chimera and Tetraploid (4N) complementation assays and analysis of mid-gestation embryos
To generate chimeric embryos, 10–12 Rosa26-EGFP labeled ES cells were injected in to B6D2F1 × B6D2F1 E3.5 wild-type blastocysts and surgically implanted into 2.5 d.p.c. pseudo-pregnant Swiss Webster female mice following standard procedures. E9.5, E13.5 or E14.5 embryos were harvested, dissected and imaged under a fluorescence dissecting scope and scored for contribution of TKO ESCs to the developing embryo based on EGFP signal. Tetraploid complementation assay was performed as described (Wernig et al., 2007) and briefly explained in the supplemental experimental procedures.
Embryoid body formation and teratoma assays
Embryoid bodies were formed from ESCs following standard hanging-drop methods as explained previously (Dawlaty et al., 2011). EBs were maintained in FBS media –LIF for 10 days (or 15 days) before use for analysis. Teratoma assays and histological analyses of tissues and EBs were performed exactly as explained before (Dawlaty et al., 2013). Hematoxylin and eosin-stained sections of teratomas were examined and scored for the presence or absence of tissue types from the three germ layers.
Quantification of 5hmC and 5mC
Combined liquid chromatography-tandem mass spectrometry with multiple reaction monitoring (LC-MS/MS-MRM) was applied to quantify 5hmC and 5hmC levels in DNA extracted from ESCs or EBs as described previously (Le et al., 2011; Dawlaty et al., 2013).
Microarray Analysis
RNA was extracted from day 10 embryoid bodies using Qiagen RNAeasy Kit, labeled with Cy3 and hybridized to Agilent arrays. Two technical replicas of WT mice (2 samples in total) and two technical replicas of two independent TKO EBs (4 TKO samples in total) were hybridized to a Mouse GE 8×60K Microarray. Arrays were normalized by quantile normalization. All probes for the same gene were summarized by mean before assaying for differential expression by a moderated t test corrected for FDR, as implemented by the limma package in Bioconductor. The cut-off used to select differentially expressed genes was fold change (FC) > 2 and adjusted p-value < 0.01. Gene ontology analyses were performed using Ingenuity Pathway Analysis software.
454 DNA bisulfite sequencing
Deep DNA bisulfite sequencing was performed as described previously (Grönniger et al., 2010). Genomic DNA from wt and TKO ESCs and wt and TKO embryoid bodies was bisulfite-treated using the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s instructions. Treated DNA was amplified with sequence-specific primers (described in supplemental experimental procedures) containing cell type-specific barcodes and standard 454 linker sequences. PCR products of 200–300 bp were gel-extracted using the peqGOLD extraction kit (Peqlab). For sequencing, equimolar amounts of all amplicons were combined in a single tube and processed on a GS Junior sequencer (Roche) according to the manufacturer’s instructions. Sequence reads were aligned and displayed as color coded heat maps.
MeDIP-seq, Mapping of sequencing data and calculation of methylation levels
MeDIP-seq was performed as described (Bocker et al., 2012) but significantly adapted and scaled down to smaller starting amounts according to the protocol by Taiwo and colleagues (Taiwo et al., 2012). 2 μg of genomic DNA isolated from day 10 EBs was sonicated to 300bp fragments using a Covaris S220 ultrasonicator (Covaris). One μg of purified adapter ligated DNA was used for pooled immunoprecipitations using a polyclonal 5mC-specific antibody (Active Motif, 39791). Details of MeDIP, library preparation and data analysis are outlined in supplemental experimental procedures. Briefly, in order to quantitatively analyze sequencing reads, position-wise read counts were normalized according to the 5mC levels of each sample DNA, as determined by mass spectroscopy. Differences in methylation levels for each gene in different samples were calculated by subtracting the normalized average read counts in TKO samples from the respective WT values. If the difference was equal or bigger than 0.25 the region was considered hypermethylated in TKO. If the difference was equal or smaller than −0.25, the region was considered as hypomethylated.
Supplementary Material
Highlights (max 85 characters with spaces for each line).
Combined loss of all three Tet enzymes restricts normal differentiation of ESCs
Tet null ESC contribute poorly to developing embryos and cannot support development
Tet loss causes promoter hypermethylation and deregulation of developmental genes
Acknowledgments
We thank Kibibi Ganz, Dongdong Fu, Ruth Flannery, Linyu Shi, Hui Yang, Ping Xu and Raaji Alagappan for help with animal husbandry, histology and blastocyst injections. We are also grateful to Tayyeb Adil, John Cassady, Thorold Theunissen and Jianlong Wang for plasmids, Alex Yoon for help with mass spectrometry, Tanja Musch for 454 sequencing and the DKFZ Genomics and Proteomics Core Facility for Illumina sequencing services. M.M.D is a Damon Runyon Postdoctoral Fellow. B.E.P is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. A.W.C. is supported by a Croucher scholarship. T.L. is supported by UCLA Molecular, Cellular and Neurobiology Training Grant, UCLA Mental Retardation Training Grant, and Eugene V. Cota-Robles Fellowship. Work in F.L.’s lab was supported by a grant from the Deutsche Forschungsgemeinschaft (SPP 1463). R.J. is funded by NIH grants 5-RO1-HDO45022 and 5-R37-CA084198 and the Simons Foundation. R.J. is an advisor to Stemgent and cofounder of Fate Therapeutics.
Footnotes
Accession numbers
The MeDIP sequencing data (accession number GSE55049) and gene expression array data sets (accession number GSE55574) have been deposited at the Gene Expression Omnibus (GEO) database, http://www.ncbi.nim.nih.gov/geo
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bocker MT, Tuorto F, Raddatz GUN, Musch T, Yang FC, Xu M, Lyko F, Breiling A. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nature Communications. 2012;3:818–12. doi: 10.1038/ncomms1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Developmental Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 Is Dispensable for Maintaining Pluripotency and Its Loss Is Compatible with Embryonic and Postnatal Development. Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönniger E, Weber B, Heil O, Peters N, Stäb F, Wenck H, Korn B, Winnefeld M, Lyko F. Aging and Chronic Sun Exposure Cause Distinct Epigenetic Changes in Human Skin. PLoS Genet. 2010;6:e1000971. doi: 10.1371/journal.pgen.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Dietmann S, Murakami K, Down TA, Leitch HG, Surani MA. Synergistic Mechanisms of DNA Demethylation during Transition to Ground-State Pluripotency. Stem Cell Reports. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-Wide Reprogramming in the Mouse Germ Line Entails the Base Excision Repair Pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martínez JA, Pape UJ, Jacobsen SE, Peters B, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proceedings of the National Academy of Sciences. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 Regulate 5-Hydroxymethylcytosine Production and Cell Lineage Specification in Mouse Embryonic Stem Cells. Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T, Kim KP, Fan G, Faull KF. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Analytical Biochemistry. 2011;412:203–209. doi: 10.1016/j.ab.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai C, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011 doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 Loss Leads to Increased Hematopoietic Stem Cell Self-Renewal and Myeloid Transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano MM, Bell DWD, Haber DAD, Li EE. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell. 1999;99:11–11. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes & Development. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, Mcloughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011:1–4. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. DNA Methylation Is Dispensable for the Growth and Survival of the Extraembryonic Lineages. Current Biology. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011:1–24. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter JOR. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nature Communications. 2011;2:241–248. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA Methylation: Mechanisms, Genomics, and Biological Functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, Beck S, Butcher LM. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc. 2012 Mar 8;7(4):617–36. doi: 10.1038/nprot.2012.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.