Abstract
Background
Epidemiological studies have evaluated the association between Apolipoprotein E (APOE) gene ε2/ε3/ε4 polymorphism and glaucoma susceptibility. However, the published data are still inconclusive. The aim of the present study is to evaluate the impact of APOE gene ε2/ε3/ε4 polymorphism on glaucoma risk by using meta-analysis.
Methods
A comprehensive literature search of PubMed, EMBASE, Cochrane, Elsevier Science Direct and CNKI databases was conducted to identify relevant articles, with the last report up to January 5, 2014. Pooled odds ratio (OR) and 95% confidence interval (CI) were used to assess the strength of association by using the fixed or random effect model.
Results
Fifteen separate studies including 2,700 cases and 2,365 controls were included in the meta-analysis. We did not detect a significant association between APOE gene ε2/ε3/ε4 polymorphism and glaucoma in overall population (P > 0.0083). In Asians, we detected an association of the ε4ε4 genotype with elevated risk for glaucoma (OR = 5.22, 95% CI = 1.85-14.68, P = 0.002), mainly for primary open angle glaucoma (OR = 4.98, 95% CI = 1.75-14.20, P = 0.003).
Conclusions
The meta-analysis suggests that APOE gene ε4ε4 may be associated with elevated risk for primary open angle glaucoma in Asians. However, more epidemiologic studies based on larger sample size, case–control design and stratified by ethnicity as well as types of glaucoma are suggested to further clarify the relationship between APOE gene ε2/ε3/ε4 polymorphism and genetic predisposition to glaucoma.
Keywords: Glaucoma, APOE, Genetic, Meta-analysis
Background
Glaucoma, a group of diseases causing optic neuropathy, is characterized by optic nerve head changes and visual field loss, and is the second leading cause of irreversible blindness across the world [1,2]. In 1995, the World Health Organization reported that 5.1 million people were bilaterally blind from glaucoma [1]. Glaucoma is expected to affect 79.6 million people worldwide by the year 2020 [2]. Its morbidity and prevalence make it a significant public health problem [1]. However, to date, the pathogenesis of glaucoma remains largely unknown, with both genetic and environmental factors contributing to the pathophysiology [1,3]. Over the last decade, the application of molecular genetic techniques to the study of glaucoma has been accelerated greatly, and the accumulating evidence supports an important role for genetics in determining risk for this disease [3].
Apolipoprotein E (APOE), which is a major apolipoprotein in the central nervous system, plays an important role in the uptake and redistribution of cholesterol within neuronal network [4]. APOE gene is located on human chromosome 19q13.2 and is polymorphic [5,6]. There are three common APOE isoforms E2, E3 and E4, encoded by different alleles (ε2, ε3 and ε4), which vary significantly in structure and functions [7]. APOE gene ε2/ε3/ε4 polymorphism has been associated with several neurodegenerative diseases such as Alzheimer's and Parkinson's diseases [8,9]. Therefore, it is possible that this polymorphism may be associated with glaucoma risk [10]. Recently, a number of studies have been conducted to investigate the association of APOE gene ε2/ε3/ε4 polymorphism with glaucoma risk [11-25]. However, the results are controversial with some reports showing positive association [13,15,18-21,24,25] while others showed no association [11,12,14,16,17,22,23]. The inconsistent conclusion may be due to the relatively small size of subjects, since small sample sized association studies could be very limited for efficient assessment of the association.
Integration of these studies may provide improved statistical power to detect the significance. Meta-analysis is a statistical procedure for combining the results of several studies to produce a single estimate of the major effect with enhanced precision [26]. Recently, two studies assessed the association of APOE gene ε2/ε3/ε4 polymorphism with primary open angle glaucoma (POAG) by using meta-analysis [27,28]. Their meta-analyses suggest that this polymorphism is not associated with POAG susceptibility. However, the results of their meta-analyses should be interpreted with caution due to some potential limitations. Therefore, in this study, we conducted a meta-analysis to derive a more precise estimation of the association between APOE gene ε2/ε3/ε4 polymorphism and the risk for glaucoma.
Methods
Search strategy and inclusion criteria
This meta-analysis was reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) Statement, issued in 2009 (Additional file 1: Table S1). Electronic databases, including PubMed, Excerpta Medica Database (EMBASE), Cochrane, Elsevier Science Direct and China National Knowledge Infrastructure (CNKI) databases, were searched for identification of the studies on APOE gene polymorphisms and glaucoma published up to January 5, 2014. Search terms included ‘glaucoma’ and ‘apolipoprotein OR APOE OR Apo E OR ApoE’. There was no language restriction. Review articles and original papers were searched by hand for additional eligible studies. The following criteria were used to select the eligible studies: (a) evaluation of the association between APOE gene ε2/ε3/ε4 polymorphism and glaucoma risk; (b) an unrelated case–control study in which family members were excluded; (c) sufficient published data for estimating an odds ratio (OR) with 95% confidence interval (CI). When authors reported two or more publications on the same patient population, only the largest study was selected. Additionally, when a study reported the results on different subpopulations, we treated them as a separate study.
Data extraction
Information was carefully extracted from all eligible publications independently by two investigators. Disagreement was resolved by discussion between the two investigators. The following data were collected from each study: the first author’s name, publication year, source of publication, ethnicity, sample size (numbers of cases and controls), types of glaucoma, and allele as well as genotype frequencies. Authors were contacted for further information when necessary.
Statistical analysis
Pooled OR with 95% CI was used to assess the strength of association between APOE gene ε2/ε3/ε4 polymorphism and glaucoma risk. The significance of the pooled OR was determined by Z test. Genotype ε3ε3 is the most common genotype [29]. Thus, genotype ε3ε3 or allele ε3 is designated as reference category in the present study. We estimated the risk of the ε2 and ε4 alleles, compared with the ε3 allele. We also performed genotypic analyses (ε2 carriers versus ε3ε3, ε2ε2 versus ε3ε3, ε4 carriers versus ε3ε3 and ε4ε4 versus ε3ε3). ε2 carriers were defined as patients with the ε2ε2 and ε2/ε3 genotypes. ε4 carriers were defined as patients with the ε3ε4 and ε4ε4 genotypes. Genotype ε2ε4 was excluded from the genotypic analyses because of the opposite effect between ε2 and ε4 alleles [15,30]. In addition, we performed subgroup analyses by ethnicity (Cauasians and Asians) and types of glaucoma (POAG) when the data were available. Subgroup analyses were also performed by excluding those studies which did not fulfill Hardy-Weinberg equilibrium (HWE). We carried out sensitivity analysis by excluding one study at a time to explore whether the results were influenced by a specific study. Heterogeneity (between-study inconsistency) was investigated and measured using Cochran’s Q statistic and I2 statistic [31,32]. A P value greater than 0.10 indicated a lack of heterogeneity among studies, so the fixed effect model (Mantel-Haenszel method) was used to calculate pooled OR [33]. Otherwise, the random effect model (DerSimonian-Laird method) was used [34]. Publication bias was detected using Egger’s linear regression test by visual inspection of funnel plot [35]. A Chi square-test was used to evaluate the deviation from HWE in controls. All analyses were done with the software Review Manager (v4.2; Oxford, England) and Stata statistical software (v10.0; StataCorp, College Station, TX, USA), using two-sided P values. P values were Bonferroni adjusted to account for multiple testing. In overall analyses, P values below 0.0083 (0.05/6) were considered statistically significant, and, in subgroup analyses, P values below 0.0042 (0.05/12) were considered statistically significant.
Results
Characteristics of studies
Characteristics of studies investigating the association of APOE gene ε2/ε3/ε4 polymorphism with glaucoma risk are presented in Table 1[11-24]. The study selection process is shown in Figure 1. There were 666 articles relevant to the searching terms (PubMed: 51; EMBASE: 95; Cochrane: 0; Elsevier Science Direct: 513; CNKI: 7). Nineteen studies examined the association between APOE gene polymorphisms and glaucoma. Three studies did not explore the ε2/ε3/ε4 polymorphism and were excluded [36-38]. One study was excluded due to duplicate report [39], and another study was excluded due to unavailable data [25]. Krumbiegel et al. [11] provided data on two different subpopulations (German and Italian), and, thus, each subpopulation was treated as a separate study. Finally, a total of fifteen separate studies including 2,700 cases and 2,365 controls were included in the present meta-analysis [11-24].
Table 1.
Characteristics of studies included in the meta-analysis of apolipoprotein E gene ε2/ε3/ε4 polymorphism and glaucoma*
| Author/Year [Reference] | Ethnicity | Sample size (Case/control) | Glaucoma type |
Number of cases |
Number of controls |
HWE P-value | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ε2 | ε3 | ε4 | ε2 | ε3 | ε4 | |||||
| Krumbiegel 2010 [11] |
Caucasian (German) |
801(459/342) |
PEXG |
79 |
740 |
99 |
51 |
547 |
86 |
0.926 |
| Krumbiegel 2010 [11] |
Caucasian (Italian) |
323(133/190) |
PEXG |
21 |
222 |
23 |
23 |
317 |
40 |
0.024 |
| Saglar 2009 [12] |
Caucasian |
194(75/119) |
POAG |
13 |
126 |
11 |
10 |
204 |
24 |
0.832 |
| Al-Dabbagh 2009 [13] |
Asian |
230(100/130) |
POAG,PACG |
0 |
181 |
19 |
0 |
249 |
11 |
0.969 |
| Jia 2009 [14] |
Asian |
376(176/200) |
POAG |
34 |
278 |
40 |
35 |
329 |
36 |
0.897 |
| Yuan 2007 [15] |
Asian |
162(105/57) |
POAG,PACG |
48 |
81 |
81 |
18 |
76 |
20 |
<0.0001 |
| Zetterberg 2007 [16] |
Caucasian |
429(242/187) |
POAG |
50 |
376 |
58 |
42 |
289 |
43 |
0.923 |
| Hu 2007 [17] |
Asian |
219(142/77) |
POAG |
17 |
229 |
38 |
11 |
129 |
14 |
0.409 |
| Tamura 2006 [18] |
Asian |
105(28/77) |
POAG |
3 |
42 |
11 |
12 |
135 |
7 |
0.079 |
| Lam 2006 [19] |
Asian |
700(400/300) |
POAG |
79 |
674 |
47 |
50 |
495 |
55 |
0.065 |
| Mabuchi 2005 [20] |
Asian |
489(310/179) |
POAG |
16 |
567 |
37 |
18 |
302 |
38 |
0.104 |
| Jünemann 2004 [21] |
Caucasian |
128(96/32) |
POAG |
19 |
146 |
27 |
9 |
40 |
15 |
0.012 |
| Lake 2004 [22] |
Caucasian |
504(155/349) |
POAG |
28 |
229 |
53 |
56 |
534 |
108 |
0.316 |
| Ressiniotis 2004 [23] |
Caucasian |
212(137/75) |
POAG |
35 |
199 |
40 |
16 |
114 |
20 |
NA# |
| Vickers 2002 [24] | Caucasian | 193(142/51) | POAG | 27 | 206 | 51 | 15 | 75 | 12 | 0.202 |
*HWE: Hardy-Weinberg equilibrium; PEXG: pseudoexfoliation glaucoma; POAG: primary open angle glaucoma; PACG: primary angle closure glaucoma; NA: not available. #Only allele frequency was extracted from the study.
Figure 1.
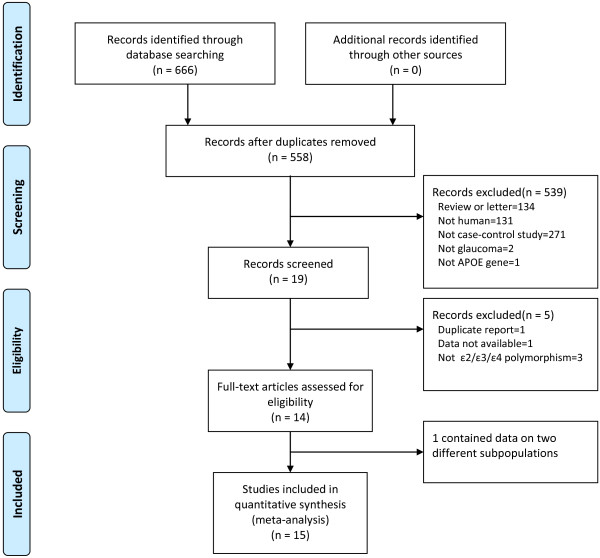
Flow diagram of the study selection process.
The allele and genotype frequencies of APOE gene ε2/ε3/ε4 polymorphism were extracted from fourteen separate studies [11-22,24]. Only allele frequency was extracted from the study by Ressiniotis et al. [23]. The results of HWE test for the distribution of the genotype in control population are shown in Table 1. The genotype distributions in eleven separate studies were in agreement with HWE. The genotype distributions in 3 separate studies were not in agreement with HWE. It was unavailable for the study by Ressiniotis et al. to perform HWE test. The fifteen studies consisted of eight Caucasian studies and seven Asian studies. Of these studies, two studies were conducted in patients with pseudoexfoliation glaucoma (PEXG), eleven studies were conducted in patients with primary open angle glaucoma (POAG), and two studies were conducted in patients with POAG and primary angle closure glaucoma (PACG).
Quantitative synthesis
The results of the meta-analysis on the association between APOE gene ε2/ε3/ε4 polymorphism and risk of glaucoma are shown in Table 2. By pooling all the studies, APOE gene ε2/ε3/ε4 polymorphism was not associated with glaucoma risk (P > 0.0083). We detected significant between-study heterogeneity for the contrasts of ε2 versus ε3, ε2 carriers versus ε3ε3, ε4 versus ε3 and ε4 carriers versus ε3ε3 in overall population (P < 0.1). We carried out subgroup analyses by excluding those studies which did not fulfill HWE, and found similar results (P > 0.0042). Significant between-study heterogeneity for the contrasts of ε2 carriers versus ε3ε3, ε4 versus ε3 and ε4 carriers versus ε3ε3 was detected in HWE population (P < 0.1).
Table 2.
Meta-analysis of the association between apolipoprotein E gene ε2/ε3/ε4 polymorphism and glaucoma*
| Groups |
Sample size |
Number of Studies |
Test of association |
Test of heterogeneity |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95 % CI) | Z | P-value | Model | χ2 | P-value | I2(%) | |||
| Overall |
ε2 vs ε3 |
4765 |
4201 |
15 |
1.08(0.88-1.32) |
0.74 |
0.46 |
R |
20.83 |
0.08 |
37.6 |
| |
ε2 carriers vs ε3ε3 |
2001 |
1804 |
14 |
1.02(0.79-1.33) |
0.18 |
0.86 |
R |
21.18 |
0.05 |
43.3 |
| |
ε2ε2 vs ε3ε3 |
1688 |
1541 |
14 |
1.11(0.54-2.28) |
0.28 |
0.78 |
F |
3.51 |
0.83 |
0.0 |
| |
ε4 vs ε3 |
4931 |
4364 |
15 |
1.14(0.86-1.51) |
0.92 |
0.36 |
R |
58.45 |
<0.0001 |
76.0 |
| |
ε4 carriers vs ε3ε3 |
2146 |
1955 |
14 |
1.15(0.82-1.59) |
0.81 |
0.42 |
R |
50.80 |
<0.0001 |
74.4 |
| |
ε4ε4 vs ε3ε3 |
1704 |
1549 |
14 |
1.66(0.99-2.78) |
1.92 |
0.05 |
F |
14.05 |
0.23 |
21.7 |
| Controls in HWE |
ε2 vs ε3 |
3994 |
3588 |
11 |
1.03(0.87-1.22) |
0.36 |
0.72 |
F |
11.02 |
0.27 |
18.3 |
| |
ε2 carriers vs ε3ε3 |
1790 |
1597 |
11 |
0.93(0.71-1.22) |
0.54 |
0.59 |
R |
15.50 |
0.08 |
41.9 |
| |
ε2ε2 vs ε3ε3 |
1525 |
1359 |
11 |
1.34(0.62-2.90) |
0.73 |
0.46 |
F |
2.05 |
0.92 |
0.0 |
| |
ε4 vs ε3 |
4112 |
3722 |
11 |
1.10(0.83-1.47) |
0.69 |
0.49 |
R |
33.90 |
0.0002 |
70.5 |
| |
ε4 carriers vs ε3ε3 |
1900 |
1723 |
11 |
1.04(0.77-1.42) |
0.26 |
0.79 |
R |
29.86 |
0.0009 |
66.5 |
| |
ε4ε4 vs ε3ε3 |
1533 |
1367 |
11 |
1.41(0.80-2.50) |
1.18 |
0.24 |
F |
10.35 |
0.32 |
13.1 |
| Caucasian |
ε2 vs ε3 |
2516 |
2342 |
8 |
1.08(0.89-1.31) |
0.74 |
0.46 |
F |
7.70 |
0.36 |
9.1 |
| |
ε2 carriers vs ε3ε3 |
996 |
963 |
7 |
1.07(0.84-1.36) |
0.55 |
0.58 |
F |
6.65 |
0.35 |
9.8 |
| |
ε2ε2 vs ε3ε3 |
821 |
813 |
7 |
0.92(0.41-2.09) |
0.20 |
0.84 |
F |
2.74 |
0.60 |
0.0 |
| |
ε4 vs ε3 |
2606 |
2468 |
8 |
0.96(0.81-1.13) |
0.51 |
0.61 |
F |
7.89 |
0.34 |
11.3 |
| |
ε4 carriers vs ε3ε3 |
1079 |
1076 |
7 |
0.93(0.75-1.14) |
0.73 |
0.46 |
F |
4.62 |
0.59 |
0.0 |
| |
ε4ε4 vs ε3ε3 |
823 |
822 |
7 |
0.92(0.47-1.77) |
0.26 |
0.79 |
F |
5.52 |
0.36 |
9.3 |
| Asian |
ε2 vs ε3 |
2249 |
1859 |
7 |
1.07(0.71-1.62) |
0.32 |
0.75 |
R |
13.05 |
0.02 |
61.7 |
| |
ε2 carriers vs ε3ε3 |
1005 |
841 |
7 |
0.95(0.56-1.63) |
0.18 |
0.86 |
R |
14.41 |
0.01 |
65.3 |
| |
ε2ε2 vs ε3ε3 |
867 |
728 |
7 |
2.08(0.45-9.63) |
0.94 |
0.35 |
F |
0.04 |
0.98 |
0.0 |
| |
ε4 vs ε3 |
2325 |
1896 |
7 |
1.53(0.83-2.84) |
1.36 |
0.17 |
R |
48.44 |
<0.0001 |
87.6 |
| |
ε4 carriers vs ε3ε3 |
1067 |
879 |
7 |
1.56(0.77-3.13) |
1.24 |
0.21 |
R |
45.75 |
<0.0001 |
86.9 |
| |
ε4ε4 vs ε3ε3 |
881 |
727 |
7 |
5.22(1.85-14.68) |
3.13 |
0.002 |
F |
2.95 |
0.71 |
0.0 |
| POAG (Caucasian) |
ε2 vs ε3 |
1454 |
1404 |
6 |
1.02(0.80-1.30) |
0.13 |
0.90 |
F |
6.95 |
0.22 |
28.1 |
| |
ε2 carriers vs ε3ε3 |
522 |
549 |
5 |
0.99(0.72-1.37) |
0.05 |
0.96 |
F |
5.92 |
0.20 |
32.5 |
| |
ε2ε2 vs ε3ε3 |
426 |
457 |
5 |
0.79(0.26-2.44) |
0.41 |
0.68 |
F |
0.56 |
0.76 |
0.0 |
| |
ε4 vs ε3 |
1522 |
1478 |
5 |
1.04(0.84-1.28) |
0.34 |
0.74 |
F |
6.56 |
0.26 |
23.8 |
| |
ε4 carriers vs ε3ε3 |
581 |
613 |
5 |
1.01(0.76-1.33) |
0.05 |
0.96 |
F |
3.59 |
0.47 |
0.0 |
| |
ε4ε4 vs ε3ε3 |
430 |
463 |
5 |
1.24(0.55-2.77) |
0.52 |
0.60 |
F |
3.05 |
0.38 |
1.6 |
| POAG (Asian) |
ε2 vs ε3 |
2091 |
1859 |
7 |
0.96(0.75-1.22) |
0.34 |
0.73 |
F |
6.27 |
0.28 |
20.2 |
| |
ε2 carriers vs ε3ε3 |
953 |
841 |
7 |
0.78(0.48-1.27) |
1.00 |
0.32 |
R |
10.68 |
0.06 |
53.2 |
| |
ε2ε2 vs ε3ε3 |
827 |
728 |
7 |
2.08(0.45-9.63) |
0.94 |
0.35 |
F |
0.04 |
0.98 |
0.0 |
| |
ε4 vs ε3 |
2149 |
1896 |
7 |
1.45(0.82-2.55) |
1.29 |
0.20 |
R |
37.92 |
<0.0001 |
84.2 |
| |
ε4 carriers vs ε3ε3 |
1000 |
879 |
7 |
1.45(0.76-2.76) |
1.13 |
0.26 |
R |
36.86 |
<0.0001 |
83.7 |
| ε4ε4 vs ε3ε3 | 838 | 727 | 7 | 4.98(1.75-14.20) | 3.01 | 0.003 | F | 3.01 | 0.70 | 0.0 | |
*POAG: primary open angle glaucoma; HWE: Hardy-Weinberg equilibrium; vs: versus; OR: odds ratio; CI: confidence interval; F: fixed effect model; R: random effect model.
When stratified by ethnicity, no association was found between APOE gene ε2/ε3/ε4 polymorphism and risk of glaucoma in Caucasians (P > 0.0042). No significant between-study heterogeneity was found in Caucasians (P > 0.1). In Asians, we detected an association of the ε4ε4 genotype with elevated risk for glaucoma (OR = 5.22, 95%CI = 1.85-14.68, P = 0.002). But we did not find significant association of APOE gene ε2/ε3/ε4 polymorphism with glaucoma in other models (P > 0.0042). Significant between-study heterogeneity was found for the contrasts of ε2 versus ε3, ε2 carriers versus ε3ε3, ε4 versus ε3 and ε4 carriers versus ε3ε3 in Asians (P < 0.1).
In subgroup analysis by types of glaucoma, we also detected an association of the ε4ε4 genotype with elevated risk for POAG in Asians (OR = 4.98, 95% CI = 1.75-14.20, P = 0.003), but not for POAG in Caucasians (P > 0.0042). In POAG population (Asians), we detected significant between-study heterogeneity for the contrasts of ε2 carriers versus ε3ε3, ε4 versus ε3 and ε4 carriers versus ε3ε3 (P < 0.1).
Potential publication bias and sensitivity analysis
For most of comparisons, the shapes of the funnel plots did not reveal any evidence of obvious asymmetry (funnel plots not shown), and these results were further supported by analysis via Egger’s linear regression test (Table 3). We only found publication bias for the contrasts of ε4 carriers versus ε3ε3 in overall population and HWE population, ε4 versus ε3 as well as ε4 carriers versus ε3ε3 in Asians and POAG (Asians) subgroup (P < 0.05). Additionally, Egger’s test was not applied in the contrast of ε2ε2 versus ε3ε3 in Asians and the contrasts of ε2ε2 versus ε3ε3 as well as ε4 carriers versus ε3ε3 in POAG (Asians) subgroup due to the rare frequency of homozygous mutation.
Table 3.
Egger’s linear regression test to measure the funnel plot asymmetry*
| Groups |
Y axle intercept:
a
(95 % CI) |
|||||
|---|---|---|---|---|---|---|
| Overall | Controls in HWE | Caucasian | Asian | POAG (Caucasian) | POAG (Asian) | |
| ε2 vs ε3 |
−0.66(−3.09-1.77) |
−0.93(−3.39-1.54) |
−0.22(−3.69-3.23) |
−1.06(−6.79-4.66) |
0.03(−5.81-5.87) |
−1.68(−4.48-1.13) |
| ε2 carriers vs ε3ε3 |
−0.03(−2.36-2.29) |
−1.38(−4.14-1.38) |
0.55(−2.73-3.83) |
−0.50(−5.90-4.90) |
1.14(−4.80-7.08) |
−2.07(−7.60-3.46) |
| ε2ε2 vs ε3ε3 |
−0.94(−5.04-3.15) |
−0.94(−5.04-3.15) |
−3.15(−8.03-1.73) |
NA |
−2.28(−16.07-11.52) |
NA |
| ε4 vs ε3 |
2.81(−0.76-6.39) |
3.21(−0.14-6.56) |
−0.45(−3.63-2.73) |
7.63(0.14-15.12) |
−1.60(−6.29-3.08) |
7.26(2.85-11.66) |
| ε4 carriers vs ε3ε3 |
3.49(0.56-6.41) |
3.23(0.16-6.31) |
0.38(−2.34-3.11) |
7.73(3.29-12.18) |
0.35(−4.70-5.40) |
6.54(3.07-10.01) |
| ε4ε4 vs ε3ε3 | −2.15(−5.52-1.22) | −2.18(−6.29-1.93) | −2.35(−6.03-1.32) | 2.01(−1.14-5.15) | −2.26(−6.69-2.17) | NA |
*POAG: primary open angle glaucoma; HWE: Hardy-Weinberg equilibrium; vs: versus; CI: confidence interval; NA: not available.
We carried out sensitivity analysis by excluding one study at a time to explore whether the positve result of POAG (Asians) subgroup were influenced by a specific study. Sensitivity analysis indicated that a significant variation in combined ORs by excluding the study by Al-Dabbagh et al. (OR = 4.01, 95% CI = 1.29-12.46, P = 0.02) or the study by Yuan et al. (OR = 4.04, 95% CI = 1.30-12.50, P = 0.02) or the study by Tamura et al. (OR = 4.62, 95% CI = 1.54-13.90, P = 0.006).
Discussion
Recently, two studies assessed the association of APOE gene ε2/ε3/ε4 polymorphism with POAG by using meta-analysis [27,28]. Their meta-analyses suggest that this polymorphism is not associated with POAG susceptibility. However, the results of their meta-analyses should be interpreted with caution due to some limitations. In the study by Song et al. [27], the authors did not include all available published studies. Five eligible studies were not include in the meta-analysis [13,15,17,21,23]. Meanwhile, two studies contained overlapping data [19,39], and only the largest study [19] should be selected for the analysis. In another study by Wang et al. [28], there were many mistakes in the data extraction (see Supplementary Table two in their article). For example, the authors confused the cases and controls for the study by Zetterberg et al. [16]. The data provided by the authors for the study by Vickers et al. [24] were not in line with the data provided by Vickers et al. in their original publication. Close inspection of the data in the study by Jia et al. [14] revealed that the number of the ε2, ε3 and ε4 alleles in cases should be 34, 278 and 40, but not 34, 280 and 38 in the study by Wang et al.. These limitations distorted the results of their meta-analysis. In the present study, we systemically reviewed all available published studies and performed a meta-analysis to derive a more precise estimation of the association between APOE gene ε2/ε3/ε4 polymorphism and susceptibility to glaucoma. Our meta-analysis included fifteen separate studies involving 2,700 cases and 2,365 controls. In Asians, we detected an association of the ε4ε4 genotype with elevated risk for glaucoma, mainly for POAG. Thus, the ε4ε4 genotype may be associated with elevated risk for POAG in Asians.
Glaucoma is a term describing a group of ocular disorders with multi-factorial etiology united by a clinically characteristic intraocular pressure-associated optic neuropathy [1,40]. This disease often goes undetected. In the United States, glaucoma is responsible for 9-12% of blindness, and impacts up to three million Americans, but only half of those are aware they have it [41]. Glaucoma is a neurodegenerative disease, which is often associated with high intraocular pressure [10,42]. In this disease, there are cell bodies (retinal ganglion cells) and nerve fibers (optic nerve axons) that are vulnerable to degeneration and amenable to protection. Neurodegeneration occurs in the retinal ganglion neuronal cell bodies, their axons in the optic nerve, the lateral geniculate nucleus, optic radiation, and visual cortex [43,44]. APOE plays a key role in lipid metabolism, cholesterol transport, as well as protein synthesis, and is also involved in numerous other functions, including tissue repair, cell growth and differentiation, and immune response and regulation [4,45]. APOE gene ε2/ε3/ε4 polymorphism has been associated with a number of neurodegenerative diseases such as Alzheimer's and Parkinson's diseases [8,9], raising the possibility that this polymorphism might predispose to neurodegeneration of the retinal ganglion cells and the optic nerve axons in glaucoma. In the study, we found that the ε4ε4 genotype is associated with elevated risk for POAG in Asians. The extracellular accumulation of insoluble fibrillar peptides in brain parenchyma and vessel walls as amyloid is the hallmark of neurodegenerative diseases [46]. Amyloid β-peptide (Aβ) deposition in senile plaques and cerebral vessels is a neuropathological feature of Alzheimer’s disease [47]. There is evidence that the amount of histologically identified Aβ in vessels and plaques in the cerebral cortex of patients with Alzheimer’s disease is a direct function of their APOE genotype, and subjects with ε4ε4 genotype have an increased amyloid deposits in vessels and density of strongly Aβ-immunoreactive plaques than those with ε3ε3 genotype [47]. Numerous similarities exist between glaucoma and Alzheimer's diseases [10], and, thus, the evidence supports our results. However, the exact function of APOE gene ε2/ε3/ε4 polymorphism remains largely unknown, and studies of the functional implications of this polymorphism are still needed in the future. Additionally, we found that the ε4ε4 genotype is associated with elevated risk for POAG in Asians, but not in Caucasians, which indicated that the genetic background of Asians and Caucasians is different.
Some limitations of this meta-analysis should be acknowledged. A first consideration is that we found publication bias for some contrasts. A second consideration is that significant between-study heterogeneity was found in some comparisons, which may be distorting the results of meta-analysis. In the subgroup analysis by ethnicity, the heterogeneity disappeared among Caucasians. The heterogeneity may have resulted from differences in the subjects’ genetic backgrounds. Additionally, different types of glaucoma may contribute to the heterogeneity. But most of studies did not provide glaucoma subtype information (e.g., normal tension glaucoma or high tension glaucoma). Third, though we detected an association of the ε4ε4 genotype with elevated risk for glaucoma (with POAG and PCAG combined) in Asians, only two studies were conducted in patients with PACG (109 patients). Thus, further studies exploring the association between APOE gene ε2/ε3/ε4 polymorphism and PACG risk are still needed. Fourth, sensitivity analysis indicated that a significant variation in combined ORs by excluding some studies. Finally, our results are based on unadjusted estimates, which may be distorting the results of meta-analysis. A more precise analysis stratified by environmental factors could be performed if individual data were available.
Conclusions
In summary, the present meta-analysis suggests that APOE gene ε4ε4 may be associated with elevated risk for POAG in Asians. However, more epidemiologic studies based on larger sample size, case–control design and stratified by ethnicity as well as types of glaucoma are suggested to further clarify the relationship between APOE gene ε2/ε3/ε4 polymorphism and genetic predisposition to glaucoma.
Abbreviations
APOE: Apolipoprotein E; Aβ: Amyloid β-peptide; CI: Confidence interval; CNKI: China National Knowledge Infrastructure; EMBASE: Excerpta Medica Database; HWE: Hardy-Weinberg equilibrium; OR: Odds ratio; PACG: Primary angle closure glaucoma; PEXG: Pseudoexfoliation glaucoma; POAG: Primary open angle glaucoma; PRISMA: Preferred reporting items for systematic reviews and meta-analysis.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
S-LL, YW, Y-FZ, B-YZ and Z-YG conceived and designed the study. S-LL, YW, Y-FZ, B-YZ and Z-YG took part in literature review and data extraction. S-LL and YW were involved in software used and data analysis. S-LL, YW, Y-FZ, B-YZ and Z-YG wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by Anhui Provincial Natural Science Foundation (1408085MH158).
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
PRISMA Checklist.
Contributor Information
Yong Wang, Email: wangyongayyk@163.com.
Yan-Feng Zhou, Email: zhouyanfengay@163.com.
Bing-Ying Zhao, Email: zhaobingyingay@163.com.
Zheng-Yu Gu, Email: guzhengyuay@163.com.
Shou-Ling Li, Email: shouling_li@163.com.
References
- Coleman AL, Brigatti L. The glaucomas. Minerva Med. 2001;15(5):365–379. [PubMed] [Google Scholar]
- Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;15(3):262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz TE, Fingert JH, Wang K, Kuehn MH, Knudtson KL, Alward WL, Boldt HC, Russell SR, Folk JC, Casavant TL, Braun TA, Clark AF, Stone EM, Sheffield VC. A genome-wide association study for primary open angle glaucoma and macular degeneration reveals novel Loci. PLoS One. 2013;15(3):e58657. doi: 10.1371/journal.pone.0058657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws SM, Hone E, Gandy S, Martins RN. Expanding the association between the APOE gene and the risk of Alzheimer's disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem. 2003;15(6):1215–1236. doi: 10.1046/j.1471-4159.2003.01615.x. [DOI] [PubMed] [Google Scholar]
- Singh PP, Singh M, Mastana SS. APOE distribution in world populations with new data from India and the UK. Ann Hum Biol. 2006;15(3):279–308. doi: 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- Eisenberg DT, Kuzawa CW, Hayes MG. Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history. Am J Phys Anthropol. 2010;15(1):100–111. doi: 10.1002/ajpa.21298. [DOI] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Sastre I, Recuero M, García MA, Aldudo J, Vázquez J, Valdivieso F. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998;15(2):105–108. doi: 10.1016/S0014-5793(97)01543-3. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;15(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Benjamin R, Leake A, Edwardson JA, McKeith IG, Ince PG, Perry RH, Morris CM. Apolipoprotein E genes in Lewy body and Parkinson's disease. Lancet. 1994;15(8912):1565. doi: 10.1016/s0140-6736(94)92961-0. [DOI] [PubMed] [Google Scholar]
- Gupta N, Yücel YH. Glaucoma as a neurodegenerative disease. Curr Opin Ophthalmol. 2007;15(2):110–114. doi: 10.1097/ICU.0b013e3280895aea. [DOI] [PubMed] [Google Scholar]
- Krumbiegel M, Pasutto F, Mardin CY, Weisschuh N, Paoli D, Gramer E, Weber BH, Kruse FE, Schlötzer-Schrehardt U, Reis A. Apolipoprotein E genotypes in pseudoexfoliation syndrome and pseudoexfoliation glaucoma. J Glaucoma. 2010;15(8):561–565. doi: 10.1097/IJG.0b013e3181ca76c4. [DOI] [PubMed] [Google Scholar]
- Saglar E, Yucel D, Bozkurt B, Ozgul RK, Irkec M, Ogus A. Association of polymorphisms in APOE, p53, and p21 with primary open-angle glaucoma in Turkish patients. Mol Vis. 2009;15:1270–1276. [PMC free article] [PubMed] [Google Scholar]
- Al-Dabbagh NM, Al-Dohayan N, Arfin M, Tariq M. Apolipoprotein E polymorphisms and primary glaucoma in Saudis. Mol Vis. 2009;15:912–919. [PMC free article] [PubMed] [Google Scholar]
- Jia LY, Tam PO, Chiang SW, Ding N, Chen LJ, Yam GH, Pang CP, Wang NL. Multiple gene polymorphisms analysis revealed a different profile of genetic polymorphisms of primary open-angle glaucoma in northern Chinese. Mol Vis. 2009;15:89–98. [PMC free article] [PubMed] [Google Scholar]
- Yuan HP, Xiao Z, Yang BB. A study on the association of apolipoprotein E genotypes with primary open-angle glaucoma and primary angle-closure glaucoma in northeast of China. Zhonghua Yan Ke Za Zhi. 2007;15(5):416–420. [PubMed] [Google Scholar]
- Zetterberg M, Tasa G, Palmér MS, Juronen E, Teesalu P, Blennow K, Zetterberg H. Apolipoprotein E polymorphisms in patients with primary open-angle glaucoma. Am J Ophthalmol. 2007;15(6):1059–1060. doi: 10.1016/j.ajo.2007.01.031. [DOI] [PubMed] [Google Scholar]
- Hu Y. Master thesis. Shantou: Shantou University; 2007. The APOE gene and its interactions with SNPs of other genes in primary open angle glaucoma and age-related macular degeneration. [Google Scholar]
- Tamura H, Kawakami H, Kanamoto T, Kato T, Yokoyama T, Sasaki K, Izumi Y, Matsumoto M, Mishima HK. High frequency of open-angle glaucoma in Japanese patients with Alzheimer's disease. J Neurol Sci. 2006;15(1–2):79–83. doi: 10.1016/j.jns.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Lam CY, Fan BJ, Wang DY, Tam PO, Yung Tham CC, Leung DY, Ping Fan DS, Chiu Lam DS, Pang CP. Association of apolipoprotein E polymorphisms with normal tension glaucoma in a Chinese population. J Glaucoma. 2006;15(3):218–222. doi: 10.1097/01.ijg.0000212217.19804.a7. [DOI] [PubMed] [Google Scholar]
- Mabuchi F, Tang S, Ando D, Yamakita M, Wang J, Kashiwagi K, Yamagata Z, Iijima H, Tsukahara S. The apolipoprotein E gene polymorphism is associated with open angle glaucoma in the Japanese population. Mol Vis. 2005;15:609–612. [PubMed] [Google Scholar]
- Jünemann A, Bleich S, Reulbach U, Henkel K, Wakili N, Beck G, Rautenstrauss B, Mardin C, Naumann GO, Reis A, Kornhuber J. Prospective case control study on genetic assocation of apolipoprotein epsilon2 with intraocular pressure. Br J Ophthalmol. 2004;15(4):581–582. doi: 10.1136/bjo.2003.020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake S, Liverani E, Desai M, Casson R, James B, Clark A, Salmon JF. Normal tension glaucoma is not associated with the common apolipoprotein E gene polymorphisms. Br J Ophthalmol. 2004;15(4):491–493. doi: 10.1136/bjo.2003.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressiniotis T, Griffiths PG, Birch M, Keers S, Chinnery PF. The role of apolipoprotein E gene polymorphisms in primary open-angle glaucoma. Arch Ophthalmol. 2004;15(2):258–261. doi: 10.1001/archopht.122.2.258. [DOI] [PubMed] [Google Scholar]
- Vickers JC, Craig JE, Stankovich J, McCormack GH, West AK, Dickinson JL, McCartney PJ, Coote MA, Healey DL, Mackey DA. The apolipoprotein epsilon4 gene is associated with elevated risk of normal tension glaucoma. Mol Vis. 2002;15:389–393. [PubMed] [Google Scholar]
- Chiras D, Tzika K, Kokotas H, Oliveira SC, Grigoriadou M, Kastania A, Dima K, Stefaniotou M, Aspiotis M, Petersen MB, Kroupis C, Kitsos G. Development of novel LOXL1 genotyping method and evaluation of LOXL1, APOE and MTHFR polymorphisms in exfoliation syndrome/glaucoma in a Greek population. Mol Vis. 2013;15:1006–1016. [PMC free article] [PubMed] [Google Scholar]
- Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;15(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Song Q, Chen P, Liu Q. Role of the APOE ε2/ε3/ε4 Polymorphism in the Development of Primary Open-Angle Glaucoma: Evidence from a Comprehensive Meta-Analysis. PLoS One. 2013;15(11):e82347. doi: 10.1371/journal.pone.0082347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhou M, Huang W, Chen S, Zhang X. Lack of association of apolipoprotein E (Apo E) ε2/ε3/ε4 polymorphisms with primary open-angle glaucoma: a meta-analysis from 1916 cases and 1756 controls. PLoS One. 2013;15(9):e72644. doi: 10.1371/journal.pone.0072644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthopoulos PG, Hamodrakas SJ, Bagos PG. Apolipoprotein E polymorphisms and type 2 diabetes: a meta-analysis of 30 studies including 5423 cases and 8197 controls. Mol Genet Metab. 2010;15(3):283–291. doi: 10.1016/j.ymgme.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Kesäniemi YA, Ehnholm C, Miettinen TA. Intestinal cholesterol absorption efficiency in man is related to apoprotein E phenotype. J Clin Invest. 1987;15(2):578–581. doi: 10.1172/JCI113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;15(1):101–129. doi: 10.2307/3001666. [DOI] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;15(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;15(4):719–748. [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;15(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Br Med J. 1997;15(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin B, Brézin AP, Valtot F, Dascotte JC, Béchetoille A, Garchon HJ. Apolipoprotein E-promoter single-nucleotide polymorphisms affect the phenotype of primary open-angle glaucoma and demonstrate interaction with the myocilin gene. Am J Hum Genet. 2002;15(6):1575–1581. doi: 10.1086/340733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressiniotis T, Griffiths PG, Birch M, Keers SM, Chinnery PF. Apolipoprotein E promoter polymorphisms do not have a major influence on the risk of developing primary open angle glaucoma. Mol Vis. 2004;15:805–807. [PubMed] [Google Scholar]
- Nowak A, Przybylowska-Sygut K, Gacek M, Kaminska A, Szaflik JP, Szaflik J, Majsterek I. Neurodegenerative Genes Polymorphisms of the -491A/T APOE, the -877T/C APP and the Risk of Primary Open-angle Glaucoma in the Polish Population. Ophthalmic Genet. 2013. DOI: 10.3109/13816810.2013.838277. [DOI] [PubMed]
- Fan BJ, Wang DY, Fan DS, Tam PO, Lam DS, Tham CC, Lam CY, Lau TC, Pang CP. SNPs and interaction analyses of myocilin, optineurin, and apolipoprotein E in primary open angle glaucoma patients. Mol Vis. 2005;15:625–631. [PubMed] [Google Scholar]
- Casson RJ, Chidlow G, Wood JP, Crowston JG, Goldberg I. Definition of glaucoma: clinical and experimental concepts. Clin Experiment Ophthalmol. 2012;15(4):341–349. doi: 10.1111/j.1442-9071.2012.02773.x. [DOI] [PubMed] [Google Scholar]
- Valenti DA. Alzheimer's disease and glaucoma: imaging the biomarkers of neurodegenerative disease. Int J Alzheimers Dis. 2011;15:793931. doi: 10.4061/2010/793931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. Neurodegeneration and neuroprotection in glaucoma: development of a therapeutic neuroprotective vaccine: the Friedenwald lecture. Invest Ophthalmol Vis Sci. 2003;15(4):1407–1411. doi: 10.1167/iovs.02-0594. [DOI] [PubMed] [Google Scholar]
- Yoles E, Schwartz M. Degeneration of spared axons following partial white matter lesion: implications for optic nerve neuropathies. Exp Neurol. 1998;15(1):1–7. doi: 10.1006/exnr.1998.6811. [DOI] [PubMed] [Google Scholar]
- Yücel Y, Gupta N. Glaucoma of the brain: a disease model for the study of transsynaptic neural degeneration. Prog Brain Res. 2008;15:465–478. doi: 10.1016/S0079-6123(08)01132-1. [DOI] [PubMed] [Google Scholar]
- Saadat M. Apolipoprotein E (APOE) Polymorphisms and Susceptibility to Breast Cancer: A Meta-Analysis. Cancer Res Treat. 2012;15(2):121–126. doi: 10.4143/crt.2012.44.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R, Ghetti B. Characterization of amyloid deposits in neurodegenerative diseases. Methods Mol Biol. 2011;15:241–258. doi: 10.1007/978-1-61779-328-8_16. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;15(20):9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.


