Abstract
Background
Esophageal cancer ranks eighth among frequent cancers worldwide. Our aim was to investigate whether and at which neoplastic stage promoter hypermethylation of CAV1 is involved in human esophageal carcinogenesis.
Methods
Using real-time quantitative methylation-specific PCR (qMSP), we examined CAV1 promoter hypermethylation in 260 human esophageal tissue specimens. Real-time RT-PCR and qMSP were also performed on OE33 esophageal cancer cells before and after treatment with the demethylating agent, 5-aza-2’-deoxycytidine (5-Aza-dC).
Results
CAV1 hypermethylation showed highly discriminative ROC curve profiles, clearly distinguishing esophageal adenocarcinomas (EAC) and esophageal squamous cell carcinomas (ESCC) from normal esophagus (NE) (EAC vs. NE, AUROC = 0.839 and p < 0.0001; ESCC vs. NE, AUROC = 0.920 and p < 0.0001). Both CAV1 methylation frequency and normalized methylation value (NMV) were significantly higher in Barrett’s metaplasia (BE), low-grade and high-grade dysplasia occurring in BE (D), EAC, and ESCC than in NE (all p < 0.01, respectively). Meanwhile, among 41 cases with matched NE and EAC or ESCC, CAV1 NMVs in EAC and ESCC (mean = 0.273) were significantly higher than in corresponding NE (mean = 0.146; p < 0.01, Student’s paired t-test). Treatment of OE33 EAC cells with 5-Aza-dC reduced CAV1 methylation and increased CAV1 mRNA expression.
Conclusions
CAV1 promoter hypermethylation is a frequent event in human esophageal carcinomas and is associated with early neoplastic progression in Barrett’s esophagus.
Keywords: CAV1, Hypermethylation, EAC, ESCC
Background
Esophageal cancer ranks eighth among frequent cancers worldwide, with estimated over 480,000 new cases diagnosed and 400,000 deaths globally in 2008 [1]. There are two major histologic subtypes in esophageal cancer: esophageal adenocarcinoma (EAC), which is more prevalent in Western countries, with a rapid recent rate of increase in incidence; and esophageal squamous cell carcinoma (ESCC), which occurs at high frequencies in many developing countries, especially in Asia, and including China [2]. Since esophageal cancer exhibits highly aggressive behavior, with rapid progression to death [3,4], it is essential to gain a better understanding of the molecular events underlying these tumors In order to make further inroads into survival, it is important to discover novel early detection biomarkers and targets for chemoprevention or therapy.
Caveolae, which are small (50–100 nanometer) invaginations of the plasma membrane in many vertebrate cell types, are vital subcellular structures that regulate endocytosis, vesicular traffic, and signal transduction [5]. Caveolin-1 (CAV1), a cytoplasmic 22-kDa scaffold protein, is an essential component of caveolae [6]. In recent years, several studies have reported downregulation of CAV1 protein levels in several malignancies, including ovarian, breast [7-10], prostate [11], oral squamous cell [12] and lung cancer [13]. Prade et al. also showed that the majority of EACs exhibit loss of CAV1 expression in tumor vs. matched normal esophageal epithelia [14]. These results suggest that reduced CAV1 expression may represent a general characteristic of tumors, and that CAV1 may inhibit tumor formation. Aberrant methylation of promoter CpG islands upstream of tumor suppressor genes is now well-established as a major mechanism of gene inactivation in tumorigenesis [15], including in ESCC and EAC [16-23]. Several of these aberrantly methylated genes appear to represent useful prognostic markers, as they precede and predict the progression of BE to EAC. Aberrant promoter methylation of CAV1 is associated with inactivation of its expression in breast and colorectal cancers [24-27]. Therefore, we hypothesized that CAV1 was inactivated via promoter hypermethylation in human esophageal cancers, and that hypermethylation of CAV1 constituted an early event in the genesis of EAC.
Methods
Tissue samples
In the current study, 67 normal esophagi (NE), 60 Barrett’s metaplasias without dysplasia (BE), 19 low-grade (LGD) and 21 high-grade (HGD) dysplasias occurring in BE (D), 67 EACs, and 26 ESCCs were examined. Outcome data were derived from a comprehensive database maintained by the institution’s cancer registry and from patients’ medical records at the University of Maryland and Baltimore Veterans Affairs Medical Centers. All patients provided written informed consent under a protocol approved by the Institutional Review Boards at the University of Maryland and Baltimore Veterans Affairs Medical Centers, where all esophagogastroduodenoscopies were performed. Biopsies were taken using a standardized biopsy protocol, as previously described [17]. Research tissues were obtained from grossly apparent Barrett’s epithelium or from mass lesions in patients manifesting these changes at endoscopic examination, and histology was confirmed using parallel aliquots obtained at endoscopy. All biopsy specimens were stored in liquid nitrogen before DNA/RNA extraction. Clinicopathologic characteristics are summarized in Table 1.
Table 1.
Clinicopathologic characteristics and methylation status of Caveolin 1 in human esophageal tissues
| |
Number of samples |
Age (year) mean |
NMV |
Methylation status (cutoff 0.2) |
||||
|---|---|---|---|---|---|---|---|---|
| Clinical characteristics | Mean | p | Frequency | UM | M | p | ||
|
Barrett’s segment |
|
|
|
|
|
|
|
|
| Short-segment ( <3 cm ) |
14 |
62.3 |
0.327 |
>0.05§ |
71.4% |
4 |
10 |
> 0.05‡ |
| Long-segment ( > = 3 cm ) |
16 |
62.8 |
0.439 |
|
81.3% |
3 |
13 |
|
|
Histology |
|
|
|
|
|
|
|
|
| Normal esophagus |
67 |
64.4 |
0.134 |
|
25.4% |
50 |
17 |
|
| Barrett’s metaplasia |
60 |
63.7 |
0.374 |
< 0.01*/§ |
81.7% |
11 |
49 |
* < 0.01† |
| Dysplasia in Barrett’s esophagus |
40 |
65.3 |
0.254 |
< 0.01*/§/ < 0.01$/§ |
60.0% |
16 |
24 |
* < 0.01†/ $ < 0.05† |
| Low-grade dysplasia |
19 |
65.3 |
0.269 |
< 0.01*/§/ < 0.05$/§ |
57.9% |
8 |
11 |
* < 0.01†/ $ < 0.05† |
| High-grade dysplasia |
21 |
65.2 |
0.240 |
< 0.01*/§/ < 0.01$/§ |
61.9% |
8 |
13 |
* < 0.01†/ $ > 0.05† |
| EAC |
67 |
65.1 |
0.294 |
< 0.01*/§/ < 0.01$/§ |
65.7% |
23 |
44 |
* < 0.01†/ $ < 0.05† |
| Well differentiation |
10 |
66.2 |
0.344 |
>0.05§§ |
80.0% |
2 |
8 |
> 0.05‡ |
| Moderate differentiation |
24 |
66.1 |
0.29 |
|
58.3% |
10 |
14 |
|
| Poor differentiation |
22 |
65.5 |
0.297 |
|
63.6% |
8 |
14 |
|
| Unknown |
11 |
61 |
0.252 |
|
72.7% |
3 |
8 |
|
| ESCC |
26 |
62.5 |
0.326 |
< 0.01*/§ |
80.8% |
5 |
21 |
* < 0.01‡ |
| Well differentiation |
3 |
61.7 |
0.307 |
>0.05§§ |
100.0% |
0 |
3 |
> 0.05‡ |
| Moderate differentiation |
11 |
62.7 |
0.381 |
|
90.9% |
1 |
10 |
|
| Poor differentiation |
5 |
64.2 |
0.256 |
|
80.0% |
1 |
4 |
|
| Unknown |
7 |
61.1 |
0.299 |
|
57.1% |
3 |
4 |
|
|
Stage of EAC patients |
|
|
|
|
|
|
|
|
| I |
7 |
63 |
0.358 |
>0.05§§ |
71.4% |
2 |
5 |
> 0.05‡ |
| II |
15 |
65.2 |
0.286 |
|
73.3% |
4 |
11 |
|
| III |
25 |
64.6 |
0.284 |
|
56.0% |
11 |
14 |
|
| IV |
7 |
66.3 |
0.242 |
|
28.6% |
5 |
2 |
|
|
Lymph node metastasis in EAC patients |
|
|
|
|
|
|
|
|
| Negative |
25 |
64.9 |
0.314 |
>0.05§ |
64.0% |
9 |
16 |
> 0.05† |
| Positive |
25 |
64.6 |
0.276 |
|
56.0% |
11 |
14 |
|
|
Smoking status of EAC patients |
|
|
|
|
|
|
|
|
| Never |
6 |
58.5 |
0.325 |
>0.05§§ |
83.3% |
1 |
5 |
> 0.05‡ |
| Former |
24 |
68.5 |
0.276 |
|
62.5% |
9 |
15 |
|
| Current |
13 |
60.8 |
0.303 |
|
53.8% |
6 |
7 |
|
|
Alcohol consumption of EAC patients |
|
|
|
|
|
|
|
|
| Never |
16 |
65.3 |
0.285 |
>0.05§§ |
68.8% |
5 |
11 |
> 0.05‡ |
| Former |
15 |
63 |
0.302 |
|
66.7% |
5 |
10 |
|
| Current | 10 | 65.7 | 0.315 | 60.0% | 4 | 6 | ||
EAC: esophageal adenocarcinoma; ESCC: esophageal squamous cell carcinoma; NMV: normalized methylation value; UM, unmethylated; M, methylated; §Student's t test; *comparisons made to normal esophagus; *comparisons made to Barrett’s metaplasia; §§Kruskal-Wallis test; †Chi-square for independence test; ‡Fisher's exact test.
Cell lines
The EAC (OE33) cell line was obtained from collaborators at the University of Michigan (Dr. David Beer). These cells were cultured in 47.5% RPMI 1640, 47.5% F-12 supplemented with 5% fetal bovine serum.
DNA and RNA extraction
Genomic DNA was extracted from biopsies and cultured cells using a DNeasy Tissue Kit (Qiagen, Valencia, CA). Total RNA was isolated cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA). DNAs and RNAs were stored at -80°C before analysis.
Bisulfite treatment and real-time quantitative methylation-specific PCR
DNA was treated with bisulfite to convert unmethylated cytosines to uracils prior to qMSP, as described previously [27]. Promoter methylation levels of CAV1 were determined with the ABI 7900 Sequence Detection System (Applied Biosystems, Foster City, CA), using primers and probes as described previously [27]. A standard curve was generated using serial dilutions of CpGenome Universal Methylated DNA (CHEMICON, Temecula, CA). The normalized methylation value (NMV) was defined as follows: NMV = (CAV1-S/CAV1-FM)/(ACTB-S/ACTB-FM), where CAV1-S and CAV1-FM represent the methylation levels of CAV1 in sample and universal methylated DNAs, respectively, while ACTB-S and ACTB-FM correspond to ß-Actin in sample and universal methylated DNAs, respectively [21].
Real-time quantitative RT-PCR
To determine CAV1 mRNA levels, one-step real-time quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) was performed using a Qiagen QuantiTect Probe RT-PCR Kit (Qiagen, Hilden, Germany) and the ABI 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). Primers and probes were the same as previously reported [27]. ß-Actin was used for normalization of data. A standard curve was generated using serial dilutions of qPCR Reference Total RNA (Clontech, Mountainview, CA). The normalized mRNA value (NRV) was calculated according to the following formula for relative expression of target mRNA: NRV = (TarS/TarC)/(ACTB-S/ACTB-C), where TarS and TarC represent levels of mRNA expression for the target gene in sample and control mRNAs, respectively, whereas ACTB-S and ACTB-C correspond to amplified ß-Actin levels in sample and control mRNAs, respectively [21].
5-Aza-dC treatment of esophageal cancer cell lines
To determine whether CAV1 inactivation was due to promoter hypermethylation in esophageal cancer, OE33 EAC cells were subjected to 5-Aza-dC (Sigma, St. Louis, MO) treatment as previously described [21]. Briefly, 1 × 105 cells/ml were seeded onto a 100 mm dish and grown for 24 h. Then, 1 ul of 5 mM 5-Aza-dC per ml of cells was added every 24 hours for 6 days. DNA and RNA were harvested on day 4.
Data analysis and statistics
Receiver-operator characteristic (ROC) curve analysis [28] was performed using NMVs for the 67 EAC, 26 ESCC and 67 NE by Analyse-it software (Version 1.71, Analyse-it Software, Leeds, UK). Using this approach, the area under the ROC curve (AUROC) yielded optimal sensitivity and specificity to distinguish normal from malignant esophageal tissues, and corresponding NMV thresholds were calculated for CAV1. The cutoff value determined from this ROC curve was applied to determine the frequency of CAV1 methylation in each tissue type included in the present study. For all other tests, Statistica (version 6.1; StatSoft, Inc., Tulsa, OK) was used. Differences with p < 0.05 were deemed significant.
Results and discussion
CAV1 promoter hypermethylation in different esophageal tissues
Promoter hypermethylation of the CAV1 gene was analyzed in 67 NE, 60 BE, 40 D (19 LGD and 21 HGD), 67 EAC and 26 ESCC samples. All assays in this study were performed in duplicate format, and data showed reproducible and concordant results. CAV1 promoter hypermethylation showed highly discriminative ROC curve profiles, which clearly distinguished both EAC (p < 0.0001) and ESCC (p <0.0001) from NE. AUROC of EAC vs. NE and ESCC vs. NE were 0.839 and 0.920, respectively. ROC curves with corresponding AUROCs for CAV1 of EAC vs. NE and ESCC vs. NE are displayed in Figure 1.
Figure 1.
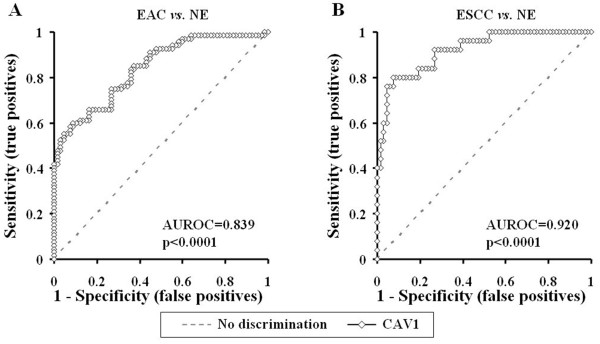
Receiver-operator characteristic (ROC) curve analysis of normalized methylation value (NMV). ROC curve analysis of CAV1 NMVs of normal esophagus (NE) vs. esophageal adenocarcinoma (EAC) (A), and NE vs. esophageal squamous cell carcinoma (ESCC) (B). The area under the ROC curve (AUROC) conveys this biomarker’s accuracy in distinguishing NE from EAC and from ESCC in terms of its sensitivity and specificity.
The cutoff NMV for CAV1 (0.2) was chosen from ROC curves to maximize sensitivity and specificity. Mean NMV and frequency of CAV1 hypermethylation for each tissue type are shown in Table 1. The mean NMV of CAV1 was significantly higher in ESCC (0.326), EAC (0.294), D (0.254), HGD (0.240), LGD (0.269) and BE (0.374) than in NE (0.134; p < 0.01, Student’s t-test). The frequency of CAV1 hypermethylation was increased in BE (81.7%), D (60%), and EAC (65.7%) vs. NE (25.4%; p < 0.01, p < 0.01 and p < 0.01, respectively; Chi-square for independence test). CAV1 was hypermethylated in 21 (80.8%) of 26 ESCCs. There were no significant differences between EAC and ESCC in mean CAV1 NMV (0.294 vs. 0.326; p > 0.05) or hypermethylation frequency (65.7% vs. 80.8%, p > 0.05). Among 41 cases with matched NE and T (EAC or ESCC), CAV1 NMVs in T (mean = 0.273) were significantly higher than in corresponding NE (mean = 0.146; p < 0.01, Student’s paired t-test; Figure 2A and B). Among 15 cases with corresponding NE, BE and EAC, one (No.8) was methylated only in EAC, three (No.7, 13 and 17) were methylated only in BE, five (No.1, 6, 9, 14 and 28) were methylated in both BE and EAC, and the remaining six were methylated in NE, BE and EAC simultaneously (Figure 2C). In addition, both CAV1 mean NMV and hypermethylation frequency were significantly higher in BE (0.374, 81.7%) than in D (0.254, 60%) and EAC (0.294, 65.7%; p < 0.01 and p < 0.05, respectively; Figure 3).
Figure 2.
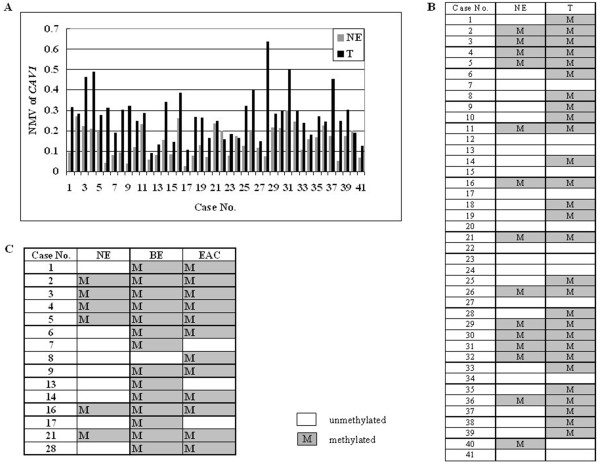
Methylation status of CAV1 in matched esophageal tissue samples. A, NMVs of CAV1 in 41 patients with matched NE and esophageal cancer (T, EAC or ESCC). B, methylation status of CAV1 in 41 cases with corresponding NE and T. C, methylation status of CAV1 in 15 cases with corresponding NE, BE and EAC.
Figure 3.
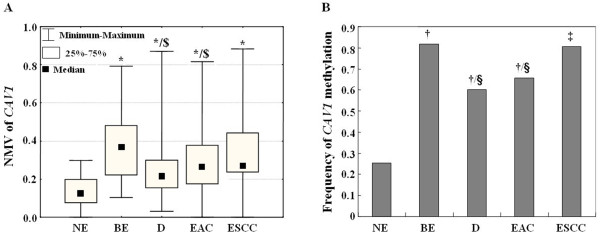
Methylation profiles of CAV1 in different esophageal tissue samples. A, The mean NMV of CAV1 was significantly higher in ESCC, EAC, D, and BE than in NE, and in BE than in D and EAC. B. The frequency of CAV1 hypermethylation was significantly higher in BE, D, EAC and ESCC than in NE, and in BE than in D and EAC. NE: normal esophagus; BE: Barrett’s metaplasia; D: Dysplasia in BE; EAC: esophageal adenocarcinoma; ESCC: esophageal squamous cell carcinoma; *: Student’s t test, p < 0.01 (comparisons made to NE); $: Student’s t test, p < 0.01 (comparisons made to BE); †: Chi-square for independence test, p < 0.01 (comparisons made to NE); ‡: Fisher’s exact test, p < 0.01 (comparisons made to NE); §: Chi-square for independence test, p < 0.05 (comparisons made to BE).
No significant associations were observed between CAV1 promoter hypermethylation and patient age, survival (data not shown), smoking or alcohol consumption status, BE segment length, tumor stage or lymph node metastasis, histologic tumor differentiation, or histologic type of esophageal carcinoma (Table 1).
CAV1 methylation and mRNA levels in OE33 cells after 5-Aza-dC treatment
OE33 cells were subjected to demethylation by 5-Aza-dC treatment. On day 4 after 5-Aza-dC treatment, the NMV of CAV1 was diminished, while CAV1 mRNA levels were increased (Figure 4).
Figure 4.
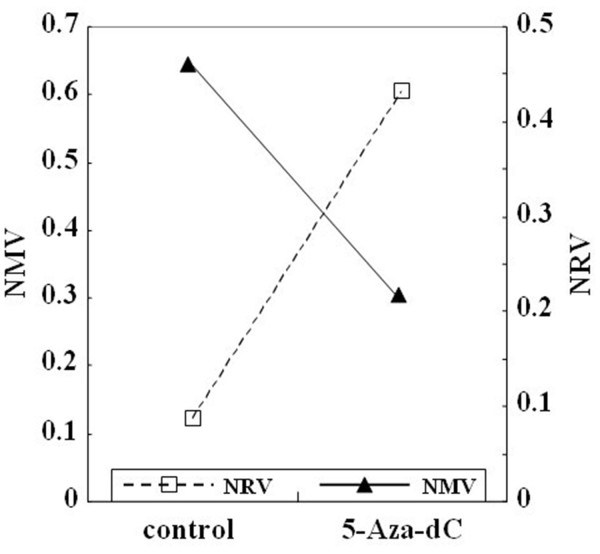
CAV1 methylation and mRNA expression in OE33 cells after treatment with 5-aza-2’-deoxycytidine (5-Aza-dC). After 5-Aza-dC treatment, the NMV of CAV1 was diminished, while the normalized mRNA value (NRV) of CAV1 was increased.
CAV1 has already been previously reported to have tumor suppressor activity via. inhibiting cell proliferation and/or metastesis in several human cancers [29-32]. CAV1 down-regulation has been reported in many types of cancer, including breast, lung, oral and esophagus [9,10,12-14]. These results suggest that low expression of CAV1 may represent a general characteristic or even a requirement of transformed cells in many kinds of carcinogenesis. Potential mechanisms underlying this suppression of expression include posttranscriptional and epigenetic changes, such as aberrant DNA methylation [26,27,33]. In the current study, we systematically investigated hypermethylation of the CAV1 gene promoter in primary human esophageal lesions of differing histological types and grades. Our results demonstrate that CAV1 promoter hypermethylation occurs frequently in both human EAC and ESCC (Table 1). CAV1 NMVs in T were significantly higher than those in corresponding NE (p < 0.01, Student’s paired t-test) in 41 cases with corresponding NE and T (Figure 2). Moreover, hypermethylation of the CAV1 promoter was significantly more frequent in premalignant lesions, such as BE and D, as well as in EAC, than in NE (Table 1). There was no significant association between CAV1 promoter hypermethylation and histological subtype of esophageal carcinoma (EAC vs. ESCC). These results suggest that hypermethylation of CAV1 may represent an early epigenetic event in these subjects, that the frequency of this epigenetic event increases during esophageal carcinogenesis, and that this event is highly prevalent in human esophageal cancers.
Barrett’s carcinogenesis is a multistep process comprising genetic and epigenetic alterations in tumor suppressor genes, cell cycle-regulatory genes, and genes essential for cell–cell adhesion [34,35]. Progressive accumulation of gene alterations is postulated to underly the transition of normal squamous epithelium to BE [36]. Many previous studies, focused on promoter hypermethylation of candidate genes for esophageal carcinomas, have shown staged growth in methylation frequency from nondysplastic esophageal squamous cell mucosa to BE and finally to EAC [15,37,38]. Interestingly, it has been suggested that CAV1 acts as a tumor modulator in a tissue type- and stage-dependent manner by binding several different proteins involved in different signal transduction [6,39-42]. Recently, we reported a preponderance of hypomethylation over hypermethylation events during the epigenomic program of BE pre-progression by comparing global DNA methylation profiles of two groups of BE patients, termed ‘progressors’ and ‘non-progressors’ [16]. Although the frequency of CAV1 hypermethylation in this study increased in parallel with esophageal carcinoma progression, the mean NMV and frequency of CAV1 hypermethylation were higher in BE than in D and EAC, and these differences were significant by Student’s t test (i.e., there was an inverse correlation between CAV1 hypermethylation and Barrett’s-associated esophageal neoplastic progression) (Figure 3). Taken together, these data suggested that the CAV1 promoter is relatively hypomethylated in EAC and D vs. BE, implying that, at least in part, this event represents an early part of the temporal program of Barrett’s-associated esophageal neoplastic progression. Two previous studies demonstrated that expression of CAV1 was elevated in ESCC compared to corresponding normal tissues, and its elevation was associated with malignant progression and poor survival [43,44]. These inconsistent results may have been due to different analytic methods used, ethnic groups studied, and smaller sample sizes in the previous studies.
In accordance with previous findings [11,45,46], we observed that methylation of CAV1 in EAC cell lines was associated with silenced or reduced expression of CAV1 mRNA. In this study, reversal of methylation and restoration of CAV1 expression were induced in OE33 cells by 5-Aza-dC treatment (Figure 4). Restoration of CAV1 mRNA expression due to 5-Aza-dC treatment is consistent with the interpretation that DNA hypermethylation silences the CAV1 gene. Although 5-Aza-dC or its derivatives have shown potential as therapeutic anticancer drugs [47-49], relatively hypomethylation of CAV1 in EAC and D vs. BE in the current study, and together with previous data on the re-expression of CAV1 in advance cancerf, would make CAV1 not an ideal molecular target for anti-cancer therapy involving demethylation in EAC patients.
Conclusions
The current study indicates that hypermethylation of the CAV1 promoter, leading to gene silencing, is a common event in human esophageal cancer and occurs early during Barrett’s-associated EAC. These results provide a basis for further research on CAV1 as a potential biomarker for the early diagnosis, classification, stratification and prognostication of esophageal cancers.
Abbreviations
5-Aza-dC: 5-aza-2’-deoxycytidine; BE: Barrett’s esophagus; CAV1: Caveolin-1; EAC: Esophageal adenocarcinomas; ESCC: Esophageal squamous cell carcinomas; NE: Normal esophagus; NMV: Normalized methylation value; qMSP: Real-time quantitative methylation-specific PCR; ROC: Receiver-operator characteristic.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZJ and SJM designed the study. ZJ, LW and ZC wrote the main manuscript text. YC, YG, XF, SC, HY, WW, ZZ, MD, XZ, JL, XF and YM analyzed and interpreted the data. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Authors’ information
Corresponding author: Zhe Jin, Department of Pathology, The Shenzhen University School of Medicine, 3688 Nanhai Ave, Rm 703, Nanshan, Shenzhen, Guangdong, People’s Republic of China 518060; Phone: 086-0755-86671904; Fax: 086-0755-86671906; email address: zhejin1995@yahoo.com Co-Correspondence: Stephen J. Meltzer, Division of Gastroenterology, Department of Medicine, Johns Hopkins University School of Medicine. 1503 E. Jefferson Street, Rm. 112, Baltimore, MD, USA 21231; Phone: 01-410-502-6071; Fax: 01-410-502-1329; email address: smeltzer@jhmi.edu
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Zhe Jin, Email: zhejin1995@yahoo.com.
Liang Wang, Email: liangwang@szu.edu.cn.
Ziyi Cao, Email: yszhdbhh@126.com.
Yulan Cheng, Email: yulancheng@jhmi.edu.
Yan Gao, Email: gaosandy101@sina.com.
Xianling Feng, Email: xianling_feng@szu.edu.cn.
Si Chen, Email: chensijnu@126.com.
Huimin Yu, Email: yuhuimin@szu.edu.cn.
Wenjing Wu, Email: wenjing2102@gmail.com.
Zhenfu Zhao, Email: Zhzhf1973@sina.com.
Ming Dong, Email: loving_jean@hotmail.com.
Xiaojing Zhang, Email: apple4328@sina.com.
Jie Liu, Email: ljljz@yahoo.com.
Xinmin Fan, Email: xinminfan1962@yahoo.com.
Yuriko Mori, Email: ymori3@jhmi.edu.
Stephen J Meltzer, Email: smeltzer@jhmi.edu.
Acknowledgements
This study supported by National Nature Science Foundation of China grant No. 81172282, the Shenzhen Peacock Plan KQCX20130621101141669, the Planned Science and Technology Project of Shenzhen No. GJHS20120621142654087, the Key Laboratory Project of Shenzhen No. ZDSY20130329101130496, Natural Science Foundation of SZU grants 201108 and T201202 to Z Jin; NIH grant DK087454to SJ Meltzer; National Nature Science Foundation of China grant 81171921 and the Science and Technology Bureau of ShenZhen City grant JC201006010727A, JCYJ20120613165853326 and GJHS20120621154321244 to L Wang.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Guo MZ. The value of epigenetic markers in esophageal cancer. Front Med China. 2010;4:378–384. doi: 10.1007/s11684-010-0230-3. [DOI] [PubMed] [Google Scholar]
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- Kleinberg L, Gibson MK, Forastiere AA. Chemoradiotherapy for localized esophageal cancer: regimen selection and molecular mechanisms of radiosensitization. Nat Clin Pract Oncol. 2007;4:282–294. doi: 10.1038/ncponc0796. [DOI] [PubMed] [Google Scholar]
- Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Ann Med. 2004;36:584–595. doi: 10.1080/07853890410018899. [DOI] [PubMed] [Google Scholar]
- Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- Razani B, Lisanti MP. Caveolin-deficient mice: insights into caveolar function human disease. J Clin Invest. 2001;108:1553–1561. doi: 10.1172/JCI200114611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Deng X, Li Q, Wang M, An W, Deli A, Gao Z, Xie Y, Dai Y, Cong YS. Down-regulation of the cavin family proteins in breast cancer. J Cell Biochem. 2012;113:322–328. doi: 10.1002/jcb.23358. [DOI] [PubMed] [Google Scholar]
- Ferraldeschi R, Latif A, Clarke RB, Spence K, Ashton G, O’Sullivan J, Evans DG, Howell A, Newman WG. Lack of caveolin-1 (P132L) somatic mutations in breast cancer. Breast Cancer Res Treat. 2012;132:1185–1186. doi: 10.1007/s10549-012-1981-0. [DOI] [PubMed] [Google Scholar]
- Patani N, Martin LA, Reis-Filho JS, Dowsett M. The role of caveolin-1 in human breast cancer. Breast Cancer Res Treat. 2012;131:1–15. doi: 10.1007/s10549-011-1751-4. [DOI] [PubMed] [Google Scholar]
- Bachmann N, Haeusler J, Luedeke M, Kuefer R, Perner S, Assum G, Paiss T, Hoegel J, Vogel W, Maier C. Expression changes of CAV1 and EZH2, located on 7q31 approximately q36, are rarely related to genomic alterations in primary prostate carcinoma. Cancer Genet Cytogenet. 2008;182:103–110. doi: 10.1016/j.cancergencyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Han SE, Park KH, Lee G, Huh YJ, Min BM. Mutation and aberrant expression of Caveolin-1 in human oral squamous cell carcinomas and oral cancer cell lines. Int J Oncol. 2004;24:435–440. [PubMed] [Google Scholar]
- Kopantzev EP, Monastyrskaya GS, Vinogradova TV, Zinovyeva MV, Kostina MB, Filyukova OB, Tonevitsky AG, Sukhikh GT, Sverdlov ED. Differences in gene expression levels between early and later stages of human lung development are opposite to those between normal lung tissue and non-small lung cell carcinoma. Lung Cancer. 2008;62:23–34. doi: 10.1016/j.lungcan.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Prade E, Tobiasch M, Hitkova I, Schaffer I, Lian F, Xing X, Tanzer M, Rauser S, Walch A, Feith M, Post S, Röcken C, Schmid RM, Ebert MP, Burgermeister E. Bile acids down-regulate caveolin-1 in esophageal epithelial cells through sterol responsive element-binding protein. Mol Endocrinol. 2012;26:819–832. doi: 10.1210/me.2011-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JS, Guo M, Montgomery EA, Thompson RE, Cosby H, Hicks L, Wang S, Herman JG, Canto MI. DNA promoter hypermethylation of p16 and APC predicts neoplastic progression in Barrett’s esophagus. Am J Gastroenterol. 2009;104:2153–2160. doi: 10.1038/ajg.2009.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, Jin Z, Yang J, Mori Y, Song JH, Kumar S, Sato M, Cheng Y, Olaru AV, Abraham JM, Verma A, Meltzer SJ. Epigenomic program of Barrett’s-associated neoplastic progression reveals possible involvement of insulin signaling pathways. Endocr Relat Cancer. 2012;19:L5–L9. doi: 10.1530/ERC-11-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, Mori Y, Olaru AV, Paun BC, Yang J, Kan T, Ito T, Hamilton JP, Selaru FM, Agarwal R, David S, Abraham JM, Wolfsen HC, Wallace MB, Shaheen NJ, Washington K, Wang J, Canto MI, Bhattacharyya A, Nelson MA, Wagner PD, Romero Y, Wang KK, Feng Z, Sampliner RE, Meltzer SJ. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res. 2009;69:4112–4115. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Mori Y, Hamilton JP, Olaru A, Sato F, Yang J, Ito T, Kan T, Agarwal R, Meltzer SJ. Hypermethylation of the somatostatin promoter is a common, early event in human esophageal carcinogenesis. Cancer. 2008;112:43–49. doi: 10.1002/cncr.23135. [DOI] [PubMed] [Google Scholar]
- Jin Z, Hamilton JP, Yang J, Mori Y, Olaru A, Sato F, Ito T, Kan T, Cheng Y, Paun B, David S, Beer DG, Agarwal R, Abraham JM, Meltzer SJ. Hypermethylation of the AKAP12 promoter is a biomarker of Barrett’s-associated esophageal neoplastic progression. Cancer Epidemiol Biomarkers Prev. 2008;17:111–117. doi: 10.1158/1055-9965.EPI-07-0407. [DOI] [PubMed] [Google Scholar]
- Jin Z, Cheng Y, Olaru A, Kan T, Yang J, Paun B, Ito T, Hamilton JP, David S, Agarwal R, Selaru FM, Sato F, Abraham JM, Beer DG, Mori Y, Shimada Y, Meltzer SJ. Promoter hypermethylation of CDH13 is a common, early event in human esophageal adenocarcinogenesis and correlates with clinical risk factors. Int J Cancer. 2008;123:2331–2336. doi: 10.1002/ijc.23804. [DOI] [PubMed] [Google Scholar]
- Jin Z, Olaru A, Yang J, Sato F, Cheng Y, Kan T, Mori Y, Mantzur C, Paun B, Hamilton JP, Ito T, Wang S, David S, Agarwal R, Beer DG, Abraham JM, Meltzer SJ. Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer. Clin Cancer Res. 2007;13:6293–6300. doi: 10.1158/1078-0432.CCR-07-0818. [DOI] [PubMed] [Google Scholar]
- Clement G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett’s oesophagus patients at risk for malignant transformation. J Pathol. 2006;208:100–107. doi: 10.1002/path.1884. [DOI] [PubMed] [Google Scholar]
- Tischoff I, Hengge UR, Vieth M, Ell C, Stolte M, Weber A, Schmidt WE, Tannapfel A. Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett’s adenocarcinoma. Gut. 2007;56:1047–1053. doi: 10.1136/gut.2006.111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TF, Su TH, Yeh KT, Chang JY, Lin TH, Chen JC, Yuang SS, Chang JG. Mutational, epigenetic and expressional analyses of caveolin-1 gene in cervical cancers. Int J Oncol. 2003;23:599–604. [PubMed] [Google Scholar]
- Chen ST, Lin SY, Yeh KT, Kuo SJ, Chan WL, Chu YP, Chang JG. Mutational, epigenetic and expressional analyses of caveolin-1 gene in breast cancers. Int J Mol Med. 2004;14:577–582. [PubMed] [Google Scholar]
- Lin SY, Yeh KT, Chen WT, Chen HC, Chen ST, Chang JG. Promoter CpG methylation of caveolin-1 in sporadic colorectal cancer. Anticancer Res. 2004;24:1645–1650. [PubMed] [Google Scholar]
- Mori Y, Cai K, Cheng Y, Wang S, Paun B, Hamilton JP, Jin Z, Sato F, Berki AT, Kan T, Ito T, Mantzur C, Abraham JM, Meltzer SJ. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology. 2006;131:797–808. doi: 10.1053/j.gastro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Chen RS, Song YM, Zhou ZY, Tong T, Li Y, Fu M, Guo XL, Dong LJ, He X, Qiao HX, Zhan QM, Li W. Disruption of xCT inhibits cancer cell metastasis via the caveolin-1/beta-catenin pathway. Oncogene. 2009;28:599–609. doi: 10.1038/onc.2008.414. [DOI] [PubMed] [Google Scholar]
- Song Y, Xue L, Du S, Sun M, Hu J, Hao L, Gong L, Yeh D, Xiong H, Shao S. Caveolin-1 knockdown is associated with the metastasis and proliferation of human lung cancer cell line NCI-H460. Biomed Pharmacother. 2012;66:439–447. doi: 10.1016/j.biopha.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Trimmer C, Sotgia F, Lisanti MP, Capozza F. Cav1 inhibits benign skin tumor development in a two-stage carcinogenesis model by suppressing epidermal proliferation. Am J Transl Res. 2013;5:80–91. [PMC free article] [PubMed] [Google Scholar]
- Trimmer C, Bonuccelli G, Katiyar S, Sotgia F, Pestell RG, Lisanti MP, Capozza F. Cav1 suppresses tumor growth and metastasis in a murine model of cutaneous SCC through modulation of MAPK/AP-1 activation. Am J Pathol. 2013;182:992–1004. doi: 10.1016/j.ajpath.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zhang XL, Lisanti MP. Sequence and detailed organization of the human caveolin-1 and -2 genes located near the D7S522 locus (7q31.1). Methylation of a CpG island in the 5’ promoter region of the caveolin-1 gene in human breast cancer cell lines. FEBS Lett. 1999;448:221–230. doi: 10.1016/S0014-5793(99)00365-8. [DOI] [PubMed] [Google Scholar]
- Jankowski JA, Harrison RF, Perry I, Balkwill F, Tselepis C. Barrett’s metaplasia. Lancet. 2000;356:2079–2085. doi: 10.1016/S0140-6736(00)03411-5. [DOI] [PubMed] [Google Scholar]
- Tannapfel A. Molecular findings in Barrett’s epithelium. Dig Dis. 2004;22:126–133. doi: 10.1159/000080311. [DOI] [PubMed] [Google Scholar]
- Jankowski JA, Wright NA, Meltzer SJ, Triadafilopoulos G, Geboes K, Casson AG, Kerr D, Young LS. Molecular evolution of the metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Pathol. 1999;154:965–973. doi: 10.1016/S0002-9440(10)65346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuester D, Dar AA, Moskaluk CC, Krueger S, Meyer F, Hartig R, Stolte M, Malfertheiner P, Lippert H, Roessner A, El-Rifai W, Schneider-Stock R. Early involvement of death-associated protein kinase promoter hypermethylation in the carcinogenesis of Barrett’s esophageal adenocarcinoma and its association with clinical progression. Neoplasia. 2007;9:236–245. doi: 10.1593/neo.06802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuester D, El-Rifai W, Peng D, Ruemmele P, Kroeckel I, Peters B, Moskaluk CA, Stolte M, Monkemuller K, Meyer F, Schulz HU, Hartmann A, Roessner A, Schneider-Stock R. Silencing of MGMT expression by promoter hypermethylation in the metaplasia-dysplasia-carcinoma sequence of Barrett’s esophagus. Cancer Lett. 2009;275:117–126. doi: 10.1016/j.canlet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005;288:C494–C506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- Navarro A, Anand-Apte B, Parat MO. A role for caveolae in cell migration. Faseb J. 2004;18:1801–1811. doi: 10.1096/fj.04-2516rev. [DOI] [PubMed] [Google Scholar]
- Goetz JG, Lajoie P, Wiseman SM, Nabi IR. Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Rev. 2008;27:715–735. doi: 10.1007/s10555-008-9160-9. [DOI] [PubMed] [Google Scholar]
- Burgermeister E, Liscovitch M, Rocken C, Schmid RM, Ebert MP. Caveats of caveolin-1 in cancer progression. Cancer Lett. 2008;268:187–201. doi: 10.1016/j.canlet.2008.03.055. [DOI] [PubMed] [Google Scholar]
- Kato K, Hida Y, Miyamoto M, Hashida H, Shinohara T, Itoh T, Okushiba S, Kondo S, Katoh H. Overexpression of caveolin-1 in esophageal squamous cell carcinoma correlates with lymph node metastasis and pathologic stage. Cancer. 2002;94:929–933. doi: 10.1002/cncr.10329. [DOI] [PubMed] [Google Scholar]
- Ando T, Ishiguro H, Kimura M, Mitsui A, Mori Y, Sugito N, Tomoda K, Mori R, Harada K, Katada T, Ogawa R, Fujii Y, Kuwabara Y. The overexpression of caveolin-1 and caveolin-2 correlates with a poor prognosis and tumor progression in esophageal squamous cell carcinoma. Oncol Rep. 2007;18:601–609. [PubMed] [Google Scholar]
- Das M, Das S, Lekli I, Das DK. Caveolin induces cardioprotection through epigenetic regulation. J Cell Mol Med. 2012;16:888–895. doi: 10.1111/j.1582-4934.2011.01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, Nakagawa T, Hatanaka M, Takeuchi T, Matsumoto E, Takenaka H, Shimizu A. Quantification of caveolin isoforms using quantitative real-time RT-PCR, and analysis of promoter CpG methylation of caveolin-1alpha in human T cell leukemia cell lines. Int J Mol Med. 2006;18:489–495. [PubMed] [Google Scholar]
- Lemaire M, Momparler LF, Bernstein ML, Marquez VE, Momparler RL. Enhancement of antineoplastic action of 5-aza-2’-deoxycytidine by zebularine on L1210 leukemia. Anticancer Drugs. 2005;16:301–308. doi: 10.1097/00001813-200503000-00009. [DOI] [PubMed] [Google Scholar]
- Momparler RL. Epigenetic therapy of cancer with 5-aza-2’-deoxycytidine (decitabine) Semin Oncol. 2005;32:443–451. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Mirza S, Sharma G, Pandya P, Ralhan R. Demethylating agent 5-aza-2-deoxycytidine enhances susceptibility of breast cancer cells to anticancer agents. Mol Cell Biochem. 2010;342:101–109. doi: 10.1007/s11010-010-0473-y. [DOI] [PubMed] [Google Scholar]


