Abstract
Zoosporic parasites have received increased attention during the last years, but it is still largely unnoted that these parasites can themselves be infected by hyperparasites. Some members of the Chytridiomycota, Blastocladiomycota, Cryptomycota, Hyphochytriomycota, Labyrinthulomycota, Oomycota, and Phytomyxea are hyperparasites of zoosporic hosts. Because of sometimes complex tripartite interactions between hyperparasite, their parasite-host, and the primary host, hyperparasites can be difficult to detect and monitor. Some of these hyperparasites use similar mechanisms as their parasite-hosts to find and infect their target and to access food resources. The life cycle of zoosporic hyperparasites is usually shorter than the life cycle of their hosts, so hyperparasites may accelerate the turnaround times of nutrients within the ecosystem. Hyperparasites may increase the complexity of food webs and play significant roles in regulating population sizes and population dynamics of their hosts. We suggest that hyperparasites lengthen food chains but can also play a role in conducting or suppressing diseases of animals, plants, or algae. Hyperparasites can significantly impact ecosystems in various ways, therefore it is important to increase our understanding about these cryptic and diverse organisms.
Keywords: hyperparasites, ecology, food web, parasite, zoospores, eDNA
Introduction
“So, naturalists observe, a flea
Has smaller fleas that on him prey;
And these have smaller still to bite ‘em,
And so proceed ad infinitum.”
Jonathan Swift, On Poetry: a rhapsody (1733)
Parasites belonging to all taxonomic groups have gained increasing attention in ecological research during recent years. It is widely recognised that the number of species of parasites are more numerous than organisms with a non-parasitic lifestyle (Lafferty et al., 2008). Also it is widely accepted that many parasites can themselves be hosts for other parasites. Such parasites of parasites are usually called “hyperparasites”; a term which is used without any reference to the phylogeny of the host or the parasite or whether the relationship is obligately or facultatively parasitic. Novel methodological tools and an increasing interest in parasites and their ecology have led to more targeted sampling approaches. This has shown that especially microbial parasites which have until now been rarely detected are abundant and diverse (Lefèvre et al., 2008; Jones et al., 2011; Hartikainen et al., 2014). It is very difficult—or in many cases impossible—to isolate and identify them because of their generic morphology, and because such parasites are often restricted to only a few host cells which makes them difficult to detect even with state of the art molecular methods. Hence, it is no surprise that microbial hyperparasites are not well understood. Some species of hyperparasites are endoparasites and difficult to see in the light microscope without special staining methods. Although zoosporic parasites of primary producers have been the focus of recent studies (Powell, 1993; Ibelings et al., 2004; Kagami et al., 2007; Marano et al., 2011; Neuhauser et al., 2011a), our knowledge about zoosporic hyperparasites and their microbial hosts remains anecdotal. In this article we focus on zoosporic hyperparasites with zoosporic hosts, their abundance and relationships between parasites and their hosts and their possible roles in ecological processes.
In two of the early works focusing on microbial hyperparasites, Karling (1942a,b) documented and discussed examples of hyperparasitism among zoosporic true fungi (Table 1). Although his study focused primarily on hyperparasites among the zoosporic true fungi, Karling was aware of hyperparasites among other microbial groups such as stramenopiles or plasmodiophorids (Table 2). Sparrow's monograph about aquatic phycomycetes contains still the most comprehensive references to zoosporic hyperparasites (Sparrow, 1960). Although hyperparasitism among true fungi has been the focus of numerous research projects, for instance in the form of biological control of plant diseases (e.g., Vinale et al., 2008), hyperparasitism involving heterotrophic stramenopiles and zoosporic true fungi has been rare (Boosalis, 1964; Barnett and Binder, 1973; Adams, 1990). Zoosporic hyperparasites have been described in the fungal groups Chytridiomycota, Blastocladiomycota, and Cryptomycota (Opisthokonts, for examples see Table 1). Within the heterokonts the groups Hyphochytriomycota, Oomycota, Labyrinthulomycota, and Phytomyxea contain hyperparasitic species (Table 2). These groups belong to various supergroups in the tree of life (Baldauf, 2003; Adl et al., 2012), but these microorganisms interact together in the same ecosystems. Because of their morphological similarity and their similarity in size they can have ecologically similar functions and are in food web studies often treated as “trophic species” (Powell, 1993; Marano et al., 2011). Many of the known hosts belong to common genera which are frequently observed in many soil and fresh water ecosystems using both baiting procedures and molecular analysis of environmental samples (Sparrow, 1960; Powell, 1993; Barr, 2001; Dick, 2001; Lozupone and Klein, 2002; Shearer et al., 2007; Lefèvre et al., 2008; Marano et al., 2011). It is very likely that zoosporic hyperparasites are as abundant on “rarer” hosts. This is of ecological importance because zoosporic true fungi and heterotrophic stramenopiles can be among the predominant groups in some ecosystems (Lefèvre et al., 2008; Freeman et al., 2009; Marano et al., 2011). Because of the large number of species of zoosporic parasites, hyperparasites, and their associated hosts, it is likely that there are many additional taxa that await discovery.
Table 1.
Selected hyperparasitic Opistokonts (Chytridiomycota, Cryptomycota, Blastocladiomycota).
| Hyperparasite | Trophic mode | Parasite (=Host of hyperparasite) | Host (=Host of parasite) | References |
|---|---|---|---|---|
| Cryptomycota | Chytridiomycota | |||
| Rozella marina | Biotroph | Chytridium polysiphoniae | Parasite, red algae | Sparrow, 1960; Held, 1981 |
| Rozella parva | Biotroph | Zygorhizidium affluens | Canter, 1965; Beakes et al., 1988 | |
| Rozella rhizophlyctii | Biotroph | Rhizophlyctis rosea | Facultative parasite | Karling, 1960; Held, 1981 |
| Biotroph | Rhizophydium globosum | Parasite, Diatoms, algae | Sparrow, 1960; Held, 1981 | |
| Rozella polyphagi | Biotroph | Polyphagus laevis | Parasite, Euglena | Sparrow, 1960; Held, 1981 |
| Biotroph | Polyphagus euglenae | Parasite, Euglena | Powell, 1984 | |
| Rozella endochytrium | Biotroph | Endochytrium operculatum | Facultative parasite, algae | Sparrow, 1960; Held, 1981 |
| Rozella cladochytrii | Biotroph | Cladochytrium replicatum | Facultative parasite, green algae | Sparrow, 1960; Held, 1981 |
| Cryptomycota | Blastocladiomycota | |||
| Rozella allomycis | Biotroph | Allomyces arbuscula | Facultative parasite, insect cadaver | Held, 1981 |
| Biotroph | Allomyces macrogynus | Held, 1974 | ||
| Cryptomycota | Oomycota | |||
| Rozella rhipidii-spinosi | Biotroph | Araiospora spinosa | Facultative parasite | Sparrow, 1960; Held, 1981 |
| Rozella apodiae-brachynematis | Biotroph | Apodachlya brachynema | Facultative parasite | Sparrow, 1960; Held, 1981 |
| Rozella achlyae | Biotroph | Achlya flagellata | Facultative parasite | Sparrow, 1960; Held, 1981 |
| Dictyuchus anomalus | Parasite, fish | |||
| Rozella cuculus | Biotroph | Pythium intermedium | Parasite, plant | Sparrow, 1960; Held, 1981 |
| P. monospermum | Parasite, nematode | Held, 1981 | ||
| Rozella laevis | Biotroph | Pythium gracile | Parasite, green algae | Sparrow, 1960; Held, 1981 |
| Rozella barrettii | Biotroph | Phytophthora cactorum | Parasite, plant | Sparrow, 1960; Held, 1981 |
| Rozella pseudomorpha | Biotroph | Lagenidium rabenhorstii | Parasite, green algae | Sparrow, 1960; Held, 1981 |
| Chytridiomycota | Chytridiomycota | |||
| Dictyomorpha dioica | Biotroph | Achlya flagellata | Mullins and Barksdale, 1965 | |
| Chytridium parasiticum | Biotroph | Septosperma rhizophydii | Parasite, chytrid | Karling, 1960 |
| Rhizophydium parasiticum | Rhizophlyctis rosea | Facultative parasite, chitin | Karling, 1960; Sparrow, 1960 | |
| Chytridiomyces verrucocsa | ||||
| Rhizophydium carpophilum | Synchytrium fulgens | Parasite, plant | Karling, 1960 | |
| S. macrosporum | Parasite, plant | |||
| S. linariae | Parasite, plant | |||
| Phlyctochytrium synchytrii | Synchytrium endobioticum | Parasite, plant | Karling, 1942a | |
| Septosperma rhizophydii | Rhizophydium macrosporum | Facultative parasite | Karling, 1960 | |
| Septosperma anomala | Phlyctidium bumelleriae | Parasite, Xanthophyceae | Karling, 1960 | |
| Chytridiomycota | Oomycota | |||
| Rhizophydium pythii | Biotroph | Pythium monospermum | Parasite, nematode | Sparrow, 1960 |
| Rhizidiomyces japonicus | Phytophthora megasperma | Parasite, plant | Sneh et al., 1977 | |
| Phytophthora erythroseptica | Parasite, plant | Wynn and Epton, 1979 | ||
| Canteriomyces stigeoclonii | Phytophthora megasperma | Parasite, plant | Sneh et al., 1977 | |
| Blastocladiomycota | Oomycota | |||
| Catenaria anguillulae | Facultative | Phytophthora cinnamomii | Parasite, plant | Daft and Tsao, 1984 |
| Phytophthora parasitica | Parasite, plant |
Hyperparasites and hosts are sorted by taxon. Higher ranks are given in bold.
Table 2.
Selected hyperparasitic Heterokonts (Oomycota, Hyphochytridiomycota, Phytomyxea).
| Hyperparasite | Trophic mode | Parasite (=Host of hyperparasite) | Host (=Host of parasite) | References |
|---|---|---|---|---|
| Oomycota | Oomycota | |||
| Olpidiopsis incrassata | Saprolegnia ferax | Parasite, fish | Slifkin, 1961 | |
| Olpidiopsis karlingiae | Rhizophlyctis rosea | Facultative Parasite | Karling, 1960 | |
| Pythiella vernalis | Pythium aphanidermatum | Parasite, plant | Pires-Zottarelli et al., 2009 | |
| Pythium gracile | Parasite, green algae | Blackwell, 2010 | ||
| Pythiella pythii | Pythium dictyosporum | Parasite, green algae | Blackwell, 2010 | |
| Pythium proliferum | Rhizophlyctis rosea | Facultative Parasite | Karling, 1960 | |
| Pythium monospermum | Phytophthora megasperma | Parasite, plant | Humble and Lockwood, 1981 | |
| Pythium oligandrum | Pythium irregulare | Parasite, plant | Ribeiro and Butler, 1995; Benhamou et al., 1999 | |
| Pythium mamillatum | Parasite, plant | |||
| Pythium paroecandrum | Parasite, plant | |||
| Pythium aphanidermatum | Parasite, plant | |||
| Pythium sylvaticum | Parasite, plant | |||
| Pythium ultimum | Parasite, plant | |||
| Hyphochytridiomycota | Oomycota | |||
| Hyphochytrium catenoides | Facultative | Pythium myriostylum | Parasite, plant | Ayers and Lumsden, 1977 |
| Aphanomyces euteiches | Parasite, plant | Ayers and Lumsden, 1977; Sneh et al., 1977 | ||
| Phytophthora erythroseptica | Parasite, plant | Wynn and Epton, 1979 | ||
| Phytophthora megasperma | Parasite, plant | Humble and Lockwood, 1981 | ||
| Phytomyxea | Oomycota | |||
| Sorodiscus cokeri | Biotroph | Pythium proliferum | Facultative Parasite | Goldie-Smith, 1951 |
| Pythium graminicolum | Facultative Parasite, moss | Goldie-Smith, 1951 | ||
| Pythium catenulatum | Facultative Parasite, plant | Goldie-Smith, 1951 | ||
| Pythium elongatum | Facultative Parasite | Goldie-Smith, 1951 | ||
| Pythium irregulare | Parasite, plant | Goldie-Smith, 1951 | ||
| Pythium undulatum | Parasite, plant | Goldie-Smith, 1951 | ||
| Woronina polycystis | Biotroph | Saprolegnia ferax | Parasite, fish | Goldie-Smith, 1954 |
| Woronina pythii | Biotroph | Pythium proliferum | Facultative Parasite | Goldie-Smith, 1956a |
| Pythium aphanidermatum | Parasite, plant | Goldie-Smith, 1956a | ||
| Pythium debaryanum | Parasite, plant | Goldie-Smith, 1956a | ||
| Pythium irregulare | Parasite, plant | Goldie-Smith, 1956a | ||
| Pythium monospermum | Parasite, nematode | Goldie-Smith, 1956a | ||
| Pythium pulchrum | Goldie-Smith, 1956a | |||
| Pythium ultimum | Parasite, plant | Goldie-Smith, 1956a |
Hyperparasites and hosts are sorted by taxon. Higher ranks are given in bold.
Zoospores
Zoospores are a shared morphological feature of the hosts and hyperparasites discussed here. Zoospores are motile propagules which permit rapid dispersal. Zoospores can sense environmental gradients which they use to identify and find potential hosts (Tyler, 2002). There are different types of zoospores (Lange and Olson, 1983), which have distinguishing features, allowing observers to determine and categorize the organisms. The most important feature is the type of flagellation. Zoospores can generally be grouped into (1) uniflagellate with posteriorly directed whiplash flagellum, (2) uniflagellate with an anteriorly directed tinsel flagellum, (3) biflagellate, heterokont, with one posteriorly directed whiplash flagellum and one anteriorly directed tinsel flagellum and (4) biflagellate, isokont, two whiplash flagellae, often of different lengths, with the shorter one anteriorly directed and the longer one posteriorly directed.
Despite their relatively simple morphology many zoosporic hyperparasites form functionally and developmentally distinct types of zoospores during their life cycle (Sparrow, 1960). A variety of names are used for different types of zoospores in different taxonomic groups, but generally one type of zoospore is formed in zoosporangia following mitosis and can be either haploid or diploid, while another type of zoospore is formed by meiosis and is haploid (Lange and Olson, 1983). The different types of zoospores can serve different functions during the parasite life cycle—such as rapid propagation and dispersal or primary infection and population establishment after periods of hibernation (e.g., Neuhauser et al., 2011b). Despite variable modes of formation and complex parasite life cycles which can result in periods where one type of zoospore is predominantly formed, the main unifying feature of all types of zoospores is that they are small, single-celled, motile propagules. Within food webs zoospores provide a rapid energy source for a variety of organisms at higher trophic levels (Gleason et al., 2011), so it is not surprising that zoospores are often treated as trophic species.
Mechanisms used by hyperparasites to access food resources
Zoosporic hyperparasites use a large variety of mechanisms to attack their hosts. Hyperparasites can grow epibiotically on the surface of their host only entering the host cell with specialized structures such as chytrid rhizoids (Figures 1A,C). Hyperparasites also grow endobiotically this means completely submerged in their hosts (Figures 1B,D). The parasite hosts of hyperparasites can be ectoparasites (Figures 1A,B) growing epibiotically on the primary host or endoparasites (Figures 1C,D) growing endobiotically inside the primary host. Hyperparasites which are infecting ectoparasites only have to overcome the defense mechanisms of their host, and often use infection strategies that are very similar to those of zoosporic parasites (Sparrow, 1960; Marano et al., 2012). On the other hand, hyperparasites which are parasites of endoparasites may have to overcome two barriers of defense—they have to enter the parasite-host and their host to get access to food resources. Most of the described zoosporic hyperparasites are parasites of ectoparasites (e.g., most Rozella species, Wornina spp.). We hypothesize that ectoparasites are easier accessible for hyperparasites with only one line of defense to break. We also hypothesize that our knowledge about zoosporic hyperparasites of endoparasites is biased by the fact that zoosporic endoparasites are a poorly studied group themselves. Therefore, most of the examples discussed here are from zoosporic hyperparasites of parasites which are not completely submerged inside their host or from endoparasitic hyperparasites of epibiotic hosts (Figures 1A–C).
Figure 1.
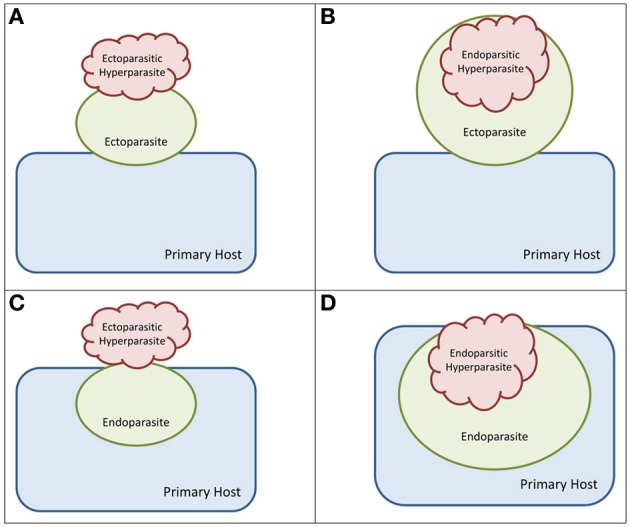
Types of hyperparasitism. Blue—primary host, green—parasite, red—hyperparasite. (A) epibiotic hyperparasite of ectoparasite. This type can be found for example in the interaction of the hyperparasite Rhizophydium parasiticum (Chytridiomycota), its and its (facultative) parasites host Rhizophlyctis rosea. (B) Endobiotic hyperparasite of ectoparasite host. This is the most commonly described mode of hyperparasitsm seen in many Rozella species (Cryptomycota) or Woronina spp. (Phytomyxea). (C) Epibiotic hyperparasite of endoparasite host. E.g., Rhizophyidum carpophilum (Chytridiomycota) on Olpidiopsis sp. (oomycetes) and Synchytrium sp. (chytrid). (D) Endobiotic hyperparasite of endoparasite host. E.g., the hyperparasitic chytrid Phlyctochytrium synchytrii in the plant pathogen Synchytrium endobioticum.
An example of an epibiotic infection (Figure 1A) is the parasitic relationship between the two chytrids Chytriomyces verrucosus and Rhizophlyctis rosea (Karling, 1960). The chemotactic zoospores of R. rosea are attracted to the host cell where they encyst. The zoospore then germinates and a germ tube penetrates the host zoosporangium. Inside the host, an endobiotic rhizoidal system develops supplying the epibiotic zoosporangium (having since formed from the body of the zoospore) with nutrients. Epibiotic parasites can also be found in the stramenopiles (Sneh et al., 1977): zoospores of the hyphochytriomycete Rhizidomyces japonicus attach to the surface of oospores of Phytophthora megasperma (Oomycetes) where thalli grow externally around the oospore and produce zoosporangia. The oomycete Pontisma lagenidioides which is a parasite of the green alga Chaetomorpha media can be infected by Labyrinthula sp. (Raghukumar, 1987).
Endobiotic parasites grow entirely submerged within their host. An example is Rozella allomycis (Rozellida/Cryptomycota) and its host Allomyces arbuscula (Blastocladiomycota) (Held, 1973, 1974). In this case, the infection process is relatively well studied and is described in more detail here to exemplify the infection process of most known endobiotic zoosporic hyperparasites. Substances produced by the host attract the chemotactic zoospores of the parasite toward the host. Once the zoospore attaches to the surface of the host cell it forms a so-called cyst, which produces a germ tube. The germ tube then grows into the host cell through the cell wall while the protoplast of Rozella is pushed into the host cell by fluid pressure produced from a vacuole in the cyst. Subsequently the parasite grows inside the host cell. In the case of Rozella allomycis the host cell is then transformed into the parasite sporangium. Other known endobiotic parasites are Rozella polyphagi (Rozellida/Cryptomycota), which parasitizes the chytrid parasite Polyphagus euglenae (Powell, 1984) and the endobiotic parasite Catenaria allomycis (Blastocladiomycota), which infects Allomyces javanicus (Sykes and Porter, 1980; Powell, 1982). Catenaria anguillulae, a member of the Blastocladiomycota, is an endobiotic parasite of the plant pathogenic oomycetes Phytophthora cinnamomi and P. parasitica (Daft and Tsao, 1984), while Hyphochytrium catenoides (Hyphochytriomycota) colonizes oospores of Pythium myriostylum (Ayers and Lumsden, 1977). Another parasite of Pythium spp. is Woronina pythii (Phytomyxea), which infects both vegetative hyphae and reproductive structures of Pythium (Dylewski and Miller, 1983).
Interactions are slightly different between hyphal forming zoosporic organisms, such as oomycetes. Here interactions between hyphae can be observed, and these interactions are different from the endo- and epibiotic parasitic interactions discussed above. Two distinct mechanisms appear to be involved in interactions between this parasite and its hosts: (1) hyperparasitism; mediated by hyphal interactions, and (2) antibiosis; causing metabolic and developmental changes prior to contact between hyphae of the parasite and host (Adams, 1990; Benhamou et al., 1999). An example of direct interactions between the organisms is the interaction between hyphae of the well-known hyperparasite Pythium oligandrum (Oomycota) and hyphae of its oomycete hosts (e.g., P. ultimum, P. aphanidermatum, Phytophthora megasperma) (Benhamou et al., 1999). Hyphae of the parasite can adhere to the surface of the host sometimes coiling around the host hyphae. Penetration of the host cells by infection pegs may follow, leading to digestion of the host cytoplasm. When the interaction is initiated by antibiosis (without contact with the host) the parasite can release soluble substances which cause biochemical changes within the host cells. Then the parasite can release extracellular enzymes, which digest the host cells.
Biodiversity and host range of hyperparasites
DNA sequences assigned to putative parasite and hyperparasite taxa of zoosporic fungi are widespread (e.g., Lara et al., 2010; Jones et al., 2011; Lara and Belbahri, 2011; Nagano and Nagahama, 2012). But molecular methods are often biased by the selection of primers and sampling methods (Hartikainen et al., 2014; Neuhauser et al., 2014) and the assignment of environmental DNA sequences to described species is only as good as the available reference datasets. Data on zoosporic microorganisms are sparse, and many of the “unknown” sequences are probably from common species which to date have no reference record in public data bases (e.g., Nagy et al., 2011; Karpov et al., 2013). Reliable reference sequences of many zoosporic hyperparasites are generally rare. One reason is that many of the known zoosporic hyperparasites are biotrophic parasites which cannot be grown without their hosts. The hosts themselves are often biotrophic parasites as well, making it very hard to isolate, identify and sequence the hyperparasites. Therefore, targeted studies to detect and characterize hyperparasites and their hosts are needed. Such targeted approaches could include baiting experiments combined with microscopic observation or DNA and RNA based screenings of various environments. Despite being very time consuming baiting and isolation experiments are highly valuable because they will allow to understand how hyperparasites interact with their hosts, to describe their life cycle, and to analyze interactions with their hosts. Baiting experiments with oospores of the oomycetes parasites Phytophthora megasperma, P. cactorum, Pythium sp. and Aphanomyces euteiches, revealed that those baits quickly became infected by different hyperparasites (Sneh et al., 1977). Another approach for characterizing zoosporic hyperparasites would be to implement a combination of DNA and RNA isolation methods combined with specific primers and to then visualize the respective organisms using specific FISH (Fluorescence in situ hybridization) probes (Not et al., 2002; Jones et al., 2011; Marano et al., 2012). Such targeted molecular probing techniques are a powerful tool to identify unknown organisms. When attempting to detect hyperparasites by this approach, however, mainly free living stages (zoospores) will be detected and the sampling is largely limited to aquatic environments because the background fluorescence in soil or sediment samples tends to be high (Wagner and Haider, 2012).
Hyperparasites, their hosts and the primary hosts are complex systems. Most studies about zoosporic hyperparasites base their evidence on laboratory studies of dual cultures of one host infected by one parasite or the host range of a single parasite (e.g., Karling, 1960; Sparrow, 1960; Held, 1981). Although to date we can only estimate how those interactions might occur in natural environments like sediment or soil (Gleason et al., 2012), simultaneous infections by different species are likely—especially for abundant parasite hosts for which more than one species of hyperparasite is known (for examples see Tables 1, 2). Similarly, unrelated or distantly related hyperparasites may infect the same hosts individually or simultaneously. An excellent example of this phenomenon was described by Karling (1960) who observed simultaneous infection of Rhizophlyctis rosea with four hyperparasites. He studied infections of the facultative parasite R. rosea with Chytriomyces verrucosa (Chytridiomycota). Karling noted that numerous sporangia of R. rosea were also infected with Rozella rhizophlyctii (Rozellida/Cryptomycota) and Olpidiopsis karlingiae (Oomycota). In addition to this, the large sporangia of R. rosea were infected by a fourth species, Pythium proliferum (Oomycota), which was itself densely parasitized by Woronina pythii (Phytomyxea). Although R. rosea is a facultative parasite, this example shows the extent to which hyperparasites can occur in nature when studied in detail.
On the other hand not all hyperparasites are host specific. Studies on the range of host specificity indicate that some species of hyperparasites in the Oomycota and Phytomyxea can infect several species of hosts (Goldie-Smith, 1951; Dylewski and Miller, 1983). Rozella allomycis only infects two susceptible hosts: Allomyces arbuscula and A. macrogynus (Held, 1974), while Olpidiopsis incrassata infects six species of Saprolegnia and three species of Isoachlya (Slifkin, 1961). Other parasites such as Woronina pythii have a broad host spectrum and can infect more than 40 species of oomycetes (Dylewski and Miller, 1983). Pythium oligandrum also infects a wide range of fungal and stramenopilous host (Ribeiro and Butler, 1995). These studies highlight the importance of isolating and characterizing species for understanding and characterizing hyperparsite biodiversity and host range. Culture based methods and well defined voucher isolates are also needed to provide a groundwork for DNA barcoding studies (del Campo et al., 2014) or for food web analyses (Hrcek et al., 2011) which form the basis for a more holistic understanding of hyperparasites and their ecological roles.
Size control of host populations by hyperparasites
Like all parasites, hyperparasites can impact population size and fitness of their hosts (Sieber and Hilker, 2011; Allen and Bokil, 2012; Preston et al., 2014). Some hyperparasites can infect persistent structures of their hosts, for example oospores, resistant sporangia, or resting spores (Gleason et al., 2010). Such resting stages are recalcitrant substrates and can survive in a dormant state in dried soil for long periods of time (Goldie-Smith, 1956b; Bruckart et al., 2011) where they accumulate, forming a “spore bank” of zoosporic parasites. But when these resting stages are infected by hyperparasites the pathogen pressure can potentially be reduced. This could explain the finding that zoosporic hyperparasites can be linked to suppressive soil properties (Weller et al., 2002) as they have the ability to reduce the viable pathogen load in soil. The presence of hyperparasites contributes to controlling their hosts in the environment, hinting at the important role of these parasites in balancing diversity and abundance of their hosts, consequently resulting in stable ecosystems.
Hyperparasites are already widely used as biological control agents to control the population size of plant pathogens. The best known example is the oomycete Pythium oligandrum which is used to control other Pythium spp. and oomycetes (Ikeda et al., 2012). Hyperparasites have a huge potential to control diseases if they can be systematically accumulated in the environment. But so far not many hyperparasites can be grown in the lab in big enough quantities that permit use as biocontrol agent. There are known hyperparasites of important plant pathogens which have not been explored as biocontrol agents because of this reason. Oospores of the potato pathogen Phytophthora erythroseptica, for example, were found to be infected with Hyphochytriun catenoides and Rhizidiomyces japonicus in waterlogged soils in England (Wynn and Epton, 1979). Given the global importance of Phytophthora spp. as existing and emerging plant pathogens (Brasier et al., 2004; Fry, 2008; Fisher et al., 2012), identifying hyperparasites that naturally control the abundance and survival of these parasites would be beneficial.
There have been observations of such effects in control of population sizes by hyperparasites in fresh water ecosystems. Populations of Zygorhizidium affluens (Chytridiomycota) are frequent parasites of populations of the diatom Asterionella formosa in freshwater lakes (Canter, 1965; Beakes et al., 1988). The growth of the parasite population follows the growth of the host population (Chave, 2013) resulting in a “chytrid epidemic.” Sporangia and resting spores of Z. affluens can be infected by the hyperparasite Rozella parva (Canter, 1965). Both a decline in the A. formosa populations and an increase in the R. parva populations as the growing season progresses would, in theory, result in a decrease in Z. affluens populations. Another example is Polyphagus euglenae, a parasite of Euglena viridis and E. gracilis and its hyperparasite Rozella polyphagi (Powell, 1984), in which an infection with the hyperparasite R. polyphagi is known to decrease the population size of its host. Blooms of toxic cyanobacteria are common in freshwater environments (Sønstebø and Rohrlack, 2011). These cyanobacteria can be parasitized by zoosporic true fungi (Canter, 1972) that have the potential to control the sizes of such toxic algal blooms. Parasites of cyanobacteria can be infected by hyperparasites, a fact which was noted, but not analyzed in any detail. A reduction in the numbers of zoosporic parasites may result in an increase in growth of the (toxic) algal blooms (Canter, 1972). However, such tripartite interactions should be the subject of future studies: hyperparasites may impact the population sizes of parasitic, zoosporic true fungi that are parasites of organisms which can be damaging to the environment. The need to study the ecological role of hyperparasites may be even more significant as cyanobacteria and microalgae are gaining increasing importance as sustainable second generation biofuels (Stephens et al., 2010). Microalgal cultures are prone to get contaminated with a wide range of bacteria and eukaryotes which potentially impact on the yield (Stephens et al., 2010; Lakaniemi et al., 2012). Especially in such semi-controlled systems a control of detrimental parasites with hyperparasites could be a successful approach to increase productivity and energy yield.
Food webs
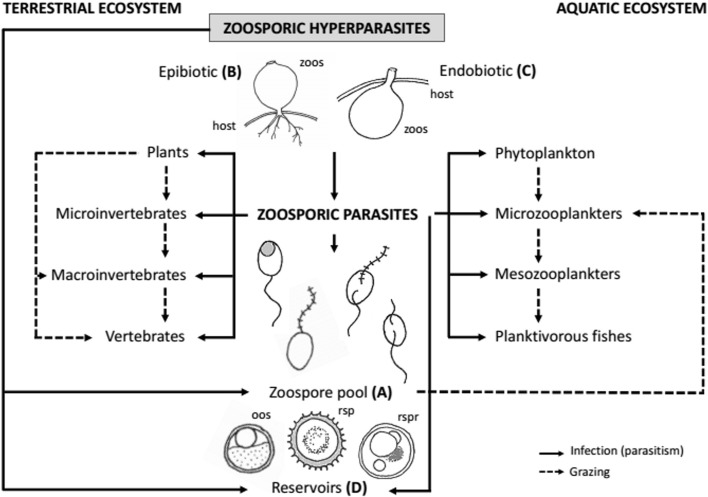
The presence of hyperparasites in food webs affect predators and grazers alike (Figure 2) (Hatcher et al., 2006; Morozova et al., 2007). By infecting resistant structures of their hosts, zoosporic parasites and hyperparasites release recalcitrant carbon, which is then potentially made available as food for protistan and metazoan predators rather than being deposited through sedimentation (Figure 2D). When zoospores are released, some will find new utilizable substrates, some will encyst, but many may provide food for grazing zooplankton and filter feeding animals (Figure 2A) (Kagami et al., 2007; Miki et al., 2011). The sizes of the mouth parts of grazing zooplankters determines the maximum size of zoopores that can be ingested (Kagami et al., 2007). For example, species of Daphnia are known to digest zoospores of any species smaller than 5 μm in diameter. The sizes of zoospores of hyperparasites tend to be smaller than those of the hosts (Sparrow, 1960; Held, 1981). This is clearly exemplified by the parasitic relationship between the fish parasite Achlya flagellata and its hyperparasite Dictyomorpha dioica (Mullins and Barksdale, 1965). The zoospores of A. flagellata are 8.5–10.5 μm in diameter while those of D. dioica are 3.5 μm in diameter (Mullins and Barksdale, 1965). The smaller size of the hyperparasite zoospores may enable zooplankton to graze on them or make their ingestion by zooplankters more likely, so that they ultimately provide better food resources for zooplankton than parasite zoospores. The population sizes of key species of grazing zooplankters, such as Daphnia, may be impacted by a decrease or increase in the total supply of zoospores which are a good food source (Kagami et al., 2007). This in turn will impact the population sizes of planktonivorous fish and other macroinvertebrates which feed on zooplankton.
Figure 2.
Possible links of a hypothesized food web in which zoosporic parasites and hyperparasites are involved. In food webs zoosporic hyperparasites can either contribute the zoospore pool (Zoospore pool, A) which is used as food source by grazers in terrestrial and acquatic ecosystems. At the same time epibiotic sporangia of hyperparasites (Epibiotic, B) can serve as food source for larger grazers. The sporangia of epibiotic hyperparasites (Endobiotic, C) are more difficult to access as food sources for grazers. Some zoosporic hyperparasites use resting stages (Reservoirs, D) as substrate. Hosts of hyperparasites can be parasites of microscopic eukaryotes, but also parasites of plants or animals. This allows for a rapid cycling of nutrients from organisms higher up in the food web towards small grazers (trophic upgrading). References: zoos, zoosporangium; host, zoosporic host; oos, oospore; rsp, resting spore; rspr, resting sporangium.
Because of the high nutritional value of zoospores, we would expect populations of Daphnia magna to increase with the onset of the chytrid epidemic. Daphnia magna also feeds on zoospores of Batrachochytrium dendrobatidis (Chytridiomycota), which is a serious pathogen of amphibians (Buck et al., 2011). It was suggested that the consumption of zoospores of B. dendrobatidis by D. magna may prevent the transmission of this fungus (Buck et al., 2011). If a crash occurs in populations of D. magna, when the total zoospore food supply rapidly decreases, the rate of transmission of amphibian chytridiomycosis could increase because fewer individuals of D. magna would be present to feed on zoospores of B. dendrobatidis. Thus, more zoospores would be available to spread chytridiomycosis through the populations of amphibians. In adult frogs B. dendrobatidis prevalence is highest during late summer and winter, while infection takes place from late spring to early summer (Russell et al., 2010; Sapsford et al., 2013). This coincides with the breakdown of the chytrid epidemics. We would expect many other biotic and abiotic factors to affect population dynamics here, but the availability of zoospores as food in the spring can be decisive for the pathogen load of B. dendrobatidis later in the year by influencing the numbers of predators feeding on zoospores.
It is important to establish the roles of zoosporic hyperparasites as well as parasites in the structure and function of aquatic food webs. Structure includes species richness, trophic levels, links, trophic chain length, and connectance (Dunne et al., 2005, 2013). Function includes the total amount, rate, and efficiency of carbon transfer, and effects on stability of the food web. Adding parasites to food webs results in an increased complexity (Lafferty et al., 2008; Thieltges et al., 2013). Adding links to food webs, such as parasites, hyperparasites, and both of their associated niches, might also add to the stability of a particular web (Hudson et al., 2006; Lafferty et al., 2006, 2008). Parasites with life cycles involving ontogenetic niche shifts—such as hyperparasites—impact food web structures more and potentially negatively because specialized life cycle stages are more prone to secondary extinction than generalist stages (Preston et al., 2014). Such ontogenetic effects can be found in zoosporic hyperparasites: different types of zoospores, or zoospores formed by different species can have considerably different swimming patterns (Lange and Olson, 1983) or serve different purposes like long or short distance dispersal (Neuhauser et al., 2011a). Consequently different zoospores will attract predators occupying different niches and will therefore enter the food web at different trophic levels. Because of the anecdotal nature of the available data it is not yet possible to include zoosporic hyperparasites into mathematical food web models to allow for more realistic estimates of population dynamics and energy flow and their impact on food web stability. However, it can be expected that once our knowledge about zoosporic hyperparasites increases, we will also be able to show that, like zoosporic true fungi, zoosporic hyperparasites are diverse, abundant, and important links for energy transfer (Grami et al., 2011; Niquil et al., 2011). Zoosporic true fungal parasites result in a significant reduction in the loss of algal carbon though sedimentation into the detritus pool, allowing carbon transfer from zoospores to grazing protists and metazoans. This contributes to longer carbon path lengths, higher levels of activity and specialization, lower recycling, and increased stability of aquatic food webs (Grami et al., 2011; Ulanowicz et al., 2014).
Hyperparasites tend to have shorter life cycles than their hosts, so they produce biomass in the form of zoospores more quickly. Some of them produce primarily zoospores, such as Rozella, which, instead of forming its own zoosporangium, uses the host sporangium to reproduce (Held, 1981; Powell, 1984). This outsourcing of energy consuming biomass production allows for faster life cycles and hyperparasites such as Rozella are therefore likely to increase and accelerate the energy flow between trophic levels (Figure 2C). On the other hand epibiotic parasites have zoosporangia that are formed on the surface of their host. Consequently, both their zoospores and the zoosporangia are likely to enter the food web contributing different types of energy for predators with different size preferences for their food (Figure 2B). Since food webs that include zoosporic hyperparasites have additional links, we suggest they could be more efficient, and therefore would support a larger population of grazing zooplankton species. This hypotheses needs to be tested quantitatively.
Conclusion and future prospects
Many hyperparasites have been discovered during research with the host species. However, it is vital that such efforts are intensified to provide the basis for the development of more rapid tools for species discovery and characterization. Although emerging techniques such as single cell genomic approaches provide a quantum leap in identifying and characterizing active cells in the environment, such methods will initially not account for the complex life cycles of zoosporic hyperparasites. To understand the life cycles, and consequently the ecological function of hyperparasites, time consuming studies involving targeted sampling and probing approaches are still needed. Even the sparse information available on hyperparasites highlights their potential in many ecosystem processes. Zoosporic hyperparasites may increase the turn-around time of certain nutrients in food webs due to their often rapid life cycles. They may play a role in trophic upgrading, as well as in the stability and complexity of food web dynamics. Hyperparasites also may play a role in the natural regulation of their host population sizes, which are also parasites. Regulation of population sizes of parasites will have an impact on their host population sizes. This may result in fine-tuning the magnitudes of patterns of energy flow in food webs and impact overall biodiversity as well as population dynamics. In summary, it is likely that zoosporic hyperparasites play a vital part of every ecosystem; hence more focused research on these important organisms is needed.
Author contributions
Frank H. Gleason and Sigrid Neuhauser drafted the initial version of the manuscript. Agostina V. Marano, Télesphore Sime-Ngando, Martin Kirchmair, Brooke K. Sullivan and Osu Lilje critically revised this draft and contributed intellectual content to the final version.
Conflict of interest statement
The reviewer, Hicham El Alaoui, declares that despite being affiliated to the same institution and department as the author, Télesphore Sime-Ndando, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Sigrid Neuhauser gratefully acknowledges funding by the Austrian Science Fund (FWF) through an Erwin Schrödinger research grant (J3175-B20). Frank H. Gleason thanks the University of Sydney Library for the use of its resources, Elayna Truszewski, Department of Biological Sciences, Macquarie University for her editorial assistance with preparation of this manuscript.
References
- Adams P. B. (1990). The potential of mycoparasites for biological control of plant diseases. Annu. Rev. Phytopathol. 28, 59–72 10.1146/annurev.py.28.090190.000423 [DOI] [PubMed] [Google Scholar]
- Adl S. M., Simpson A. G. B., Lane C. E., Lukes J., Bass D., Bowser S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–493 10.1111/j.1550-7408.2012.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen L. J. S., Bokil V. A. (2012). stochastic models for competing species with a shared pathogen. Math. Biosci. Eng. 9, 461–485 10.3934/mbe.2012.9.461 [DOI] [PubMed] [Google Scholar]
- Ayers W., Lumsden R. (1977). Mycoparasitism of oospores of Pythium and Aphanomyces species by Hyphochytrium catenoides. Can. J. Microbiol. 23, 38–44 10.1139/m77-005 [DOI] [Google Scholar]
- Baldauf S. (2003). The deep roots of eukaryotes. Science 300, 1703–1706 10.1126/science.1085544 [DOI] [PubMed] [Google Scholar]
- Barnett H., Binder F. (1973). The fungal host-parasite relationship. Annu. Rev. Phytopathol. 11, 273–292 10.1146/annurev.py.11.090173.001421 [DOI] [Google Scholar]
- Barr D. J. S. (2001). Chytridiomycota, in Systematics and Evolution, eds McLaughlin D. J., McLaughlin E. G., Lemke P. A. (Berlin; Heidelberg: Springer; ), 93–112 10.1007/978-3-662-10376-0_5 [DOI] [Google Scholar]
- Beakes G. W., Canter H. M., Jaworski G. H. (1988). Zoospore ultrastructure of Zygorhizidium affluens and Z. planktonicum, two chytrids parasitizing the diatom Asterionella formosa. Can. J. Bot. 66, 1054–1067 10.1139/b88-151 [DOI] [Google Scholar]
- Benhamou N., Rey P., Picard K., Tirilly Y. (1999). Ultrastructural and cytochemical aspects of the interaction between the mycoparasite Pythium oligandrum and soilborne plant pathogens. Phytopathology 89, 506–517 10.1094/PHYTO.1999.89.6.506 [DOI] [PubMed] [Google Scholar]
- Blackwell W. H. (2010). The enigmatic genus Pythiella (Oomycota). Phytologia 92, 304–311 [Google Scholar]
- Boosalis M. G. (1964). Hyperparasitism. Annu. Rev. Phytopathol. 2, 363–376 10.1146/annurev.py.02.090164.002051 [DOI] [Google Scholar]
- Brasier C. M., Kirk S. A., Delcan J., Cooke D. E., Jung T., Man In't Veld W. A. (2004). Phytophthora alni sp. nov. and its variants: designation of emerging heteroploid hybrid pathogens spreading on Alnus trees. Mycol. Res. 108, 1172–1184 10.1017/S0953756204001005 [DOI] [PubMed] [Google Scholar]
- Bruckart W. L., Eskandari F. M., Widmer T. L. (2011). Synchytrium solstitiale: reclassification based on the function and role of resting spores. Mycologia 103, 775–778 10.3852/10-286 [DOI] [PubMed] [Google Scholar]
- Buck J. C., Truong L., Blaustein A. R. (2011). Predation by zooplankton on Batrachochytrium dendrobatidis: biological control of the deadly amphibian chytrid fungus? Biodivers. Conserv. 20, 3549–3553 10.1007/s10531-011-0147-4 [DOI] [Google Scholar]
- Canter H. M. (1965). Studies on British chytrids. J. R. Microsc. Soc. 84, 549–557 10.1111/j.1365-2818.1965.tb02155.x [DOI] [Google Scholar]
- Canter H. M. (1972). A guide to the fungi occurring on planktonic blue-green algae, in Taxonomy and Biology of Blue-green Algae, ed Desikachary T. V. (Madras: University of Madras, Centre for Advance study in Botany; ), 145–158 [Google Scholar]
- Chave J. (2013). The problem of pattern and scale in ecology: what have we learned in 20 years? Ecol. Lett. 16, 4–16 10.1111/ele.12048 [DOI] [PubMed] [Google Scholar]
- Daft G. C., Tsao P. H. (1984). Parasitism of Phytophthora cinnamomi and P. parasitica spores by Catenaria anguillulae in a soil environment. Trans. Br. Mycol. Soc. 82, 485–490 10.1016/S0007-1536(84)80013-3 [DOI] [Google Scholar]
- del Campo J., Sieracki M. E., Molestina R., Keeling P., Massana R., Ruiz-Trillo I. (2014). The others: our biased perspective of eukaryotic genomes. Trends Ecol. Evol. 29, 252–259 10.1016/j.tree.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick M. W. (2001). Straminipilous Fungi: Systematics of the Peronosporomycetes Including Accounts of the Marine Straminipilous protists, the Plasmodiophorids and Similar Organisms. Dordrecht: Kluwer Academic Publishers; 10.1007/978-94-015-9733-3 [DOI] [Google Scholar]
- Dunne J. A., Brose U., Williams R. J., Martinez N. D. (2005). Modeling food-web dynamics: complexity-stability implications, in Aquatic Food Webs: An Ecosystem Approach, eds Belgrano A., Scharler U. M., Dunne J., Ulanowicz R. E. (Oxford: Oxford University Press; ), 117–129 10.1093/acprof:oso/9780198564836.003.0011 [DOI] [Google Scholar]
- Dunne J. A., Lafferty K. D., Dobson A. P., Hechinger R. F., Kuris A. M., Martinez N. D., et al. (2013). Parasites affect food web structure primarily through increased diversity and complexity. PLoS Biol. 11:e1001579 10.1371/journal.pbio.1001579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dylewski D. P., Miller C. E. (1983). Systematic and host range studies of Woronina pythii (Plasmodiophoromycetes) and host, Pythium species, from axenic culture. Mycologia 75, 412–422 10.2307/3792683 [DOI] [Google Scholar]
- Fisher M. C., Henk D. A., Briggs C. J., Brownstein J. S., Madoff L. C., McCraw S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194 10.1038/nature10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman K., Martin A., Karki D., Lynch R., Mitter M., Meyer A., et al. (2009). Evidence that chytrids dominate fungal communities in high-elevation soils. Proc. Natl. Acad. Sci. U.S.A. 106, 18315–18320 10.1073/pnas.0907303106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry W. (2008). Phytophthora infestans: the plant (and R gene) destroyer. Mol. Plant Pathol. 9, 385–402 10.1111/j.1364-3703.2007.00465.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason F. H., Crawford J. W., Neuhauser S., Handerson L. E., Lilje O. (2012). Resource seeking strategies of zoosporic true fungi in heterogeneous soil habitats at the microscale level. Soil Biol. Biochem. 45, 79–88 10.1016/j.soilbio.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason F. H., Küpper F. C., Amon J. P., Picard K., Gachon C. M., Marano A. V., et al. (2011). Zoosporic true fungi in marine ecosystems: a review. Mar. Freshw. Res. 62, 383–393 10.1071/MF10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason F. H., Schmidt S. K., Marano A. V. (2010). Can zoosporic true fungi grow or survive in extreme or stressful environments? Extremophiles 14, 417–425 10.1007/s00792-010-0323-6 [DOI] [PubMed] [Google Scholar]
- Goldie-Smith E. (1956a). A new species of Woronina, and Sorodiscus cokeri emended. J. Elisha Mitchell Sci. Soc. 72, 348–356 [Google Scholar]
- Goldie-Smith E. K. (1951). A new species of Sorodiscus on Pythium. J. Elisha Mitchell Sci. Soc. 67, 108–121 [Google Scholar]
- Goldie-Smith E. K. (1954). The position of Woronina polycystis in the Plasmodiophoraceae. Am. J. Bot. 41, 441–448 10.2307/2438854 [DOI] [Google Scholar]
- Goldie-Smith E. K. (1956b). Maintenance of stock cultures of aquatic fungi. J. Elisha Mitchell Sci. Soc. 72, 158–166 [Google Scholar]
- Grami B., Rasconi S., Niquil N., Jobard M., Saint-Béat B., Sime-Ngando T. (2011). Functional effects of parasites on food web properties during the spring diatom bloom in Lake Pavin: a linear inverse modeling analysis. PLoS ONE 6:e23273 10.1371/journal.pone.0023273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartikainen H., Ashford O. S., Berney C., Okamura B., Feist S. W., Baker-Austin C., et al. (2014). Lineage-specific molecular probing reveals novel diversity and ecological partitioning of haplosporidians. ISME J. 8, 177–186 10.1038/ismej.2013.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher M. J., Dick J. T. A., Dunn A. M. (2006). How parasites affect interactions between competitors and predators. Ecol. Lett. 9, 1253–1271 10.1111/j.1461-0248.2006.00964.x [DOI] [PubMed] [Google Scholar]
- Held A. A. (1973). Encystment and germination of the parasitic chytrid Rozella allomycis on host hyphae. Can. J. Bot. 51, 1825–1835 10.1139/b73-234 [DOI] [Google Scholar]
- Held A. A. (1974). Attraction and attachment of zoospores of the parasitic chytrid Rozella allomycis in response to host-dependent factors. Arch. Microbiol. 95, 97–114 10.1007/BF02451752 [DOI] [PubMed] [Google Scholar]
- Held A. A. (1981). Rozella and Rozellopsis: naked endoparasitic fungi which dress-up as their hosts. Bot. Rev. 47, 451–515 10.1007/BF02860539 [DOI] [Google Scholar]
- Hrcek J., Miller S. E., Quicke D. L., Smith M. (2011). Molecular detection of trophic links in a complex insect host–parasitoid food web. Mol. Ecol. Resour. 11, 786–794 10.1111/j.1755-0998.2011.03016.x [DOI] [PubMed] [Google Scholar]
- Hudson P. J., Dobson A. P., Lafferty K. D. (2006). Is a healthy ecosystem one that is rich in parasites? Trends Ecol. Evol. 21, 381–385 10.1016/j.tree.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Humble S. J., Lockwood J. (1981). Hyperparasitism of oospores of Phytophthora megasperma var. sojae. Soil Biol. Biochem. 13, 355–360 10.1016/0038-0717(81)90076-6 [DOI] [Google Scholar]
- Ibelings B. W., de Bruin A., Kagami M., Rijkeboer M., Brehm M., Donk E. V. (2004). Host parasite interactions between freshwater phytoplankton and chytrid fungi (Chytridiomycota). J. Phycol. 40, 437–453 10.1111/j.1529-8817.2004.03117.x [DOI] [Google Scholar]
- Ikeda S., Shimizu A., Shimizu M., Takahashi H., Takenaka S. (2012). Biocontrol of black scurf on potato by seed tuber treatment with Pythium oligandrum. Biol. Control 60, 297–304 10.1016/j.biocontrol.2011.10.016 [DOI] [Google Scholar]
- Jones M. D., Forn I., Gadelha C., Egan M. J., Bass D., Massana R., et al. (2011). Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474, 200–203 10.1038/nature09984 [DOI] [PubMed] [Google Scholar]
- Kagami M., de Bruin A., Ibelings B. W., van Donk E. (2007). Parasitic chytrids: their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 578, 113–129 10.1007/s10750-006-0438-z15134249 [DOI] [Google Scholar]
- Karling J. S. (1942a). Parasitism among the chytrids. Am. J. Bot. 29, 24–35 10.2307/2436540 [DOI] [Google Scholar]
- Karling J. S. (1942b). A synopsis of Rozella and Rozellopsis. Mycologia 34, 193–208 10.2307/375481117486963 [DOI] [Google Scholar]
- Karling J. S. (1960). Parasitism Among the Chytrids. II Chytriomyces verrucosus sp. nov. and Phlyctochytrium synchytrii. Bull. Torrey Bot. Club 87, 326–336 10.2307/2482628 [DOI] [Google Scholar]
- Karpov S. A., Mikhailov K. V., Mirzaeva G. S., Mirabdullaev I. M., Mamkaeva K. A., Titova N. N., et al. (2013). Obligately phagotrophic aphelids turned out to branch with the earliest-diverging fungi. Protist 164, 195–205 10.1016/j.protis.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Lafferty K. D., Allesina S., Arim M., Briggs C. J., de Leo G., Dobson A. P., et al. (2008). Parasites in food webs: the ultimate missing links. Ecol. Lett. 11, 533–546 10.1111/j.1461-0248.2008.01174.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty K. D., Dobson A. P., Kuris A. M. (2006). Parasites dominate food web links. Proc. Natl. Acad. Sci. U. S. A. 103, 11211–11216 10.1073/pnas.0604755103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakaniemi A.-M., Hulatt C. J., Wakeman K. D., Thomas D. N., Puhakka J. A. (2012). Eukaryotic and prokaryotic microbial communities during microalgal biomass production. Bioresour. Technol. 124, 387–393 10.1016/j.biortech.2012.08.048 [DOI] [PubMed] [Google Scholar]
- Lange L., Olson L. W. (1983). The fungal zoospore. Its structure and biological significance, in Zoosporic Plant Pathogens, ed Buczacki S. T. (London: Academic Press; ), 1–42 [Google Scholar]
- Lara E., Belbahri L. (2011). SSU rRNA reveals major trends in oomycete evolution. Fungal Divers. 49, 93–100 10.1007/s13225-011-0098-9 [DOI] [Google Scholar]
- Lara E., Moreira D., López-García P. (2010). The environmental clade LKM11 and Rozella form the deepest branching clade of fungi. Protist 161, 116–121 10.1016/j.protis.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Lefèvre E., Roussel B., Amblard C., Sime-Ngando T. (2008). The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS ONE 3:e2324 10.1371/journal.pone.0002324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Klein D. (2002). Molecular and cultural assessment of chytrid and Spizellomyces populations in grassland soils. Mycologia 94, 411–420 10.2307/3761775 [DOI] [PubMed] [Google Scholar]
- Marano A., Pires-Zottarelli C., Barrera M., Steciow M., Gleason F. (2011). Diversity, role in decomposition, and succession of zoosporic fungi and straminipiles on submerged decaying leaves in a woodland stream. Hydrobiologia 659, 93–109 10.1007/s10750-009-0006-4 [DOI] [Google Scholar]
- Marano A. V., Gleason F. H., Baerlocher F., Pires-Zottarelli C. L. A., Lilje O., Schmidt S. K., et al. (2012). Quantitative methods for the analysis of zoosporic fungi. J. Microbiol. Methods 89, 22–32 10.1016/j.mimet.2012.02.003 [DOI] [PubMed] [Google Scholar]
- Miki T., Takimoto G., Kagami M. (2011). Roles of parasitic fungi in aquatic food webs: a theoretical approach. Freshw. Biol. 56, 1173–1183 10.1111/j.1365-2427.2010.02562.x [DOI] [Google Scholar]
- Morozova A. Y., Robin C., Franc A. (2007). A simple model for the dynamics of a host-parasite-hyperparasite interaction. J. Theor. Biol. 249, 246–253 10.1016/j.jtbi.2007.05.041 [DOI] [PubMed] [Google Scholar]
- Mullins J. T., Barksdale A. W. (1965). Parasitism of the chytrid Dictyomorpha dioica. Mycologia 57, 352–359 10.2307/3756864 [DOI] [Google Scholar]
- Nagano Y., Nagahama T. (2012). Fungal diversity in deep-sea extreme environments. Fungal Ecol. 5, 463–471 10.1016/j.funeco.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Nagy L. G., Petkovits T., Kovács G. M., Voigt K., Vágvölgyi C., Papp T. (2011). Where is the unseen fungal diversity hidden? A study of Mortierella reveals a large contribution of reference collections to the identification of fungal environmental sequences. New Phytol. 191, 789–794 10.1111/j.1469-8137.2011.03707.x [DOI] [PubMed] [Google Scholar]
- Neuhauser S., Kirchmair M., Bulman S., Bass D. (2014). Cross-kingdom host shifts of phytomyxid parasites. BMC Evol. Biol. 14:33 10.1186/1471-2148-14-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser S., Kirchmair M., Gleason F. H. (2011a). The ecological potentials of Phytomyxea (plasmodiophorids) in aquatic food webs. Hydrobiologia 659, 23–35 10.1007/s10750-010-0508-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser S., Kirchmair M., Gleason F. H. (2011b). Ecological roles of the parasitic phytomyxids (plasmodiophorids) in marine ecosystems–a review. Mar. Freshw. Res. 62, 365–371 10.1071/MF10282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niquil N., Kagami M., Urabe J., Christaki U., Viscogliosi E., Sime-Ngando T. (2011). Potential role of fungi in plankton food web functioning and stability: a simulation analysis based on Lake Biwa inverse model. Hydrobiologia 659, 65–79 10.1007/s10750-010-0308-6 [DOI] [Google Scholar]
- Not F., Simon N., Biegala I. C., Vaulot D. (2002). Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat. Microb. Ecol. 28, 157–166 10.3354/ame02815718359836 [DOI] [Google Scholar]
- Pires-Zottarelli C., Santos A. D. S., Milanez A., Cipriano M. (2009). Occurrence of Pythiella vernalis from Pythium aphanidermatum on hydroponic culture of Lepidium sativum in Brazil. Summa Phytopathol. 35, 325–326 10.1590/S0100-54052009000400012 [DOI] [Google Scholar]
- Powell M. J. (1982). Ultrastructure of the host-parasite interface between Allomyces javanicus and its endoparasite Catenaria allomycis. Bot. Gaz. 143, 176–187 10.1086/337286 [DOI] [Google Scholar]
- Powell M. J. (1984). Fine structure of the unwalled thallus of Rozella polyphagi in its host Polyphagus euglenae. Mycologia 76, 1039–1048 10.2307/3793019 [DOI] [Google Scholar]
- Powell M. J. (1993). Looking at mycology with a Janus face: a glimpse at Chytridiomycetes active in the environment. Mycologia 85, 1–20 10.2307/3760471 [DOI] [Google Scholar]
- Preston D., Jacobs A., Orlofske S., Johnson P. J. (2014). Complex life cycles in a pond food web: effects of life stage structure and parasites on network properties, trophic positions and the fit of a probabilistic niche model. Oecologia 174, 953–965 10.1007/s00442-013-2806-5 [DOI] [PubMed] [Google Scholar]
- Raghukumar C. (1987). Fungal parasites of marine algae from Mandapam (South India). Dis. Aquat. Org. 3, 137–145 10.3354/dao003137 [DOI] [Google Scholar]
- Ribeiro W. R., Butler E. (1995). Comparison of the mycoparasites Pythium periplocum, P. acanthicum and P. oligandrum. Mycol. Res. 99, 963–968 10.1016/S0953-7562(09)80757-0 [DOI] [Google Scholar]
- Russell D. M., Goldberg C. S., Waits L. P., Rosenblum E. B. (2010). Batrachochytrium dendrobatidis infection dynamics in the Columbia spotted frog Rana luteiventris in north Idaho, USA. Dis. Aquat. Org. 92, 223–230 10.3354/dao02286 [DOI] [PubMed] [Google Scholar]
- Sapsford S. J., Alford R. A., Schwarzkopf L. (2013). Elevation, Temperature, and Aquatic connectivity all influence the infection dynamics of the amphibian chytrid fungus in adult frogs. PLoS ONE 8:e82425 10.1371/journal.pone.0082425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer C. A., Descals E., Kohlmeyer B., Kohlmeyer J., Marvanová L., Padgett D., et al. (2007). Fungal biodiversity in aquatic habitats. Biodivers. Conserv. 16, 49–67 10.1007/s10531-006-9120-z [DOI] [Google Scholar]
- Sieber M., Hilker F. M. (2011). Prey, predators, parasites: intraguild predation or simpler community modules in disguise? J. Anim. Ecol. 80, 414–421 10.1111/j.1365-2656.2010.01788.x [DOI] [PubMed] [Google Scholar]
- Slifkin M. K. (1961). Parasitism of Olpidiopsis incrassata on members of the Saprolegniaceae. I. Host range and effects of light, temperature, and stage of host on infectivity. Mycologia 53, 183–193 10.2307/3756236 [DOI] [Google Scholar]
- Sneh B., Humble S., Lockwood J. (1977). Parasitism of oospores of Phytophthora megasperma var. sojae, P. cactorum, Pythium sp., and Aphanomyces euteiches in soil by Oomycetes, Chytridiomycetes, Hyphomycetes, Actinomycetes, and bacteria. Phytopathology 67, 622–628 10.1094/Phyto-67-622 [DOI] [Google Scholar]
- Sønstebø J. H., Rohrlack T. (2011). Possible implications of chytrid parasitism for population subdivision in freshwater cyanobacteria of the genus Planktothrix. Appl. Environ. Microbiol. 77, 1344–1351 10.1128/AEM.02153-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow F. (1960). Aquatic Phycomycetes. Ann Arbor: University of Michigan Press [Google Scholar]
- Stephens E., Ross I. L., Mussgnug J. H., Wagner L. D., Borowitzka M. A., Posten C., et al. (2010). Future prospects of microalgal biofuel production systems. Trends Plant Sci. 15, 554–564 10.1016/j.tplants.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Sykes E. E., Porter D. (1980). Infection and development of the obligate parasite Catenaria allomycis on Allomyces arbuscula. Mycologia 72, 288–300 10.2307/3759252 [DOI] [Google Scholar]
- Thieltges D. W., Amundsen P. A., Hechinger R. F., Johnson P. T., Lafferty K. D., Mouritsen K. N., et al. (2013). Parasites as prey in aquatic food webs: implications for predator infection and parasite transmission. Oikos 122, 1473–1482 10.1111/j.1600-0706.2013.00243.x [DOI] [Google Scholar]
- Tyler B. M. (2002). Molecular basis of recognition between Phytophthora pathogens and their hosts. Annu. Rev. Phytopathol. 40, 137–167 10.1146/annurev.phyto.40.120601.125310 [DOI] [PubMed] [Google Scholar]
- Ulanowicz R. E., Holt R. D., Barfield M. (2014). Limits on ecosystem trophic complexity: insights from ecological network analysis. Ecol. Lett. 17, 127–136 10.1111/ele.12216 [DOI] [PubMed] [Google Scholar]
- Vinale F., Sivasithamparam K., Ghisalberti E. L., Marra R., Woo S. L., Lorito M. (2008). Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 40, 1–10 10.1016/j.soilbio.2007.07.00224293705 [DOI] [Google Scholar]
- Wagner M., Haider S. (2012). New trends in fluorescence in situ hybridization for identification and functional analyses of microbes. Curr. Opin. Biotechnol. 23, 96–102 10.1016/j.copbio.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Weller D. M., Raaijmakers J. M., Gardener B. B. M., Thomashow L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348 10.1146/annurev.phyto.40.030402.110010 [DOI] [PubMed] [Google Scholar]
- Wynn A. R., Epton H. A. S. (1979). Parasitism of oospores of Phytophthora erythroseptica in soil. Trans. Br. Mycol. Soc. 73, 255–259 10.1016/S0007-1536(79)80109-6 [DOI] [Google Scholar]