Significance
Acidic soils compose significant land areas in the tropics and subtropics. On acid soils, aluminum (Al) toxicity inhibits root growth, leading to reduced crop yields. Rice is the most Al-tolerant cereal crop. We examined the natural variation in rice Nramp aluminum transporter (OsNRAT1), which encodes a transporter mediating Al uptake from the root tip cell wall into the cell, where it is sequestered in the vacuole. We identified tolerant and sensitive NRAT1 alleles that exhibited lower NRAT1 expression and reduced Al uptake in the sensitive allele, resulting in a significant reduction in rice Al tolerance. These findings indicate that the NRAT1 transporter plays a major role in rice Al tolerance and open the door for using NRAT1 to improve Al tolerance in other cereal species.
Keywords: aluminum transport, cell wall aluminum
Abstract
Aluminum (Al) toxicity is a major constraint for crop production on acid soils which compose ∼40% of arable land in the tropics and subtropics. Rice is the most Al-tolerant cereal crop and offers a good model for identifying Al tolerance genes and mechanisms. Here we investigated natural variation in the rice Nramp aluminum transporter (NRAT1) gene encoding a root plasma membrane Al uptake transporter previously hypothesized to underlie a unique Al tolerance mechanism. DNA sequence variation in the NRAT1 coding and regulatory regions was associated with changes in NRAT1 expression and NRAT1 Al transport properties. These sequence changes resulted in significant differences in Al tolerance that were found to be associated with changes in the Al content of root cell wall and cell sap in 24 representative rice lines from a rice association panel. Expression of the tolerant OsNRAT1 allele in yeast resulted in higher Al uptake than did the sensitive allele and conferred greater Al tolerance when expressed in transgenic Arabidopsis. These findings indicate that NRAT1 plays an important role in rice Al tolerance by reducing the level of toxic Al in the root cell wall and transporting Al into the root cell, where it is ultimately sequestered in the vacuole. Given its ability to enhance Al tolerance in rice and Arabidopsis, this work suggests that the NRAT1 gene or its orthologs may be useful tools for enhancing Al tolerance in a wide range of plant species.
As much as 40% of the world’s potentially arable lands are highly acidic (1). At low soil pH values (pH < 5.0), the toxic aluminum (Al) species Al3+ is released from soil clay minerals, damaging and stunting root systems and resulting in significant reductions in crop yields due to drought stress and nutritional deficiencies (2). Therefore, the development of cultivars exhibiting elevated levels of Al tolerance is important for improving crop yields on acid soils, particularly in developing countries where food security is quite tenuous.
Plants have evolved both Al tolerance and Al avoidance mechanisms to cope with Al stress (2). Al tolerance involves internal mechanisms that allow plants to deal with Al toxicity in the root cell wall and/or to detoxify Al3+ that enters root cells by forming nontoxic organic acid (OA)–Al complexes in the cytosol and/or by sequestering the Al in subcellular compartments, such as vacuoles (3–5). The primary Al avoidance mechanism involves exclusion of Al from the growing root tip via the exudation of Al-chelating OAs into the rhizosphere, where the OAs form nontoxic OA–Al complexes which do not enter the root. Over the past decade, some key cellular/molecular components for both the Al tolerance and exclusion mechanisms have been identified in plants (6–12).
Rice is the most Al-tolerant of the cereal crop species (13–15). This is surprising, given the aquatic origin of the species and the fact that the majority of rice is grown in flooded paddies, where soil pH is effectively neutral and, thus, Al3+ toxicity is not a problem. Genetic and molecular analysis of rice Al tolerance indicates that this superior tolerance is due to the functioning of multiple Al tolerance mechanisms and genes that confer tolerance to Al in both the root cell wall and the root symplasm (6–8). Probably the most unique of these putative tolerance mechanisms involves NRAT1, which is a member of the Nramp family of metal transporters and functions as a plasma membrane-localized Al uptake transporter in cells of the root apex (the site of Al toxicity). Because the majority of the Al in the root resides in the cell wall (16, 17), the transport of cell wall Al into the root cell may function to reduce toxic levels of Al in the wall. It has been hypothesized that the Al transported into the cytoplasm by NRAT1 is subsequently sequestered in the vacuole by the tonoplast membrane Al transporter OsALS1 (5).
Our previous rice genome-wide association (GWA) study identified a significant marker–trait association on chromosome 2 in close vicinity to OsNRAT1, and a total of 11 distinct OsNRAT1 haplotypes were identified based on genotypic analysis of this region across the entire rice diversity panel (373 accessions). One haplotype was found to be unique to the eight Al-sensitive aus accessions in the diversity panel, and this haplotype explained 40% of the phenotypic variation for Al tolerance within the aus subpopulation (18). In the current study, we found that sequence variations in the OsNRAT1promoter and coding regions both play an important role in regulating Al tolerance in rice. Somewhat surprisingly, we found here that overexpression of OsNRAT1 confers enhanced Al tolerance in transgenic Arabidopsis, a plant species that does not possess an NRAT1 homolog and normally employs an Al exclusion mechanism to deal with Al toxicity (19). These findings suggest that OsNRAT1 or its orthologs in other cereal species may prove to be useful tools for improving aluminum tolerance in crop species.
Results
NRAT1 Expression Analysis.
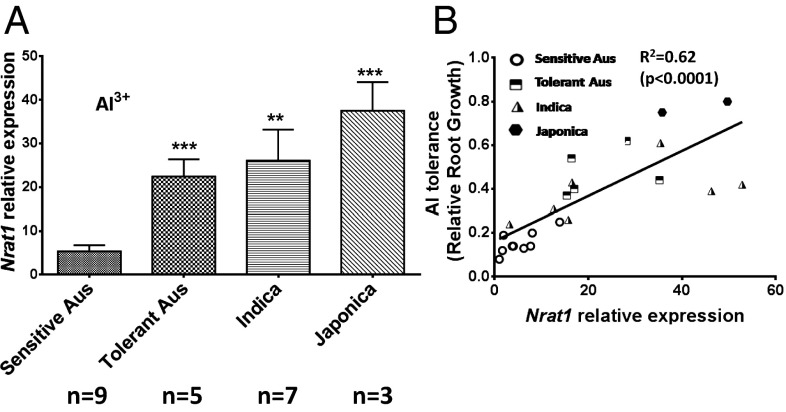
To evaluate the possible role of NRAT1 natural variation in differential rice Al tolerance, 24 rice lines from the GWA panel, including nine Al-sensitive aus accessions, five Al-tolerant aus accessions, and ten indica and japonica accessions that are as tolerant or more tolerant than the tolerant aus lines, were selected for further study. Within the 24 lines, the level of NRAT1 transcript abundance in the sensitive aus lines was significantly lower than in Al-tolerant aus and was also lower than in all of the indica and japonica lines under both ±Al growth conditions (Fig. 1A and Fig. S1 A–C). More detailed analysis of these data demonstrated that there is a strong positive association between the level of NRAT1 expression and Al tolerance across all 24 lines (R2 = 0.62) (Fig. 1B).
Fig. 1.
NRAT1 expression in 24 diverse rice accessions. (A) NRAT1 expression determined using quantitative real-time PCR with RNA from the root tips (1 cm) of 24 diverse rice lines grown for 3 d on nutrient solution containing an Al3+ activity of 160 µM. NRAT1 expression for each line is presented in relation to NRAT1 expression in the most sensitive aus line, NSF-317, whose expression was set to 1. The rice gene OsHistoneH3:5′ was used as the endogenous calibrator for each single real-time qRT-PCR experiment. The rice lines used for the NRAT1 expression analysis are classified into the following categories: sensitive aus, tolerant aus, indica, and japonica. Values are the mean ± SE. Asterisks indicate significant differences by t tests (**P < 0.01, ***P < 0.001). (B) Regression analysis for Al tolerance measured as relative root growth as a function of root NRAT1 expression for each of the 24 rice lines.
NRAT1 Haplotypes for the Promoter and Coding Regions.
To identify possible causative elements responsible for the observed variation in NRAT1 expression, we sequenced and analyzed the putative NRAT1 promoter (2,061-bp region upstream of the start codon) in each of the 24 rice accessions. Additionally, to look for sequence variation that could be associated with NRAT1 transport function, NRAT1 cDNA was also sequenced and analyzed in the 24 lines (Table 1). Haplotype analysis based on the promoter and cDNA sequence yielded a total of 14 SNPs and one indel representing five haplotypes for the promoter and coding regions (Table 1). Eight of the nine sensitive aus lines share one haplotype across the 15 polymorphisms (haplotype 1), whereas the other Al-sensitive aus accession, Kasalath, uniquely carries haplotype 2, which differs from the other eight sensitive aus lines only at polymorphisms M1 and M10. Additionally, all of the tolerant indica and japonica lines, as well as two of the tolerant aus lines (Ca 902/B/21 and Goria), share a common haplotype (haplotype 5) (Table 1). The five tolerant aus lines (Ca 902/B/21, Goria, Karkati 87, DM59, and P 737) carry three different haplotypes: Ca 902/B/21 and Goria carry haplotype 3, which is a modified mixture of haplotype 5 and haplotypes 1 and 2, with a unique SNP (M14) in the coding region. Karkati 87 carries haplotype 4, which is identical to the haplotype found in the tolerant indica and japonica lines, except for a unique SNP (M11) in the coding region. Finally, lines DM59 and P 737 have the same haplotype (haplotype 5) as all of the indica and japonica lines (Table 1). The sequence variation in the coding region can be translated into three different amino acid sequences among these 24 lines (Table S3). With regards to the NRAT1 promoter region, five SNPs (M2–M6) distinguish the sensitive aus haplotypes (1, 2) from the tolerant aus, indica, and japonica haplotypes (3–5) (Table S1). We had previously determined Nrat1 haplotypes using both exonic and intronic DNA sequence (18) and when the intron SNPs from that analysis were combined with the exon SNPs determined here, a larger number of haplotypes are seen (Table S1). The primary difference observed when intron SNPs are included is that different indica and japonica Nrat1 haplotypes are generated (Table S1).
Table 1.
SNPs and haplotype for Nrat1 promoter and coding regions
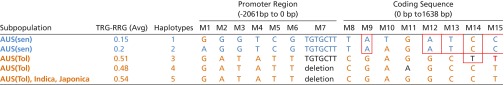 |
Genotypes of the 24 diverse lines assayed by sequencing a 3.7-kb region, including the 2.1-kb promoter region (M1 to M7) and a 1,638-bp coding sequence (M8 to M15). Fifteen natural mutation sites were identified (M1 to M15) based on comparison of the Nipponbare NRAT1 sequence, including 14 SNPs and one 7-bp insertion (M7). A total of five haplotypes were identified among the 24 diverse rice lines. Mutation sites marked with a red rectangle result in amino acid alterations. Also shown in the column labeled TRG-RRG is the Al tolerance for each group, based on the RRG of the total root system (TRG, total root growth).
Eight SNPs (M8–M15) were identified in the NRAT1 coding region (Table 1). Of these eight SNPs, five are unique to the sensitive aus lines, and four of these unique SNPs (M9, M12, M13, and M15) cause missense mutations (18). SNP M14, which is unique to the two haplotype 3 tolerant aus lines (Ca 902/B/21 and Goria), causes a different missense mutation (Table 1 and Table S2).
NRAT1 Expression in Yeast Causes Al Hypersensitivity due to Al Uptake.
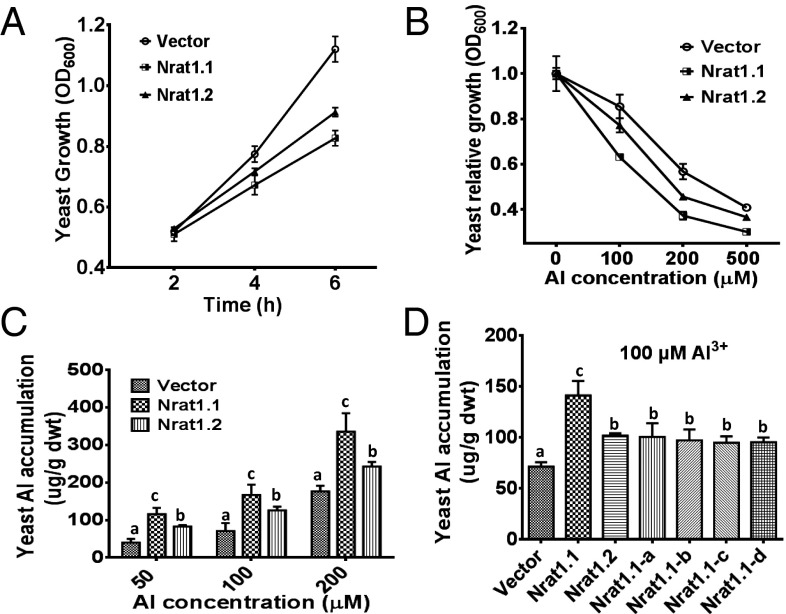
To investigate whether the altered amino acid sequences in NRAT1 affect its ability to transport Al, the Al-tolerant and Al-sensitive NRAT1 alleles (tolerant NRAT1.1 allele from the japonica reference genome, Nipponbare, haplotype 5; sensitive NRAT1.2 allele from aus lines, haplotypes 1 and 2; Table 1 and Table S2) were cloned into a yeast expression vector and transformed into yeast cells. The expression of NRAT1.1 and NRAT1.2 in yeast had no effect on cell growth under control (−Al) conditions, indicating that NRAT1 is not toxic to yeast cells (Fig. 2). However, both the NRAT1.1 and the NRAT1.2 expressing yeast genotypes were significantly more sensitive to Al stress than wild-type (WT) yeast transformed with the empty vector at all tested Al concentrations and treatment durations (Fig. 2 A and B). The hypersensitive NRAT1.1 and NRAT1.2 genotypes accumulated significantly higher levels of Al than the empty vector expression yeast did (Fig. 2C). Yeast has no mechanism to sequester toxic Al in its vacuole, which explains why the NRAT1.1 and NRAT1.2 genotypes were more sensitive to Al than yeast expressing the empty vector. The NRAT1.1 genotype accumulated more Al and was more sensitive to Al than the NRAT1.2 genotype, suggesting that the four missense mutations (M9, M12, M13, and M15) in the NRAT1.2 allele lead to reduced Al uptake via the NRAT1 transporter encoded by the sensitive NRAT1 haplotype (Fig. 2C). Although the NRAT1.3 genotype from two tolerant aus lines (Table 1 and Table S2) contains an altered amino acid due to SNP M14, it displayed a similar phenotype of yeast Al sensitivity and enhanced yeast Al accumulation to that of the yeast expressing NRAT1.1, indicating that this amino acid is not critical for full NRAT1 transport function.
Fig. 2.
Al sensitivity and Al uptake in yeast expressing the tolerant and sensitive NRAT1 alleles. (A) Growth of yeast expressing the empty vector (WT) and the tolerant NRAT1.1 or sensitive NRAT1.2 alleles after 2, 4, and 6 h of growth in low pH, low magnesium (LPM) medium plus 50 µM Al3+. (B) Growth of the WT, NRAT1.1, and NRAT1.2 expressing yeast lines after 6 h of growth in LPM media containing 0, 100, 200, or 500 µM Al3+. (C) Al3+ content of the WT, NRAT1.1, and NRAT1.2 expressing yeast lines after 15 h of growth in LPM media containing 50, 100, or 200 µM Al3+. (D) Al3+ uptake for yeast cells expressing the empty vector, the tolerant NRAT1.1 allele, the sensitive NRAT1.2 allele, and the single amino acid mutants, grown on LPM medium containing 100 µM Al3+ for 15 h. For data in all four panels, values are means ± SE. Histograms with different letters were significantly different determined by t tests (P < 0.01).
To test the effects on yeast Al transport and Al sensitivity of each of the individual missense mutations in NRAT1.2, single point mutations corresponding to each of the NRAT1.2 missense mutations were generated in the NRAT1.1 background through DNA sequence fragment substitution (Fig. S2A). Yeast lines expressing these single NRAT1 mutations displayed a similar growth and Al accumulation phenotype to yeast expressing the sensitive NRAT1.2 allele (Fig. 2D and Fig. S2 B and C), indicating that each of the mutated amino acids in NRAT1 is of equal importance for full NRAT1 Al transport function.
Cell Wall Al Content Analysis in Rice.
It was previously shown that when OsNRAT1 was knocked out in rice, this resulted in increased Al accumulation in the cell wall and decreased Al in the cell sap of root apices, and these responses were associated with increased Al sensitivity (6). These findings led to the suggestion that NRAT1 plays a role in partitioning Al between the rice root cell wall and symplasm as an Al resistance mechanism.
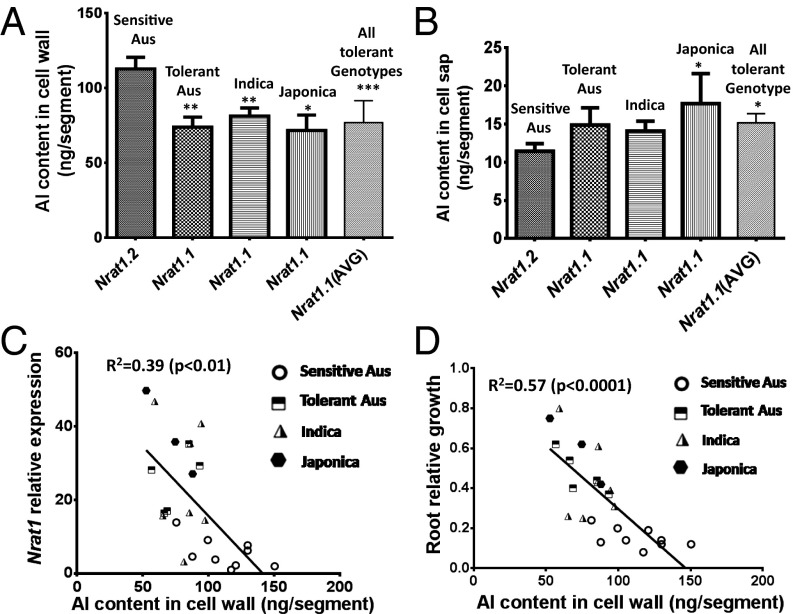
To investigate the role of NRAT1 variation in Al partitioning in cells of the rice root tip, we measured the Al content in the cell wall and cell sap of root apical cells for the 24 lines (Table S4). The lines harboring the tolerant NRAT1 allele (NRAT1.1; tolerant aus, indica, and japonica lines) accumulated significantly lower levels of Al in their root tip cell wall (Fig. 3A) and significantly higher levels of Al in the cell sap than did the Al-sensitive aus lines carrying the sensitive NRAT1.2 allele (Fig. 3B and Table S4). Cell wall Al content was negatively correlated with the level of NRAT1 expression (P < 0.001) and Al tolerance [relative root growth (RRG %)] (P < 0.0001) (Fig. 3 C and D). It has been well documented that most of the root Al (∼90%) occurs in the cell wall, presumably due in part to the cell wall’s cation exchange capacity (16, 17). For the smaller amount of Al that is transported into the root symplasm, most of this symplastic Al is believed to be sequestered in the vacuole, which occupies most (∼90%) of the volume of the root cell symplasm (5). Thus, the majority of the cell sap Al quite likely resides in the root cell vacuole. It is of note that the decrease in root cell wall Al content in the more tolerant rice lines expressing the tolerant NRAT1.1 allele is greater than the increase in cell sap Al in these same lines. There are two possible explanations for this. First, the techniques used to quantify root tip cell wall and symplastic Al involve the disruption of the root symplasm by freeze-thawing, following by centrifugation of the cell sap containing the symplastic Al away from the cell wall. It is possible that during these procedures, some of the symplastic Al released from the ruptured cell binds to the cell wall, thus causing an overestimation of cell wall Al and an underestimation of symplastic Al. Second, it is possible that in addition to sequestration in the vacuole of the Al transported across the plasma membrane from the cell wall, some of the transported Al could move radially into the root where it is loaded into the xylem and then transported to the shoot for storage in a manner similar to what occurs in the Al accumulators, hydrangea and buckwheat (4, 5).
Fig. 3.
Root Al content, Al tolerance, and NRAT1 expression in the 24 diverse rice accessions. (A) Cell wall Al content for the Al-sensitive aus lines carrying the Al-sensitive NRAT1.1 allele compared with the rice lines carrying the Al-tolerant NRAT1.1 allele (tolerant aus, indica, japonica, and the mean for all lines expressing the tolerant NRAT1.1 allele).(B) Cell sap Al content for the same five groupings of rice lines examined in A. (C) The relationship between NRAT1 expression and cell wall Al content. (D) The relationship between Al tolerance [relative root growth (RRG)] and cell wall Al content. Regression analysis was conducted on the data for each individual rice line from the 24 line diversity panel. Values are the mean ± SE. In A and B, asterisks indicate significant differences as determined by t tests (*P < 0.05, **P < 0.01, ***P < 0.001).
It has been suggested that the tonoplast-localized ABC transporter aluminum sensitive 1 (OsALS1) might be responsible for the sequestration of Al3+ from the cytosol into the rice root vacuole (5). To evaluate the possible involvement of OsALS1 in the variation in Al tolerance associated with NRAT1, the level of root tip OsALS1 expression was measured in the 24 lines in response to Al3+ treatment. As depicted in Fig. S3 A–C, root tip OsALS1 expression in the sensitive aus lines was significantly lower than in the tolerant aus, indica, and japonica lines, and OsALS1 expression in these lines was also significantly and positively associated with NRAT1expression (P < 0.01).
Overexpression of Rice NRAT1 Confers Enhanced Al Tolerance in Transgenic Arabidopsis.
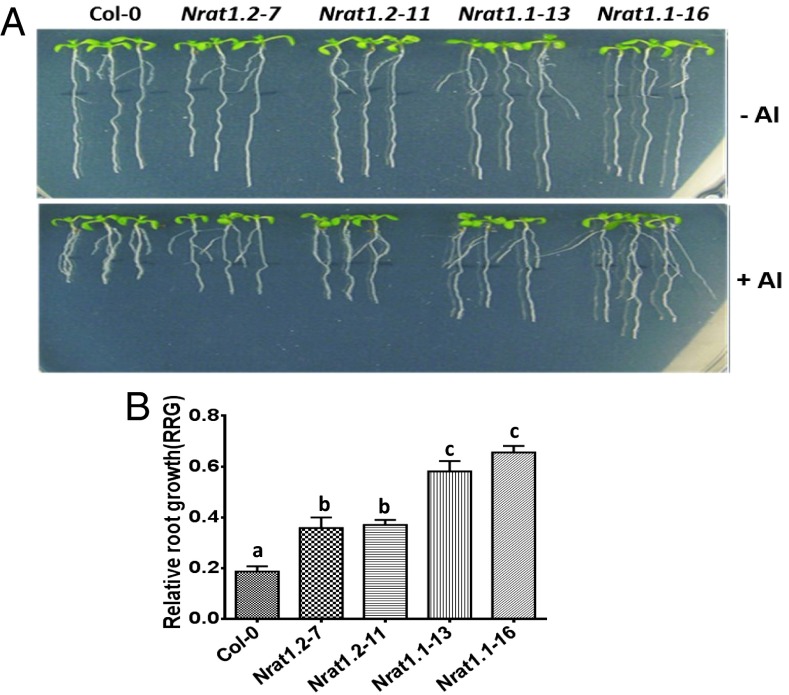
To determine if the rice NRAT1 gene could be used for improving Al tolerance in other plant species, NRAT1.1 and NRAT1.2 under the control of the CaMV 35S promoter were individually transformed into Arabidopsis (Col-0). Seven independent transgenic lines were generated for each of the NRAT1 alleles and four of these lines, NRAT1.1–13, NRAT1.1–16, NRAT1.2–7, and NRAT1.2–11, with the comparable and highest levels of the transgene expression were chosen for further study (Fig. S4). No significant differences were observed in root growth between the WT and transgenic lines under control (−Al) conditions, whereas the transgenic lines overexpressing either NRAT1.1 or NRAT1.2 exhibited significantly enhanced Al tolerance (Fig. 4 A and B). Among the transgenic lines, the lines expressing the tolerant NRAT1.1 allele conferred greater Al tolerance than did the transgenic lines expressing the sensitive NRAT1.2 allele (Fig. 4 A and B), suggesting that the rate and/or efficiency of Al uptake mediated by NRAT1.1 is higher than the same transport properties for NRAT1.2, which is consistent with the yeast Al uptake results (Fig. 2).
Fig. 4.
Overexpression of rice NRAT1 confers enhanced Al tolerance in transgenic Arabidopsis. (A) Al tolerance (root growth in +Al media/root growth in −Al media) of Arabidopsis WT (Col-0) and NRAT1 overexpressing transgenic lines grown on nutrient agarose containing 0 or 150 μM Al. (B) Quantification of Al tolerance as RRG (root growth [+Al]/Root growth [–Al]) for the lines depicted visually in A. Values in B are the mean ± SE. Histograms with different letters were significantly different as determined by t tests (P < 0.01).
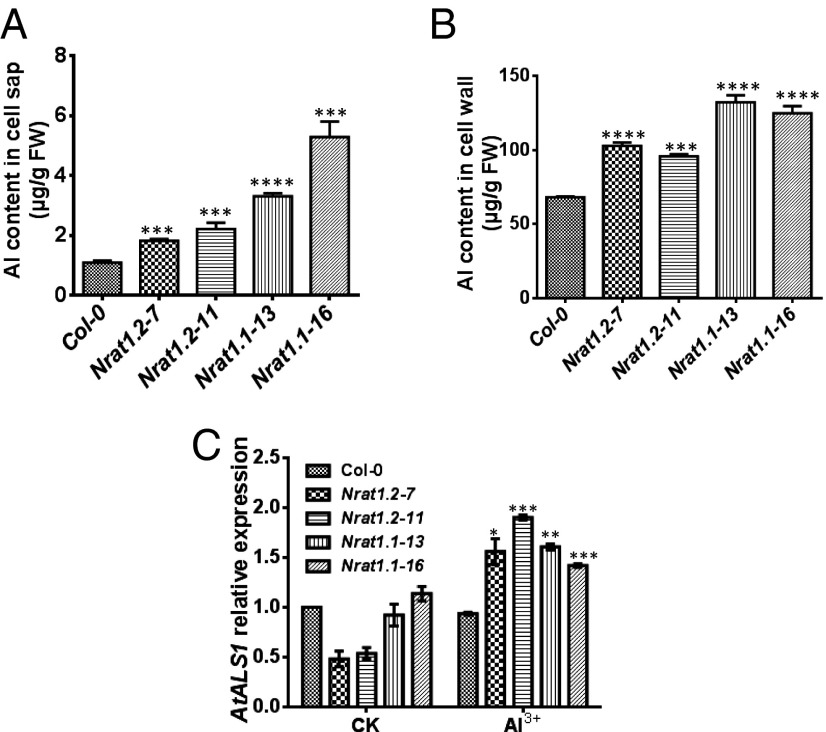
In the transgenic Arabidopsis lines overexpressing NRAT1, we were surprised to see that unlike in rice, the Al content of both the cell sap and cell wall was significantly higher in the more Al-tolerant transgenic lines compared with WT plants (Fig. 5 A and B). It was notable, however, that the increase in root cell sap Al in the transgenic Arabidopsis plants correlated closely with the increase in Al tolerance in these lines compared with WT plants (R2 = 0.89) (compare Figs. 4B and 5A), whereas there was no correlation between cell wall Al content and increased Al tolerance in the different transgenic lines (Figs. 4B and 5B). We currently do not have an explanation for the anomalous Arabidopsis root cell wall results.
Fig. 5.
Al concentration in the root cell wall and cell sap, and AtALS1 expression in WT and NRAT1-expressing transgenic Arabidopsis lines. (A) Al concentration in the root cell sap in WT and NRAT1-expressing transgenic Arabidopsis lines. (B) Al concentration in the root cell wall in WT and NRAT1-expressing transgenic Arabidopsis lines. (C) Quantitative real-time PCR analysis of AtALS1 expression in WT and NRAT1-expressing transgenic Arabidopsis lines grown in control (−Al) and +Al media. For A–C, values are mean ± SE for three biological replicates. Asterisks indicate significant difference: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
There currently is no direct evidence for a functional homolog of OsNRAT1 in Arabidopsis, but it has been speculated that the tonoplast-localized AtALS1, which is similar in sequence to OsALS1, might be responsible for the sequestration of Al from the cytosol into the vacuole in Arabidopsis. Here we found that overexpression of rice NRAT1 resulted in enhanced expression of the endogenous AtALS1 under Al stress compared with that in WT plants, whereas there was no difference in expression under control conditions (Fig. 5C). This might be consistent with the involvement of AtALS1 with the transport of the increased cytosolic Al facilitated by overexpression of OsNRAT1 into the vacuole. However, the enhanced AtALS1 expression in the transgenic lines was not found to be associated with Al content in cell sap or Al tolerance (R2 = 0.07 and R2 = 0.15, respectively).
As mentioned above, Arabidopsis primarily employs a root tip Al exclusion mechanism based on AtALMT1 and AtMATE1-mediated root malate and citrate exudation to cope with Al stress (19, 20). To test if this exclusion mechanism might possibly be up-regulated when NRAT1 was expressed in Arabidopsis, we examined the expression of AtALMT1 and AtMATE in the WT and the NRAT1 overexpression lines. As seen in Fig. S5, there were no significant differences in AtMATE and AtALMT1 expression under either control or +Al treatment between WT and the NRAT1 overexpression lines, suggesting that enhanced OA exudation was not responsible for the increased Al3+ tolerance in the NRAT1 transgenic lines.
Discussion
It has been previously shown that promoter sequence variation influences the expression of other plant Al tolerance genes. For instance, the size of a tourist-like miniature inverted repeat transposable element (MITE) in the promoter of the sorghum Al tolerance gene, SbMATE, is positively correlated with multidrug and toxic compound extrusion (SbMATE) expression (10), where increase in MITE size results in a greater number of sequence repeats in the MITE. Also, in barley, a 1-kb insertion in the promoter of the barley Al tolerance gene, aluminum-activated citrate transporter (HvAACT1), enhances HvAACT1 expression and alters the localization of HvAACT1 expression in roots so that it is expressed in the root tip epidermis, which is a novel feature of Al tolerance for this allele (21).
In this study, we identified five SNPs (M2–M6) in the NRAT1 promoter unique to the sensitive aus lines that might be involved in the regulation of NRAT1 expression, as the lines harboring these five SNPs all exhibit low NRAT1 expression (Fig. 1A). None of these five SNPs are localized in sequence motifs similar to any previously identified Al responsive cis-elements (7, 22); thus, they may be associated with novel cis-acting elements that regulate the expression of the NRAT1 Al tolerance gene. However, currently, we do not have any direct functional evidence that these SNPs are causative elements responsible for the elevated NRAT1 expression in rice. Further functional promoter studies are required to confirm/identify these SNPs as causative elements underlying elevated NRAT1 expression in the more tolerant rice lines. In addition to these five SNPs in the promoter region, we identified four SNPs in the coding region that were exclusively present in the nine sensitive aus lines and cause missense mutations (Table 1 and Table S2) (21). In this report we did conclusively demonstrate that each of these four SNPs in the sensitive aus allele significantly affects NRAT1’s Al transport activity (Fig. 2 A–C), and each of them is equally important for the fully functional NRAT1.1 found in the more Al-tolerant rice lines (Fig. 2D and Fig. S2 B and C). Taken together, our findings indicate that natural variation in the NRAT1 coding region functionally contributes to the observed differences in Al tolerance in diverse rice lines and that variation in the NRAT1 promoter also possibly contributes to this variation in Al tolerance via elevated expression.
The cell wall is a major target for Al accumulation and Al toxicity in higher plants (23, 24). The root, and more specifically, the root tip, is the primary site of Al toxicity, and the majority (∼90%) of the root Al resides in the cell wall (16, 17). Al accumulation has been suggested to have a number of deleterious effects within the cell wall. For example, Al can increase the rigidity of the cell wall by cross-linking pectin residues which inhibits cell wall loosening needed for root elongation, leading to inhibition of root growth (25). Additionally, expansins, which are wall proteins that loosen the cell wall during the process of cellular expansion and growth, have been shown to be very sensitive to Al (26). In fact, Al3+ ions have been reported to be the most potent inhibitor of expansin activity (26). A previous study indicated that OsNRAT1 could affect the Al content of the cell wall and cell sap of the rice root tip (6). Our study showed that the sensitive aus rice lines that contain the aus-specific, sensitive NRAT1 allele maintain significantly higher Al levels in the cell wall and lower levels in the cell sap compared with cell wall and cell sap Al content in the root tips of tolerant aus, indica, and japonica lines that harbor one of the tolerant NRAT1 alleles (Table S1 and Fig. 3A). In order for a plasma membrane-localized Al uptake transporter to be effective in Al tolerance, it must work in concert with other Al transporters that mediate the transport of the cytoplasmic Al to endomembrane compartments. The root cell vacuole is the likely site of symplasmic Al sequestration because it occupies most of the volume of root cells. Huang et al. (4) recently hypothesized that OsALS1, which is located in the tonoplast membrane, may be the transporter that mediates vacuolar Al sequestration. Here we found that root tip OsALS1 expression in the sensitive aus lines was significantly lower than in the tolerant aus, indica, and japonica lines (Fig. S3 A and B), and the variation in NRAT1 expression in these lines was significantly and positively correlated with the variation in OsALS1 expression (Fig. S3C). However, as this transporter has not been shown to mediate the uptake of Al or any other metal, we cannot rule out the possibility that other currently unidentified vacuolar Al transport mechanisms might be involved in Al sequestration in vacuole. Furthermore, we were intrigued by the observation that the decrease in root cell wall Al in tolerant compared with sensitive rice lines was larger than the associated increase in cell sap Al. As we suggested in Results, this could be an artifact of the techniques used to fractionate root cell wall and symplastic Al. Alternatively, it could be that additional transport processes are involved that move a portion of the absorbed Al from the root tip to the xylem, where it is transported to the shoot for sequestration.
The phenotype of enhanced Al tolerance for the Arabidopsis NRAT1 overexpression lines contrasted with the increased sensitivity to Al seen when NRAT1 was overexpressed in rice (27) and yeast (Fig. 2). It was speculated that the hypersensitive phenotype of the rice NRAT1 overexpression lines was due to entry of Al into the cytoplasm in excess of the capacity of the tonoplast-localized Al transporter, OsALS1, to move this excess Al into the vacuole (5). Because yeast lacks a functional homolog of OsALS1 to sequester Al into the vacuole, overexpression of NRAT1 in yeast also could result in hyperaccumulation of Al in the cytosol, leading to the observed enhanced Al toxicity of the NRAT1-expressing yeast (Fig. 2). In WT Arabidopsis, high levels of Al content in the cytosol of root cells should not normally occur due to the lack of a functional OsNRAT1 homolog and the dependence of Arabidopsis on a root tip Al exclusion mechanism mediated by Al-activated root malate and citrate exudation. Thus, we were surprised to see significantly increased Al tolerance when OsNRAT1 was expressed in transgenic Arabidopsis plants. It is possible that in OsNRAT1-expressing transgenic Arabidopsis plants, a coordinated enhancement of AtALS1 expression leads to increased capacity to bring Al into the cytosol and immediately sequester the cytosolic Al in the vacuole (Figs. 4 and 5C). We speculate that a high level of Al in the cytosol may be required for enhanced AtALS1 expression, based on the findings in Fig. 5C where we observed that Al exposure alone did not increase AsALS1 expression in roots of WT Arabidopsis. However, in roots of transgenic Arabidopsis expressing either OsNRAT1 allele, Al exposure resulted in a significant increase in AtALS1 expression. Thus, the presence of the transgenic NRAT1 in Arabidopsis roots could be working coordinately with the endogenous AtALS1 to play a role in the significantly enhanced Al tolerance in transgenic NRAT1-expressing Arabidopsis plants.
As addressed in Results, we were surprised that there was an apparent increase in both cell sap and cell wall Al in roots of transgenic Arabidopsis overexpressing the rice NRAT1 (Fig. 5 A and B). However, only the sequential increases in cell sap Al in the different transgenic lines were highly correlated with the similar sequential increases in Al tolerance for these four transgenic lines, whereas the increase in cell wall Al to fairly similar levels in all four transgenic lines showed little correlation with the variation in Al tolerance. We speculate that this may be in part due to technical difficulties in accurately quantifying root cell wall Al, and some of the Al could actually have originally come from the root symplasm. If this occurs, the problem could be exacerbated in Arabidopsis because dicot roots generally have a much higher cation exchange capacity than do roots of cereals and other monocots and thus bind considerably more Al in their root cell walls (28). Thus, it is possible that the overestimation of the binding of Al released from the root symplasm by the cell wall could be exacerbated by the greater Al binding by the Arabidopsis cell wall. Finally, although Al content in the root cell sap was significantly and positively associated with Al tolerance between the WT and the transgenic lines, the levels of enhanced AtALS1 expression were not well correlated with Al tolerance in these lines (compare Figs. 4B and 5C), suggesting the possibility that other currently unknown vacuolar Al transporter(s) might also be involved in Al sequestration in the Arabidopsis root cell vacuole.
Unlike many other crop plants, rice appears to use both symplastic and apoplastic Al tolerance mechanisms to cope with Al stress. Coordinated plasma membrane/tonoplast Al transport systems in rice appear to be a major contributor to rice’s superior level of Al tolerance compared with other cereal crops. This notion is supported by the fact that even the most Al-sensitive aus lines which have a functionally deficient OsNRAT1 transporter are still more Al-tolerant than other cereal species, including maize, sorghum, and wheat (15). In the current study, through the introduction of the rice plasma membrane-localized NRAT1 into Arabidopsis plants, it appears that a coordinated root plasma membrane/tonoplast Al transport and sequestration system was established, which allowed the transgenic Arabidopsis plants to combine a rice-like internal Al tolerance mechanism with the plant’s normal root Al exclusion mechanism (Fig. 4 A and B and Fig. S5D). This raises an interesting evolutionary question about why a functional OsALS1 homolog in Arabidopsis would have maintained the capacity for Al ion sequestration. Because of the lack of a functional OsNRAT1 to transport Al into the cytoplasm, AtALS1 should not normally operate as a tonoplast Al transporter. One explanation would be that AtALS1 is a multifunctional tonoplast transporter, capable of sequestering other critical ion(s) in addition to Al. This would explain why the addition of OsNRAT1 would enable transgenic Arabidopsis plants to combine a rice-like internal Al tolerance mechanism with the plant’s normal root Al exclusion mechanism. This pyramiding of the two types of tolerance mechanisms allows for greater root growth at higher concentrations of Al, significantly increasing the level of Arabidopsis Al tolerance. We are now investigating whether the transgenic expression of OsNRAT1 in cereal species such as wheat that depends exclusively on root tip Al exclusion will significantly increase Al tolerance. We also are looking for homologs of OsNat1 and OsALS1 in maize, sorghum, and wheat to see if there are possibilities to enhance this coordination of tolerance mechanisms in other cereal species via marker-assisted plant breeding.
Materials and Methods
See Supporting Information for details concerning materials and methods.
Plant Materials and Growth Conditions.
Rice seedlings were germinated and grown hydroponically as described in ref. 15.
Yeast Al Tolerance and Uptake Analysis.
The NRAT1 coding sequences were amplified by PCR from cDNAs generated from each of the 24 rice diversity lines and cloned into the yeast expression vector, pYES2. Single amino acid NRAT1 mutants were generated via partial DNA fragment substitution. The resulting constructs were then transformed into the yeast DY4741 cell line. Al tolerance was measured by the growth of each of the yeast genotypes at indicated Al concentrations or Al treatment duration. For the measurement of Al content, yeast cells were harvested and washed with deionized water and then digested with 2 N HCl. The concentration of Al was determined by inductively coupled plasma mass spectrometry (ICP-MS).
Root Cell Sap Preparation and Al Determination.
After Al treatment, the first 1 cm of root tip segments were cut and washed with dH2O and then centrifuged to remove apoplastic solution. The root cell sap solution was obtained by freezing and thawing the samples, followed by centrifuging. The residual cell wall was washed with 70% (vol/vol) ethanol and then digested in 2 N HCl. Al content was determined by ICP-MS.
Sequence and Haplotype Analysis.
The NRAT1 coding and promoter sequences were amplified from cDNA and genomic DNA, respectively, by PCR.
Supplementary Material
Acknowledgments
We thank Eric Craft, Michael Rutzke, and Shree Giri for conducting the ICP-MS experiments; Adam Famoso for providing rice Al tolerance data; Sandra Harrington for providing rice seeds used in the experiments; and Jon Shaff and Eric Craft for carrying out the rice seed increases in the greenhouse. The work was supported by US Department of Agriculture’s Agricultural Food Research Institute Grant 2009-02273.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318975111/-/DCSupplemental.
References
- 1.Uexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171(1):1–15. [Google Scholar]
- 2.Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol. 2004;55(1):459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- 3.Ma JF, Hiradate S, Nomoto K, Iwashita T, Matsumoto H. Internal detoxification mechanism of Al in hydrangea (identification of Al form in the leaves) Plant Physiol. 1997;113(4):1033–1039. doi: 10.1104/pp.113.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen R, Ma JF, Kyo M, Iwashita T. Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta. 2002;215(3):394–398. doi: 10.1007/s00425-002-0763-z. [DOI] [PubMed] [Google Scholar]
- 5.Huang CF, Yamaji N, Chen Z, Ma JF. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012;69(5):857–867. doi: 10.1111/j.1365-313X.2011.04837.x. [DOI] [PubMed] [Google Scholar]
- 6.Xia J, Yamaji N, Kasai T, Ma JF. Plasma membrane-localized transporter for aluminum in rice. Proc Natl Acad Sci USA. 2010;107(43):18381–18385. doi: 10.1073/pnas.1004949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaji N, et al. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell. 2009;21(10):3339–3349. doi: 10.1105/tpc.109.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CF, et al. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell. 2009;21(2):655–667. doi: 10.1105/tpc.108.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki T, et al. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37(5):645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 10.Magalhaes JV, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 2007;39(9):1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- 11.Yokosho K, Yamaji N, Ma JF. An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J. 2011;68(6):1061–1069. doi: 10.1111/j.1365-313X.2011.04757.x. [DOI] [PubMed] [Google Scholar]
- 12.Tovkach A, et al. Transposon-mediated alteration of TaMATE1B expression in wheat confers constitutive citrate efflux from root apices. Plant Physiol. 2013;161(2):880–892. doi: 10.1104/pp.112.207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foy CD. Plant adaptation to acid, aluminum‐toxic soils. Commun Soil Sci Plant Anal. 1988;19(7-12):959–987. [Google Scholar]
- 14.Ma JF, et al. Response of rice to Al stress and identification of quantitative trait Loci for Al tolerance. Plant Cell Physiol. 2002;43(6):652–659. doi: 10.1093/pcp/pcf081. [DOI] [PubMed] [Google Scholar]
- 15.Famoso AN, et al. Development of a novel aluminum tolerance phenotyping platform used for comparisons of cereal aluminum tolerance and investigations into rice aluminum tolerance mechanisms. Plant Physiol. 2010;153(4):1678–1691. doi: 10.1104/pp.110.156794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor GJ, et al. Direct measurement of aluminum uptake and distribution in single cells of Chara corallina. Plant Physiol. 2000;123(3):987–996. doi: 10.1104/pp.123.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang YC, Yamamoto Y, Matsumoto H. Accumulation of aluminium in the cell wall pectin in cultured tobacco (Nicotiana tabacum L.) cells treated with a combination of aluminium and iron. Plant Cell Environ. 1999;22(8):1009–1017. [Google Scholar]
- 18.Famoso AN, et al. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 2011;7(8):e1002221. doi: 10.1371/journal.pgen.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Magalhaes JV, Shaff J, Kochian LV. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009;57(3):389–399. doi: 10.1111/j.1365-313X.2008.03696.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, et al. A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance. Plant J. 2012;71(2):327–337. doi: 10.1111/j.1365-313X.2012.04994.x. [DOI] [PubMed] [Google Scholar]
- 21.Fujii M, et al. Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun. 2012;3:713. doi: 10.1038/ncomms1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsutsui T, Yamaji N, Feng Ma J. Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 2011;156(2):925–931. doi: 10.1104/pp.111.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabuchi A, Matsumoto H. Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiol Plant. 2001;112(3):353–358. doi: 10.1034/j.1399-3054.2001.1120308.x. [DOI] [PubMed] [Google Scholar]
- 24.Ma JF, Shen R, Nagao S, Tanimoto E. Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 2004;45(5):583–589. doi: 10.1093/pcp/pch060. [DOI] [PubMed] [Google Scholar]
- 25.Horst WJ, Wang Y, Eticha D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: A review. Ann Bot (Lond) 2010;106(1):185–197. doi: 10.1093/aob/mcq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosgrove DJ. Characterization of long-term extension of isolated cell walls from growing cucumber hypocotyls. Planta. 1989;177(1):121–130. doi: 10.1007/BF00392162. [DOI] [PubMed] [Google Scholar]
- 27.Xia J, Yamaji N, Ma JF. Further characterization of an aluminum influx transporter in rice. Plant Signal Behav. 2011;6(1):160–163. doi: 10.4161/psb.6.1.14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allan DL, Jarrell WM. Proton and copper adsorption to maize and soybean root cell walls. Plant Physiol. 1989;89(3):823–832. doi: 10.1104/pp.89.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.