Significance
Ribonucleotide reductases (RNR) play a critical role in supplying cellular deoxynucleotide pools. Nucleotide reduction by class Ia RNR requires a diferric-tyrosyl radical cofactor, which is a target of anticancer agents. How this essential cofactor is assembled in vivo is not well understood. We show here that a conserved protein complex composed of the Fe-S–requiring Dre2 and the diflavin-requiring Tah18, previously shown to donate electrons for Fe-S cluster assembly for proteins found in the cytosol and nucleus, also is required for RNR cofactor assembly. Deficiency in this complex leads to activation of both the DNA-damage checkpoint and the iron regulon, linking iron homeostasis to maintenance of genome stability. These findings may provide new insights into development of RNR-targeted therapeutics.
Keywords: iron cofactor, iron regulon, dNTP pool, genome stability, S phase
Abstract
Eukaryotic ribonucleotide reductases (RNRs) require a diferric-tyrosyl radical (FeIII2-Y•) cofactor to produce deoxynucleotides essential for DNA replication and repair. This metallocofactor is an important target of RNR-based therapeutics, although mechanisms of in vivo cofactor assembly, inactivation, and reactivation are poorly understood. Here, we demonstrate that the conserved Fe-S protein–diflavin reductase complex, Dre2–Tah18, plays a critical role in RNR cofactor biosynthesis. Depletion of Dre2 affects both RNR gene transcription and mRNA turnover through the activation of the DNA-damage checkpoint and the Aft1/Aft2-controlled iron regulon. Under conditions of comparable RNR protein levels, cells with diminishing Dre2 have significantly reduced ability to make deoxynucleotides. Furthermore, the kinetics and levels of in vivo reconstitution of the RNR cofactor are severely impaired in two conditional tah18 mutants. Together, these findings provide insight into RNR cofactor formation and reveal a shared mechanism underlying assembly of the FeIII2-Y• cofactor in RNR and the Fe-S clusters in cytosolic and nuclear proteins.
Ribonucleotide reductase (RNR) converts NDPs to dNDPs by using radical-based chemistry and supplies the essential building blocks for DNA replication and repair (1–3). Class Ia RNR, conserved from bacteria to human, is composed of α and β subunits that form active quaternary structure(s) (α2)3(β2)m (m = 1 or 3) in eukaryotes (4–7). The α subunit contains the catalytic and allosteric sites that control overall activity and substrate specificity. The β subunit houses a di-iron center that generates and maintains a tyrosyl radical (Y•), which is essential to initiate nucleotide reduction in the catalytic site of the α subunit via a long-range radical transfer pathway (8, 9). In this study we focus on the mechanism by which the requisite diferric-tyrosyl radical (FeIII2-Y•) cofactor is generated in the β subunit of yeast RNR.
The Saccharomyces cerevisiae RNR holoenzyme is proposed to have an (α2)3ββ′ configuration, in which α, β, and β′ are encoded by RNR1, RNR2, and RNR4, respectively. A fourth gene, RNR3, encodes an isoform of subunit α that normally is repressed and is inducible by genotoxic stress. The active form of yeast β2 is a heterodimer (ββ′) (10, 11). Only β is capable of iron binding and cofactor assembly, and consequently there is a maximum of one Y• per ββ′. However, β′ is essential to maintain β in a conformation competent for iron binding both in vivo and in vitro (12–14).
Eukaryotic cells tightly control their RNR activity to maintain an adequately sized and balanced dNTP pool that ensures high-fidelity DNA synthesis. The levels and activities of S. cerevisiae RNR are regulated by both the cell cycle and environmental signals including genotoxic stress and low iron availability. Cells in S phase have increased expression of the α subunit and redistribution of ββ′ from the nucleus to the cytoplasm, where the α subunit resides (15). In response to DNA damage, an activated Mec1–Rad53–Dun1 checkpoint kinase cascade increases RNR levels by phosphorylation-dependent removal of Crt1, the transcriptional repressor of RNR2/3/4 (16). Checkpoint kinase-mediated phosphorylation also leads to degradation of two negative regulators of RNR: Sml1 that binds and inhibits subunit α (17, 18) and Dif1 that facilitates nuclear sequestration of ββ′ (19, 20). Another negative regulator of RNR is the nuclear WD40 protein Wtm1, which binds and retains ββ′ in the nucleus (21, 22). Under iron deficiency, mRNAs of RNR2 and RNR4 and, to a much greater extent, of WTM1 are degraded in a Cth1/Cth2-dependent fashion as part of a metabolic remodeling process to conserve and optimize utilization of iron (23). Cth1/Cth2 belong to the iron regulon, a group of genes controlled by transcriptional factors Aft1 and Aft2 that are activated upon iron depletion (24).
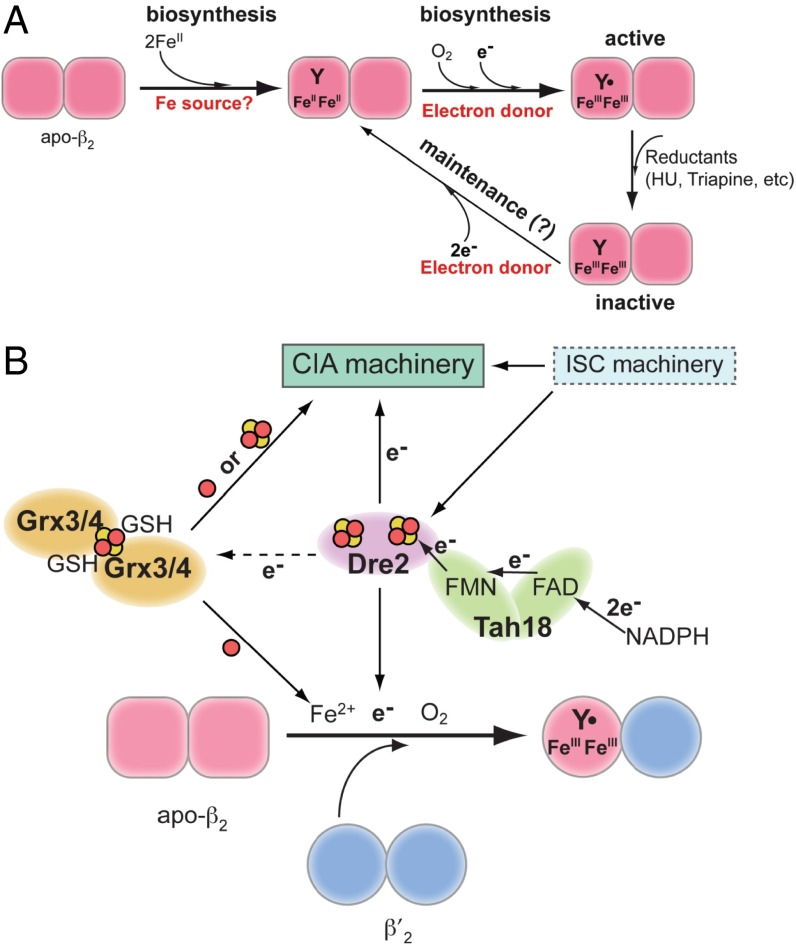
An additional layer of RNR regulation, given that the level of Y• of the FeIII2-Y• cofactor is directly correlated with nucleotide reduction activity, involves the assembly and maintenance of this essential cluster. The cellular machinery required for these processes has been explored only recently (13, 25). The metallo-cofactor can be generated in vitro by self-assembly from apo-β2, FeII, and O2, with FeII supplying the required reducing equivalent (26, 27) (Eq. 1 and Fig. 1A). However, the self-assembly process is inefficient in general, pointing to the importance of a biosynthesis pathway for controlled cofactor assembly (28). The Y• in cells also can be destroyed rapidly by endogenous reductants or exogenous reducing agents such as hydroxyurea (HU) and triapine (29, 30, and thus must be repaired to restore RNR activity (Fig. 1A).
Fig. 1.
The role of Dre2–Tah18 in RNR cofactor biosynthesis. (A) The proposed pathways for biosynthesis and maintenance of the FeIII2-Y• cofactor of class Ia RNR. The biosynthetic pathway requires delivery of two FeII/β and a reducing equivalent to carry out the four-electron reduction of O2 to H2O (Eq. 1); the other three electrons come from the two FeII and Tyr residue to form the FeIII2-Y•. The maintenance pathway may use the same source of reducing equivalents to convert the inactive FeIII2-Y cluster to FeII2-Y, which subsequently forms FeIII2-Y• in the presence of O2 via the biosynthesis pathway. (B) A model depicting the central role of the Dre2–Tah18 complex as a source of reducing equivalent for cluster assembly in RNR and cytosolic and nuclear Fe-S cluster-containing proteins. Assembly of FeIII2-Y• in β is facilitated by β′, which stabilizes β in a conformation that allows iron binding. Grx3/4 functions in intracellular iron trafficking and is required for most iron-requiring pathways, including the biosynthesis of Fe-S clusters in the mitochondria (ISC) and cytosol (CIA) and FeIII2-Y• assembly in RNR (25). Electrons from NADPH are transferred via FAD and FMN, the two flavin cofactors in Tah18, to the Fe-S cluster(s) in Dre2, which subsequently deliver the electrons to proteins in the CIA pathway and β in RNR. Dre2–Tah18 also may provide reducing equivalents to facilitate iron release from the [2Fe2S]-GSH2 cluster in the Grx3/Grx4 dimer.
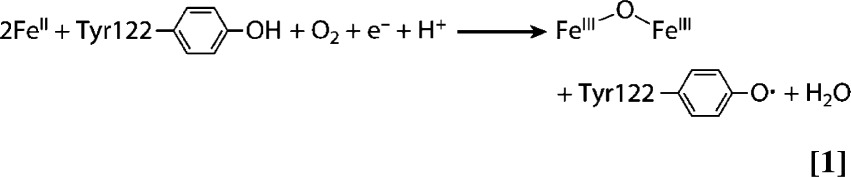 |
The key issues of in vivo RNR cofactor assembly are loading of FeII to apo-β2 and delivery of the obligatory reducing equivalent (Eq. 1). In S. cerevisiae, two cytosolic monothiol glutaredoxins, Grx3 and Grx4, which form a dimer with a [2Fe2S]-GSH2 cluster at the subunit interface, recently have been proposed to play an important role in the delivery of iron to Fe-S, heme, and di-iron–requiring proteins (25, 31, 32). Depletion of Grx3/4 in yeast cells reduced iron loading into ββ′ and impaired the ability of RNR to make dNTPs; both effects are consistent with a role in providing iron to RNR (25).
Our previous study also led us to propose that the Fe-S cluster protein Dre2 is a conduit of the reducing equivalent required for RNR cluster assembly and for reduction of the Fe-S cluster in Grx3/Grx4 for iron delivery (13) (Fig. 1B). This role was supported by a synthetic growth defect between dre2 and grx3/4 mutants and by the finding that depletion of Dre2 in yeast cells causes hypersensitivity to the Y•-quenching reagent HU and a decrease in both Y• content and RNR activity (13). However, these studies were complicated by the instability of ββ′ in Dre2-deficient cells.
Recently Dre2 has been shown to form a complex with the diflavin reductase Tah18 (33, 34) and to supply reducing equivalents to the early steps of the cytosolic Fe-S assembly (CIA) pathway (33). Thus, it is possible that together Dre2–Tah18 donate the electron for RNR cluster assembly (Fig. 1B). This hypothesis is appealing, because we recently have shown that FeIII2-Y• maintenance of the Escherichia coli NrdB (β2) is facilitated by a [2Fe-2S]-ferredoxin encoded by yfaE, which resides in the same operon as nrdA (α) and nrdB (β) (35). Although the S. cerevisiae ferredoxin-ferredoxin reductase (Fd-Fre) orthologs Yah1-Arh1 are localized exclusively in the mitochondria (36, 37), the Dre2–Tah18 pair has emerged as their cytoplasmic counterparts (33).
In this work, we have characterized the pleiotropic effects of Dre2–Tah18 deficiency on RNR including Cth1/2-mediated RNR2/RNR4 mRNA degradation and activation of the DNA-damage checkpoint leading to RNR induction and activation. Furthermore, using genetic manipulations, we have developed methods of circumventing the variability of ββ′ levels to determine the effect of Dre2–Tah18 deficiency on RNR cofactor assembly. We have found that the low ββ′ levels in Dre2-depleted cells can be partially suppressed by an increase in intracellular manganese levels. Upon controlling for variability in ββ′ levels, depletion of Dre2 causes a significant decrease in Y• content and RNR activity. Moreover, we took advantage of a GalRNR4 Δcrt1 system in which β is constitutively overexpressed because of the removal of transcriptional repression and in which reconstitution of Y• and ββ′ activity can be monitored over a time course upon induction of β′ by turning on the GAL promoter. Under these conditions, we found that two tah18 conditional mutants exhibit significant defects in both the kinetics and the maximum levels of Y• and ββ′ activity reconstitution relative to the WT control. Together, our findings support the model that Dre2–Tah18 functions in RNR cluster assembly and raise the intriguing perspective that the same protein pair functions as a donor of reducing equivalents to two different types of cytosolic iron clusters: the Fe-S cluster in CIA and the di-iron cluster in RNR.
Results
GalDRE2 Mutant Has Lower Y• and β Levels Even in the Absence of the RNR2/RNR4 Transcription Repressor Crt1.
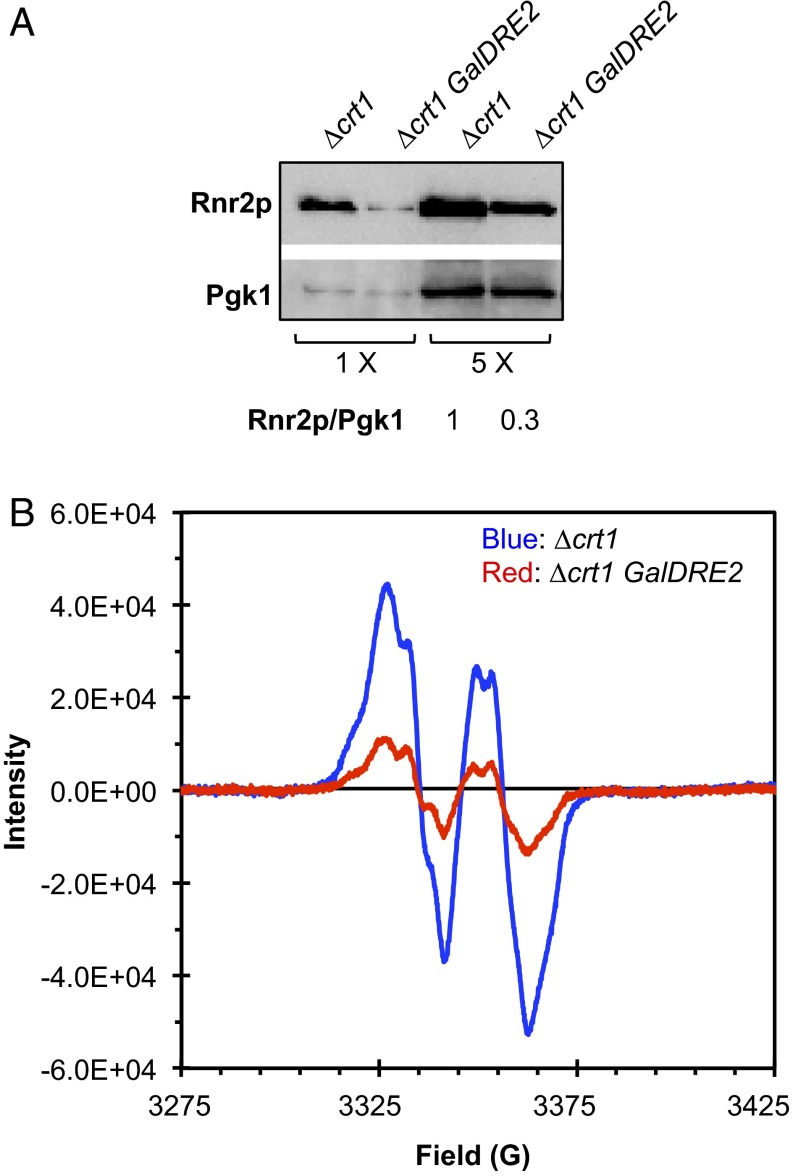
Because DRE2 is essential for cell viability, the downstream effects of Dre2 deficiency can be investigated by replacing the native DRE2 promoter with the glucose-repressible GAL1 promoter to allow transcriptional shut-off. We have shown previously that Dre2 depletion in GalDRE2 cells led to concurrent decreases in levels of Y•, ββ′ activity, and ββ′ proteins (13). To determine whether the decrease in ββ′ protein levels is mediated transcriptionally or posttranscriptionally, we constructed a Δcrt1GalDRE2 double mutant in which CRT1, the major transcriptional repressor of RNR2 and RNR4, was removed. The protein levels of β and β′ in Δcrt1GalDRE2 cells were still threefold lower than in the Δcrt1 single mutant, (Fig. 2A), suggesting that the decrease in ββ′ levels in GalDRE2 cells is mediated by a posttranscriptional mechanism. Moreover, the Y• content of Δcrt1 GalDRE2 cells is 5.3-fold lower than that of Δcrt1 (Fig. 2B). Thus, after correction for the difference in ββ′ protein levels, Dre2 depletion in Δcrt1 cells resulted in a twofold decrease in Y•/ββ′ ratio.
Fig. 2.
The GalDRE2 mutant has lower Y• and ββ′ levels even in the absence of the RNR2/RNR4 transcription repressor Crt1. Cells from a galactose-containing plate (GAL on) were inoculated into glucose-containing liquid (GAL off) and grown at 30 °C for 24 h to reach log phase before being harvested for EPR and Western blotting. (A) Comparison of Rnr2 (β) levels in Δcrt1 and Δcrt1GalDRE2 cells by Western blot with Pgk1 as a loading control. Rnr2/Pgk1 ratios were quantified based on signal intensity. (B) Whole-cell EPR spectra of Δcrt1 (blue, 8.4 × 109 cells/mL) and GalDRE2 Δcrt1 (red, 9.3 × 109 cells/mL).
The Decrease of ββ′ Levels in GalDRE2 Mutant Is Mediated by CTH2 and CTH1.
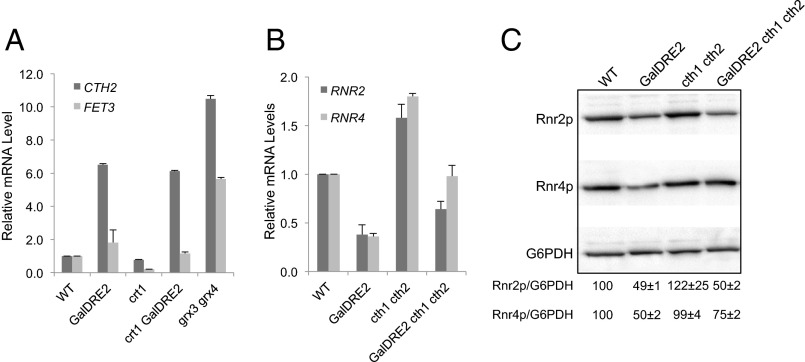
A recently discovered mechanism of posttranscriptional regulation of RNR2 and RNR4 is targeted mRNA turnover mediated by Cth1 and Cth2 (23), two homologous proteins that bind to specific AU-rich elements in the 3′ UTRs of many mRNAs including those of RNR2 and RNR4 (38). In response to iron deficiency, cells activate transcription of CTH1 transiently and of CTH2 persistently, which target specific mRNA degradation leading to metabolic reprogramming of iron utilization and iron storage (39, 40). To determine whether CTH2 is induced in Dre2-depeleted cells, we performed reverse transcription and quantitative real-time PCR (RT-qPCR) to compare CTH2 mRNA levels in WT and GalDRE2 mutant cells under GAL promoter-off conditions. CTH2 mRNA is ∼6.5-fold higher in GalDRE2 cells than in WT cells (Fig. 3A). A similar increase in CTH2 mRNA also was observed in Δcrt1 GalDRE2 relative to Δcrt1 cells, suggesting that induction of CTH2 is caused by depletion of Dre2 instead of by removal of CRT1. Unlike CTH2, the mRNA level of FET3, another member of the iron regulon, is induced only slightly in GalDRE2 (Fig. 3A).
Fig. 3.
The decrease in ββ′ levels in the GalDRE2 mutant is mediated by CTH1/CTH2. (A) CTH2 is induced in the GalDRE2 mutant. Levels of CTH2 and FET3 mRNAs in WT, GalDRE2, Δcrt1, and Δcrt1GalDRE2 cells were determined by reverse transcription and RT-qPCR. Signals of CTH2 and FET3 were normalized against that of ACT1 in each strain, and the resulting ratios in WT cells were arbitrarily defined as onefold. Mutant Δgrx3Δgrx4 was included as a positive control for iron regulon activation (41). (B) Comparison of RNR2 and RNR4 mRNA levels in WT, GalDRE2, Δcth1Δcth2, and GalDRE2Δcth1Δcth2 cells by RT-qPCR as described in A. (C) Comparison of Rnr2 (β) and Rnr4 (β′) protein levels in WT, GalDRE2, Δcth1Δcth2, and GalDRE2Δcth1Δcth2 cells by Western blot with G6PDH as a loading control. Rnr2/G6PDH ratios were quantified, and the average and SD from triplicates are shown.
Concurrent with an increase in CTH2 levels, we observed a 2.5-fold decrease in RNR2 and RNR4 mRNA levels in GalDRE2 cells (Fig. 3B). The decrease in RNR2 and RNR4 transcripts appeared to be mediated mainly by CTH1/CTH2 because deletion of both genes in GalDRE2 cells restores RNR2 and RNR4 mRNA levels to ∼70% and 100% of those in WT cells (Fig. 3B). Interestingly, although Rnr4 (β′) protein in GalDRE2 cells was restored to close to WT levels by removal of CTH1/CTH2, no significant increase in Rnr2 (β) protein level was observed in Δcth1Δcth2GalDRE2 cells relative to GalDRE2 cells (Fig. 3C). The discrepancy between transcript and protein levels of RNR2 likely reflects decreased stability of apo-β in Dre2-depleted cells.
Dre2 Depletion Activates both the DNA-Damage Checkpoint and Aft1/Aft2-Dependent CTH2 Transcription and Thereby Exerts Complex Effects on RNR.
We noted that induction of CTH2 in Dre2-depleted cells is less robust (sixfold) than in the Δgrx3Δgrx4 mutant (∼10.5-fold), which activates transcription of many genes of the iron regulon including FET3 (41) (Fig. 3A). CTH2 transcription can be activated by both Aft1 and Aft2 (41, 42). Moreover, an endogenously tagged Cth2-GFP fusion protein has been shown to become more abundant in cells under HU-caused replicational stress (43), suggesting that CTH2 expression may be subjected to other regulation in addition to Aft1/Aft2.
To determine whether the increased CTH2 transcription in GalDRE2 mutants is mediated by Aft1, Aft2, or the DNA replication checkpoint, we compared CTH2 mRNA levels by RT-qPCR in WT cells, GalDRE2 cells, and GalDRE2 cells lacking AFT1, AFT2, or the checkpoint kinase DUN1. The CTH2 mRNA level was unaffected in Δdun1 and was slightly lower in Δaft1 and Δaft2 cells (Fig. 4A). The increase of CTH2 mRNA in the GalDRE2 mutant was independent of DUN1, because the CTH2 level in the Δdun1GalDRE2 mutant cells was comparable to that in GalDRE2 cells. In contrast, CTH2 mRNA levels decreased by 20% in the Δaft1GalDRE2 cells and 50% in Δaft2GalDRE2 double mutants relative to the GalDRE2 single mutant (Fig. 4A). In keeping with the decrease of CTH2, RNR2 mRNA was restored from 43% in the GalDRE2 single mutant to ∼80% in both Δaft1GalDRE2 and Δaft2GalDRE2 double mutants relative to the WT strain (Fig. 4A). Together, these data indicate that Aft1/Aft2-medidated induction of CTH2 is responsible for the decrease of RNR2 mRNA levels in the GalDRE2 mutant.
Fig. 4.
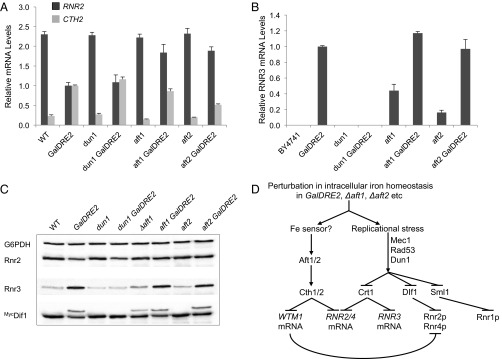
The GalDRE2 mutant exhibits checkpoint activation. (A) Comparison by RT-qPCR of RNR2 and CTH2 mRNA levels in WT, GalDRE2 single mutants, and GalDRE2 double mutants with Δdun1, Δaft1, and Δaft2 as described in Fig. 3A. The RNR2/ACT1 and CTH2/ACT1 ratios of GalDRE2 were arbitrarily defined as onefold. (B) Checkpoint-dependent RNR3 induction in the GalDRE2 mutant. RNR3 mRNA levels were determined in WT and mutant cells grown in YPD (GAL off) by RT-qPCR, with the RNR3/ACT1 ratio of GalDRE2 defined as onefold. (C) Checkpoint-mediated phosphorylation of Myc3-Dif1 in the GalDRE2 mutant, manifested as the slower mobility band on Western blot (20). (D) A scheme depicting the downstream events in cells deficient in Dre2 or Aft1/2. Cth2 is induced in Dre2-depleted cells via Aft1/Aft2 activation, leading to degradation of RNR2/RNR4 mRNAs and, to a much greater extent, of the mRNA of WTM1, which encodes a nuclear anchoring protein of Rnr2/4 (21, 22). Moreover, the DNA-damage checkpoint is activated in cells lacking Dre2 or Aft1/Aft2, causing phosphorylation-dependent removal of three negative regulators of RNR: Crt1 that represses transcription of RNR2/3/4, Sml1 that inhibits Rnr1 (α), and Dif1 that imports Rnr2/4 (ββ′) into the nucleus away from α.
We also observed an increase in RNR3 mRNA levels in the GalDRE2 mutant by RT-qPCR analysis. In contrast to CTH2, RNR3 induction in GalDRE2 cells was clearly DUN1-dependent and Aft1/Aft2-independent, because it was abolished in the Δdun1GalDRE2 but unchanged in Δaft1GalDRE2 and Δaft2GalDRE2 double mutants relative to the GalDRE2 single mutant (Fig. 4B). Interestingly, RNR3 also was moderately induced in Δaft1 and Δaft2 single mutants. Consistent with the increased RNR3 mRNA levels, Rnr3 protein levels also were higher in GalDRE2 and Δaft1 mutant cells than in the WT control (Fig. 4C). Induction of RNR3 is a signature of activation of the Mec1–Rad53–Dun1 checkpoint kinase cascade (16, 44) (Fig. 4D). To determine further whether GalDRE2 cells have a constitutively activated DNA-damage response, we monitored phosphorylation status of Dif1, the DNA-damage–regulated nuclear import facilitator of ββ′ that is phosphorylated in a DUN1-dependent manner in response to genotoxic stress (19, 20). A 3Myc-Dif1 from the GalDRE2 and Δaft1 mutants exhibited a phosphorylation-specific slow mobility shift on SDS/PAGE, which is undetectable in the Δdun1GalDRE2 double mutant (Fig. 4C). Therefore, GalDRE2, Δaft1, and, to a lesser degree, Δaft2 mutants all have a constitutively activated checkpoint. No further increase in RNR3 levels or Dif1 slow mobility shift was seen in the Δaft1GalDRE2 and Δaft2GalDRE2 double mutants relative to the GalDRE2 single mutant, suggesting that the signal leading to RNR3 induction in GalDRE2 and Δaft1/Δaft2 is of the same nature, likely a perturbation in intracellular iron homeostasis (Fig. 4D).
Depletion of Dre2 Decreases ββ′ Activity.
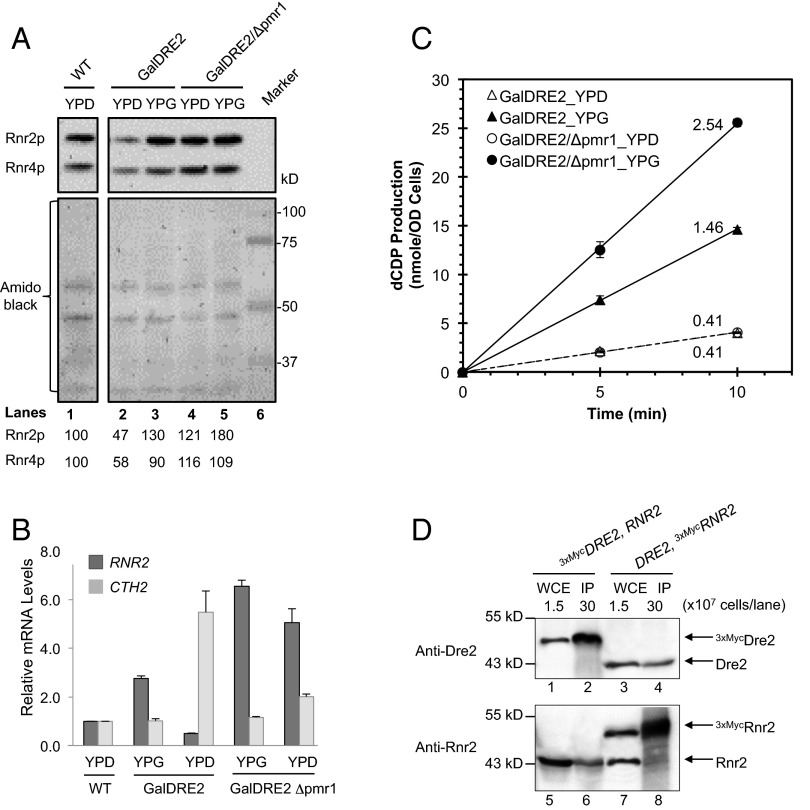
Considering the pleiotropic effects of Dre2 depletion on ββ′ function, including transcription, mRNA turnover, and subcellular localization (Fig. 4D), we directed our effort toward identifying conditions under which the ββ′ protein levels remain relatively unchanged before and after GalDRE2 shut-off so that the effect on ββ′ activity can be measured without concerns for drastically varying ββ′ levels. Manganese is known to be able to occupy the iron-binding site of class Ia RNR, resulting in catalytically inactive β2 (45). A possible effect of increased cytosolic manganese levels is to form Mn-β that stabilizes the β protein. Because increasing intracellular manganese levels are known to down-regulate manganese uptake in WT cells (46), we chose the Δpmr1 mutant that is defective in manganese transport from the cytosol to Golgi (47) and that apparently is deficient in the negative feedback control of manganese uptake. As a result, Δpmr1 cells have ∼10-fold elevated intracellular manganese levels (48, 49). To assess whether the increased intracellular manganese would affect β protein levels, we constructed the Δpmr1GalDRE2 double mutant. The β protein levels of the Δpmr1GalDRE2 cells remain comparable in GAL-repressed [yeast extract/peptone/dextrose (YPD)] and GAL-induced [yeast extract/peptone/Gal (YPG)] growth conditions (121% and 180%, respectively) but in GalDRE2 single mutant varied by threefold in the YPD (47%) and the YPG (130%) conditions (Fig. 5A). The changes in β protein levels were mirrored in changes in RNR2 mRNA levels. Consistent with the notion that CTH2 induction leads to RNR2 mRNA degradation, we found that the levels of CTH2 transcript in Δpmr1GalDRE2 double mutant were much lower than those in GalDRE2 single mutant under GAL-repressed (YPD) conditions (Fig. 5B).
Fig. 5.
Depletion of Dre2 causes decrease in ββ′ activity. (A) Comparison of Rnr2 protein levels in GalDRE2 Δpmr1 and GalDRE2 cells under GAL-on (YPG) and GAL-off (YPD) conditions. Total protein extract from an equal number of cells was loaded for each sample. The protein blot was probed with anti-Rnr2 and anti-Rnr4 (Upper) and stained with amino black (Lower) as a control for loading. Relative Rnr2 and Rnr4 signals are shown. (B) Removal of PMR1 in GalDRE2 cells abolished induction of CTH2 and decrease of RNR2 under GAL-off conditions. Relative levels of RNR2 and CTH2 mRNAs were determined by RT-qPCR and normalized against ACT1 mRNA signals; the resulting ratios in WT cells were arbitrarily defined as onefold. (C) Comparison of ββ′ activities of GalDRE2 Δpmr1 and GalDER2 cells under GAL-on (YPG) and GAL-off (YPD) conditions. The ββ′ activity of each sample was assayed in permeabilized cells in the presence of an excess of α as previously described (13). The ββ′ activities for GalDRE2_YPD, GalDRE2_YPG, GalDRE2/Δpmr1_YPD, and GalDRE2/Δpmr1_YPG are 0.41, 1.46, 0.41, and 2.54 nmol dCDP/min in OD600 cells, respectively. (D) Coimmunoprecipitation of Dre2 and Rnr2 (β). Whole-cell extracts (WCE) of the 3MycDRE2 (AXY1767) and 3MycRNR2 (MHY340) strains were incubated a with anti-Myc monoclonal antibody 9E10. The immunoprecipitates (IP) were brought down with Protein A beads, and the protein blots were probed with polyclonal anti-Dre2 (lanes 1–4) and anti-Rnr2 (lanes 5–8) antibodies. The WCE lanes were loaded with lysates of 1.5 × 107 cells, and the IP lanes contained immunoprecipitates from lysates of 3 × 108 cells.
Taking advantage of the comparable protein levels of β and β′ in the Δpmr1GalDRE2 cells in the GAL-repressed and GAL-induced states (Fig. 5A), we measured and compared the ββ′ activity of Δpmr1GalDRE2 cells grown in YPD and YPG by a permeabilized cell-based RNR activity assay (13). The activity of ββ′ in Δpmr1GalDRE2 cells is 6.5-fold lower under Dre2-repressed (YPD) conditions than in Dre2-induced (YPG) conditions (Fig. 5C, open circles versus filled circles). Under GAL-induced conditions, the ββ′ activity of Δpmr1GalDRE2 cells is ∼40% higher than in GalDRE2 cells (Fig. 5C, filled circles versus filled triangles), as is consistent with ∼40% higher protein levels of β (Fig. 5A, lanes 3 and 5). Under Dre2-repressed conditions the ββ′ activity of Δpmr1GalDRE2 cells is as low as that of GalDRE2 cells (Fig. 5C, open circles and open triangles), although Δpmr1GalDRE2 has 2.5-fold more of ββ′ protein (Fig. 5A, lanes 1 and 3). Collectively, these results indicate that Dre2 is required for the formation of catalytically active ββ′.
The proposal that Dre2 serves as the electron donor for cluster assembly in β requires a transient interaction between them. We thus performed reciprocal immunoprecipitation experiments in the strains containing either N-terminally epitope-tagged 3xMycDRE2 or 3xMycRNR2 under their respective native promoters. The anti-Myc immunocomplex from 3xMycDRE2 cells brings down not only 3xMycDre2 but also Rnr2 (Fig. 5D, lanes 2 and 6, respectively). Conversely, we detected both 3xMycRnr2 and Dre2 in the anti-Myc immunocomplex from 3xMycRNR2 cells (Fig. 5D, lanes 8 and 4, respectively). Together, these results indicate that Dre2 and β can exist in the same protein complex in vivo, as is consistent with the model that Dre2 is involved in delivering the reducing equivalent to β for its cofactor assembly.
Conditional Mutants of tah18 Exhibit Slow S-Phase Progression and Synthetic Growth Defect with GalRNR4.
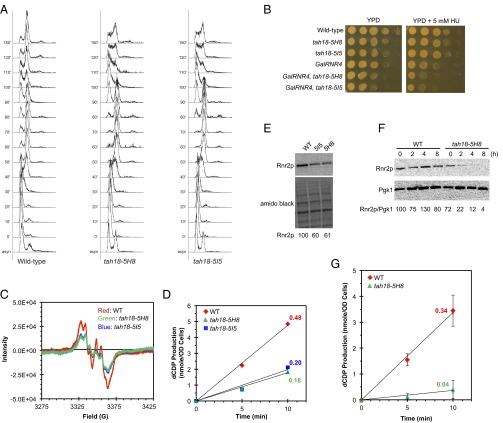
The diflavin reductase Tah18 forms a stable complex with Dre2 to transfer electrons from NADPH to Dre2’s Fe-S clusters. We inferred that inactivation of Tah18 would have effects on RNR similar to that of Dre2 depletion. TAH18 is essential for viability. A previous study has identified two temperature-sensitive (ts) mutant alleles of TAH18, tah18-5H8, and tah18-5I5 (34). We characterized the original tah18 ts strains on HU-containing plates and found that tah18-5I5 was hypersensitive to HU at the permissive temperature 25 °C (Fig. S1). Moreover, both tah18 ts mutants exhibited defects in cell-cycle progression upon release from α factor-mediated G1 arrest: a prolonged S phase for tah18-5I5 and a delay at the G1/S boundary for tah18-5H8 (Fig. S2).
HU sensitivity and defects in G1/S transition or S-phase progression are characteristics of mutants deficient in DNA replication or cellular dNTP pools. To investigate whether the RNR cluster formation is defective in the tah18 ts mutants, we used whole-cell EPR to measure and compare Y• contents in the mutants and their isogenic WT strain. The EPR spectra of these strains had a very high background (Fig. S3) relative to the W303 and S288C strains (11, 13) and thus impeded accurate quantitation. To circumvent this problem, we moved the two tah18 ts mutant alleles into the W303 strain background by crossings with a W303 WT parental strain six times. The outcrossed mutants retained the ts and cell-cycle delay phenotypes but no longer showed obvious HU sensitivity, suggesting that background mutations in the original strains contribute to the growth defect on HU. Interestingly, both the tah18-5I5 and tah18-5H8 mutants showed a delay in S-phase progression after being released from G1 (Fig. 6A). Moreover, both tah18-5I5 and tah18-5H8 enhance HU sensitivity of the GalRNR4 mutant in which the chromosomal RNR4 promoter was replaced by the glucose-repressible GAL1 promoter, on a glucose-containing plate (Fig. 6B), suggesting exacerbation of RNR deficiency when both Rnr4 (β′) and Tah18 are compromised. The synthetic growth defect between GalRNR4 and tah18 ts mutants is reminiscent of the synthetic defect observed between Δrnr4 and GalDRE2 (13).
Fig. 6.
The tah18 ts mutants exhibit S-phase defects and a concurrent decrease in Y• content and β protein levels. (A) The defect in S-phase progression of tah18 ts mutants. WT and tah18 mutant cells from log-phase cultures were synchronized in G1 at 25 °C before being shifted to 37 °C for 1 h and released into the cell cycle at 37 °C. Cells were taken at the indicated time points for analysis of DNA content by flow cytometry. (B) Synthetic growth defect in tah18 ts mutants and GalRNR4. Both the WT and tah18 ts mutant strains were grown in galactose-containing medium (GAL-on) to log phase at 23 °C. Tenfold serial dilutions of each culture, starting at 106 cells, were dot-plated on glucose-containing plates (YPD, GAL off) or YPD containing 5 mM HU and were incubated at 23 °C for 2.5 d before being imaged. (C) Measurement of Y• in WT, tah18-5H8, and tah18-5I5 mutants by whole-cell EPR analysis. Cells from log-phase culture at 25 °C were washed in ice-cold PBS, resuspended in PBS with 30% glycerol at a density of ∼6.5 × 109 cells/mL, and packed into EPR tubes. (D) Measurement of ββ′ activity by CDP reduction in permeabilized cells of WT, tah18-5H8, and tah18-5I5 mutant strains in the presence of an excess of α. The ββ′ activities for WT, tah18-5H8, and tah18-5I5 are 0.48, 0.18, and 0.20 nmol dCDP/min in OD600 cells, respectively. (E) Comparison of Rnr2 protein levels in WT, tah18-5H8, and tah18-5I5 mutant cells grown at 25 °C. The protein blot was probed with anti-Rnr2 and stained with amido black as a control for loading. Relative Rnr2 signals were quantified. (F) Comparison of Rnr2 protein levels in WT and tah18-5H8 cells at 37 °C. Cells were grown to log phase at 25 °C, shifted to 37 °C at time 0, and harvested at the indicated time points for protein extraction and Western blotting. (G) Measurement of ββ′ activities by CDP reduction in permeabilized WT and tah18-5H8 cells 3 h after cells were shifted to 37 °C, in the presence of an excess of α. The ββ′ activities for WT and tah18-5H8 are 0.34 and 0.04 nmol dCDP/min in⋅OD600 cells, respectively.
Concurrent Decreases in ββ′ Activity and Proteins Levels in the tah18 ts Mutants.
As anticipated, the outcrossed tah18 ts mutants have a much cleaner EPR background and thus allow quantitative comparison of the Y• content in WT and mutants. Both tah18-5I5 and tah18-5H8 mutants have ∼50% of Y• content seen in the WT strain even at the permissive temperature 25 °C (Fig. 6C). Consistent with the low Y• levels, the activities of ββ′ of the two tah18 ts mutant cells were ∼40% of activities of the WT cells when normalized by cell numbers (Fig. 6D). However, when probing for β protein, we found that the two tah18 ts mutants also had a much lower β levels (Fig. 6E). The protein levels of β in the tah18-5H8 mutant exhibited a further and dramatic decline when the cells were shifted from 25 °C to the nonpermissive temperature 37 °C, dropping to ∼20% of the WT levels after 2 h at 37 °C and becoming undetectable after 4 h (Fig. 6F). Consistent with the decrease in β protein levels, RNR activity of tah18-5H8 dropped to 10% of the WT levels 3 h after being shifted to 37 °C (Fig. 6G). Thus, as in Dre2-depleted cells, the concurrent decrease in protein levels and activity of ββ′ complicated the assessment of the effect of inactivating Tah18 on RNR.
Inactivation of Tah18 at Nonpermissive Temperature Impairs Formation of Y• and Reconstitution of ββ′ Activity upon Induction of β′ in GalRNR4 Cells.
To circumvent the effect of Tah18 inactivation on β protein stability, we searched for conditions under which the β protein level is constant and not rate-limiting for measurement of Y• content and ββ′ activity. We have shown previously that induction of β′ in GalRNR4 cells leads to rapid and efficient FeIII2-Y• formation up to fourfold of the level in WT cells (13). GalRNR4 cells under GAL-repressed conditions accumulate five- to 10-fold more β protein than WT cells because activation of the Mec1-Rad53-Dun1 checkpoint leads to the removal of the transcriptional repressor Crt1. Upon induction of β′, formation of the active ββ′ and replenishment of cellular dNTP pools gradually diminishes the checkpoint signaling, and the levels of both β and Y• eventually return to those of WT cells because of Crt1-mediated negative feedback regulation (13, 16).
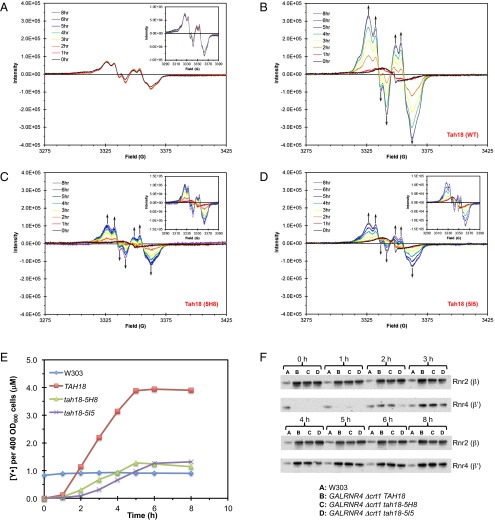
We capitalized on the inducible GalRNR4 system and the conditional tah18 ts mutant alleles to investigate whether Tah18 is required for the Y• cofactor formation upon β′ induction by generating GalRNR4 tah18 double mutants. To remove Crt1-mediated negative feedback, we deleted CRT1 in GalRNR4 TAH18 (expressing WT Tah18) and GalRNR4 tah18-ts strains so that β is constitutively overexpressed and consequently is not rate-limiting for reconstitution of Y• and ββ′ activity upon induction of β′. Both tah18-5H8 and tah18-5I5 mutants exhibited lower Tah18 protein levels that dwindled quickly when shifted to the nonpermissive temperature 30 °C (Fig. S4). As expected, induction of β′ at 30 °C in the GalRNR4 Δcrt1 TAH18 cells led to a time-dependent increase of Y• signal, reaching a plateau that was approximately fourfold higher than that of a WT W303 strain (Fig. 7 A, B, and E). In contrast, formation of Y• signal in GalRNR4 Δcrt1 tah18-5H8 cells (Fig. 7C) and GalRNR4 Δcrt1 tah18-5I5 cells (Fig. 7D) upon β′ induction at 30 °C occurred at a much slower pace and reached a plateau that was only 25% of the level in GalRNR4 Δcrt1 TAH18 cells (Fig. 7E).
Fig. 7.
Inactivation of Tah18 impairs reconstitution of ββ′ activity upon induction of β′ in Δcrt1 GalRNR4 cells. (A–D) EPR spectra showing changes in Y• content at different time points after β′ induction in TAH18 WT cells (B), in ts mutants tah18-5H8 (C), and in tah18-5I5 cells (D), all in the GalRNR4 Δcrt1 background. EPR spectra of a WT W303 strain (A) are shown as controls. (E) Comparison of changes in Y• content over an 8-h time course of β′ induction. Signals of Y• were normalized against cell number (OD600) at different time points. The Y• level remained unchanged in W303, which has RNR4 under its endogenous promoter. (F) Comparison of Rnr2 (β) and Rnr4 (β′) protein levels during the time course of β′ induction. Western blots show constitutive and comparable Rnr2 (β) levels and induction of Rnr4 (β′) at different time points in TAH18, tah18-5H8, and tah18-5I5 cells, all in the GalRNR4Δcrt1 background.
The β protein of the GalRNR4 Δcrt1 TAH18, GalRNR4 Δcrt1 tah18-5H8, and GalRNR4 Δcrt1 tah18-5I5 cells remained at a constitutive level that is much higher relative to the W303 WT strain over the time course of the experiments as a result of loss of Crt1-mediated transcriptional repression (Fig. 7F). Under such conditions, induced expression of β′ protein becomes the rate-limiting step in ββ′ formation and cluster assembly so that the level of Y• formation would be proportional to the level of β′ protein induced. The time course and levels of induced β′ protein were comparable in GalRNR4 Δcrt1 TAH18 cells and GalRNR4 Δcrt1 tah18-5H8 cells. In both strains, β′ levels became detectable at 2 h and plateaued at 5 h (Fig. 7E), as was consistent with the time course of Y• appearance and increase that reached a plateau at 5 h (Fig. 7E). On the other hand, β′ induction and Y• formation in GalRNR4 Δcrt1 tah18-5I5 cells occurred at a slower pace, becoming detectable at 3 h and reaching a plateau at 6 h (Fig. 7 E and F). Nevertheless, upon reaching the plateau (6 h for GalRNR4 Δcrt1 TAH18 and GalRNR4 Δcrt1 tah18-5H8 cells and 8 h for GalRNR4 Δcrt1 tah18-5I5 cells), the levels of β and β′ proteins were comparable in all three strains, but the Y• levels of GalRNR4 Δcrt1 tah18-5H8 cells and GalRNR4 Δcrt1 tah18-5I5 cells were only ∼25% of that of GalRNR4 Δcrt1 TAH18 cells. As we have shown previously (13), there is a good correlation between the Y• signal determined by EPR and the ββ′ activity measured by the permeabilized cell-based RNR activity assay in these strains (e.g., ββ′ activities at 2 h after β′ induction are shown in Fig. S5). Thus these findings strongly support our model that Tah18 is required for de novo formation of the FeIII2-Y• cofactor in ββ′.
Discussion
In this work, we demonstrated that Dre2–Tah18, a protein complex recently identified as a donor of reducing equivalents to the CIA machinery (33), also plays a critical role in formation of the FeIII2-Y• cofactor in RNR. Our efforts to determine the contribution of Dre2–Tah18 to RNR function were complicated by the decrease in ββ′ levels in dre2 and tah18 mutant cells. Our studies to understand the molecular basis for the decrease in ββ′ associated with Dre2–Tah18 inactivation thus have unexpectedly unveiled important regulatory mechanisms linking RNR stability with iron limitation and activation of the DNA-damage checkpoint.
First, we found that depletion of Dre2 induces CTH2 transcription in an Aft1/Aft2-dependent manner. Previous studies have suggested that Aft1/2 sense cellular iron levels by responding to deficiency in the mitochondrial iron-sulfur cluster (ISC) assembly process (50). Because mitochondrial ISC is not affected by Dre2 deficiency (51), it was unclear how Aft1/Aft2 became activated in Dre2-depleted cells. One possible explanation is based on our model that Dre2–Tah18 supplies electrons to Grx3/Grx4 for their function in the delivery of iron for the assembly of all iron-requiring cofactors, including those in the mitochondria (Fig. 1B). This notion is supported by synthetic lethality between grx3/4 and dre2 mutants (13) and by interactions between Dre2 and Grx3 in both yeast and human (52, 53). Moreover, we show that the CTH2 transcript is induced to a much greater extent in the grx3/4 mutant than in GalDRE2 mutant (Fig. 3A). Thus, it is possible that Dre2 depletion may cause deficiency in Grx3/4 activity, which could be sensed directly or indirectly by Aft1/Aft2, leading to transcriptional induction of CTH2. We noted that, unlike CTH2, another member of the iron regulon FET3 is induced in the grx3/4 but not much in GalDRE2 mutant, perhaps reflecting a differential degree of activation of Aft1/Aft2 in these mutants.
Second, we found that Dre2-depleted cells have an activated DNA-damage checkpoint resulting in transcriptional induction of RNR3 as well as RNR2/RNR4 through phosphorylation-mediated removal of repressor Crt1 from its target promoters. Thus, the apparent static level of ββ′ would result from the opposing effects of checkpoint-mediated transcriptional induction and Cth1/Cth2-mediated mRNA turnover of RNR2/RNR4. Consistent with this notion, Dre2-depleted cells have higher Rnr3 levels but lower ββ′ levels than WT cells. Cth1/Cth2 promote mRNA turnover not only of RNR2/RNR4 but also of WTM1, which acts to prevent nuclear release of ββ′ and its colocalization with α (21, 22). The apparent paradoxical down-regulation of both ββ′ and its negative regulator Wtm1 suggests that, in an effort to optimize the use of limited iron, yeast cells prioritize nucleus-to-cytoplasm redistribution and iron loading of existing ββ′ proteins over the synthesis of more apo proteins.
Third, the findings in GalDRE2 and Δaft1 mutants of RNR3 induction and Dif1 phosphorylation, two downstream events mediated by checkpoint kinase Dun1, indicate that the Mec1–Rad53–Dun1 checkpoint cascade can be activated in mutants defective in Fe-S cluster synthesis or cellular iron homeostasis. Fe-S cluster–binding domains have been found in an increasing number of nuclear proteins involved in DNA replication and repair including DNA primase, DNA helicases, and DNA polymerases (54). The importance of Dre2–Tah18 in DNA replication also was supported by the synthetic lethality between a ts mutant of POL3 encoding the Fe-S cluster containing DNA polymerase δ and dre2 and tah18 mutant alleles (55). Our findings show that the Dre2–Tah18 complex is required for assembly of the di-iron enzyme RNR cluster and for cellular supplies of dNTP. As such, deficiencies in the CIA pathway or proper distribution of intracellular iron utilization would impact many aspects of DNA replication and repair directly, leading to checkpoint activation.
Our finding that removal of Cth1/Cth2 only partially restores the decrease of ββ′ levels in Dre2-depleted cells suggests additional, unidentified regulatory mechanism(s), likely instability of the apo-ββ′ proteins. Therefore, our focus was to identify strains with increased ββ′ levels so that cofactor FeIII2-Y• could be monitored in cells lacking Dre2 and Tah18. We achieved this goal by increasing intracellular manganese levels in the conditional strain GalDRE2 via Δpmr1 and by keeping β constitutively overexpressed in the GalRNR4 tah18 ts mutant via Δcrt1. The results of our studies using these strategies strongly support the requirement of Dre2–Tah18 in RNR cluster assembly, either by delivering the obligatory reducing equivalent for RNR cluster formation in β (Eq. 1) or by being involved indirectly in iron delivery. The finding of coimmunoprecipitation between Dre2 and Rnr2 (β) (Fig. 5D) is consistent with the proposed role of Dre2 in electron delivery. Because Dre2 also has been shown to interact with Grx3/Grx4, we further postulate that Dre2–Tah18 might provide the reducing equivalents to allow Fe2+ transfer from the [2Fe2S]-(GSH)2 cluster at the Grx3/4 dimer interface to apo-ββ′.
The active cluster in both the di-iron– and Fe-S cluster–requiring proteins can form by self-assembly with varying degrees of efficiency in vitro (28, 56). Both require carefully controlled delivery of reducing equivalents. Thus, in both cases biosynthesis and perhaps maintenance pathways may have evolved to ensure highly efficient construction of an essential cofactor. The central role of Dre2–Tah18 in the assembly of the Fe-S cluster in many cytosolic and nuclear proteins, including enzymes involved in DNA replication and repair, complicated experimental designs to obtain evidence for our model (Fig. 1B). Our results together with previous studies by the Lill group (25) suggest that the CIA machinery and RNR cluster assembly share the same sources of iron, in the form of [2Fe2S]-(GSH)2 from Grx3/Grx4, and also the same source of reducing equivalents from Dre2–Tah18. The point of bifurcation of the CIA and the RNR cluster assembly processes remains to be unraveled.
The pathway for FeIII2-Y• assembly is likely conserved between S. cerevisiae and human despite the differences in structures of the two β2 subunits (heterodimer versus homodimer) (1). The mammalian Grx3, PICOT, recently has been shown to be required for multiple pathways in iron homeostasis, including biogenesis of the Fe-S cluster and hemoglobin maturation (31). The Dre2–Tah18 complex may function as the cytosolic equivalent of the mitochondrial Fd-Fre pair Yah1-Arh1 to deliver electrons to multiple and divergent pathways of iron cofactor biogenesis. The human counterparts of Dre2 and Tah18, CIAPIN1 and NDOR1, respectively, recently have been shown to function in their place in yeast cells in Fe-S cluster assembly in Leu1, a substrate of the CIA machinery (33, 51). It remains to be determined if CIAPIN1-NDOR1 are involved in the assembly of the FeIII2-Y• cluster of RNR and the Fe-S cluster of CIA in mammalian cells. Discovering the cellular machinery required for FeIII2-Y• assembly and repair would provide still another tier to the multilayered RNR regulation and would provide new insights into development of RNR-targeted therapeutics.
Experimental Procedures
Yeast Strains, Plasmids, and Growth Conditions.
Yeast strains and plasmids used in this study are listed in Tables S1 and S2, respectively. Growth of yeast strains and genetic manipulations were as described (57). GalDRE2 and GalRNR4 strains were constructed by replacing sequences between nucleotides −50 and −1 of each endogenous promoter with the GAL1 promoter (58). AXY1664, AXY1668, and AXY1696 were constructed by integrating an N-terminally Flag-tagged TAH18 (WT) or tah18-5H8 and tah18-5I5 mutants into the tah18::KanMX4 locus. Cell-cycle synchronization and FACS analysis were as described (59).
Protein Analysis.
Yeast protein extracts were prepared by trichloroacetic acid precipitation (10) or alkaline treatment (60) for Western blotting and by lysis buffer B for immunoprecipitation (20). Antibodies used for immunoprecipitation and Western blotting were anti-Rnr2/3/4, as previously described (61), anti-G6PDH (Sigma-Aldrich), and monoclonal 9E10 (anti-Myc; Covance). Signals from protein blots were recorded and quantitated using ChemiDoc MP (Bio-Rad).
RNA Extraction, Reverse Transcription, and RT-qPCR.
Total RNA was extracted from 2 × 108 cells by using a hot-phenol method (62). Total RNA (10 μg) was treated with 10 units of RNase-free DNase I (New England Biolabs) for 30 min at 37 °C to remove contaminating DNA. First-strand cDNA synthesis was carried out by M-MuLV reverse transcriptase (New England Biolabs) on aliquots of 1 μg RNA with a random primer mix. The single-stranded cDNA products were used in qPCR on a Bio-Rad CFX96 real-time PCR detection system based on SYBR Green fluorescence. Sequences of oligo pairs are listed in Table S3.
Whole-Cell EPR Spectroscopy and ββ′ Activity Assays in Permeabilized Cells.
Whole-cell EPR spectroscopy, preparation of permeabilized yeast cells, and measurement of RNR activity were performed as described previously (13).
Supplementary Material
Acknowledgments
We thank Drs. A. Dancis, S. Puig, R. Lill, and L. Vernis for sharing of yeast strains, antibodies, and plasmids. This work was supported by National Institutes of Health Grants R01GM29595 (to J.S.), R01CA125574 (to M.H.), and R01GM81393 (to J.S. and M.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405204111/-/DCSupplemental.
References
- 1.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Stubbe J, van Der Donk WA. Protein Radicals in Enzyme Catalysis. Chem Rev. 1998;98(2):705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 3.Hofer A, Crona M, Logan DT, Sjöberg BM. DNA building blocks: Keeping control of manufacture. Crit Rev Biochem Mol Biol. 2012;47(1):50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J Biol Chem. 2006;281(38):27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine-5′-diphosphate inhibit ribonucleotide reductases. Proc Natl Acad Sci USA. 2007;104(36):14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairman JW, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18(3):316–322. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: Refinements and consequences. Biochemistry. 2003;42(6):1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- 8.Minnihan EC, Nocera DG, Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc Chem Res. 2013;46(11):2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stubbe J, Nocera DG, Yee CS, Chang MC. Radical initiation in the class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103(6):2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 10.An X, Zhang Z, Yang K, Huang M. Cotransport of the heterodimeric small subunit of the Saccharomyces cerevisiae ribonucleotide reductase between the nucleus and the cytoplasm. Genetics. 2006;173(1):63–73. doi: 10.1534/genetics.105.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlstein DL, et al. The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry. 2005;44(46):15366–15377. doi: 10.1021/bi051616+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voegtli WC, Ge J, Perlstein DL, Stubbe J, Rosenzweig AC. Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer. Proc Natl Acad Sci USA. 2001;98(18):10073–10078. doi: 10.1073/pnas.181336398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: Requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins. J Biol Chem. 2011;286(48):41499–41509. doi: 10.1074/jbc.M111.294074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortigosa AD, et al. Determination of the in vivo stoichiometry of tyrosyl radical per betabeta’ in Saccharomyces cerevisiae ribonucleotide reductase. Biochemistry. 2006;45(40):12282–12294. doi: 10.1021/bi0610404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao R, et al. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci USA. 2003;100(11):6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang MX, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94(5):595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20(13):3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2(3):329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 19.Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell. 2008;32(1):70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Huang M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol. 2008;28(23):7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YD, Elledge SJ. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 2006;20(3):334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, et al. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc Natl Acad Sci USA. 2006;103(5):1422–1427. doi: 10.1073/pnas.0510516103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanvisens N, Bañó MC, Huang M, Puig S. Regulation of ribonucleotide reductase in response to iron deficiency. Mol Cell. 2011;44(5):759–769. doi: 10.1016/j.molcel.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan CD, Kaplan J. Iron acquisition and transcriptional regulation. Chem Rev. 2009;109(10):4536–4552. doi: 10.1021/cr9001676. [DOI] [PubMed] [Google Scholar]
- 25.Mühlenhoff U, et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12(4):373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollinger JM, Jr, et al. Mechanism of assembly of the tyrosyl radical-dinuclear iron cluster cofactor of ribonucleotide reductase. Science. 1991;253(5017):292–298. doi: 10.1126/science.1650033. [DOI] [PubMed] [Google Scholar]
- 27.Atkin CL, Thelander L, Reichard P, Lang G. Iron and free radical in ribonucleotide reductase. Exchange of iron and Mössbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J Biol Chem. 1973;248(21):7464–7472. [PubMed] [Google Scholar]
- 28.Cotruvo JA, Stubbe J. Class I ribonucleotide reductases: Metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem. 2011;80:733–767. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y, Wong J, Lovejoy DB, Kalinowski DS, Richardson DR. Chelators at the cancer coalface: Desferrioxamine to Triapine and beyond. Clin Cancer Res. 2006;12(23):6876–6883. doi: 10.1158/1078-0432.CCR-06-1954. [DOI] [PubMed] [Google Scholar]
- 30.Aye Y, Long MJ, Stubbe J. Mechanistic studies of semicarbazone triapine targeting human ribonucleotide reductase in vitro and in mammalian cells: Tyrosyl radical quenching not involving reactive oxygen species. J Biol Chem. 2012;287(42):35768–35778. doi: 10.1074/jbc.M112.396911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haunhorst P, et al. Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol Biol Cell. 2013;24(12):1895–1903. doi: 10.1091/mbc.E12-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Mapolelo DT, Randeniya S, Johnson MK, Outten CE. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry. 2012;51(8):1687–1696. doi: 10.1021/bi2019089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Netz DJ, et al. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat Chem Biol. 2010;6(10):758–765. doi: 10.1038/nchembio.432. [DOI] [PubMed] [Google Scholar]
- 34.Vernis L, et al. A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS ONE. 2009;4(2):e4376. doi: 10.1371/journal.pone.0004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C-H, Jiang W, Krebs C, Stubbe J. YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry. 2007;46(41):11577–11588. doi: 10.1021/bi7012454. [DOI] [PubMed] [Google Scholar]
- 36.Lange H, Kaut A, Kispal G, Lill R. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc Natl Acad Sci USA. 2000;97(3):1050–1055. doi: 10.1073/pnas.97.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Saxena S, Pain D, Dancis A. Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J Biol Chem. 2001;276(2):1503–1509. doi: 10.1074/jbc.M007198200. [DOI] [PubMed] [Google Scholar]
- 38.Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120(1):99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 39.Martínez-Pastor M, Vergara SV, Puig S, Thiele DJ. Negative feedback regulation of the yeast CTH1 and CTH2 mRNA binding proteins is required for adaptation to iron deficiency and iron supplementation. Mol Cell Biol. 2013;33(11):2178–2187. doi: 10.1128/MCB.01458-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puig S, Vergara SV, Thiele DJ. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008;7(6):555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ojeda L, et al. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281(26):17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 42.Courel M, Lallet S, Camadro JM, Blaiseau PL. Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol Cell Biol. 2005;25(15):6760–6771. doi: 10.1128/MCB.25.15.6760-6771.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tkach JM, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14(9):966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4(5):740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 45.Atta M, Nordlund P, Aberg A, Eklund H, Fontecave M. Substitution of manganese for iron in ribonucleotide reductase from Escherichia coli. Spectroscopic and crystallographic characterization. J Biol Chem. 1992;267(29):20682–20688. [PubMed] [Google Scholar]
- 46.Culotta VC, Yang M, Hall MD. Manganese transport and trafficking: Lessons learned from Saccharomyces cerevisiae. Eukaryot Cell. 2005;4(7):1159–1165. doi: 10.1128/EC.4.7.1159-1165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNaughton RL, et al. Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc Natl Acad Sci USA. 2010;107(35):15335–15339. doi: 10.1073/pnas.1009648107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddi AR, Culotta VC. Regulation of manganese antioxidants by nutrient sensing pathways in Saccharomyces cerevisiae. Genetics. 2011;189(4):1261–1270. doi: 10.1534/genetics.111.134007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu XF, Culotta VC. The requirement for yeast superoxide dismutase is bypassed through mutations in BSD2, a novel metal homeostasis gene. Mol Cell Biol. 1994;14(11):7037–7045. doi: 10.1128/mcb.14.11.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen OS, Hemenway S, Kaplan J. Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron export: Evidence that Yfh1p affects Fe-S cluster synthesis. Proc Natl Acad Sci USA. 2002;99(19):12321–12326. doi: 10.1073/pnas.192449599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol Cell Biol. 2008;28(18):5569–5582. doi: 10.1128/MCB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito Y, et al. PICOT is a molecule which binds to anamorsin. Biochem Biophys Res Commun. 2011;408(2):329–333. doi: 10.1016/j.bbrc.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 53.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320(5882):1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 54.White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Curr Opin Struct Biol. 2012;22(1):94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Chanet R, Heude M. Characterization of mutations that are synthetic lethal with pol3-13, a mutated allele of DNA polymerase delta in Saccharomyces cerevisiae. Curr Genet. 2003;43(5):337–350. doi: 10.1007/s00294-003-0407-2. [DOI] [PubMed] [Google Scholar]
- 56.Malkin R, Rabinowitz JC. The reconstitution of clostridial ferredoxin. Biochem Biophys Res Commun. 1966;23(6):822–827. doi: 10.1016/0006-291x(66)90561-4. [DOI] [PubMed] [Google Scholar]
- 57.Burke D, Sawson D, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2000. [Google Scholar]
- 58.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 59.Gasch AP, et al. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12(10):2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16(9):857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen HH, Ge J, Perlstein DL, Stubbe J. Purification of ribonucleotide reductase subunits Y1, Y2, Y3, and Y4 from yeast: Y4 plays a key role in diiron cluster assembly. Proc Natl Acad Sci USA. 1999;96(22):12339–12344. doi: 10.1073/pnas.96.22.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.