Abstract
Rice (Oryza sativa) is one of the most important cereal grains in the world today and serves as a staple food source for more than half of the world’s population. Research into when, where, and how rice was brought into cultivation and eventually domesticated, along with its development into a staple food source, is thus essential. These questions have been a point of nearly continuous research in both archaeology and genetics, and new information has continually come to light as theory, data acquisition, and analytical techniques have advanced over time. Here, we review the broad history of our scientific understanding of the rice domestication process from both an archaeological and genetic perspective and examine in detail the information that has come to light in both of these fields in the last 10 y. Current findings from genetics and archaeology are consistent with the domestication of O. sativa japonica in the Yangtze River valley of southern China. Interestingly, although it appears rice was cultivated in the area by as early 8000 BP, the key domestication trait of nonshattering was not fixed for another 1,000 y or perhaps longer. Rice was also cultivated in India as early as 5000 BP, but the domesticated indica subspecies currently appears to be a product of the introgression of favorable alleles from japonica. These findings are reshaping our understanding of rice domestication and also have implications for understanding the complex evolutionary process of plant domestication.
Keywords: Oryza rufipogon, Oryza nivara, domestication gene
Archaeological Evidence for Rice Domestication and Development of Rice Agriculture
Given the broad importance of domesticated rice as a food source, its origin and development from the wild species Oryza rufipogon have driven much of the interest and research in archaeology in East and South Asia during the last century. An early focus was the geographic origin of domesticated rice. Several areas were proposed, including India (1), South China (2), the Yangtze River area in China (3), the so-called “belt region” with its great diversity of Oryza species along the southern slope of the Himalayas (4), and coastal swamp habitats in Southeast Asia (5). However, there were comparatively few serious studies on the chronology of rice domestication, which was presumed to have occurred about 10,000 y ago, probably because previous research showed that the origins of agriculture in the other parts of world, such as western Asia, took place ∼10,000 y ago (6). During the last 10 y, research into rice origins and dispersal has benefitted, as has domestication research in other regions of the world (papers in this volume), from the generation of a considerable amount of new empirical data from archaeological sites, itself driven by the application of new methodological procedures that can better detect Oryza and follow its early history. For example, the recent widespread use of flotation (7–9) in East and South Asia has resulted in the retrieval of rich macrobotanical remains of rice grains and husks from some important sites (10–15). Phytolith analysis has also proved useful for identifying microscopic remains of plants to the genus level, including in very early (e.g., Pleistocene) deposits where grains and husks are not present. This advance has allowed identification of presumably domesticated, or at least cultivated, rice occurring beyond the areas of wild Oryza distribution with enough accuracy to separate the two major subspecies of Oryza sativa (indica and japonica) from each other (16–18).
Many scholars now accept that the Yangtze River area in China is the place where rice was originally domesticated as a consequence of these newer findings (11, 19–23). However, as discussed elsewhere in this paper, whether indica and japonica had single or multiple origins is a question under active research in the genetic and archaeological arenas, and there is little consensus of opinion with regard to the available genetic evidence (24). The resolution of this question depends to a large extent on archaeological research, which has revealed separate cultivation origins for indica and japonica. Current arguments in archaeology are also focused on fundamental questions of when rice cultivation began in China and how long the domestication process took. Cultivation and subsequent domestication are increasingly seen as being considerably more separated in time than once thought, as a horizon of what’s termed “predomestication cultivation” sometimes lasting thousands of years is being increasingly documented in the Old World (see Introduction in this volume), and this also appears true for rice. Moreover, recent studies suggest that there is no clear boundary line between hunting-gathering and agriculture and that the transformation between the two is not a revolutionary change but rather a slow process of qualitative and quantitative shifts that may have taken thousands of years (6, 11). These questions, in turn, are related to theories of agricultural origins in China and around the world that are currently of great interest in anthropology and archaeology (25). Accordingly, we now focus on new archaeobotanical data bearing on these issues and the subsequent spread of rice into Korean Peninsula, the Japanese archipelago, and India.
Archaeobotanical Data from China.
In recent years, flotation of archaeological sediments for recovery of macrobotanical remains of plants has been carried out on more than 100 archaeological sites throughout China, and numerous charred plant remains have been retrieved for study (Fig. 1). They are from a variety of crop species including rice (10, 11). The earliest rice remains recovered in China have been reported from three archaeological sites: Xianrendong and Diaotonghuan in Jiangxi Province (26) and Shangshan in Zhejiang Province (27). The cultural remains of these sites are dated to around 10,000 BP (all dates are in calibrated years), although it should be noted that the cultural deposits in the Xianrendong cave site have a very long sequence; the lower layer was recently dated to about 20,000 BP (28). This information suggests that a new date might also be needed for the rice remains found in the upper layers of this site.
Fig. 1.
Locations of the sites with early rice remains in the Yangtze Rivers areas: (1) Yuchanyan; (2) Pengtoushan; (3) Bashidang; (4) Jiahu; (5) Xianrendong/Diaotonghuan; (6) Shangshan; (7) Kuahuqiao; (8) Xiaohuangshan; (9) Hemudu/Tianluoshan.
Shangshan is an early Neolithic settlement with house and pit features, and artifactual remains that include pottery and stone tools. The cultural assemblage recorded can be divided into two periods: the Shangshan culture dated to ∼11,000–9000 BP, and the Kuahuqiao culture dated to 8000–7000 BP (29). Although more than 400 soil samples were floated, only 10 charred rice grains and a few rice spikelet bases were recovered. Most of these belong to the Kuahuqiao cultural horizon. A few rice grains were recovered from the Shangshan culture horizon. Rice remains were also found by other methods. For example, rice husks can be easily identified in the paste of pottery sherds, and they were commonly found in sherds dating to both of the two periods. Heaps of rice husks were also found in burnt soil blocks from the early period layers of the site. The combined evidence indicates that Shangshan people intensively exploited rice.
The Shangshan rice was believed to be in an early stage of domestication, based on grain size and morphological characteristics (e.g., length-to-width ratios) (27). However, others note that grain size and shape may exhibit considerable variability in wild and domesticated populations, some of which is probably influenced by plant responses to environmental factors and therefore may not be a reliable indicator of early domestication (30, 31). In light of the abundance of rice husks in the pottery and other site contexts, it seems that Shangshan people had a high demand for rice, and the cultivation of rice may have begun at that time (11). Further work is required to unequivocally establish rice cultivation; if it occurred, it could be interpreted as being primarily an attempt to improve the yield of wild rice.
In China, the time around 8,000 y ago appears to have been critical for agricultural origins, not only for rice in the Yangtze River area, but also for millet agriculture in the Yellow River system of northern China. For example, several archaeological sites exhibiting the characteristics of early rice farming have all been dated to around 8000 BP. They are Pengtoushan and Bashidang in Hunan Province (32), Kuahiao and Xiaohuangshan in Zhejiang Province (33, 34), and Jiahu in Henan Province (35).
Jiahu was a permanent village dated by dozens of radiocarbon determinations to a period between 9000 and 7800 BP. Flotation work was carried out on a total of 125 soil samples, and a large number of charred plant remains were recovered, including several hundred rice grains (36). Other plant remains include soybean (Glycine sp.), water chestnut (Trapa sp.), lotus roots (Nelumbo nucifera), and acorn (Quecus sp.). Zhao’s research on the Jiahu rice indicates that it may well be domesticated, as its grain phenotypic characteristics, including size and shape, are much like modern domesticated rice. A discussion has ensued about these characteristics (37, 38). For example, it has been suggested that Jiahu rice might belong to a wild rice species because the grains are remarkably small (31). Alternatively, it has also been argued that the Jiahu rice grains are not small but characterized by a great variation in size, based on measurements of hundreds of rice grains recently recovered from Jiahu (38). Another factor that should be considered in establishing the status of the Jiahu rice is the abundance of weedy grasses, which may represent weeds of cultivation (e.g., Digitaria and Echinochloa spp.). Furthermore, the location of the Jiahu site is far from the natural distribution of wild rice today. All of these factors indicate that domesticated rice and rice agriculture were established at the site 8,000 y ago.
It should also be noted that the rice remains at Jiahu were accompanied by a large amount of wild food resources, such as lotus and water chestnut, along with fish bones and shells. Quantitative analysis of the plant remains showed that rice was not the dominant plant in the remains (36). It appears that rice did not play a dominant role in the subsistence of the Jiahu people and that the overall subsistence economy was a mixture of plant cultivation, fishing, and other wild resource procurement. This type of mixed subsistence is a pattern coming to light in other regions of the world where early agriculture was established.
The discovery of the Hemudu site in the 1970s resounded throughout the world. Because of the waterlogged condition of the site, organic materials were well preserved (39). A huge number of plant remains have been recovered, among which the most noticeable were rice. Some scholars accordingly suggested that the Hemudu people might have had a very mature agricultural economy based on rice (40). However, little was known about the phenotypic characteristics of the rice that could reveal if they were morphologically still wild or domesticated. Another unresolved problem was whether the rice was indeed the main food resource at Hemudu, especially considering that a significant number of other edible wild plant remains were found during the excavation.
The Tianluoshan site, discovered in 2004, afforded a chance to answer these questions. Its location is only 7 km from Hemudu, and the cultural deposits of the two sites are almost identical (41). A systematic sampling strategy was applied during the excavation to recover plant remains, including water screening and flotation. More than 200 soil samples have been processed thus far, and a tremendous number of plant remains have been found, including rice, water chestnut, acorns, bottle-gourd, Euryale ferox, Ziziphus jujube, Diospyros sp., and various weed seeds. The most important study of the Tianluoshan data are a systemic analysis of the rice spikelet bases carried out by Fuller et al. (42). The results show that the Tianluoshan rice consists of a high proportion of shattering, WT spikelets, which suggests that the process of rice domestication was not yet complete in the Hemudu period, i.e., sometime about 6500 BP. However, the quantitative analysis of the plant remains suggested that rice was one of the most important food resources at Tianluoshan and that the Tianluoshan people probably engaged in rice farming activities. Nonetheless, rice farming did not replace hunting-gathering as the dominant economy of the Tianluoshan residents or even probably the Hemudu culture. Wild resources such as acorns were still important foods at that time.
Liu et al. (38) published additional research that analyzed the morphological characteristics of both grains and spikelet bases from early occupations at Shangshan, Kuahuqiao, and Hemudu. They also presented new measurements of rice grains recovered from Jiahu. They argued based on all of the data that the process of rice domestication began during the early Holocene at about 9000 BP with the “management of phenotypically wild plants.” Fuller et al. (37) counter argued that the new metric data of Liu et al. were still insufficient to determine the wild or domesticated status for rice remains from the early sites. Analyses of spikelet bases unearthed from the early sites in the lower Yangtze River region, such as Kuahuqiao, Luojiajiao (Majiabang period), and Tianluoshan, show few signs of domestication.
Loss of shattering is a key characteristic of domesticated cereal crops that distinguishes them from their wild ancestors (31). Furthermore, domestication is a long and slow process in which the proportion of shattering forms should be high in the beginning and then decrease gradually until nonshattering types dominate populations. Before or during this transition, wild or cultivated rice that shattered at maturity may have been harvested at an immature stage to prevent resource loss. Therefore, the study and detection of “immature rice” is a strong conceptual advance and will become a key for understanding the process of rice domestication, as emphasized by Fuller. However, the accurate identification of immature rice is still at issue (43). Broadly speaking, the distinctive characteristics of immaturity, shattering, and nonshattering states based on spikelet bases has the potential to be reliable, whereas characters relating to grain morphometrics and their diagnostic power for domestication need further study.
As can be seen, major questions relating to when rice cultivation ensued and how it can be identified, together with how domestication should be determined, currently surround issues of rice cultivation in China. On the basis of current data, two significant patterns in agricultural origins and development can be identified. First, as shown by the percentages of nonshattering grains through time, the fixation of nonshattering grains under cultivation was a slow process that occurred over a few thousand years. The comprehensive study of the early sites (e.g., Shangshan) suggests that a predomestication cultivation horizon may have existed for rice before domestication and that cultivation probably began 9,000–8,000 y ago. Second, the transition to rice agriculture from hunting and gathering was not a clear-cut revolutionary change but a slower evolutionary process. During this long-term process, hunting and gathering gradually waned, whereas rice agriculture slowly achieved a dominant position and finally became the major subsistence practice. This slow process is a pattern emerging in many other areas of the world.
Archaeobotanical Data from Korea, Japan, and India.
Korea.
Oryza is not native to the Korean peninsula; thus, rice research focuses on diffusion from China and possible routes by which the plant may have spread. It was conventionally thought that rice farming spread from China to the Korean peninsula during the Early Mumun period (3400-2800 BP). Some studies suggested that rice may have arrived in Korea as early as the Chulmun period (7500-4000 BP) (13). Recent archaeobotanical studies have clarified this issue.
Macrobotanical evidence now indicates that Chulmun subsistence, especially the Middle-Late Chulmun, was clearly based on an agricultural economy. However, Chulmun agriculture was a dry-land farming system based not on rice, but apparently on millets, including both foxtail (Setaria italic) and broomcorn millet (Panicum miliaceum), legumes such as soybean (Glycine max), adzuki bean (Vigna angularis), and other crops (14). No Chulmun rice remains have been found through macrobotanical analysis. Nevertheless, rice presence is indicated by phytoliths found in pottery shards dating to the Chulmun period. Future work will address further aspects of rice at this site.
Analysis of plant remains confirmed that rice agriculture was indeed a major part of the subsistence economy of the Mumun period and also suggested that paddy rice farming occurred. This fact suggests that rice agriculture appeared in the Korean peninsula as an already developed rice farming system (13, 14), indicating that rice agriculture diffused into the Korean peninsula. The route of spread is still under active research and debate.
Japan.
As with the Korean peninsula, wild rice does not occur in Japan today and probably never did. It is therefore generally agreed that rice agriculture diffused to Japan, and it is thought this occurred during the Yayoi period, which began ∼2800 BP (44). Very similar to the situation in the Korean peninsula, early rice farming in Japan appears to have involved paddy rice farming. Although a highly productive rice agriculture probably began in the Yayoi period, it is increasingly likely that domesticated rice was introduced into the Japanese archipelago earlier, during the late Jomon period, some time around 4000 BP. Rice seed impressions on Jomon period pottery documented through scanning electronic microscopic study (45) appear to establish that domesticated rice was introduced into Japan before the Yayoi period, although it is not clear how large a part of the subsistence base rice was at that time. This rice appears to have been a part of a dry-land agriculture system, as other crops of dry-land farming have been found in sites dating to the late and middle Jomon, including various millets such as barnyard millet (Echinochloa utiliz) and legumes (soybean and adzuki beans).
It is interesting to see the similarity between the Korean peninsula and Japan regarding the early development of agriculture. First, the beginning of agriculture in these two areas was characterized by a dry-land farming system with the major crops being millets and beans. Second, rice agriculture diffused to these two areas at a relatively late time, but with a paddy rice farming system. This new farming system quickly replaced the indigenous dry-land farming system. It can be suggested this change was one of the reasons for the cultural transitions that took place; i.e., from Mumun to Chulmunin the Korean peninsula and Jomon to Yayoi in Japan. Third, sporadic rice remains were also found in the periods before the occurrence of paddy rice farming system in these two areas. This early arriving rice presumably had little impact on the indigenous dry-land farming system, but more evidence is needed to study its significance.
India.
The prehistory of indica and japonica in India presents one of the more interesting stories of domestication, long distance spread, and subsequent interactions of cultivars within a single genus of plants. Both O. rufipogon and another close wild relative, Oryza nivara, are native to India and well distributed there today, and probably were present since the Pleistocene (46). The country has a number of long archaeological sequences with good plant records including those in the Ganges River valley in the north where rice, likely wild O. rufipogon and O. nivara, is documented by 9000 BP (46, 47). It is now recognized that the Indian subcontinent was probably an independent center of agricultural origins with important regions in the Ganges plain and to the south on the Deccan Plateau. Native plants that were cultivated or domesticated before crops were introduced from elsewhere include mung bean and small-seeded grasses, among others (47). The question of an origin of indica rice in India has been under active discussion, and recent research has done much to clarify and resolve the issue. It now appears that an independent origin of cultivation of ancestral indica or proto-indica rice took place in the Ganges plains, but that the plant was completely domesticated only when domesticated japonica arrived from China and hybridized with it about 4,000 y ago (47). Indica consumption began early, by 8400 BP, and the plant was cultivated and appears to have been a staple food by 5000 BP (47).
Summary.
A subject that is as important as the origins and spread of domesticated Oryza and that is increasingly informed by multiple empirical databases from genetics and archaeology has naturally given rise to a number of controversies. Points of disagreement include how and when cultivation began and domesticated varieties emerged, how this can be documented in rice’s major center of origin in China, and when archaeological rice remains can be associated with an economy partly or fully dependent on rice as a staple food. These issues have done much to inspire and advance methodological aspects of rice domestication research and make it ever clearer that establishing accurate and feasible criteria for distinguishing domesticated and wild species is of prime importance in agricultural origins research.
Methodological techniques will continue to evolve. For example, recent work by Zhao and Gu (48) demonstrates that identification of spikelet bases has its limitations, despite the fact that pertinent qualitative features have a clear tendency of polarization between wild and domesticated rice. Wild rice spikelet bases have a shallow and round abscission scar and a small distinct vascular pore, whereas domesticated spikelet bases display irregular-shaped and deeper scars. Although this would seem to make them easy to distinguish, the irregular morphology of domesticated rice spikelet bases and their small size can make microscopic measurements very difficult. Applying qualitative criteria alone may introduce perceptual differences and variability in how different scholars evaluate the same features and make repeatability and comparisons between sites difficult. Ongoing examinations of other traits such as rice embryo characteristics (48) may provide additional data on domestication that will independently, or in combination with other criteria, provide more precise identifications of wild and domesticated rice. Productive discussions of theoretical issues are also dependent on such advances.
Genetic Evidence for Rice Domestication
Just as physical remains provide evidence of the presence and transformation of wild rice into domesticated rice in the archaeological record, the genomes of wild and domesticated rice preserve a record of the evolutionary forces they have been subject to over time. These records include information ranging from the possible geographical origin(s) of extant domesticated rice as a whole to the origin and assembly of individual alleles that combine to create the domestication phenotype (e.g., loss of shattering, increase in grain size and number, change in grain color and plant growth stature). Combined with complimentary information from archaeology and ethnobotany, this information can be essential to understanding the process of rice domestication. What has genetic information from wild and domesticated rice yielded to date? Lately, the answer seems to be that genetics has told us a constantly changing story. Although this is not necessarily surprising, given the rapid advances in genetic technology over the last 20 y, it has had the effect of both expanding our understanding of the origin and evolution of domesticated rice and stirring strong feelings about the history of one of the world's most important food crops.
The two major Oryza sativa subspecies are differentiated by a number of morphological and physiological characters, along with a substantial (although incomplete) sterility barrier (49, 50). All genetic analyses have confirmed the distinctiveness of the subspecies and further confirmed the existence of recognized subgroups within these groups (i.e., temperate and tropical japonica along with aus) (51). The consistent genetic and phenotypic distinctiveness has long been considered an indicator that indica and japonica might have distinct origins. However, genetic distinctiveness alone is not enough to establish independent origins. Multiple origins of a single domesticated species is most convincingly demonstrated when varieties or subspecies within the species show genetic affiliations to existing wild populations with distinct geographic and genetic provenance. What, then, prevents us from quickly identifying the genetic and geographic source populations that gave rise to domesticated rice, given its obvious socio-economic importance and the extensive genetic resources for the Oryza system? After all, the likely geographic origin of the extant maize lineage was pinpointed more than 10 y ago (52, 53), whereas arguments for and against multiple origins of rice have appeared in the literature on a nearly annual basis.
There are three main complicating factors that have made the history of domesticated rice difficult to read based on the patterns in its genome. These factors include (i) a paucity of genetic markers; (ii) a paucity of samples from the wild relative of domesticated rice; and (iii) difficulty resolving the relationship between gene genealogies for domestication genes and gene genealogies for neutral genes. Below, we will describe the progress of rice domestication genetics with these three factors in mind and discuss how the field might be advanced in the future.
Neutral Markers.
Many authors have documented evidence consistent with independent origins of the japonica and indica subspecies based on molecular markers ranging from allozymes to retrotransposons (54–57). However, to a large extent, the recent exchange of papers identifying a single vs. multiple origins of domesticated Asian rice began in 2006, with the publication by Londo et al. showing that phylogeographic evidence was consistent with separate origins of indica and japonica (58). This paper surveyed only three sequenced loci, but its strength lay in the relatively broad sampling of both domesticated and wild Asian rice (203 O. sativa and 129 O. rufipogon), with a strong effort to sample across the range of the wild species. In particular, this paper included many field-collected samples of O. rufipogon from China; these samples were particularly important because they include one of the potential geographic origins of domesticated rice. The phylogenetic patterns detected in this paper suggested two likely origins of domesticated rice, with indica originating in eastern India and japonica originating in southern China.
The possibility that the O. sativa subspecies had unique origins was subsequently bolstered by the analysis of Caicedo et al. (59). Although this paper did not include a phylogeographical component (it was focused on the effects of selection on the patterns of genetic variation in domesticated rice), the large number of sequenced regions (111 gene fragments) provided a strongly supported phylogenetic tree indicating that indica and japonica were not just genetically distinct but also that each subspecies was more closely related to a separate set of O. rufipogon than to the other subspecies. In contrast to the Londo et al. analysis, however, this analysis included a relatively small number of O. rufipogon samples (21 total), the majority of which were from China or Nepal. Indeed, the phylogenetic tree presented in the paper would seem to indicate that all subspecies of domesticated Asian rice arose from wild populations similar to those found in modern-day China. Overall, the lack of samples from other countries makes it impossible to draw conclusions about the geographic origin of rice (and, indeed, the paper's authors do not try to). A complementary study published in the same year, using 22 sequence-based markers, also showed patterns consistent with independent origins of the two subspecies (60). The level of sampling, however, was still too low (at 30 samples) to pinpoint a possible geographical origin of either subspecies.
Shortly thereafter, an analysis using 60 microsatellites conducted by Gao and Innan suggested “nonindependent” origins of the two subspecies (61). The authors of this paper took the interesting approach of evaluating evidence for bottlenecks at corresponding loci in the genomes of the two subspecies. The logic was that the stochastic nature of a domestication bottleneck would reduce diversity at some neutral loci to a greater extent than other loci, but that levels of diversity at a given locus would not be correlated across subspecies if they were domesticated independently (assuming an absence of parallel selective pressures acting on these loci and limited gene flow between independent origins). The authors found a significant, positive correlation when they compared the subspecies, consistent with a single origin or extensive recent gene flow. The samples used in this study included 92 individuals, 35 of which were O. rufipogon, all sampled from within China (61, 62). Whether this wild reference population was the most appropriate comparison for indica varieties that may or may not have been domesticated in that geographical area is not addressed in the paper.
Domestication Genes.
This succession of papers coincided with the first set of domestication genes being cloned and characterized in Asian domesticated rice, both of which controlled shattering (sh4, qSH1) (63, 64). Although the qSH1 domestication allele was confined to a subset of japonica varieties, analyses of sh4 revealed that the mutations associated with the nonshattering phenotype had a single origin and that a single allele was now distributed across japonica and indica (despite the sterility barrier between them) (65, 66). The single origin of a gene underlying this major domestication trait prompted researchers to consider possibilities of interactive domestication scenarios such as the “snowballing model” and the “combination model” proposed by Sang and Ge (65, 67). These models attempt to reconcile the divergence at neutral loci with the similarity at domestication loci through various scenarios involving gene flow, although multiple origins were still favored based on the deep divergence between the subspecies (57, 68). Following the shattering genes, the Rc gene, underlying a change in pericarp color from red in wild rice to white in domesticated rice, was cloned and its origin was characterized in a diverse collection of more than 400 rice cultivars (69, 70). Much like sh4, the rc allele (causing white pericarps) was found to be common across both indica and japonica. The survey also clearly indicated that the domestication allele originated in japonica and spread to indica.
All three of the genes mentioned thus far can be classified as domestication genes, in that they either control a trait that is critical to a domesticated condition (loss of shattering) or they are found in a large majority of domesticated varieties (white pericarps). The Waxy (Wx) gene, which was characterized at the molecular level over a number of years (71–74), is best described as an improvement or diversification gene: it is selected in response to cultural preferences in some areas of the world but not uniformly favored. The majority of indica varieties have the fully functional Wxa allele, whereas japonica varieties generally carry the Wxb allele (resulting in a stickier grain), or its derivative, the waxy allele, which results in fully glutinous rice. The waxy allele has spread out of japonica and into some indica varieties when the glutinous phenotype was favored (71). This example conforms again to an emerging pattern at this point in rice domestication genetic research: domestication and improvement alleles are either restricted to the japonica subspecies (e.g., qSH1, Wxb), or they originated there and subsequently spread to indica (e.g., rc, waxy) (75). Again, the common origin of important domestication genes might seem to indicate that O. sativa was domesticated only once, but this scenario was inconsistent with the deep genetic divergence between the subspecies based on neutral loci. The combination model, suggesting that the domestication process was initiated multiple times and that this was followed by extensive introgression of strongly selected domestication alleles, was considered to be most consistent with the data at this point (75).
These four loci represented an early view into the future of rice domestication genetics based on functional genes: following the identification of these domestication and improvement genes, new genes were identified at a rapid rate (76, 77). In general, when surveys included a broad sampling of rice varieties, the allele associated with a domesticated state showed one of three patterns: (i) the allele was unique to japonica (78, 79), (ii) the same allele was found in a subset of both japonica and indica (80, 81), or (iii) the same allele was found in the majority of japonica and indica varieties (82, 83). These results revealed the genetic complexity of rice domestication; alleles underlying domestication phenotypes can have different origins and different distributions depending on their desirability and dispersal across the range of domesticated rice. However, in every case where the survey included an evolutionary component, the origin of the domestication allele was found to be the japonica subspecies (80, 81). As a result, the phylogenies generated from domestication genes showed japonica and indica as a monophyletic group (69, 84), consistent with a single origin for allele controlling a domestication trait. In contrast, neutral loci generally recovered a polyphyletic relationship (51, 59), and analyses that included O. rufipogon showed that japonica and indica were more closely related to different populations of the wild species than to each other, consistent with multiple origins. It should be noted that the discordance in phylogenetic trees is expected under a scenario of introgression at loci controlling domestication traits (76) and illustrates the difference between gene trees and species trees (85), but the question of how to interpret the origins of indica and japonica in light of these conflicts was still an open one.
Genomic Patterns.
These common patterns for domestication genes vs. neutral loci were challenged in a publication by Molina et al. (86), where 630 gene fragments were sequenced in 20 O. rufipogon, 20 indica, and 16 japonica sampled from throughout the native range of wild and domesticated rice. When methods of demographic inference (using ∂a∂i) were applied to this dataset (87), the results indicated that a single origin for domesticated rice was significantly more likely than multiple origins. The authors found that a single origin was more likely even when they excluded regions that showed evidence of selective sweeps; i.e., gene fragments that looked the most like domestication genes were not influencing the analysis. The paper also included a reanalysis of previously published sequence data using *BEAST (88), a Bayesian approach that generates a species tree based on heterogeneous gene trees: this removes the need for concatenation and potentially provides a more accurate phylogeny. Although the smaller datasets indicated a lack of monophyly, larger datasets (>5,000 bp, five or more loci) were strongly supportive of a single origin for domesticated rice. Overall, the paper is consistent with a single origin of O. sativa in the Yangtze Valley of China, and this corresponds to the majority opinion based on archaeological research. Although it has been pointed out that the modeling component of the analysis was influenced by the assumption of no structure in O. rufipogon (89), a situation that appears to be unlikely based on subsequent studies (90, 91), this paper opened many questions about genetic research into the origins of domesticated rice (were previous phylogenetic analyses wholly inaccurate? how should the contrasting results from the same starting data be interpreted?). In addition, the study served as a harbinger of a return to a focus on data from neutral loci in addition to domestication genes.
At nearly the same time, an analysis using an almost identical sampling strategy (22 samples each from O. rufipogon, indica, and japonica from throughout the native range of wild rice) applied whole-genome resequencing to evaluate the evolutionary history of domesticated rice (92). In contrast to the methods of Molina et al. (86), He et al. (92) used a coalescent-based approach that showed strong support for separate origins of japonica and indica based on the majority of the genome, but also found that regions of low diversity (possibly indicative of a selective sweep) displayed a pattern that was most consistent with a single origin. Overall, the data were consistent with unique origins of japonica and indica accompanied by extensive gene flow between the subspecies at domestication loci. Clearly, given the similarities of these two studies, it would be optimal to see each analysis (∂a∂i vs. coalescent modeling) applied to each dataset. As it currently stands, it is difficult to say whether the different results are due to the differences in the type of data (gene fragments vs. whole genomes), the analytical methods, or the identity of the samples chosen in each case. The unique findings of Molina et al. have not yet been recovered in other studies, despite their potential importance for our understanding of rice domestication.
The large genetic datasets but relatively small sample sizes in these studies were countered in a study focusing firmly on the neutral genetic variation in O. rufipogon (90). The study used 42 gene fragments and 180 accessions (108 of which were O. rufipogon); this represented the largest combined genetic/sample size at the time (93). Analysis indicated that O. rufipogon showed strong population structure and fell into two groups; this corresponds well to previous studies. However, in a surprising finding with no precedent in the literature, the results also showed that indica was most closely related to wild rice from southern China, a location that is usually associated with the origin of japonica rice. Even more surprisingly, japonica samples showed no close affinity to either of the O. rufipogon groups. These results open many questions about the origin of domesticated Asian rice and its relationship with its closest wild relative. The results have yet to be replicated, but indicate that a greater emphasis on sampling O. rufipogon may reveal unexpected patterns.
The use of resequenced rice genomes as a tool to understand the evolutionary origins of domesticated Oryza has expanded most recently with the publication and analysis of 446 resequenced O. rufipogon genomes and 1,083 japonica and indica genomes (91). This analysis provides one of the most extensive datasets exploring the genetic structure of O. rufipogon, which is critical for understanding the evolution of domesticated rice. The analyses based on genomewide variation indicated a close relationship between japonica and O. rufipogon from far southern China (in the Pearl River valley rather than the Yangtze River valley), and a similarly close relationship between indica and O. rufipogon from eastern India. When phylogenetic analyses were conducted based on variation at 55 regions that showed evidence of selective sweeps under domestication, both indica and japonica were most closely related to populations in the Pearl River valley. The authors therefore suggest a single origin of domesticated rice in far southern China, followed by dispersal to Southeast Asia and hybridization with local O. rufipogon. This study corresponds well to archaeological evidence relating to the origin of indica, but differs in that archaeological research has not indicated the Pearl River as a location for the domestication of japonica. One item to note about this paper is that, although the dataset is very impressive, the authors did not take advantage of powerful coalescent or other model-based techniques that could yield more precise of the population history of the domesticated and wild species (e.g., population size and timing of domestication).
Summary.
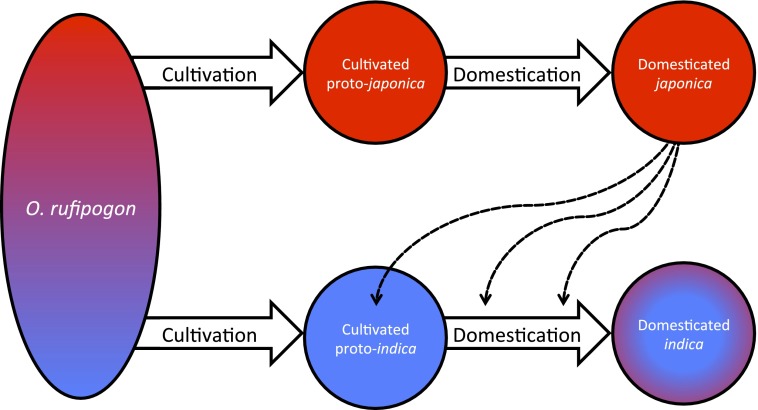
Considering these recent studies, especially in light of the factors that might complicate our understanding of rice domestication, we can see real progress in some areas. As is the case for many other biological systems, the number of genetic makers in a given study has increased by orders of magnitude, so that limited sampling of genetic variation is no longer a concern. Sample size and diversity has not increased as steadily; indeed, there has been a trend toward decreasing sample sizes with an increasing number of molecular markers. Although necessary in some circumstances, this trend can hopefully be avoided in future studies of rice domestication, because sampling sparsely from a large geographical range is bound to introduce error in the form of missing populations and diversity. Finally, the integration of patterns seen at domestication genes and neutral loci is still being resolved. Although the possibility of introgression between indica and japonica is clearly well accepted, along with the genealogical discordance this can cause across different loci, it is still unclear how to relate this directly to the origin of indica and japonica (Fig. 2). In particular, should we conclude that there were two origins of domesticated rice and that phylogenetic incongruity is caused by introgression of domestication genes (92)? Or should we conclude that there was a single origin, with indica being brought to a domesticated state through a series of hybridization events between japonica and O. rufipogon populations (91)? The former scenario would suggest that some domestication alleles might have arisen in indica, but that these alleles were replaced by more desirable alleles from japonica. The later scenario would suggest that all domestication alleles arose exclusively in japonica. Which scenario is more likely currently remains unresolved, but the answer seems much closer now than it has in the past.
Fig. 2.
Possible scenarios for the origin of japonica and indica from a genetically diverse O. rufipogon population, with genetic differentiation represented by different colors. Possible timing for the movement of domestication alleles from japonica into indica via hybridization is shown with dashed lines. Colors in the final domesticated indica circle represent the contributions from the original O. rufipogon populations (blue) and introgression from japonica (red).
Conclusions
Domestication is not a single event, but a continuum, within which there are different degrees of codependence between humans and plants (94). Rice has moved far along the continuum (becoming a major food source for humans) and has been extensively modified in this process, so that it now differs from its closest wild relative for a suite of traits that encompass life history, breeding system, morphology, and physiology. Given the exciting accumulation of new information from both archaeology and genetics about when and where Oryza sativa started along this continuum and how domestication traits eventually arose across the range of the species, how can the two fields better inform each other to answer key questions about the origin of indica and japonica?
Both fields offer unique approaches to identifying the geographical origin of rice domestication, and the bulk of studies indicate that japonica originated in the Yangtze River valley, whereas indica originated in the Ganges plains (although there are counter examples with interesting alternatives). Archaeological research indicates that appearance of one of the most iconic domestication traits (loss of shattering) was later than originally thought and that it was potentially not complete even by 6500 BP, although other traits (more difficult to detect from physical remains) may well have followed a different trajectory. Genetic approaches have shown that domestication traits are controlled by the same alleles in japonica and indica and that these alleles often originated in japonica, but that the subspecies are divergent at neutral loci. These findings from archaeology and genetics have been combined to suggest the independent origins of rice cultivation in China and India, followed by the introgression of domestication traits from japonica into proto-indica cultivated plants to result in the establishment of the domesticated indica subspecies (47). This scenario, a type of “domestication by hybridization” for which there is increasing evidence from other plant and animal domesticates (95), is consistent with both the deep divergence between the subspecies at neutral loci and allele sharing at loci controlling key domestication genes.
One question about the domestication process with strong synergistic potential for the two fields deals with the process of domestication and the preferential spread of some domestication alleles over to others. For example, while the most common allele resulting in white rice grains (rc) originated in japonica and spread to indica, there are independent mutations resulting in an identical phenotype found in both the aus subspecies of O. sativa (closely related to indica) and the African domesticate O. glaberrima (69, 96). This pattern may indicate the incipient development of domestication traits in some populations, with further spread or development stopped by the arrival of a variety with more favorable characteristics. Although genetic surveys of extant lineages can provide some indication of the frequency of this type of phenomenon, archaeological evidence of the presence of domestication traits (e.g., shattering and grain shape) are required to complete our understanding of the history of rice domestication.
Supplementary Material
Acknowledgments
This work is based on the National Evolutionary Synthesis Center (NESCent) catalysis meeting entitled “Domestication as an Evolutionary Phenomenon: Expanding the Synthesis.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Vavilov NI. Studies on the origin of cultivated plants. Bull Appl Biol. 1926;16:139–248. [Google Scholar]
- 2.Ding Y. The origin and evolution of Chinese cultivated rice. J Agriculture. 1957;8(3):243–260. [Google Scholar]
- 3.Yan W. Re-thinking the origin of rice agriculture in China. Nongye Kaogu. 1989;89(2):74–83. [Google Scholar]
- 4.Chang T. Rice. In: Simmonds NW, editor. Evolution of Crop Plants. London: Longman; 1976. pp. 98–104. [Google Scholar]
- 5.Higham C. The transition to rice cultivation in Southeast Asia. In: Price T, Gebauer A, editors. Last Hunters—First Farmers. Santa Fe, NM: School of American Research Press; 1995. pp. 127–155. [Google Scholar]
- 6.Zeder MA. The origins of agriculture in the Near East. Curr Anthropol. 2011;52(S4):S221–S236. [Google Scholar]
- 7.Watson P. In pursuit of prehistoric subsistence: A comparative account of some contemporary flotation techniques. Mid-Continental J Archaeol. 1976;1:77–100. [Google Scholar]
- 8.Crawford G. Paleoethnobotany of the Kameda Peninsula Jamon. Ann Arbor, MI: Museum of Anthropology; 1983. [Google Scholar]
- 9.Pearsall D. Paleoethnobotany: A Handbook of Procedures. San Diego: Academic Press; 1989. [Google Scholar]
- 10.Zhao Z. Paleoethnobotany and its recent advances in China. Kaogu (Archaeology) 2005;7:42–49. [Google Scholar]
- 11.Zhao Z. New Archaeobotanic data for the study of the origins of agriculture in China. Curr Anthropol. 2011;52(S4):S295–S306. [Google Scholar]
- 12.Fuller DQ. Agricultural origins and frontiers in South Asia: A working synthesis. J World Prehist. 2006;20:1–86. [Google Scholar]
- 13.Ahn S. The emergence of rice agriculture in Korea: Archaeobotanical perspectives. Archaeol Anthropol Sci. 2010;2(2):89–98. [Google Scholar]
- 14.Lee G. The transition from foraging to farming in prehistoric Korea. Curr Anthropol. 2011;52(S4):S307–S330. [Google Scholar]
- 15.Crawford G. Advances in understanding early agriculture in Japan. Curr Anthropol. 2011;52(S4):S331–S346. [Google Scholar]
- 16.Lu H, et al. Rice domestication and climatic change: Phytolith evidence from East China. Boreas. 2002;31(4):378–385. [Google Scholar]
- 17.Zheng Y, Matsui A, Fujiwara H. Phytoliths of rice detected in the Neolithic sites in the valley of the Taihu Lake in China. Environ Archaeol. 2003;8(2):177–183. [Google Scholar]
- 18.Gu Y, Zhao Z, Pearsall D. Phytolith morphology research on wild and domesticated rice species in East Asia. Quat Int. 2012;287:141–148. [Google Scholar]
- 19.Bellwood P. First Farmers: The Origins of Agricultural Societies. Malden, MA: Blackwell; 2005. [Google Scholar]
- 20.Barker G. The Agricultural Revolution in Prehistory. Oxford: Oxford Univ Press; 2006. [Google Scholar]
- 21.Crawford G. Prehistoric plant domestication in East Asia. In: Cowan C, Watson P, editors. The Origins of Agriculture: An International Perspective. London: Smithsonian Institution Press; 1992. [Google Scholar]
- 22.Nakamura S. The origin of rice cultivation in the Lower Yangtze Region, China. Archaeol Anthropol Sci. 2010;2(2):107–113. [Google Scholar]
- 23.Cohen D. The beginnings of agriculture in China: A multi-regional view. Curr Anthropol. 2011;52(S4):S273–S293. [Google Scholar]
- 24.Fuller DQ, Sato Y-I. Japonica rice carried to, not from, Southeast Asia. Nat Genet. 2008;40(11):1264–1265. doi: 10.1038/ng1108-1264. [DOI] [PubMed] [Google Scholar]
- 25.Price T, Bar-Yosef O. The origins of agriculture, new data, new ideas: An introduction to supplement 4. Curr Anthropol. 2011;52(S4):S163–S174. [Google Scholar]
- 26.Zhao Z. The middle Yangtze region in China is one place where rice was domesticated: Phytolith evidence from the Diaotonghuan cave, northern Jiangxi. Antiquity. 1998;72(278):885–897. [Google Scholar]
- 27.Jiang L, Liu L. New evidence for the origins of sedentism and rice domestication in the Lower Yangzi River, China. Antiquity. 2006;80(308):355–361. [Google Scholar]
- 28.Wu X, et al. Early pottery at 20,000 years ago in Xianrendong Cave, China. Science. 2012;336(6089):1696–1700. doi: 10.1126/science.1218643. [DOI] [PubMed] [Google Scholar]
- 29. Jiang L (2005) The Shangshan Neolithic site in Pujiang County, Zhejing: New evidence of rice civilization in the Lower Yangtze River region (in Chinese). Gudai Wenming Yanjiu Zhongxin Tongxun 7:51–55.
- 30.Fuller DQ. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann Bot (Lond) 2007;100(5):903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuller DQ, Harvey E, Qin L. Presumed domestication? Evidence for wild rice cultivation and domestication in the fifth millennium BC of the Lower Yangzte region. Antiquity. 2007;81(312):316–331. [Google Scholar]
- 32.Hunan Institue of Archaeology . Pengtoushan and Bashidang. Beijing: Kexue Chubanshe; 2006. (in Chinese) [Google Scholar]
- 33.Zhejiang Institute of Archaeology and Xiaoshan Museum . Kuahuqiao. Beijing: Kexue Chubanshe; 2004. (in Chinese) [Google Scholar]
- 34.Zhang H, Wang H, Yang W. Early Neolithic Relics Found in the Xiaohuangshan Site in Shengzhou, Zhejiang. Zhongguo Wenwu Bao; 2005. Chinese. [Google Scholar]
- 35.Henan Institute of Archaeology . Wuyang Jiahu. Beijing: Kexue Chubanshe; 1999. Chinese. [Google Scholar]
- 36.Zhao Z, Zhang J. The report of flotation work at the Jiahu site. Kaogu (Archaeology) 2009;8:84–93. [Google Scholar]
- 37.Fuller DQ, Qin L, Harvey E. Rice archaeobotany revisted: Comments on Liu et al. Antiquity. 2008 82(315):Project Gallery, online (www.antiquity.ac.uk/ProjGall/fuller1/) [Google Scholar]
- 38.Liu L, et al. Evidence for the early beginning (c. 9000 cal. BP) of rice domestication in China: A response. Holocene. 2008;17(8):1059–1068. [Google Scholar]
- 39.Liu J. The Hemudu Culture. Beijing: Cultural Relics Press; 2006. Chinese. [Google Scholar]
- 40.Ellis JR, Pashley CH, Burke JM, McCauley DE. High genetic diversity in a rare and endangered sunflower as compared to a common congener. Mol Ecol. 2006;15(9):2345–2355. doi: 10.1111/j.1365-294X.2006.02937.x. [DOI] [PubMed] [Google Scholar]
- 41.Research Center of Chinese Archaeology . Comprehensive Study of Natural Remains at the Tianluoshan Site. Beijing: Cultural Relics Press; 2011. Chinese. [Google Scholar]
- 42.Fuller DQ, et al. The domestication process and domestication rate in rice: Spikelet bases from the Lower Yangtze. Science. 2009;323(5921):1607–1610. doi: 10.1126/science.1166605. [DOI] [PubMed] [Google Scholar]
- 43.Pan Y. Immature wild rice harvesting at Kuahuqiao, China? Antiquity. 2008 82(316):Project Gallery, online (www.antiquity.ac.uk/projGall/pan/) [Google Scholar]
- 44.Imamura I. News Letter No.7. National Museum of Japanese History, Sakura, Japan; 2007. Chronology of the Yayoi period; pp. 6–7. Japanese. [Google Scholar]
- 45.Obata H. Origins of domesticated plants in Jomon period, Japan. In: Obata H, editor. Prehistoric and Ancient Cultigens in the Far East. Vol 3. Kumamoto, Japan: Univ of Kumamoto; 2008. pp. 43–94. Japanese. [Google Scholar]
- 46.Fuller DQ, Allaby RG, Stevens C. Domestication as innovation: The entanglement of techniques, technology and chance in the domestication of cereal crops. World Archaeol. 2010;42(1):13–28. [Google Scholar]
- 47.Fuller DQ. Finding plant domestication in the Indian subcontinent. Curr Anthropol. 2011;52(S4):S347–S362. [Google Scholar]
- 48.Zhao Z, Gu H. Study of identification methods of archaeological rice remains. Hunan Kaogu Jikan. 2011;8:5–68. [Google Scholar]
- 49.Oka H-I. Phylogenetic differrentiation of the cultivated rice plant. 1. Variations in respective characteristics and their combinations in rice cultivars. Jap J Breeding. 1953;3:33–43. [Google Scholar]
- 50.Oka H-I. Intervarietal variation and classification of cultivated rice. Indian J Genet Plant Breed. 1958;18:79–89. [Google Scholar]
- 51.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169(3):1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA. 2002;99(9):6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Heerwaarden J, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA. 2011;108(3):1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng C, et al. Polyphyletic origin of cultivated rice: Based on the interspersion pattern of SINEs. Mol Biol Evol. 2003;20(1):67–75. doi: 10.1093/molbev/msg004. [DOI] [PubMed] [Google Scholar]
- 55.Second G. Origin of the genetic diversity of cultivated rice (Oryza spp.): Study of the polymorphism scored at 40 isozyme loci. Jpn J Genet. 1982;57:25–57. [Google Scholar]
- 56.Wang ZY, Second G, Tanksley SD. Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet. 1992;83(5):565–581. doi: 10.1007/BF00226900. [DOI] [PubMed] [Google Scholar]
- 57.Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 2005;167(1):249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
- 58.Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA. 2006;103(25):9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caicedo AL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3(9):1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rakshit S, et al. Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice. Theor Appl Genet. 2007;114(4):731–743. doi: 10.1007/s00122-006-0473-1. [DOI] [PubMed] [Google Scholar]
- 61.Gao LZ, Innan H. Nonindependent domestication of the two rice subspecies, Oryza sativa ssp. indica and ssp. japonica, demonstrated by multilocus microsatellites. Genetics. 2008;179(2):965–976. doi: 10.1534/genetics.106.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao LZ, Zhang CH, Jia JZ. Cross-species transferability of rice microsatellites in its wild relatives and the potential for conservation genetic studies. Genet Resour Crop Evol. 2005;52(7):931–940. [Google Scholar]
- 63.Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;311(5769):1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 64.Konishi S, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312(5778):1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 65.Sang T, Ge S. Genetics and phylogenetics of rice domestication. Curr Opin Genet Dev. 2007;17(6):533–538. doi: 10.1016/j.gde.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L-B, et al. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009;184(3):708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 67.Sang T, Ge S. The puzzle of rice domestication. J Integr Plant Biol. 2007;49(6):760–768. [Google Scholar]
- 68.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA. 2004;101(34):12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sweeney MT, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3(8):e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18(2):283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olsen KM, Purugganan MD. Molecular evidence on the origin and evolution of glutinous rice. Genetics. 2002;162(2):941–950. doi: 10.1093/genetics/162.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olsen KM, et al. Selection under domestication: Evidence for a sweep in the rice waxy genomic region. Genetics. 2006;173(2):975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z-Y, et al. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995;7(4):613–622. doi: 10.1046/j.1365-313x.1995.7040613.x. [DOI] [PubMed] [Google Scholar]
- 74.Hirano HY, Eiguchi M, Sano Y. A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol Biol Evol. 1998;15(8):978–987. doi: 10.1093/oxfordjournals.molbev.a026013. [DOI] [PubMed] [Google Scholar]
- 75.Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23(11):578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 76.Gross BL, Olsen KM. Genetic perspectives on crop domestication. Trends Plant Sci. 2010;15(9):529–537. doi: 10.1016/j.tplants.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Olsen KM, Wendel JF (2013) Crop plants as models for understanding plant adaptation and diversification. Front Plant Sci, 10.3389/fpls.2013.00290. [DOI] [PMC free article] [PubMed]
- 78.Asano K, et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc Natl Acad Sci USA. 2011;108(27):11034–11039. doi: 10.1073/pnas.1019490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40(8):1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 80.Takano-Kai N, et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics. 2009;182(4):1323–1334. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kovach MJ, Calingacion MN, Fitzgerald MA, McCouch SR. The origin and evolution of fragrance in rice (Oryza sativa L.) Proc Natl Acad Sci USA. 2009;106(34):14444–14449. doi: 10.1073/pnas.0904077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan L, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40(11):1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 83.Jin J, et al. Genetic control of rice plant architecture under domestication. Nat Genet. 2008;40(11):1365–1369. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 84.Gross BL, et al. Seeing red: The origin of grain pigmentation in US weedy rice. Mol Ecol. 2010;19(16):3380–3393. doi: 10.1111/j.1365-294X.2010.04707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nichols R. Gene trees and species trees are not the same. Trends Ecol Evol. 2001;16(7):358–364. doi: 10.1016/s0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]
- 86.Molina J, et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci USA. 2011;108(20):8351–8356. doi: 10.1073/pnas.1104686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5(10):e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Drummond AJ, et al. (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29(8):1969–1973. [DOI] [PMC free article] [PubMed]
- 89.Ge S, Sang T. Inappropriate model rejects independent domestications of indica and japonica rice. Proc Natl Acad Sci USA. 2011;108(39):E755. doi: 10.1073/pnas.1111601108. author reply E756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang P, et al. Phylogeography of Asian wild rice, Oryza rufipogon: A genome-wide view. Mol Ecol. 2012;21(18):4593–4604. doi: 10.1111/j.1365-294X.2012.05625.x. [DOI] [PubMed] [Google Scholar]
- 91.Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490(7421):497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He Z, et al. Two evolutionary histories in the genome of rice: The roles of domestication genes. PLoS Genet. 2011;7(6):e1002100. doi: 10.1371/journal.pgen.1002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gross BL. Rice domestication: Histories and mysteries. Mol Ecol. 2012;21(18):4412–4413. doi: 10.1111/j.1365-294X.2012.05626.x. [DOI] [PubMed] [Google Scholar]
- 94.Zeder MA. Central questions in the domestication of plants and animals. Evol Anthropol. 2006;15(3):105–117. [Google Scholar]
- 95.Larson G, Burger J. A population genetics view of animal domestication. Trends Genet. 2013;29(4):197–205. doi: 10.1016/j.tig.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 96.Gross BL, Steffen FT, Olsen KM. The molecular basis of white pericarps in African domesticated rice: Novel mutations at the Rc gene. J Evol Biol. 2010;23(12):2747–2753. doi: 10.1111/j.1420-9101.2010.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]