Significance
Glycosylating toxins are major virulence factors of pathogenic Clostridia, including Clostridium difficile toxins A and B, which cause antibiotics-associated diarrhea and pseudomembranous colitis. Host cell uptake of these toxins is suggested to be mediated via C-terminal combined repetitive oligopeptides (CROP). Here, we identified LDL receptor-related protein 1 (LRP1) as a host cell receptor of Clostridium perfringens TpeL toxin, a recently identified clostridial glycosylating toxin, which has no CROP domain. TpeL binds to LRP1 with a C-terminal receptor-binding domain, which is conserved in all other clostridial glycosylating toxins and precedes the CROP domain. Thus, our study identifies LRP1 as TpeL receptor and suggests a two-receptor model for clostridial glycosylating toxins, which offers additional perspectives in antitoxin strategies.
Keywords: toxin receptor, receptor-mediated endocytosis
Abstract
Large glycosylating toxins are major virulence factors of various species of pathogenic Clostridia. Prototypes are Clostridium difficile toxins A and B, which cause antibiotics-associated diarrhea and pseudomembranous colitis. The current model of the toxins’ action suggests that receptor binding is mediated by a C-terminal domain of combined repetitive oligopeptides (CROP). This model is challenged by the glycosylating Clostridium perfringens large cytotoxin (TpeL toxin) that is devoid of the CROP domain but still intoxicates cells. Using a haploid genetic screen, we identified LDL receptor-related protein 1 (LRP1) as a host cell receptor for the TpeL toxin. LRP1-deficient cells are not able to take up TpeL and are not intoxicated. Expression of cluster IV of LRP1 is sufficient to rescue toxin uptake in these cells. By plasmon resonance spectroscopy, a KD value of 23 nM was determined for binding of TpeL to LRP1 cluster IV. The C terminus of TpeL (residues 1335–1779) represents the receptor-binding domain (RBD) of the toxin. RBD-like regions are conserved in all other clostridial glycosylating toxins preceding their CROP domain. CROP-deficient C. difficile toxin B is toxic to cells, depending on the RBD-like region (residues 1349–1811) but does not interact with LRP1. Our data indicate the presence of a second, CROP-independent receptor-binding domain in clostridial glycosylating toxins and suggest a two-receptor model for the cellular uptake of clostridial glycosylating toxins.
Clostridial glycosylating toxins are major pathogenicity factors that are responsible for numerous severe diseases in humans and animals. Prototypes of these toxins are Clostridium difficile toxins A and B, the causative agents of antibiotics-associated diarrhea and pseudomembranous colitis (1, 2). During recent years, morbidity and mortality of C. difficile-induced infections (CDI) largely increased (3, 4). In fact, CDI advanced to one of the most important nosocomial infections in developed countries. Other members of the family of clostridial glycosylating toxins are Clostridium sordellii lethal and hemorrhagic toxin, and the α-toxin from Clostridium novyi, which cause gas gangrene syndromes (5). All these toxins have a very similar primary structure comprising an amino acid sequence identity of 40–90% (1, 5). Recently, an ABCD model has been proposed for these toxins with an N-terminally located glycosyltransferase domain (domain A), a subsequent cysteine protease domain for autoproteolytic cleavage (domain C), a putative pore-forming and delivery domain (domain D), and a C-terminal binding domain (domain B) (6). After binding to cell surface receptors, the toxins are endocytosed in a clathrin-dependent and dynamin-dependent manner (7). At a low pH of endosomes, the toxins insert into endosomal membranes and form pores, which probably allow translocation of the glycosyltransferase (GT) and cysteine protease domains into the cytosol where inositol hexakisphosphate activates the protease for autoproteolytic cleavage and release of the GT into the cytosol (8–10). In the cytosol, Rho and/or Ras proteins are glucosylated or modified by GlcNAcylation, resulting in inhibition of these switch proteins and, eventually, in inflammation and cell death (11–13). Although autoproteolytic processing and toxin-induced glycosylation of Rho/Ras proteins are well characterized, the interaction of the toxins with cell surface receptors is still enigmatic. Cell surface-binding is suggested to be mediated by C-terminal polypeptide repeats (B domain) termed combined repetitive oligopeptides (CROP) that might recognize cell surface carbohydrate structures (14–17). Recombinant fragments of this C-terminal toxin part blocks toxin binding and cytotoxicity (18). Moreover, monoclonal antibodies raised against this toxin portion prevent cell intoxication (19). Putative receptors have been described for C. difficile toxin A, including carbohydrates, glycophospholipids, and proteins (16, 17, 20, 21). The hypothesis that the C-terminal part of clostridial glycosylating toxins is solely responsible for receptor interaction has been challenged. For example, after removal of the CROP domain of toxin A or toxin B, the toxins were still cytotoxic (10, 22). Thus, we and others hypothesized that a receptor binding site different from the CROP domain might be involved in toxin binding.
Recently, TpeL, the most recent member of the family of clostridial glycosylating toxins that is produced by Clostridium perfringens type A, B, and C strains, was described (23). TpeL exhibits 30–40% amino acid sequence identity with other clostridial glycosylating toxins and shares with them the glycosyltransferase domain, the cysteine protease domain, and the delivery domain. However, it does not possess a CROP domain. Nevertheless, TpeL intoxicates cells and kills mice (23). Toxic effects of TpeL are probably due to GlcNAcylation of Ras proteins at threonine-35 (24). In addition, TpeL-induced modification of Rac has been reported (25).
Thus, TpeL is a model toxin to unravel the interaction of clostridial glycosylating toxins with target cells. Here, we identify the low-density lipoprotein receptor-related protein 1 (LRP1) as a target molecule for binding and cell entry of TpeL. We report that the C-terminal part of TpeL binds to LRP1 and represents the receptor-binding domain. Furthermore, we show that the respective part within C. difficile toxin B, which resembles to the receptor-binding domain of TpeL, binds to cells. Our study strongly supports a two-receptor model of clostridial glycosylating toxins and offers an additional perspective in the understanding of the pathogenicity of this group of clinically important toxins.
Results
Haploid Genetic Screen Yields LRP1 as a Possible Receptor of TpeL.
Recently, we used a genetic screen based on the human haploid cell line Hap1 to identify the cell membrane receptor of C. difficile binary toxin CDT (26). Here, the same method was used to identify the cell surface receptor of TpeL. Hap1 cells rounded up in the presence of toxin, indicating the presence of a cell surface receptor. We enriched toxin-resistant clones and genomic DNA was isolated from the entire pool of toxin-resistant cells, followed by inverse PCR to amplify DNA sequences, flanking the retroviral integration sites, and parallel sequencing (Solexa). Although hits of the genetic screen, which were not linked to any cell surface receptor, were neglected, we identified the low-density lipoprotein receptor-related protein 1 (LRP1, A2MR, APR, CD91) and, strikingly, sorting nexin 17 (SNX17), a protein that regulates cell surface levels of LRP1 (27).
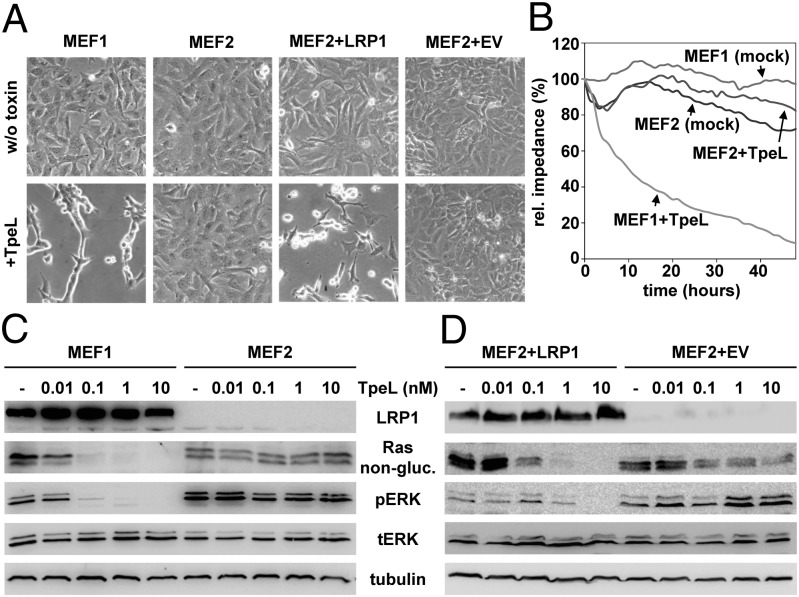
To test whether LRP1 is required for cell entry of TpeL, we used wild-type mouse embryonic fibroblasts (MEF LRP1+/+; termed MEF1) and LRP1-deficient MEFs (MEF LRP1−/−; termed MEF2) (28), respectively, and challenged both cell lines with the toxin (10 nM) for 2 h at 37 °C. Whereas MEF1 cells exhibited cell shrinking and rounding, MEF2 cells were resistant toward TpeL (Fig. 1A). Importantly, sensitivity toward TpeL was restored in MEF2 cells transfected with an LRP1-expressing plasmid (termed MEF2+LRP1), but not in MEF2 cells that were transfected with an empty vector (termed MEF2+EV) (Fig. 1A). To compare TpeL intoxication in MEF1 and MEF2 cells over a prolonged time period at a low dose of toxin (1 nM), we performed electric cell substrate impedance sensing (ECIS) measurements for continuous detection of toxin effects on cell monolayers impedance. The ECIS measurements confirmed that MEF2 but not MEF1 cells were resistant toward TpeL even after 2 d of toxin treatment (Fig. 1B).
Fig. 1.
LRP1 is essential for intoxication of MEF cells by TpeL. (A) MEF1, MEF2, MEF2+LRP1, and MEF2+EV cells were either incubated with 10 nM TpeL for 2 h at 37 °C (+TpeL) or were left untreated (w/o toxin), before microscopic analysis of cell morphology. (B) MEF1 and MEF2 cells were grown on ECIS wells and incubated either with 1 nM TpeL (+TpeL) or without toxin (mock), and decrease of impedance was measured over time. The initial resistance of each assay after toxin addition was set to 100%. (C and D) MEF1 and MEF2 (C), as well as MEF2+LRP1 and MEF2+EV cells (D), were intoxicated for 2 h with increasing concentrations of TpeL (as indicated), before immunoblot analysis of nonglycosylated Ras (Rasnon-gluc.) and phospho-ERK (pERK) levels in whole-cell lysates. Equal load of samples was verified by detection of total ERK (tERK) and tubulin levels.
To additionally prove that TpeL required LRP1 for cell uptake, we studied the glycosylation state of Ras in lysates of TpeL-treated MEF cells with an antibody that recognizes only nonglycosylated Ras protein. TpeL dose dependently decreased the amount of nonglycosylated Ras in MEF1 but not in MEF2 cells (Fig. 1C). Concomitantly, TpeL inhibited Ras downstream signaling in MEF1 but not in MEF2 cells monitored by probing the phosphorylation state of ERK1/2 (Fig. 1C). Notably, Ras glycosylation and inhibition of Ras downstream signaling by TpeL was restored in MEF2+LRP1 cells but not in MEF2+EV cells (Fig. 1D). Thus, our studies with MEF cells indicated that TpeL required LRP1 to reach its intracellular target substrates.
In line with these data, short interfering RNA-mediated knockdown of SNX17 expression decreased TpeL-induced inactivation of Ras signaling in MEF1 cells (Fig. S1). Thus, these findings confirm an essential role of SNX17 in TpeL-induced intoxication.
LRP1 Increases Binding of TpeL to the Cell Surface and Mediates Endocytic Uptake.
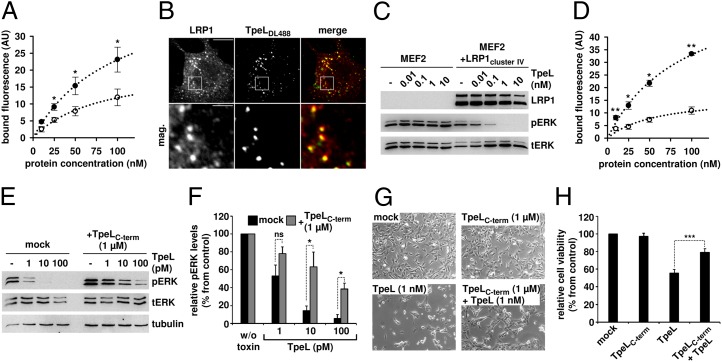
Next, we studied LRP1-dependent cell surface binding and endocytic uptake of TpeL by fluorescence-activated cell sorting (FACS). Remarkably, fluorescence-labeled TpeL bound to MEF1 and MEF2 cells with similar affinity [KD, 97.1 ± 10.3 nM (MEF1) vs. KD, 79.1 ± 13.2 nM (MEF2)] (Fig. 2A) at 4 °C. However, the TpeL-binding capacity was nearly double as high for MEF1 cells compared with MEF2 cells [Bmax: 45.5 ± 2.8 AU (MEF1) versus 21.2 ± 1.9 AU (MEF2)] (Fig. 2A). Although the data showed that the presence of LRP1 increases cell surface binding of TpeL, the study suggested additional LRP1-independent binding sites for TpeL. To investigate colocalization of TpeL with endocytic vesicles containing LRP1, we incubated fluorescence-labeled TpeL with MEF1 cells at 37 °C. As shown in Fig. 2B, colocalization of TpeL with immunolabeled LRP1 was obvious.
Fig. 2.
Prerequisites for LRP1-dependent binding and uptake of TpeL. (A and B) LRP1 increases binding of TpeL to the cell surface and mediates endocytic uptake the toxin. (A) MEF1 (filled circles) and MEF2 (open circles) cell suspensions (200,000 cells in 0.2 mL of medium) were incubated with increasing concentrations of DyLight488-labeled TpeL for 15 min at 4 °C, and cell-surface binding of TpeL was analyzed by FACS. Bound fluorescence is represented as arbitrary units (AU), n = 4, ±SEM, *P < 0.05. (B) MEF1 cells were incubated with 0.1 µg/µL DyLight488-labeled TpeL (TpeLDL488) for 30 min at 4 °C. Following an incubation of the cells for 15 min at 37 °C to induce endocytic uptake, cells were fixed, permeabilized, and endogenous LRP1 protein was immunostained before confocal fluorescence microscopy. mag., magnified area (white box). (Scale bars: Upper, 20 µm; Lower, 4 µm.) (C) Cluster IV of LRP1 is sufficient for direct interaction with TpeL. MEF2 cells and LRP1cluster IV-expressing MEF2 cells (MEF2+LRP1cluster IV) were incubated with indicated concentrations of TpeL for 2 h at 37 °C, following immunodetection of pERK and tERK in whole-cell lysates. (D–F) Binding of TpeL to LRP1 is mediated by the C-terminal part of the toxin. (D) MEF1 (filled circles) and MEF2 (open circles) cells were incubated with increasing concentrations of DyLight488-labeled TpeLC-term as described in A, and cell-surface binding of the TpeL fragment was analyzed by FACS. Bound fluorescence is represented as arbitrary units (AU), n = 3, ±SEM, *P < 0.05, **P < 0.01. (E) MEF1 cells were treated with indicated concentrations of TpeL for 2 h at 37 °C, in the absence (mock) or presence of 1 µM TpeLC-term, before immunoblot analysis for the detection of pERK levels in whole-cell lysates. Equal loading of samples was verified by detection of total ERK (tERK) and tubulin. (F) Quantification of pERK levels shown in E. Results (n = 3, ±SEM, *P < 0.05) are shown relative to pERK levels from untreated cells (w/o toxin) that were set to 100% (control). ns, not significant. (G) Microscopy images of MEF1 cells that were treated for 4 h at 37 °C with indicated concentrations of either TpeLC-term or TpeL, both proteins together, or that were left untreated (mock). (H) Confluent MEF1 cells were treated for 14 h at 37 °C with proteins as shown in G, before addition of CellTiter-Blue reagent, incubation for 2 h at 37 °C, and fluorescence detection. Results (n = 3, ±SEM, ***P < 0.001) are shown relative to the cell viability of untreated control cells (mock) that was set to 100%.
Cluster IV of LRP1 Is Sufficient for the Interaction of the TpeL Toxin with the Receptor.
LRP1 is a single-pass transmembrane protein with a very large extracellular domain (heavy chain) that contains four putative ligand binding clusters (I–IV) and a small intracellular domain (light chain). Most ligands bind to the clusters II and IV (29–31). Pseudomonas exotoxin A, which also gains entry into cells via LRP1, binds exclusively to cluster IV of the receptor (29). Therefore, we tested whether cluster IV of LRP1 is sufficient for uptake of TpeL, using LRP1-deficient MEF2 cells that ectopically express an LRP1 “minireceptor” that lacks cluster I, II, and III (termed MEF2+LRP1cluster IV). To this end, MEF2 and MEF2+LRP1cluster IV cells were treated with increasing concentrations of TpeL (0, 0.01, 0.1, 1, 10 nM) for 2 h followed by phospho-ERK1/2 immunoblot analysis. TpeL dose-dependently decreased phosphorylated ERK1/2 levels in MEF2+LRP1cluster IV cells but not in MEF2 controls (Fig. 2C).
The C Terminus of TpeL Harbors the Receptor-Binding Domain of the Toxin.
Previously, we reported that a C-terminally truncated TpeL variant, produced by C. perfringens strain MC18, is not toxic to cultured cells of various origin (24). Therefore, we assumed that the C-terminal part of TpeL harbors the receptor-binding domain. We expressed a C-terminal TpeL fragment, including amino acids 1335–1779 (denoted here as TpeLC-term) and tested binding of this fragment to MEF cells by FACS. To this end, TpeLC-term was labeled with DyLight488 and increasing concentrations were added to MEF1 or MEF2 cell suspensions. After incubation at 4 °C, where endocytic uptake is prevented, cells were washed and bound fluorescence was quantified. As shown in Fig. 2D, TpeLC-term was able to bind to both MEF1 and MEF2 cells with similar affinity [KD, 91.7 ± 27.3 nM (MEF1) vs. KD, 58.9 ± 32.1 nM (MEF2)]. However, the binding capacity of the fragment was notably higher for MEF1 cells compared with MEF2 cells [Bmax, 63.3 ± 10.9 AU (MEF1) versus 16.9 ± 4.6 AU (MEF2)]. It is important to note that similar results were observed with the full-length TpeL protein (Fig. 2A). Next, we performed competition studies, where MEF1 cells were incubated for 2 h with increasing concentrations of the toxin (0, 1, 10, and 100 pM) in the presence or absence of 1 µM TpeLC-term. As expected, excess addition of TpeLC-term decreased the uptake of the toxin and subsequent inhibition of Ras downstream signaling (Fig. 2 E and F). Supportingly, 1,000-fold excess addition of TpeLC-term efficiently reduced the cytotoxic potential of TpeL on cultured MEF1 cells, as observed microscopically (Fig. 2G) or by measuring the cell viability (Fig. 2H).
Interaction of TpeL and LRP1 in Vitro.
Human plasma contains a soluble form of LRP1 (sLRP1), consisting of the extracellular domain (heavy chain) of the receptor (32). Using a dot blot approach, interaction of sLRP1 with TpeL was verified (Fig. S2). To study the direct binding and to characterize the kinetics of the interaction between TpeL and LRP1, we performed surface plasmon resonance (SPR) spectroscopy studies with a protein corresponding to cluster IV of LRP1. Thereby, we obtained for TpeL a binding constant of 23.2 ± 0.2 nM (Fig. S3A) and for Pseudomonas exotoxin A and TpeLC-term, a binding constant of 5.2 ± 0.3 nM and 18.9 ± 0.7 nM, respectively (Fig. S3 B and C). These results confirmed direct and high-affinity binding of TpeL and its C-terminal portion to the LRP1 receptor.
Receptor-Associated Protein Prevents Intoxication of MEF1 Cells by TpeL.
Receptor-associated protein (RAP) is an ER-resident chaperone that binds tightly to multiple sites on LRP1 (33–35). Therefore, we purified RAP as a GST-fusion protein and tested its ability to competitively inhibit binding and entry of TpeL into MEF1 cells. To this end, cells were incubated with TpeL (1 and 10 nM) and an excess of GST-RAP (1 µM) at 4 °C, where the clearance of LRP1 from the plasma membrane is prevented, followed by incubation at 37 °C for 2 h to induce the uptake of the toxin. TpeL intoxication was then analyzed by probing the glycosylation state of Ras and the phosphorylation state of ERK1/2 in cell lysates. As shown in Fig. S4, 1,000-fold excess of GST-RAP, but not of GST alone, inhibited TpeL-induced intoxication of MEF1 cells. These results indicated a common binding site of RAP and TpeL at LRP1.
C. difficile Toxin B Harbors a TpeL-Like Receptor-Binding Domain That Acts Independent of the CROP Domain.
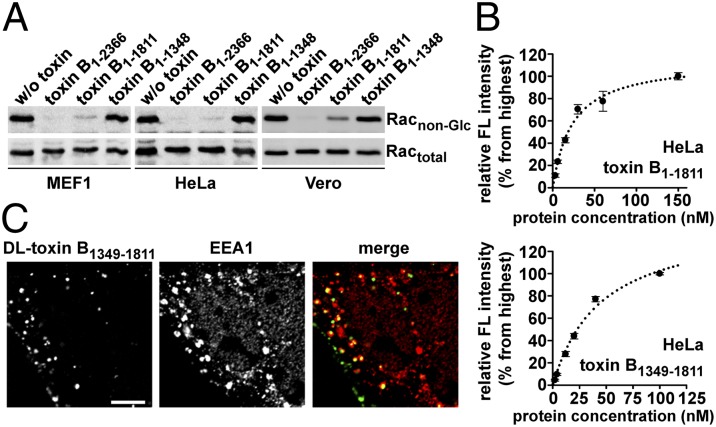
We and others showed that C. difficile toxins A and B lacking the CROP domain are still capable to enter into host cells and to cause cell rounding (10, 22), suggesting a second, CROP-independent receptor-binding site, which might follow the translocation domain and reside directly adjacent to the CROP domain (10). Therefore, we generated C-terminal truncations of toxin B either lacking the CROP domain (toxin B1–1811) or the CROP domain and a putative second receptor-binding site (toxin B1–1348). Using three cell lines (MEF1, HeLa, and Vero cells), we detected Rac glycosylation as an indication of toxin uptake, only with full-length toxin B and the CROP-deficient toxin B variant (Fig. 3A). Furthermore, we performed FACS binding studies with HeLa and Vero cells and CROP-deficient toxin B1–1811 or only with the putative receptor-binding domain (toxin B1349–1811). Notably, both proteins, toxin B1–1811 and toxin B1349–1811, exhibited saturable binding to the cell surface of HeLa (Fig. 3B) and Vero cells (Fig. S5). Coincubation of increasing concentrations of nonlabeled toxin B1349–1811 prevented binding of DyLight488-labeled toxin B1–1811 to Vero cells in a dose-dependent manner and vice versa (Fig. S6A). As expected, nonlabeled toxin B1–543 (representing the glycosyltransferase domain only) did not markedly inhibit binding of DyLight488-labeled toxin B1–1811 (Fig. S6B). Finally, we tested by confocal fluorescence microscopy, whether the toxin B fragment (toxin B1349–1811) was taken up into endocytic vesicles. We identified a dot-like intracellular distribution of toxin B1349–1811 overlapping with the endosomal marker protein EEA1 (Fig. 3C). Thus, the data indicate the presence of a functional receptor-binding domain within amino acids 1349–1811 of toxin B that acts independently of the CROP domain in binding and uptake into host cells.
Fig. 3.
Binding and uptake analysis of CROP-deficient C. difficile toxin B (toxin B1–1811) and toxin B1349–1811. (A) Monolayers of MEF1, HeLa, and Vero cells were treated overnight at 37 °C with full-length toxin B (toxin B1–2366), toxin B1–1811, toxin B1–1348 (each 10 pM), or were left without treatment (w/o toxin). Following cell lysis, Rac glucosylation was monitored by immunoblot with specific anti-Rac antibodies either recognizing only the nonmodified form of Rac (Racnon-Glc) or total Rac levels (Ractotal). (B) FACS analysis of cell surface-binding of toxin B1–1811 (Upper) or toxin B1349–1811 (Lower). HeLa cell suspensions (200,000 cells in 0.5 mL of medium) were incubated with increasing concentrations of DyLight488-labeled toxin B1–1811 or toxin B1349–1811 for 15 min at 4 °C before FACS analysis. Bound fluorescence (FL) is shown relative to the highest value that was set to 100% (n = 3, ±SEM). Dotted curve was calculated by SigmaPlot. (C) CaCo-2 cells grown as a monolayer were incubated with 70 nM DyLight488-labeled toxin B1349–1811 for 25 min at 37 °C. Subsequently, cells were fixed, permeabilized, and EEA1 was immunostained, before confocal fluorescence microscopy. (Scale bar: 5 µm.) DL, DyLight488.
LRP1 Is Not Involved in the Cellular Uptake of CROP-Deficient C. difficile Toxin A and B.
Next, we wondered whether LRP1 might be also the receptor for the second receptor-binding site in the C. difficile toxins A and B. To this end, we performed ECIS measurements with MEF1 and MEF2 cells that were treated for 10 h with CROP-deficient toxin A (toxin A1–1849) or toxin B (toxin B1–1811). After 1 h, decrease of the impedance was observed only in samples that were treated with C. difficile toxin A1–1849 or toxin B1–1811. However, MEF1 and MEF2 cells were intoxicated with similar efficiency by both toxin variants (Fig. S7). These findings argue for an LRP1-independent uptake mechanism that mediates cell entry of CROP-deficient toxin A and B.
Discussion
The main aim of the current study was to identify the host cell receptor of the TpeL toxin, which is a more recently identified member of the family of clostridial glycosylating toxins. To this end, we used a genetic screen (36), which is based on a human haploid cell library with null mutations in nonessential genes. By the same approach, we identified the host cell receptor of the C. difficile binary toxin CDT (26). Here, we found TpeL-resistant clones with retroviral insertions in the gene encoding the LRP1. We focused on this gene for two reasons. First, LRP1 is a cell-surface protein and could, thereby, serve as a putative receptor for TpeL. Second, we found TpeL-insensitive cells in our screen that carried retroviral insertions in the gene encoding SNX17, a protein known to regulate endosomal recycling and to maintain normal levels of LRP1 at the cell surface (27).
LRP1 is a cell surface receptor and belongs to the LDL receptor gene family (37). Members of this family participate in a range of biological processes, including lipid metabolism and various scavenger functions (30). A variety of ligands have been described so far that are endocytosed in an LRP1-dependent manner (e.g., apolipoprotein E, ref. 38; chylomicron remnants, ref. 39; α2-macroglobulin, ref. 40; amyloid precursor protein, ref. 41; and several proteases and protease inhibitors, refs. 42 and 43). Interestingly, LRP1 functions also as the cell entry receptor for the Pseudomonas exotoxin A and the minor-group common cold virus (44, 45). In addition, LRP1 acts in cell communication and signal transduction (30, 46). The LRP1 protein is a type I single-pass transmembrane protein with the size of 600 kDa that is processed by the furin protease (47). The N-terminal, extracellular part (denoted as heavy chain) has a size of 515 kDa and remains noncovalently attached to the 85 kDa membrane-integrated intracellular part (light chain) (30). The heavy chain of LRP1 consists of four ligand-binding clusters. With the exception of the protein RAP that acts as a molecular chaperone for LRP1 and interacts with all LRP1 clusters, most ligands bind exclusively to cluster II and/or IV (30).
Several findings that are summarized in Fig. 1 indicate that LRP1 acts as a host cell receptor for the TpeL toxin. First, LRP1-deficient MEFs were resistant toward TpeL treatment. This finding was shown by microscopic analysis of the cell morphology, by measuring the integrity of the cell monolayer with the ECIS method, and by TpeL-induced glycosylation of its target protein Ras and inactivation of Ras downstream signaling. Second, ectopic expression of LRP1 restored the sensitivity of LRP1-deficient MEF cells toward TpeL toxin. A protective effect against the TpeL toxin was also observed in wild-type MEF cells by knockdown of SNX17 expression. Thus, these results corroborated data from the genetic screen, suggesting that both proteins, LRP1 and SNX17, are crucial for the cytotoxic activity of the TpeL toxin.
The chaperone RAP binds tightly to LRP1 and is frequently used to inhibit binding of ligands to LRP1 (34, 35, 48). Accordingly, excess of RAP prevented the intoxication of MEF cells by TpeL, indicating that TpeL binds directly to LRP1 and shares a common LRP1 binding site with RAP. As shown in Fig. 2, intoxication of MEF2 cells by TpeL after expression of an LRP1 “mini-receptor” with a shortened heavy chain, containing only cluster IV, indicate that this cluster is sufficient for cell entry of the TpeL toxin. Accordingly, we used recombinantly expressed cluster IV of LRP1 for analysis of the binding kinetics of TpeL to its receptor. TpeL bound with comparable affinity (KD: 23.2 ± 0.2 nM) to cluster IV of LRP1 as Pseudomonas exotoxin A (KD: 5.2 ± 0.3 nM), a known ligand of LRP1. Binding of TpeL to LRP1 is most likely mediated via the C-terminal part of the toxin, because a toxin fragment, covering residues 1335–1779, prevented intoxication of cells. Moreover, surface plasmon resonance spectroscopy revealed the same high-affinity binding of this fragment to LRP1 cluster IV as full-length TpeL.
Binding of TpeL was also observed to LRP1-deficient MEF cells, indicating additional TpeL binding sites on MEF cells. This finding is in line with many ligands of LRP1. For example, internalization of apoE/lipoprotein particles depends on LRP1 and heparin sulfate proteoglycan (HSPG) (49). Similarly, the LRP1 ligands, MMP-2 (prometalloproteinase)/TIMP-2 (tissue inhibitor of MMPs) (50), collagenase-3 protein (51), or amyloid-β peptide (52) require LRP1 and a second receptor. In the case of collagenase-3, a two-step uptake mechanism has been suggested (51). First, collagenase-3 binds to a specific cell membrane protein, and, second, interaction of LRP1 with the collagenase-3/receptor complex results in internalization of collagenase-3. We cannot exclude a similar two-step uptake mechanism for the TpeL protein or additional cell surface molecules that mediate toxin uptake via LRP1 in a cooperative manner. However, because LRP1-deficient MEF cells are resistant toward TpeL and because of the fact that we observe direct binding between TpeL and LRP1, we conclude that LRP1 is the main host cell receptor for this toxin.
Our finding that LRP1 is a cell membrane receptor for TpeL is a major challenge for the current concept of the action of large clostridial glycosylating toxins. According to this model, receptor binding and host cell entry of clostridial glycosylating toxins is mediated by the large CROP domain at the C terminus of the toxins (14, 18, 53). By contrast, TpeL, a member of the same toxin family, exhibits cytotoxicity and high-affinity host cell binding, although it does not possess a C-terminal CROP domain (23). As shown (10, 22) and confirmed here, a C. difficile toxin B fragment, covering residues 1–1811 (lacking the CROP domain) is still capable to intoxicate target cells. This finding could only be explained by an additional receptor-binding site in C. difficile toxin B 1–1811. However, the TpeL receptor LRP1 is not involved in toxin B binding, because MEF1 and MEF2 cells were similarly sensitive toward the CROP-deficient C. difficile toxin B variant.
Also previous studies questioned the simple model of an exclusively CROP-mediated uptake of clostridial glycosylating toxins. For example, Barroso et al. (54) reported that the removal of the CROP domain of C. difficile toxin B reduced, but did not block, cytotoxicity. Furthermore, intoxication studies with C. difficile toxins revealed that the CROP domain itself exhibited weaker competition with the holotoxin than a fragment consisting of the CROP domain plus the intermediate part of the toxin (55).
Thus, all these data support the view that another region, beside the CROP domain, is essentially involved in binding and/or uptake of large clostridial glycosylating toxins, including C. difficile toxins A and B. Where is this region located? Previously, we defined a translocation domain (D or delivery domain) in the ABCD model of clostridial glycosylating toxins, residing between the cysteine protease domain and the CROP domain (6), covering residues ∼830–1851. Subsequent studies narrowed down the region involved in toxin translocation to residues 830 to ∼1500 (10). Using BLAST analysis, Borden Lacy and coworkers suggested that the region between the cysteine protease and the CROP domain of C. difficile toxins A and B may cover two domains, exhibiting two different homology profiles (56). The first region (D1, residues ∼800–1400) corresponded to the translocation domain of clostridial glycosylating toxins, whereas the second region (D2, residues ∼1400–1800) was so far without any attributed function. As discussed above, in TpeL the C-terminal region between residues 1335–1779 is involved in receptor binding. This region of TpeL is 27% identical (∼50% similar) to the region of C. difficile toxin B that lies between the translocation and the CROP domain (Fig. S8). To study a similar receptor binding function of this region in C. difficile toxin B, we expressed the fragment covering residues 1349–1811. As shown in Fig. 3 and Figs. S5 and S6, this fragment bound to HeLa and Vero cells and inhibited the binding of C. difficile toxin B1–1811 (and vice versa). Moreover, toxin B1349–1811 colocalized with the endosomal marker protein EEA1 in CaCo-2 cells, indicating that the toxin fragment not only binds to cells but also triggers endocytic uptake.
Taken together, our findings show that LRP1 is the cell entry receptor for TpeL. Moreover, our data extend the current model for the domain architecture (an updated model is presented in Fig. S9) and for the uptake of clostridial glycosylating toxins. We suggest a two-receptor model for the cell entry of clostridial glycosylating toxins. The CROP domain might primarily facilitate the accumulation of the toxins at the cell surface by interacting primarily with carbohydrates on the cell surface. The additional receptor-binding domain in these toxins might then interact with a specific cell surface protein that acts as the endocytic receptor of the toxins. A similar strategy is used by the botulinum and tetanus neurotoxins for target cell entry (57). A two-receptor model of clostridial glycosylating toxins might explain previous difficulties in receptor identification experienced with these toxins; for example, gene trapping studies are extremely difficult with two functional receptors. Based on the two-receptor model for clostridial glycosylating toxins that we suggest here, haploid genetic screens with variants of toxin A and B that lack one receptor domain are now feasible.
Materials and Methods
Generation and Cultivation of Cells Used in This Study.
Hap1 cells were grown in Iscove's Modified Dulbecco's medium supplemented with 10% (vol/vol) FCS, penicillin (100 U/mL)/streptomycin (100 µg/mL), and 0.2% MycoZap Plus-CL (Lonza). HeLa, Vero, MEF, and CaCo-2 cells were cultivated in DMEM (12 mM l-glutamine) supplemented with 10% (vol/vol) FCS, penicillin (100 U/mL)/streptomycin (100 µg/mL), 0.1 mM nonessential amino acids, and 1 mM sodium pyruvate (Vero and CaCo-2 cells only). Isolation of MEF cells from C57BL/6 LRP1+/+ (MEF1) and LRP1−/− (MEF2) littermate embryos at embryonic date 13 and stable transfection of MEF2 cells with an empty pcDNA3.1 vector (MEF2+EV) or with pcDNA3.1/LRP1 (MEF2+LRP1) is described (58, 59). To obtain MEF1 cells with reduced SNX17 expression levels, cells were transfected with an SNX17-directed shRNA expression vector (pSUPER-SNX17; kindly provided by Reinhard Fässler, Max Planck Institute of Biochemistry, Martinsried, Germany). For the expression of LRP1cluster IV in MEF2 cells, the corresponding gene fragment was first cloned into a pBABE-puro vector by using the In-Fusion PCR Cloning System (Clontech). Then, the resulting plasmid was transfected in the Platinum-E retroviral packaging cell line with PEI (polyethylenimine) (60). Two days after transfection, retrovirus-containing supernatant was collected and applied to cultured MEF2 cells together with 10 µg/mL polybrene. Following an overnight incubation at 37 °C, transduced MEF2 cells were selected for 48 h by the addition of puromycin (5 µg/mL) into the growth medium. All cells were incubated at 37 °C with 5% (vol/vol) CO2 under humidified conditions.
Cloning, Recombinant Production, or Source of Toxins and Proteins Used in This Study.
Full-length TpeL (strain JGS1495) and C. difficile toxin B (strain VPI 10463) were used in this study as recombinant, C-terminally His-tagged proteins. Cloning of the respective toxin genes into the bacterial expression vector pHIS1522, expression in the expression host Bacillus megaterium, and nickel affinity purification of the recombinant toxins was described (7, 24, 61). In this study, the same procedure was used for the recombinant production of the toxin fragments TpeL1335–1779, toxin A1–1849, toxin B1–1811, toxin B1–1348, toxin B1–543, and toxin B1349–1811. Plasmids encoding either GST alone (pGEX-2T) or the fusion protein GST-RAP (RAP; pGEX-KG-RAP; ref. 35) were transformed and expressed in Escherichia coli strain BL21(DE3), followed by the purification of the proteins with Glutathione-Sepharose 4B (GE Healthcare). DNA sequence encoding Pseudomonas exotoxin A (a kind gift from Joseph T. Barbieri, Medical College of Wisconsin, Milwaukee) was cloned in pET28a and transformed in the Escherichia coli strain BL21. The expression and purification of Pseudomonas exotoxin A was performed by nickel affinity purification. LRP1cluster IV-Fc chimera was purchased from R&D Systems (5395-L4-050).
Additional methods were applied in this study and are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Otilia Wunderlich and Sven Hornei for technical assistance, Carsten Schwan and Thilo Nölke for help in microscopy, Joseph T. Barbieri and Reinhard Fässler for sharing plasmids, and Vincent Blomen for help in data analysis. This study was supported by the Deutsche Forschungsgemeinschaft AK6/16-3 and in part by the Excellence Initiative of the German Research Foundation (SFB; GSC-4, Spemann Graduate School). J.H. was supported by grants from the National Institutes of Health (HL63762), the Bright Focus Foundation, the Ted Nash Longlife Foundation, the Consortium for Frontotemporal Dementia Research, and SFB780.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323790111/-/DCSupplemental.
References
- 1.Voth DE, Ballard JD. Clostridium difficile toxins: Mechanism of action and role in disease. Clin Microbiol Rev. 2005;18(2):247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med. 2008;359(18):1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 3.McDonald LC, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353(23):2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 4.Loo VG, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353(23):2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 5.Just I, Gerhard R. Large clostridial cytotoxins. Rev Physiol Biochem Pharmacol. 2004;152(1):23–47. doi: 10.1007/s10254-004-0033-5. [DOI] [PubMed] [Google Scholar]
- 6.Jank T, Aktories K. Structure and mode of action of clostridial glucosylating toxins: The ABCD model. Trends Microbiol. 2008;16(5):222–229. doi: 10.1016/j.tim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Papatheodorou P, Zamboglou C, Genisyuerek S, Guttenberg G, Aktories K. Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS ONE. 2010;5(5):e10673. doi: 10.1371/journal.pone.0010673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reineke J, et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446(7134):415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- 9.Egerer M, Giesemann T, Jank T, Satchell KJ, Aktories K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J Biol Chem. 2007;282(35):25314–25321. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- 10.Genisyuerek S, et al. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol Microbiol. 2011;79(6):1643–1654. doi: 10.1111/j.1365-2958.2011.07549.x. [DOI] [PubMed] [Google Scholar]
- 11.Just I, et al. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375(6531):500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 12.Selzer J, et al. Clostridium novyi α-toxin-catalyzed incorporation of GlcNAc into Rho subfamily proteins. J Biol Chem. 1996;271(41):25173–25177. doi: 10.1074/jbc.271.41.25173. [DOI] [PubMed] [Google Scholar]
- 13.Popoff MR, et al. Ras, Rap, and Rac small GTP-binding proteins are targets for Clostridium sordellii lethal toxin glucosylation. J Biol Chem. 1996;271(17):10217–10224. doi: 10.1074/jbc.271.17.10217. [DOI] [PubMed] [Google Scholar]
- 14.von Eichel-Streiber C, Sauerborn M. Clostridium difficile toxin A carries a C-terminal repetitive structure homologous to the carbohydrate binding region of streptococcal glycosyltransferases. Gene. 1990;96(1):107–113. doi: 10.1016/0378-1119(90)90348-u. [DOI] [PubMed] [Google Scholar]
- 15.Greco A, et al. Carbohydrate recognition by Clostridium difficile toxin A. Nat Struct Mol Biol. 2006;13(5):460–461. doi: 10.1038/nsmb1084. [DOI] [PubMed] [Google Scholar]
- 16.Krivan HC, Clark GF, Smith DF, Wilkins TD. Cell surface binding site for Clostridium difficile enterotoxin: Evidence for a glycoconjugate containing the sequence Gal alpha 1-3Gal beta 1-4GlcNAc. Infect Immun. 1986;53(3):573–581. doi: 10.1128/iai.53.3.573-581.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker KD, Wilkins TD. Toxin A of Clostridium difficile binds to the human carbohydrate antigens I, X, and Y. Infect Immun. 1991;59(1):73–78. doi: 10.1128/iai.59.1.73-78.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauerborn M, Leukel P, von Eichel-Streiber C. The C-terminal ligand-binding domain of Clostridium difficile toxin A (TcdA) abrogates TcdA-specific binding to cells and prevents mouse lethality. FEMS Microbiol Lett. 1997;155(1):45–54. doi: 10.1111/j.1574-6968.1997.tb12684.x. [DOI] [PubMed] [Google Scholar]
- 19.Frey SM, Wilkins TD. Localization of two epitopes recognized by monoclonal antibody PCG-4 on Clostridium difficile toxin A. Infect Immun. 1992;60(6):2488–2492. doi: 10.1128/iai.60.6.2488-2492.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pothoulakis C, et al. Rabbit sucrase-isomaltase contains a functional intestinal receptor for Clostridium difficile toxin A. J Clin Invest. 1996;98(3):641–649. doi: 10.1172/JCI118835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Na X, Kim H, Moyer MP, Pothoulakis C, LaMont JT. gp96 is a human colonocyte plasma membrane binding protein for Clostridium difficile toxin A. Infect Immun. 2008;76(7):2862–2871. doi: 10.1128/IAI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olling A, et al. The repetitive oligopeptide sequences modulate cytopathic potency but are not crucial for cellular uptake of Clostridium difficile toxin A. PLoS ONE. 2011;6(3):e17623. doi: 10.1371/journal.pone.0017623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amimoto K, Noro T, Oishi E, Shimizu M. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology. 2007;153(Pt 4):1198–1206. doi: 10.1099/mic.0.2006/002287-0. [DOI] [PubMed] [Google Scholar]
- 24.Guttenberg G, et al. Molecular characteristics of Clostridium perfringens TpeL toxin and consequences of mono-O-GlcNAcylation of Ras in living cells. J Biol Chem. 2012;287(30):24929–24940. doi: 10.1074/jbc.M112.347773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagahama M, et al. Clostridium perfringens TpeL glycosylates the Rac and Ras subfamily proteins. Infect Immun. 2011;79(2):905–910. doi: 10.1128/IAI.01019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papatheodorou P, et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT) Proc Natl Acad Sci USA. 2011;108(39):16422–16427. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Kerkhof P, et al. Sorting nexin 17 facilitates LRP recycling in the early endosome. EMBO J. 2005;24(16):2851–2861. doi: 10.1038/sj.emboj.7600756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiesberger T, et al. Cellular uptake of saposin (SAP) precursor and lysosomal delivery by the low density lipoprotein receptor-related protein (LRP) EMBO J. 1998;17(16):4617–4625. doi: 10.1093/emboj/17.16.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obermoeller-McCormick LM, et al. Dissection of receptor folding and ligand-binding property with functional minireceptors of LDL receptor-related protein. J Cell Sci. 2001;114(Pt 5):899–908. doi: 10.1242/jcs.114.5.899. [DOI] [PubMed] [Google Scholar]
- 30.Herz J, Strickland DK. LRP: A multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108(6):779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willnow TE, Orth K, Herz J. Molecular dissection of ligand binding sites on the low density lipoprotein receptor-related protein. J Biol Chem. 1994;269(22):15827–15832. [PubMed] [Google Scholar]
- 32.Quinn KA, et al. Soluble low density lipoprotein receptor-related protein (LRP) circulates in human plasma. J Biol Chem. 1997;272(38):23946–23951. doi: 10.1074/jbc.272.38.23946. [DOI] [PubMed] [Google Scholar]
- 33.Willnow TE, et al. RAP, a specialized chaperone, prevents ligand-induced ER retention and degradation of LDL receptor-related endocytic receptors. EMBO J. 1996;15(11):2632–2639. [PMC free article] [PubMed] [Google Scholar]
- 34.Bu G, Geuze HJ, Strous GJ, Schwartz AL. 39 kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J. 1995;14(10):2269–2280. doi: 10.1002/j.1460-2075.1995.tb07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herz J, Goldstein JL, Strickland DK, Ho YK, Brown MS. 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor. J Biol Chem. 1991;266(31):21232–21238. [PubMed] [Google Scholar]
- 36.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326(5957):1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 37.Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor-related protein 1: Unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88(3):887–918. doi: 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herz J, et al. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7(13):4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowal RC, Herz J, Goldstein JL, Esser V, Brown MS. Low density lipoprotein receptor-related protein mediates uptake of cholesteryl esters derived from apoprotein E-enriched lipoproteins. Proc Natl Acad Sci USA. 1989;86(15):5810–5814. doi: 10.1073/pnas.86.15.5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strickland DK, et al. Sequence identity between the alpha 2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J Biol Chem. 1990;265(29):17401–17404. [PubMed] [Google Scholar]
- 41.Kounnas MZ, et al. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell. 1995;82(2):331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- 42.Bu G, Williams S, Strickland DK, Schwartz AL. Low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor is an hepatic receptor for tissue-type plasminogen activator. Proc Natl Acad Sci USA. 1992;89(16):7427–7431. doi: 10.1073/pnas.89.16.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warshawsky I, Bu G, Schwartz AL. LRP and the receptor-mediated endocytosis of plasminogen activators. Ann N Y Acad Sci. 1994;737:70–87. doi: 10.1111/j.1749-6632.1994.tb44302.x. [DOI] [PubMed] [Google Scholar]
- 44.Kounnas MZ, et al. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A. J Biol Chem. 1992;267(18):12420–12423. [PubMed] [Google Scholar]
- 45.Hofer F, et al. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91(5):1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gotthardt M, et al. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J Biol Chem. 2000;275(33):25616–25624. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
- 47.Willnow TE, Moehring JM, Inocencio NM, Moehring TJ, Herz J. The low-density-lipoprotein receptor-related protein (LRP) is processed by furin in vivo and in vitro. Biochem J. 1996;313(Pt 1):71–76. doi: 10.1042/bj3130071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashcom JD, et al. The human alpha 2-macroglobulin receptor: Identification of a 420-kD cell surface glycoprotein specific for the activated conformation of alpha 2-macroglobulin. J Cell Biol. 1990;110(4):1041–1048. doi: 10.1083/jcb.110.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahley RW, Ji ZS. Remnant lipoprotein metabolism: Key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40(1):1–16. [PubMed] [Google Scholar]
- 50.Emonard H, et al. Low density lipoprotein receptor-related protein mediates endocytic clearance of pro-MMP-2.TIMP-2 complex through a thrombospondin-independent mechanism. J Biol Chem. 2004;279(52):54944–54951. doi: 10.1074/jbc.M406792200. [DOI] [PubMed] [Google Scholar]
- 51.Barmina OY, et al. Collagenase-3 binds to a specific receptor and requires the low density lipoprotein receptor-related protein for internalization. J Biol Chem. 1999;274(42):30087–30093. doi: 10.1074/jbc.274.42.30087. [DOI] [PubMed] [Google Scholar]
- 52.Kanekiyo T, et al. Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J Neurosci. 2011;31(5):1644–1651. doi: 10.1523/JNEUROSCI.5491-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Eichel-Streiber C, Sauerborn M, Kuramitsu HK. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutans glucosyltransferases. J Bacteriol. 1992;174(20):6707–6710. doi: 10.1128/jb.174.20.6707-6710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barroso LA, Moncrief JS, Lyerly DM, Wilkins TD. Mutagenesis of the Clostridium difficile toxin B gene and effect on cytotoxic activity. Microb Pathog. 1994;16(4):297–303. doi: 10.1006/mpat.1994.1030. [DOI] [PubMed] [Google Scholar]
- 55.Frisch C, Gerhard R, Aktories K, Hofmann F, Just I. The complete receptor-binding domain of Clostridium difficile toxin A is required for endocytosis. Biochem Biophys Res Commun. 2003;300(3):706–711. doi: 10.1016/s0006-291x(02)02919-4. [DOI] [PubMed] [Google Scholar]
- 56.Pruitt RN, Chambers MG, Ng KK, Ohi MD, Lacy DB. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc Natl Acad Sci USA. 2010;107(30):13467–13472. doi: 10.1073/pnas.1002199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rummel A. Double receptor anchorage of botulinum neurotoxins accounts for their exquisite neurospecificity. Curr Top Microbiol Immunol. 2013;364:61–90. doi: 10.1007/978-3-642-33570-9_4. [DOI] [PubMed] [Google Scholar]
- 58.Zurhove K, Nakajima C, Herz J, Bock HH, May P. Gamma-secretase limits the inflammatory response through the processing of LRP1. Sci Signal. 2008;1(47):ra15. doi: 10.1126/scisignal.1164263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willnow TE, Herz J. Homologous recombination for gene replacement in mouse cell lines. Methods Cell Biol. 1994;43(Pt A):305–334. doi: 10.1016/s0091-679x(08)60610-x. [DOI] [PubMed] [Google Scholar]
- 60.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7(12):1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 61.Yang G, et al. Expression of recombinant Clostridium difficile toxin A and B in Bacillus megaterium. BMC Microbiol. 2008;8(192):192. doi: 10.1186/1471-2180-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.