Significance
Sea stars are emblematic of the seashore. Despite this, their ability to pry open mussels and attach strongly but temporarily to rocks in their environments are poorly understood. Here we report, to our knowledge, the first sequence of a protein, Sea star footprint protein 1 (Sfp1), a primary constituent of the adhesive footprints secreted by sea star tube feet. Sfp1 is unusually large and complex compared with other marine adhesive proteins such as those of mussels. It is translated from a single mRNA and then fragmented into four subunits, which display specific domains that mediate interactions with other proteins present in the adhesive material and on the tube foot surface. After secretion, Sfp1 forms a structural scaffold and appears to provide footprints with cohesion.
Keywords: echinoderms, Asterias rubens, marine adhesion
Abstract
Sea stars adhere firmly but temporarily to various substrata as a result of underwater efficient adhesive secretions released by their tube feet. Previous studies showed that this material is mainly made up of proteins, which play a key role in its adhesiveness and cohesiveness. Recently, we solubilized the majority of these proteins and obtained 43 de novo-generated peptide sequences by tandem MS. Here, one of these sequences served to recover the full-length sequence of Sea star footprint protein 1 (Sfp1), by RT-PCR and tube foot transcriptome analysis. Sfp1, a large protein of 3,853 aa, is the second most abundant constituent of the secreted adhesive. By using MS and Western blot analyses, we showed that Sfp1 is translated from a single mRNA and then cleaved into four subunits linked together by disulphide bridges in tube foot adhesive cells. The four subunits display specific protein-, carbohydrate-, and metal-binding domains. Immunohistochemistry and immunocytochemistry located Sfp1 in granules stockpiled by one of the two types of adhesive cells responsible for the secretion of the adhesive material. We also demonstrated that Sfp1 makes up the structural scaffold of the adhesive footprint that remains on the substratum after tube foot detachment. Taken together, the results suggest that Sfp1 is a major structural protein involved in footprint cohesion and possibly in adhesive interactions with the tube foot surface. In recombinant form, it could be used for the design of novel sea star-inspired biomaterials.
For decades, nature’s adhesion and bonding techniques have fascinated laypeople and scientists alike. Fundamentally, all living organisms are glued together, with most of their components connected by adhesive bonds. There are in addition many organisms for which adhesion is vital in conjunction with food procurement, locomotion, defense, or attachment (1). Robust attachment is particularly crucial in the marine environment, where hydrodynamic forces are strong, recurrent, and directionally unpredictable. That is why adhesion is “a way of life” in the sea where bacteria, algae, and invertebrates produce highly viscous or solid adhesive secretions that are able to bond surfaces tenaciously underwater (2–4). The evolutionary background and biology of the species and the environmental constraints both influence the specific composition and properties of adhesive secretions in a particular organism. The diversity of marine adhesives is therefore huge, but, from a functional point of view, two main types of adhesion have been distinguished (5). Permanent adhesion involves the secretion of an adhesive that hardens with time and is characteristic of sessile organisms that remain in the same place throughout their life (e.g., the attachment of barnacles on rocks) (6, 7). Temporary adhesion, on the contrary, allows simultaneous adhesion and locomotion, thus enabling adult organisms to graze, hunt, or locate a mate, and larval forms to explore immersed surfaces before they metamorphose (e.g., the foot secretions of gastropod molluscs) (8–10). From a molecular point of view, most studies have focused on the permanent adhesives from mussels, tubeworms, and barnacles (7, 11, 12) because these organisms are important macrofoulers of marine structures and the characterization of their adhesives stoked, at least originally, by the need for improved antifouling strategies. Several unique adhesive proteins have been characterized and assigned specific bulk or surface functions. By comparison, temporary adhesives have been studied to a lesser extent, and, although the presence of specific proteins has been highlighted in some organisms, none has been fully characterized so far (10, 13, 14).
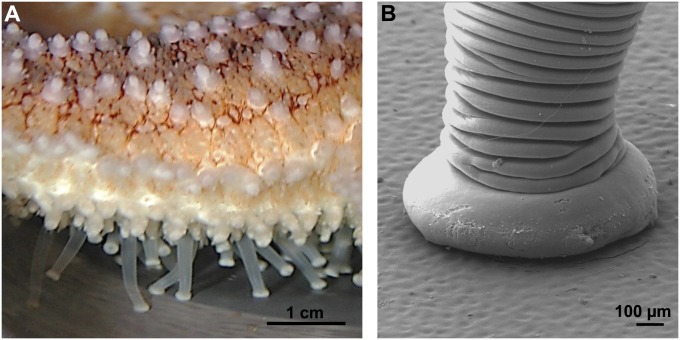
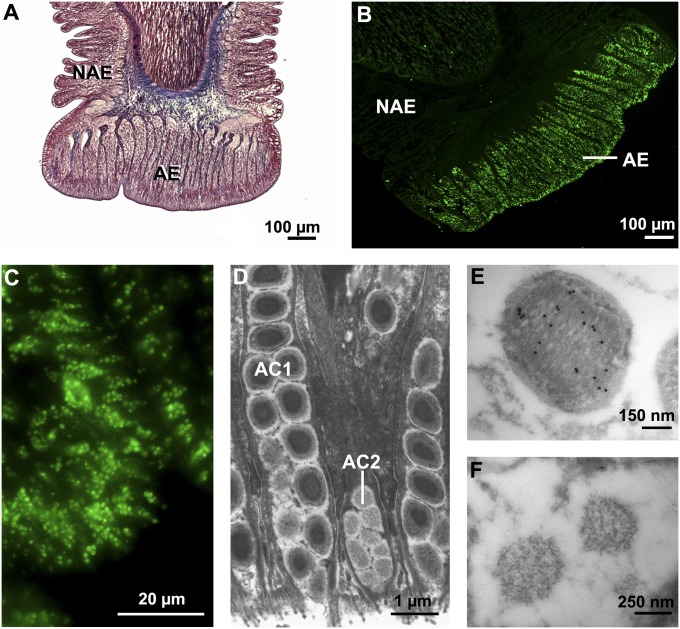
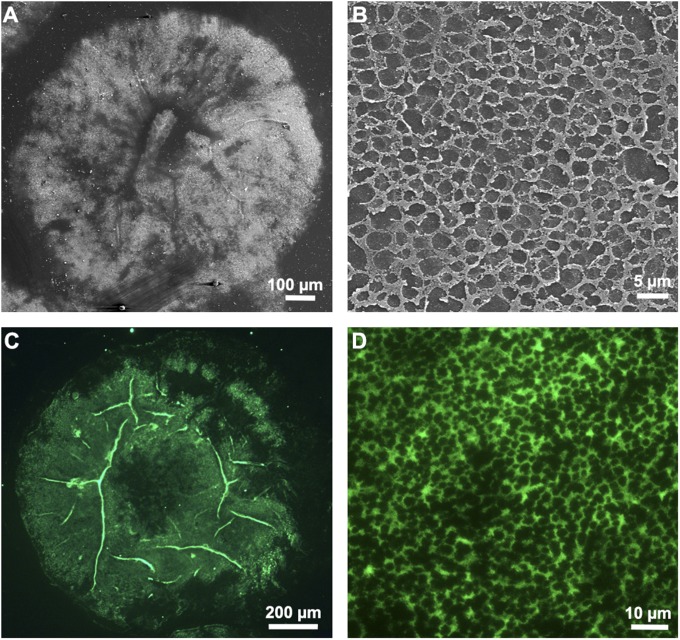
Sea stars represent an interesting model for the study of temporary adhesion (15). Indeed, attachment to underwater substrata is versatile and relies on a multitude of small appendages, the so-called tube feet (Fig. 1). In these organisms, adhesion is multifunctional: for dynamic attachment to the substratum during locomotion, vis-à-vis a static sustained attachment to withstand the action of waves, and, finally, to grip and pry open a mussel during feeding. The distal part of each tube foot, the disk, is specialized for adhesion, and its epidermis comprises two types of adhesive secretory cells (Fig. 2D), which release an adhesive material bonding the tube foot, or, more precisely, the fuzzy coat (the outermost layer of the epidermal cuticle), to the substratum surface (16, 17). Tube feet are also able to detach by releasing an as yet uncharacterized deadhesive substance from another type of secretory cells, the deadhesive cells, that enables each tube foot to disengage from its footprint on the surface of the substratum (16) (Fig. 3A). Morphological and ultrastructural investigations showed that footprints consist of a meshwork of fibrillar material, secreted by type 1 adhesive cells, deposited onto a thin homogeneous film produced by type 2 adhesive cells (17) (Fig. 3B). In previous studies, the disruption of footprints from the substratum by trypsin treatment demonstrated the crucial role of proteins for adhesion and cohesion (5, 18). In the species Asterias rubens, proteins are the main organic components of the adhesive material (20% of the footprint dry weight compared with 8% for carbohydrates and 6% for lipids) (14, 16). Recently, we solubilized the majority of these proteins by using denaturing and reducing conditions (14). Upon gel electrophoresis, they separated into 11 major protein bands in which no homolog proteins were identified by using an MS analysis and homology-database search. Proteins in these bands therefore probably correspond to novel adhesive proteins and were named Sea star footprint proteins (Sfps) (14). The labeling of two of these protein bands in lectin blots demonstrated that they contain glycoproteins with oligosaccharide moieties that comprise galactose, GalNAc, fucose, and sialic acid residues (19). Trypsin-digested Sfps were analyzed by tandem MS (MS/MS), which yielded 43 de novo-generated peptide sequences (14).
Fig. 1.
The tube feet of the sea star A. rubens. (A) Lateral view of one of the five arms showing the tube feet actively involved in locomotion. Their proximal extensible part, the stem, allows movements while their whitish distal part, the disk, makes contact with the substratum. (B) Scanning EM photograph of an attached tube foot. The disk flattens over the surface where it releases an adhesive material.
Fig. 2.
Histology, ultrastructure, and immunolabeling of tube feet in A. rubens. (A) Longitudinal section stained with Heidenhain azan trichrome showing the stem covered by a nonadhesive epidermis (NAE) and the disk presenting a thick adhesive epidermis (AE). (B) Similar section labeled with antibodies directed against Sfp1α (immunofluorescence). The adhesive epidermis of the disk is the only immunoreactive tissue. (C) At higher magnification, secretory granules of adhesive cells are clearly labeled (immunofluorescent labeling with antibodies directed against Sfp1β). (D) Transmission EM photograph showing the two types of adhesive cells enclosed in the adhesive epidermis. AC1, type 1 adhesive cell; AC2, type 2 adhesive cell. (E and F) Immunocytochemical localization of Sfp1 in the secretory granules from AC1 (E) but not from AC2 (F) (immunogold labeling with antibodies directed against Sfp1β).
Fig. 3.
Adhesive footprints left on the substratum after tube foot detachment observed in scanning EM (A, general view; B, detail) and immunolabeled with antibodies directed against Sfp1β (C, general view; D, detail). Immunoreactivity demonstrates that Sfp1 is secreted into the footprints, where it can be localized at the level of the fibrillar meshwork.
In this study, we selected one of these peptides, which was prevalent in a major protein band and lacked sequence ambiguities, and used it to obtain the full-length sequence of a first Sfp, Sfp1. This protein was then characterized in terms of structure, abundance, localization, and function by a combination of in silico and experimental analyses.
Results and Discussion
We designed degenerate oligonucleotide primers on the basis of the peptide sequence HEASGEYYR determined by MS/MS for the Sfps (14). The primers were used with the RT-PCR to amplify the cDNA corresponding to the protein that contains the peptide sequence. In parallel, we performed a tBLASTn search of the peptide sequence in the tube foot transcriptome of A. rubens. Both techniques allowed the recovery of the same 12-kb coding cDNA sequence. A search of the National Center for Biotechnology Information nonredundant database with the cDNA-deduced protein sequence did not find significant homology with any known protein. Consequently, the protein was considered to be novel and named Sfp1 (as the first sea star footprint protein). The full-length protein sequence comprises a 20-aa-long putative signal peptide followed by a proprotein sequence of 3,833 aa with a calculated molecular mass of 426 kDa.
Having determined the complete sequence of Sfp1, it was now possible to estimate its abundance from a batch trypsin digestion and MS analysis of footprint material. The calculation of the exponentially modified protein abundance index, which estimates the relative abundance of proteins in a complex mixture (20), showed that Sfp1 is the second most abundant constituent of the adhesive footprints.
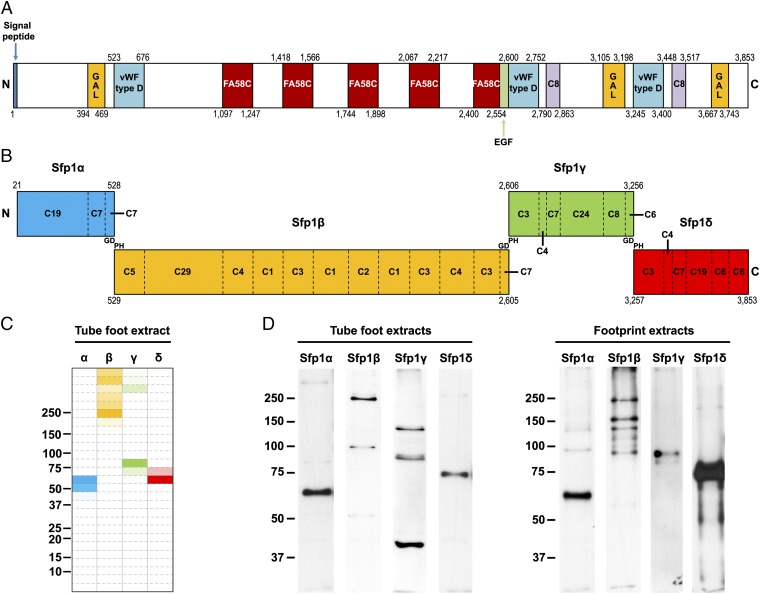
The primary sequence of Sfp1 was also interrogated by the Conserved Domain Database (CDD) of the National Center for Biotechnology Information (21), which revealed that the protein is modular with (i) five coagulation factor 5/8 C-terminal domains (also called discoidin domains), (ii) three von Willebrand factor type D domains (vWFD), (iii) three galactose binding lectin domains, (iv) two C8 domains, and (v) one calcium-binding EGF-like domain (Fig. 4A and Dataset S1). These domains are known from other studies to mediate protein–protein, protein–carbohydrate, or protein–metal interactions (22–25) and could therefore provide cohesive and adhesive interactions between Sfp1 and other glycans and/or proteins present in the adhesive material and in the outermost layer of the tube foot cuticle, respectively (14, 16, 19). To the best of our knowledge, the only other marine adhesive proteins presenting such conserved functional domains are the mussel proteins mussel foot protein 2 (mfp-2), with 11 repeats of EGF-like domains (26, 27), and proximal thread matrix protein 1 (Ptmp-1), with two von Willebrand factor type A domains (28). In mussel byssus, these domains in mfp-2, for example, reinforce interprotein interactions dominated by chelate complexes between the unusual amino acid dopa and Fe3+, leading to self-assembly through bis- or tris-dopa–iron complexes (27). One of the EGF-like motifs in mfp-2 is also able to link calcium and appears to contribute moderately to the cohesion of mussel adhesive plaques (27). Dopa is absent from sea star adhesive material (16), and calcium bridging is therefore the most likely interaction for the EGF domain of Sfp1.
Fig. 4.
Domains and subunits of the protein Sfp1. (A) Main structural domains of Sfp1 predicted by the CDD. Only specific hits as defined by CDD were considered. EGF, calcium-binding EGF-like domain; GAL, galactose binding lectin domain; FA58C, coagulation factor 5/8 C-terminal domain; vWF, von Willebrand factor. (B) The four subunits of Sfp1. Cx denotes the number of cysteine residues in each domain. (C) Detection of Sfp1 peptides in tube foot proteins extracted in a 1.5-M Tris⋅HCl buffer (pH 7.8) containing 0.5 M DTT, 6 M urea, 2% (wt/vol) SDS, and protease inhibitors. The extract was resolved by reducing SDS/PAGE and, after electrophoresis, the gel lane was entirely cut into 27 successive sections, which were analyzed by MS. In the figure, four lanes were created to highlight the distribution of peptides from the four Sfp1 subunits in the different sections of the SDS/PAGE gel. The sections are color-coded according to the abundance of peptides matching each of the four Sfp1 subunits, this abundance being calculated as: (number of peptides observed in the band/maximum number of peptides observed in any band) × 100. Full colors are therefore used for sections presenting the maximum number of peptides matching the subunit (100%). Only peptides with a peptide confidence >95 were considered. (D) Western blot analysis of the four subunits. Proteins extracted from tube feet and adhesive footprints were separated by reducing SDS/PAGE and immunolabeled with polyclonal antibodies directed against one peptide from each subunit. For C and D, molecular weight markers (in kilodaltons) are indicated on the left.
The amino acid composition of Sfp1 is very similar to that of whole footprint material (Table S1). The cysteine content (5%; Fig. 4B and Table S1) in the protein sequence is higher than average for eukaryotic proteins (1–2%) (29). In other cysteine-rich, marine adhesive proteins, such as the proteins mfp-2 from mussels (27) or the protein cement protein 20k (cp-20k) from barnacles (30), the cysteine residues are involved in intramolecular disulfide bonds providing good conformational structure for interaction with the neighboring proteins.
We compared MS/MS data obtained in our previous study of proteins extracted from footprint material and resolved by SDS/PAGE (14) to the sequence of Sfp1 to identify the corresponding gel band. Overall, 23 of the 43 de novo-generated peptides obtained for the Sfps (14) belong to Sfp1. Moreover, peptides with matches in the full-length Sfp1 were detected in all protein bands analyzed, their number varying between 1 and 92 depending on the band in question. It was clear therefore that some protein degradation had occurred in the footprint material, before or during its collection. This degradation could be a result of the tube foot detachment process, whether natural or forced. It has been suggested that the deadhesive material secreted by tube feet could act enzymatically on footprint proteins to enable their release from the tube feet (16). On the contrary, when tube feet are forcefully peeled away from the surface, their tissues can tear and release contaminating proteins including proteases (14). To bypass this problem, we extracted proteins directly from whole tube feet. They were separated by SDS/PAGE, the gel lane was entirely cut into 27 successive sections, and all the pieces were subjected to analysis by MS. Resulting MS/MS data were searched for matches with Sfp1. Matching peptides were retrieved principally from three gel sections with apparent molecular weights of ∼60 kDa, 80 kDa, and 250 kDa (Fig. 4C), and corresponded specifically to four large nonoverlapping fragments of the protein sequence (Fig. 4B and Fig. S1). This suggests that the large precursor protein corresponding to the 12-kb cDNA is processed into four fragments before secretion. To pinpoint the potential cleavage sites between these fragments, we first looked for MS-derived peptide sequences not preceded by the arginine or lysine residue typical of peptides resulting from trypsin digestion. Only two of the peptides matching Sfp1 fulfilled this requirement. The first one corresponds to the N terminus of the first fragment, following the signal peptide, and is therefore not involved in the cleavage site. The second peptide, on the contrary, corresponds to the N-terminal part of the fourth fragment, and its sequence is PHYITFDDVR. In parallel, we aligned the three regions overlapping the four fragments within the Sfp1 sequence, and revealed the presence of a conserved sequence, GDPHY (Fig. S2), precisely comprising the first 3 aa PHY of the fourth fragment’s N-terminal peptide. Interestingly, the sequence GDPH has been described as a site of cleavage in several proteins, such as mucins (31, 32), the heavy chain 3 of the preα-inhibitor (33), repulsive axon guidance molecules (34, 35), and zonadhesin (36). All these proteins are synthesized in the form of precursors which undergo cleavage between the aspartate and proline in the GDPH sequence. In these proteins, cleavage apparently is autocatalytic, occurring at low pH in the late secretory pathway, or at neutral pH in the endoplasmic reticulum (31–33). A similar mechanism could also occur during the progression of Sfp1 from rough endoplasmic reticulum cisternae, through the Golgi apparatus, to the maturing secretory granules in the tube foot adhesive cells (37). It would generate four protein fragments (namely Sfp1α, Sfp1β, Sfp1γ, and Sfp1δ) with calculated molecular weights of, respectively, 57 kDa, 231 kDa, 72 kDa, and 66 kDa (Fig. 4B). These molecular weights correspond closely to those of the gel bands from which the peptides were retrieved, demonstrating the presence of the four fragments in the tube feet.
To further confirm the cleavage of Sfp1, we raised four sets of polyclonal antibodies directed against one peptide selected in each fragment (Fig. S1) and used them in a Western blot of proteins extracted from tube feet and adhesive footprints. In both extracts, labeling was observed for protein bands with an apparent molecular weight corresponding to the four fragments (Fig. 4D). Several other bands were also observed with the antibodies directed against Sfp1β for footprint extracts, meaning that this fragment could be further processed after secretion. When the proteins were extracted from tube feet by using nonreducing buffers, labeling was observed only at the level of the entry of the stacking and running gels and, in some cases, for a protein band with a high molecular weight. When 0.5 M DTT was added to these extracts, the protein bands corresponding to the four fragments were observed again (Fig. S3). These results demonstrate that, inside the tube foot adhesive cells, the four fragments are linked by disulfide bonds and can therefore be considered as four subunits. As DTT is essential for the extraction of footprint proteins (14), it is likely that the four Sfp1 subunits are still linked together through these disulfide bonds within the adhesive footprints after secretion.
We also tested the four sets of polyclonal antibodies in immunohistochemistry and immunocytochemistry on tube foot sections as well as on footprints. In all cases, specific labeling was observed only with the antibodies directed against the Sfp1α and Sfp1β subunits. Immunoreactivity was restricted to tube foot adhesive cells (Fig. 2 B and C), and more precisely to the secretory granules enclosed in type 1 adhesive cells (Fig. 2 E and F). These ellipsoid granules (0.6 × 1 µm) have a complex ultrastructure, most of their volume being occupied by a bundle of parallel rods approximately 30 nm in diameter (37). It is at the level of these rods that we observed an extensive immunoreactivity (Fig. 2E). Within the footprints, the antibodies labeled specifically the fibrillar meshwork (Fig. 3 C and D). Antibodies therefore demonstrate the cellular origin and function of Sfp1: synthesized and packaged in type 1 adhesive cells, this protein has a bulk function and forms the structural core of the adhesive layer after release (see also ref. 17).
To our knowledge, our study is notable as the first identification and characterization of a protein involved in the temporary underwater adhesive of sea stars. The only convergence displayed by Sfp1 with the proteins characterized in the permanent adhesives from other marine organisms lies in its high cysteine content. Conversely, its multimodular organization, with four covalently cross-linked subunits each comprising several putative protein-, carbohydrate-, and metal-binding domains, appears to be unique among marine adhesives. Sfp1 could therefore possess many potential sites for self-interaction or interaction with other yet-unexplored sea star footprint proteins [including those forming the primary adhesive film directly covering the substratum (17)], and correlates well with a cohesive function at the level of the structural scaffold forming the bulk of the footprint. In addition, this complex quaternary structure of Sfp1 could allow adhesive interactions between the footprint meshwork and the outermost layer of the cuticle covering the disk epithelium. As this fuzzy coat consists mostly of proteoglycans (38), the coagulation factor 5/8 C-terminal and galactose binding lectin domains are the best candidates to promote interactions between Sfp1 and the cuticular carbohydrate residues (24, 25). Additional cross-linking possibilities between Sfp1 and other molecules within the adhesive material and cuticle could be provided by free cysteines, as proposed for the multimerization of vWFD (39) as well as by the reactive aspartate residues (anhydride) generated by Sfp1 cleavage, as proposed for preα-inhibitor and mucins (31, 40).
In A. rubens, previous investigations have clearly demonstrated that the adhesive material is released by the two types of adhesive secretory cells occurring in the disk epidermis of the tube feet (16, 17, 37). Type 2 adhesive cells are the first to release their contents and are responsible for the formation of the homogeneous film covering the substratum. Type 1 adhesive cells then start to release their contents, a prepacked array of Sfp1-containing rods that self-assemble into a fibrillar meshwork. The thickness of this meshwork layer appears to be influenced by the substratum on which it is deposited. Indeed, more fibrillar material is deposited on surfaces to which the animal adheres strongly (e.g., glass and mica) than on surfaces on which adhesion is weak (e.g., Teflon) (17). The secretion of Sfp1 therefore appears to be correlated to strong adhesion and could be triggered by sensory cells via the nerve plexus (37). In other sea star species, much less information is available. Interestingly, however, many species do not rely on strong adhesion for their activities and several species possess type 1 adhesive cells with granules lacking internal rods, or are even devoid of this cell type (5, 41). It will be interesting therefore to investigate the phylogenetic distribution of Sfp1 homologs in the class Asteroidea.
The bioadhesives of sessile marine organisms have attracted growing attention during the past 30 y and inspired numerous technological developments, including water-resistant biomedical adhesives or biomaterials (42, 43). The molecular layout of Sfp1, notably its multimodular structure, provides a relatively unexplored design paradigm especially for engineering specific adhesion between different tissues for potential application as adhesives and sealants for surgery, as tissue-engineering scaffolds, or as mucoadhesive hydrogels for drug delivery. To date, most of the bioinspired adhesive proteins and polymers have relied on dopa functionalities that require challenging and costly processing, e.g., in recombinant proteins, the enzymatic conversion of targeted tyrosines into dopa (44). The apparent absence of this posttranslationally modified amino acid in Sfp1 (14) renders it an easier target for recombinant production. Although its unusually large size could pose challenges, production or synthesis of smaller functional subunits or domains have proven useful for mimicking barnacle adhesive proteins [cp-20k (45)], and is likely to provide a suitable basis for further development of Sfp1-inspired materials.
Materials and Methods
Molecular Biology.
Total RNA was extracted from tube feet with TRI Reagent (Applied Biosystems) and reverse transcription (RT)-3′RACE was carried out by using the FirstChoice RLM-RACE kit (Ambion). Degenerate primers were designed on the basis of one of the peptide sequences obtained by MS/MS (14). The amplified fragment was used to design new primers that were used in 5′RACE to obtain the full sequence of the cDNA coding for Sfp1.
MS.
Proteins extracted from tube feet were resolved on a 4–20% (wt/vol) Mini-PROTEAN TGX precast polyacrylamide gel (Bio-Rad). The gel lane was entirely cut up into successive sections, and all pieces were digested with trypsin (Promega). The tryptic peptides were analyzed by reverse-phase HPLC–electrospray ionization MS/MS by using an Eksigent Ultra Plus nano-LC 2D HPLC system connected to a quadrupole time-of-flight TripleTOF 5600 mass spectrometer (AB SCIEX). MS/MS data were compared with the translated tube foot transcriptome using ProteinPilot Software v4.1 (AB SCIEX). For relative abundance estimation, proteins were extracted from footprints and digested without prior separation. MS/MS data were analyzed by using MASCOT Daemon software (Matrix Sciences).
Antibodies.
One peptide from each Sfp1 subunit was selected on the basis of its potential for successful synthesis and immunogenicity for polyclonal antibodies production in rabbits (Eurogentec). The antibodies were isolated from the crude serum by affinity purification by using the synthetic peptides (Eurogentec).
Western Blotting.
Adhesive footprints were collected as described previously (14). Tube feet were rapidly dissected and frozen in liquid nitrogen. Proteins from both materials were extracted in the buffers detailed in the legend of Fig. 4, centrifuged, and loaded on 8% (wt/vol) SDS/PAGE gels. After electrophoresis, the proteins were transferred to PVDF membranes. The blots were probed with the four sets of purified polyclonal antibodies, followed by HRP-conjugated anti-rabbit antibodies (Amersham) and chemiluminescence detection (Roche).
Immunohistochemistry.
Tube feet were fixed in 4% (wt/vol) paraformaldehyde (PAF) in sodium phosphate buffer (i.e., PBS solution, pH 7.4), rinsed in PBS solution, dehydrated through an ethanol series, embedded in paraffin, and sectioned longitudinally as previously described (14). The sections were probed with the four sets of purified polyclonal antibodies, followed by Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulins (Invitrogen), and observed by using a Zeiss Axioscope A1 microscope [excitation filter, band-pass (BP) 474/40 nm; beam splitter filter, farb teiler (FT) 500 nm; emission filter, BP 530/50 nm].
Footprint Immunolabeling.
Footprints were fixed in 4% (wt/vol) PAF in PBS solution and probed with the four sets of purified polyclonal antibodies as for tube foot immunohistochemistry.
Immunocytochemistry.
Tube feet were fixed on ice with 4% (wt/vol) PAF, 0.25% glutaraldehyde in PBS solution (pH 8.0), rinsed in PBS solution, dehydrated through an ethanol series, and embedded in LR White Resin (Fluka). Ultrathin (∼70 nm) longitudinal sections were probed with the four sets of purified antibodies, followed by goat anti-rabbit immunoglobulins conjugated to 15-nm gold particles (BB International). They were further stained with aqueous uranyl acetate and lead citrate and observed with a Zeiss LEO 906E transmission electron microscope.
A full description of the methods and associated references are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Benoit Hennuy (GIGA-Genomics) for the sequencing of tube foot transcriptome and Nathan Puozzo for his help in the design of Fig. 4C. This work was supported by Service Public de Wallonie–Programme Winnomat 2 (Project BIOMADH 0716661), by Communauté française de Belgique–Actions de Recherche Concertées (Project ARC AUWB-2008-08/12-UMH15), and by European Cooperation in Science and Technology Action TD0906. E.H. and P.F. are, respectively, Postdoctoral Researcher and Research Director of the Fund for Scientific Research of Belgium (F.R.S.-FNRS). This study is a contribution of the Centre Interuniversitaire de Biologie Marine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KJ472215).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400089111/-/DCSupplemental.
References
- 1.Gorb SN. Biological attachment devices: Exploring nature’s diversity for biomimetics. Phil Trans R Soc A. 2008;366(1870):1557–1574. doi: 10.1098/rsta.2007.2172. [DOI] [PubMed] [Google Scholar]
- 2.Walker G. In: Synthetic Adhesives and Sealants. Wake WC, editor. Chichester, UK: Wiley; 1987. pp. 112–135. [Google Scholar]
- 3.Smith AM, Callow JA. Biological Adhesives. Berlin: Springer-Verlag; 2006. [Google Scholar]
- 4.von Byern J, Grunwald I. Biological Adhesive Systems – From Nature to Technical and Medical Application. Berlin: Springer-Verlag; 2010. [Google Scholar]
- 5.Flammang P. In: Echinoderm Studies. Jangoux M, Lawrence JM, editors. Rotterdam: Balkema; 1996. [Google Scholar]
- 6.Silverman HG, Roberto FF. Understanding marine mussel adhesion. Mar Biotechnol (NY) 2007;9(6):661–681. doi: 10.1007/s10126-007-9053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamino K. Molecular design of barnacle cement in comparison with those of mussel and tubeworm. J Adhes. 2010;86(1):96–110. [Google Scholar]
- 8.Whittington ID, Cribb BW. Adhesive secretions in the Platyhelminthes. Adv Parasitol. 2001;48:101–224. doi: 10.1016/s0065-308x(01)48006-7. [DOI] [PubMed] [Google Scholar]
- 9.Flammang P, Santos R, Haesaerts D. In: Marine Molecular Biotechnology: Echinodermata. Matranga V, editor. Berlin: Springer-Verlag; 2005. [Google Scholar]
- 10.Aldred N, Clare AS. The adhesive strategies of cyprids and development of barnacle-resistant marine coatings. Biofouling. 2008;24(5):351–363. doi: 10.1080/08927010802256117. [DOI] [PubMed] [Google Scholar]
- 11.Lee BP, Messersmith PB, Israelachvili JN, Waite JH. Mussel-inspired adhesives and coatings. Annu Rev Mater Res. 2011;41:99–132. doi: 10.1146/annurev-matsci-062910-100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang CS, Stewart RJ. Localization of the bioadhesive precursors of the sandcastle worm, Phragmatopoma californica (Fewkes) J Exp Biol. 2012;215(2):351–361. doi: 10.1242/jeb.065011. [DOI] [PubMed] [Google Scholar]
- 13.Smith AM, Quick TJ, St Peter RLS. Differences in the composition of adhesive and non-adhesive mucus from the limpet Lottia limatula. Biol Bull. 1999;196(1):34–44. doi: 10.2307/1543164. [DOI] [PubMed] [Google Scholar]
- 14.Hennebert E, Wattiez R, Waite JH, Flammang P. Characterization of the protein fraction of the temporary adhesive secreted by the tube feet of the sea star Asterias rubens. Biofouling. 2012;28(3):289–303. doi: 10.1080/08927014.2012.672645. [DOI] [PubMed] [Google Scholar]
- 15.Hennebert E. In: Biological Adhesive Systems. von Byern J, Grunwald I, editors. Berlin: Springer-Verlag; 2010. [Google Scholar]
- 16.Flammang P, Michel A, Van Cauwenberge A, Alexandre H, Jangoux M. A study of the temporary adhesion of the podia in the sea star Asterias rubens (Echinodermata, Asteroidea) through their footprints. J Exp Biol. 1998;201(16):2383–2395. doi: 10.1242/jeb.201.16.2383. [DOI] [PubMed] [Google Scholar]
- 17.Hennebert E, Viville P, Lazzaroni R, Flammang P. Micro- and nanostructure of the adhesive material secreted by the tube feet of the sea star Asterias rubens. J Struct Biol. 2008;164(1):108–118. doi: 10.1016/j.jsb.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Thomas LA, Hermans CO. Adhesive interactions between the tube feet of a starfish, Leptasterias hexactis, and substrata. Biol Bull. 1985;169:675–688. [Google Scholar]
- 19.Hennebert E, Wattiez R, Flammang P. Characterization of the carbohydrate fraction of the temporary adhesive secreted by the tube feet of the sea star Asterias rubens. Mar Biotechnol (NY) 2011;13(3):484–495. doi: 10.1007/s10126-010-9319-6. [DOI] [PubMed] [Google Scholar]
- 20.Ishihama Y, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Marchler-Bauer A, et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer D, Girma J-P. von Willebrand factor: Structure and function. Thromb Haemost. 1993;70:99–104. [PubMed] [Google Scholar]
- 23.Rao Z, et al. The structure of a Ca2+-binding epidermal growth factor-like domain: Its role in protein-protein interactions. Cell. 1995;82(1):131–141. doi: 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 24.Baumgartner S, Hofmann K, Chiquet-Ehrismann R, Bucher P. The discoidin domain family revisited: New members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998;7(7):1626–1631. doi: 10.1002/pro.5560070717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tateno H. SUEL-Related Lectins, a lectin family widely distributed throughout organisms. Biosci Biotechnol Biochem. 2010;74(6):1141–1144. doi: 10.1271/bbb.100086. [DOI] [PubMed] [Google Scholar]
- 26.Inoue K, Takeuchi Y, Miki D, Odo S. Mussel adhesive plaque protein gene is a novel member of epidermal growth factor-like gene family. J Biol Chem. 1995;270(12):6698–6701. doi: 10.1074/jbc.270.12.6698. [DOI] [PubMed] [Google Scholar]
- 27.Hwang DS, et al. Protein- and metal-dependent interactions of a prominent protein in mussel adhesive plaques. J Biol Chem. 2010;285(33):25850–25858. doi: 10.1074/jbc.M110.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C, Lucas JM, Waite JH. Collagen-binding matrix proteins from elastomeric extraorganismic byssal fibers. Biomacromolecules. 2002;3(6):1240–1248. doi: 10.1021/bm0255903. [DOI] [PubMed] [Google Scholar]
- 29.Miseta A, Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol Biol Evol. 2000;17(8):1232–1239. doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- 30.He L-S, Zhang G, Qian P-Y. Characterization of two 20kDa-cement protein (cp20k) homologues in Amphibalanus Amphitrite. PLoS ONE. 2013;8:e64130. doi: 10.1371/journal.pone.0064130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lidell ME, Johansson MEV, Hansson GC. An autocatalytic cleavage in the C Terminus of the human MUC2 mucin occurs at the low pH of the late secretory pathway. J Biol Chem. 2003;278(16):13944–13951. doi: 10.1074/jbc.M210069200. [DOI] [PubMed] [Google Scholar]
- 32.Lidell ME, Hansson GC. Cleavage in the GDPH sequence of the C-terminal cysteine-rich part of the human MUC5AC mucin. Biochem J. 2006;399(1):121–129. doi: 10.1042/BJ20060443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thuveson M, Fries E. The low pH in trans-Golgi triggers autocatalytic cleavage of pre-α-inhibitor heavy chain precursor. J Biol Chem. 2000;275(40):30996–31000. doi: 10.1074/jbc.M002399200. [DOI] [PubMed] [Google Scholar]
- 34.Monnier PP, et al. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419(6905):392–395. doi: 10.1038/nature01041. [DOI] [PubMed] [Google Scholar]
- 35.Zhang A-S, West AP, Wyman AE, Bjorkman PJ, Enns CA. Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. J Biol Chem. 2005;280(40):33885–33894. doi: 10.1074/jbc.M506207200. [DOI] [PubMed] [Google Scholar]
- 36.Bi M, Hickox JR, Winfrey VP, Olson GE, Hardy DM. Processing, localization and binding activity of zonadhesin suggest a function in sperm adhesion to the zona pellucida during exocytosis of the acrosome. Biochem J. 2003;375(2):477–488. doi: 10.1042/BJ20030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flammang P, Demeulenaere S, Jangoux M. The role of podial secretions in adhesion in two species of sea stars (Echinodermata) Biol Bull. 1994;187(1):35–47. doi: 10.2307/1542163. [DOI] [PubMed] [Google Scholar]
- 38.Ameye L, Hermann R, Dubois P, Flammang P. Ultrastructure of the echinoderm cuticle after fast-freezing/freeze substitution and conventional chemical fixations. Microsc Res Tech. 2000;48(6):385–393. doi: 10.1002/(SICI)1097-0029(20000315)48:6<385::AID-JEMT8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 39.Mayadas TN, Wagner DD. Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc Natl Acad Sci USA. 1992;89:3531–3535. doi: 10.1073/pnas.89.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enghild JJ, et al. Chondroitin 4-sulfate covalently cross-links the chains of the human blood protein pre-α-inhibitor. J Biol Chem. 1991;266(2):747–751. [PubMed] [Google Scholar]
- 41.Santos R, Haesaerts D, Jangoux M, Flammang P. Comparative histological and immunohistochemical study of sea star tube feet (Echinodermata, Asteroidea) J Morphol. 2005;263(3):259–269. doi: 10.1002/jmor.10187. [DOI] [PubMed] [Google Scholar]
- 42.Waite JH, Andersen NH, Jewhurst S, Sun C. Mussel adhesion: Finding the tricks worth mimicking. J Adhes. 2005;81(3):297–317. [Google Scholar]
- 43.Lee H, Lee BP, Messersmith PB. A reversible wet/dry adhesive inspired by mussels and geckos. Nature. 2007;448(7151):338–341. doi: 10.1038/nature05968. [DOI] [PubMed] [Google Scholar]
- 44.Taylor SW. Chemoenzymatic synthesis of peptidyl 3,4-dihydroxyphenylalanine for structure-activity relationships in marine invertebrate polypeptides. Anal Biochem. 2002;302(1):70–74. doi: 10.1006/abio.2001.5522. [DOI] [PubMed] [Google Scholar]
- 45.Nakano M, Shen J-R, Kamino K. Self-assembling peptide inspired by a barnacle underwater adhesive protein. Biomacromolecules. 2007;8(6):1830–1835. doi: 10.1021/bm0612236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.