Significance
Biogenesis of iron–sulfur proteins is a complex process requiring a large number of accessory proteins. In eukaryotes, [2Fe-2S] clusters are synthesized in mitochondria on a scaffold protein. The cluster is then released to monothiol glutaredoxin 5 (GRX5), which was proposed to mediate the transfer of [2Fe-2S] clusters from the scaffold protein to several target proteins, but its precise molecular function remains to be clarified. By investigating the molecular recognition between human GRX5 and its partner proteins (human ISCA1 and ISCA2) and characterizing at the molecular level the cluster transfer process between them, we have shown that a switch between two conformational states of holo GRX5 drives the cluster transfer event, which occurs by a specific protein–protein recognition process.
Keywords: Fe/S protein maturation, [2Fe-2S] cluster transfer mechanism, monothiol Grxs, NMR
Abstract
Monothiol glutaredoxins play a crucial role in iron–sulfur (Fe/S) protein biogenesis. Essentially all of them can coordinate a [2Fe-2S] cluster and have been proposed to mediate the transfer of [2Fe-2S] clusters from scaffold proteins to target apo proteins, possibly by acting as cluster transfer proteins. The molecular basis of [2Fe-2S] cluster transfer from monothiol glutaredoxins to target proteins is a fundamental, but still unresolved, aspect to be defined in Fe/S protein biogenesis. In mitochondria monothiol glutaredoxin 5 (GRX5) is involved in the maturation of all cellular Fe/S proteins and participates in cellular iron regulation. Here we show that the structural plasticity of the dimeric state of the [2Fe-2S] bound form of human GRX5 (holo hGRX5) is the crucial factor that allows an efficient cluster transfer to the partner proteins human ISCA1 and ISCA2 by a specific protein–protein recognition mechanism. Holo hGRX5 works as a metallochaperone preventing the [2Fe-2S] cluster to be released in solution in the presence of physiological concentrations of glutathione and forming a transient, cluster-mediated protein–protein intermediate with two physiological protein partners receiving the [2Fe-2S] cluster. The cluster transfer mechanism defined here may extend to other mitochondrial [2Fe-2S] target proteins.
Glutaredoxins (Grxs) and glutathione (GSH) are universally distributed among all organisms, and they have been shown to play a fundamental role in iron–sulfur (Fe/S) protein biogenesis (1–5). Specifically, the [2Fe-2S]–bound forms of monothiol Grxs and a [2Fe-2S]–glutathione complex are the species suggested to be responsible for trafficking [2Fe-2S] clusters within the cell (6–9). The current working model is that in the cell monothiol Grxs receive a [2Fe-2S] cluster from the scaffold protein ISCU (where de novo synthesis of the [2Fe-2S] cluster occurs) and transfer it to specific targeting proteins, which then facilitate Fe/S cluster insertion into the final acceptor apo protein (7, 10, 11). Another possible cluster transfer mechanism, which has been proposed (8), hypothesizes the cellular presence of a [2Fe-2S](GS)4 complex, which could transiently store [2Fe-2S] clusters, facilitate cluster exchange with the cellular Fe/S cluster biosynthesis machineries, and regulate the biosynthesis of Fe/S clusters. However, a drawback of the latter model is that all of the Fe/S cellular trafficking processes will result to be protein-independent and therefore highly unspecific, thus potentially inflicting severe cellular damage.
The mitochondrial, monothiol glutaredoxin 5 protein (GRX5) belongs to the core part of the mitochondrial Fe/S cluster (ISC) assembly system (10, 12, 13), is required in the maturation of all cellular [2Fe-2S] and [4Fe-4S] proteins (11), and participates in cellular iron regulation (14). Human GRX5 in vitro binds a [2Fe-2S] cluster (15) and yeast GRX5, which in vivo and in vitro binds a [2Fe-2S] cluster (11), has been shown to interact with two highly homologous mitochondrial proteins, in humans named ISCA1 and ISCA2 (16, 17). In eukaryotes, these two proteins were reported to bind in vitro either a [2Fe-2S] cluster (18) or an iron ion (19, 20). It has been also suggested (19) that the ISCA proteins in vivo bind a Fe/S cofactor under normal circumstances and that they may only switch to an iron ion binding mode in cells either with a compromised mitochondrial ISC assembly system or upon ISCA overproduction. In eukaryotes, ISCA1 and ISCA2 were found to be required for the maturation of mitochondrial [4Fe-4S] proteins but to be dispensable for the generation of mitochondrial [2Fe-2S] proteins and for heme biosynthesis, thus performing a specific task late in the biosynthetic pathway of mitochondrial [4Fe-4S] proteins (19, 21). In this work we unraveled the molecular mechanism of the transfer of a [2Fe-2S] cluster from human GRX5 to its two partner proteins, human ISCA1 and ISCA2, showing that the structural plasticity of human GRX5 is the crucial factor for acting as a [2Fe-2S] transfer protein.
Results and Discussion
Interaction of Glutathione with Apo and Holo hGRX5.
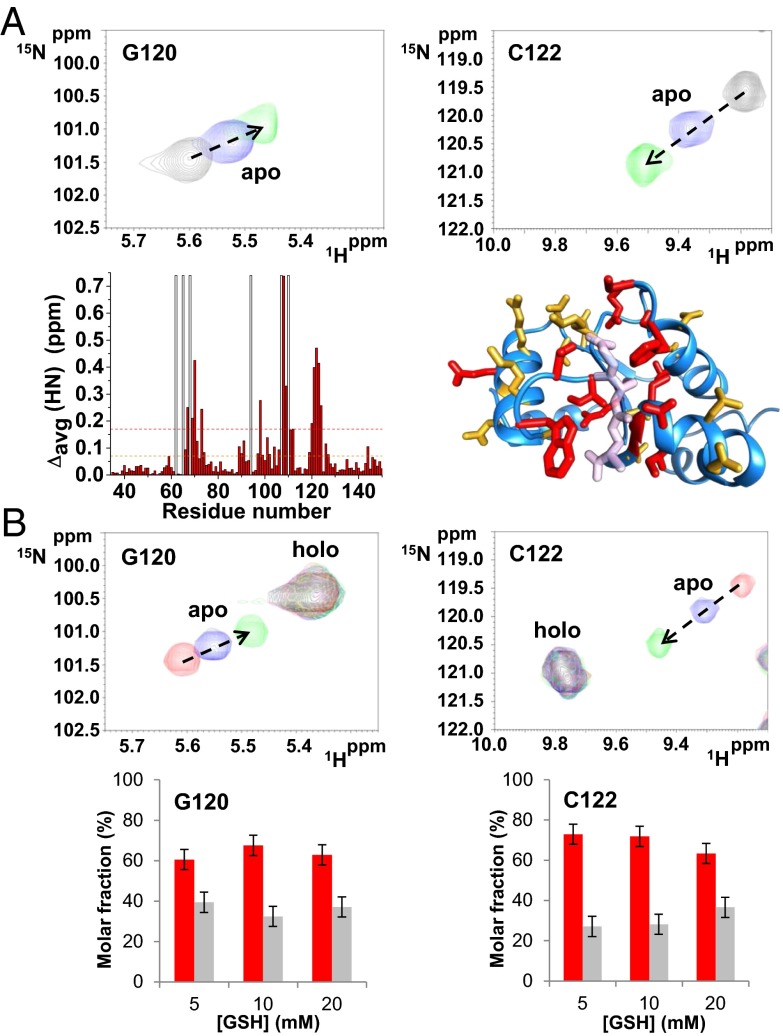
Fe/S/protein quantification, UV/visible and paramagnetic 1D 1H NMR data (Fig. S1 and SI Text) indicate that human holo GRX5 (holo hGRX5, hereafter) quantitatively binds one [2Fe-2S] cluster per two GRX5 molecules. Solution NMR interaction studies on hGRX5 titrated with GSH show that one GSH per protein molecule is involved in cluster binding (i.e., 2 GSH:2 hGRX5:1 [2Fe-2S] ratio), in agreement with the available crystallographic structures of monothiol [2Fe-2S] glutaredoxins (15, 22–24). Specifically, along the titration of apo hGRX5 with GSH the chemical shifts of a restricted set of signals of apo hGRX5 change upon increasing amounts of GSH, identifying a well-defined GSH binding site (Fig. 1A). Chemical shift mapping analysis shows that this GSH binding region matches with the GSH binding site observed in the crystallographic structure of holo hGRX5 (Fig. 1A). The chemical shifts of all signals of apo hGRX5 affected by GSH interaction move toward the corresponding chemical shifts of holo hGRX5 (Fig. 1B), indicating that the iron-bound GSH molecule of holo hGRX5 is located at the same GSH binding site identified in apo hGRX5. Finally, because the chemical shifts of holo hGRX5 in the 1H-15N heteronuclear single-quantum coherence (HSQC) map (Fig. 1B) do not change by adding large excess of GSH, the cluster binding site in holo hGRX5 is fully saturated by the two GSH molecules.
Fig. 1.
Interaction of glutathione with apo and holo hGRX5. Chemical shift changes of G120 and C122 residues induced by the addition of different GSH concentrations (5, 10, and 20 mM) in apo (A) and holo hGRX5 (B) (the latter sample contains a residual 30% of apo hGRX5) are shown. (A) Backbone weighted average chemical shift differences Δavg(HN) [i.e., ((ΔH)2 + (ΔN/5)2)/2)1/2, where ΔH and ΔN are chemical shift differences for 1H and 15N, respectively] observed upon addition of 20 mM GSH to 15N-labeled apo hGRX5. White bars indicate Pro residues and the unassigned backbone NH of Gly-68. Thresholds of 0.17 ppm (mean value of Δavg(HN) plus 1σ, red dashed line) and a lower one of 0.07 ppm (orange dashed line) were used to identify primary and secondary effects on the chemical shift changes. Residues with Δavg(HN) values greater than 0.17 ppm and 0.07 ppm are shown in red and orange, respectively, on a subunit of the tetrameric crystal structure of holo hGRX5. GSH molecule is shown in light pink. (B) Molar fractions of the apo (gray) and holo (red) forms of Gly-120 and Cys-122 residues are reported at increasing GSH concentrations. The values were obtained integrating the corresponding NH signals in the 1H-15N HSQC maps recorded on holo hGRX5 samples containing 5, 10, and 20 mM GSH concentrations. Error bars show the SD from two independent experiments.
[2Fe-2S]2+ Cluster Transfer and Protein–Protein Interaction Between hGRX5 and hISCA1.
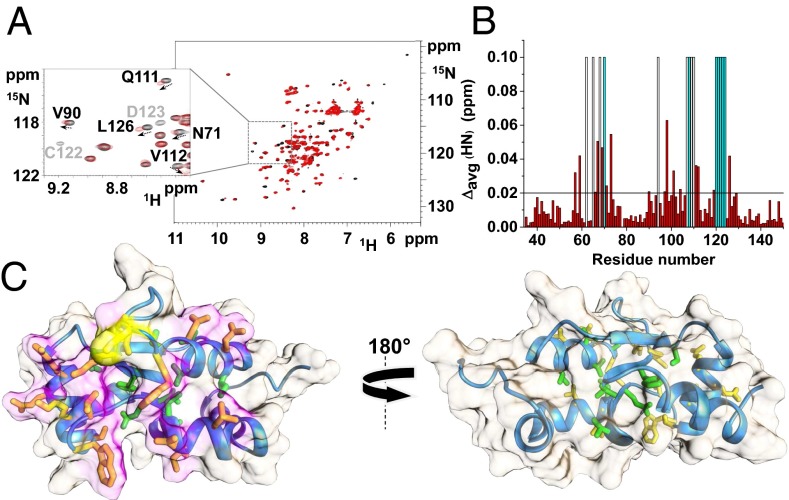
When holo hGRX5 was incubated with apo human ISCA1 (hISCA1 hereafter) in a ∼1:1 ratio, cluster transfer from hGRX5 to hISCA1 occurred. This was monitored by UV/visible spectra recorded on the isolated proteins before mixing and after reaction and separating them through nickel-affinity chromatography thanks to the presence of a His6-tag on hISCA1. The typical absorbance peaks of the oxidized [2Fe-2S]2+ cluster-bound form of hGRX5 (322, 412, and 455 nm, Fig. S1) were drastically reduced in the UV/visible spectra of hGRX5 once incubated and separated from hISCA1 (Fig. 2A), whereas the absorbance peaks, typical of the oxidized [2Fe-2S]2+ cluster-bound form of hISCA1 (322, 416, and 457 nm, holo hISCA1, Fig. S1), appeared in the spectra of hISCA1 once separated from hGRX5 after incubation (Fig. 2B). These data demonstrate the transfer of a [2Fe-2S]2+ cluster from hGRX5 to hISCA1. The presence of a [2Fe-2S] cluster bound to hISCA1, after its incubation with holo hGRX5 and its separation from the mixture, was also confirmed by the EPR spectrum of the dithionite-reduced cluster, which showed g values typical of a S = 1/2 [2Fe-2S]+ cluster as found in [2Fe-2S] ferredoxins (Fig. S1). The reverse reaction, that is, incubation of holo hISCA1 with apo hGRX5 under the same conditions, did not produce any change in the UV/visible spectra, indicating that the cluster transfer has a unique direction (Fig. 2C). 1H-15N HSQC NMR experiments of 15N-labeled hGRX5 were exploited to investigate whether the cluster transfer occurs through a specific protein–protein interaction process. To this end, apo hGRX5 was mixed with holo hISCA1 because, with these two protein forms, no cluster transfer takes place, and therefore chemical shift variations can directly monitor the protein–protein interaction event only. Chemical shift changes in the 1H-15N HSQC spectrum of 15N-labeled apo hGRX5 were observed at increasing amounts of unlabeled holo hISCA1, whereas a few signals broadened beyond detection (Fig. 3A). These spectral changes indicate that apo hGRX5 is interacting with holo hISCA1, consistently with the formation of a protein–protein adduct. The observed chemical shift variations (Fig. 3B), once mapped on the solution structure of apo hGRX5 (described below), indicate that the interacting surface involves the protein region surrounding Cys-67 (i.e., where the cluster and GSH binding occurs) (Fig. 3C). 15N backbone NMR relaxation experiments, performed on 15N-labeled apo hGRX5 in absence or in presence of two equivalents of holo hISCA1, showed an increase of the molecular tumbling value (τm) from 7.97 ± 0.63 ns (consistent with a monomeric protein state in solution; for details see SI Text) to 9.72 ± 0.7 ns. This small increase indicates the formation of a low-populated protein–protein adduct in solution. Chemical shift and heteronuclear relaxation NMR data therefore indicate the presence of an equilibrium between the protein–protein adduct and the free proteins, which is largely shifted toward the free proteins. A possible mechanism that would release the cluster free in solution to be then transferred to apo hISCA1 can be excluded because addition of GSH, even at large excess, is not able to quantitatively extract the [2Fe-2S] cluster from holo hGRX5 by competing with the two Cys protein ligands (Fig. 1B). This result indicates that the cluster in hGRX5 is stably bound and is not released in solution. Overall, the data are consistent with the cluster transfer occurring via an associative process that involves a specific protein–protein interaction event, in agreement with what was reported for homologous systems (6).
Fig. 2.
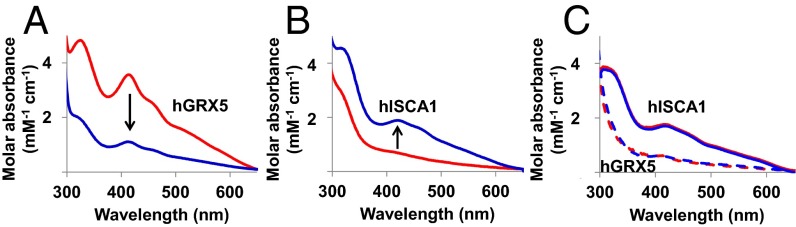
[2Fe-2S]2+ cluster transfer between hGRX5 and hISCA1 followed by UV/visible spectroscopy. UV/visible spectra of holo hGRX5 (A) and His-tagged apo hISCA1 (B) were recorded before (red) and after (blue) incubation in a ∼1:1 protein ratio and their separation through nickel-affinity chromatography. (C) Reverse titration between apo hGRX5 (dashed lines) and His-tagged, holo hISCA1 (solid lines) before (red) and after (blue) their incubation and separation.
Fig. 3.
Complex formation between hGRX5 and hISCA1. (A) 1H-15N HSQC spectrum of 15N-labeled apo hGRX5 before (black) and after (red) the addition of two equivalents of unlabeled holo hISCA1. (Inset) Shift of signals (indicated by an arrow) and beyond detection broadening effects (indicated in light gray) observed upon the addition of holo hISCA1. (B) Backbone weighted average chemical shift differences Δavg(HN) between 15N-labeled apo hGRX5 and the 2:1 unlabeled holo hISCA1/15N-labeled apo hGRX5 mixture. White bars indicate Pro residues and the unassigned backbone NHs of Gly-68 (in both apo form and mixture) and Ile-109 (in the mixture). Cyan bars indicate residues whose backbone NH signals broaden beyond detection at 2:1 protein ratio. A threshold of 0.02 ppm [mean value of Δavg(HN) plus 1σ] was used to identify meaningful chemical shift differences. (C) The meaningful chemical shift variations are mapped on the solution structure of apo hGRX5. Side chains of residues with Δavg(HN) values greater than the threshold of 0.02 ppm are shown in yellow/orange (solvent-exposed) and green (buried in the protein core). The interacting surface is in magenta and Cys-67, also showing a Δavg(HN) higher than 0.02 ppm, is in yellow. A 180° rotated view on the right shows that the interaction specifically involves residues on one side of the protein, where the GSH/[2Fe-2S] cluster binding region is located.
Solution Structure Characterization of Apo and Holo hGRX5.
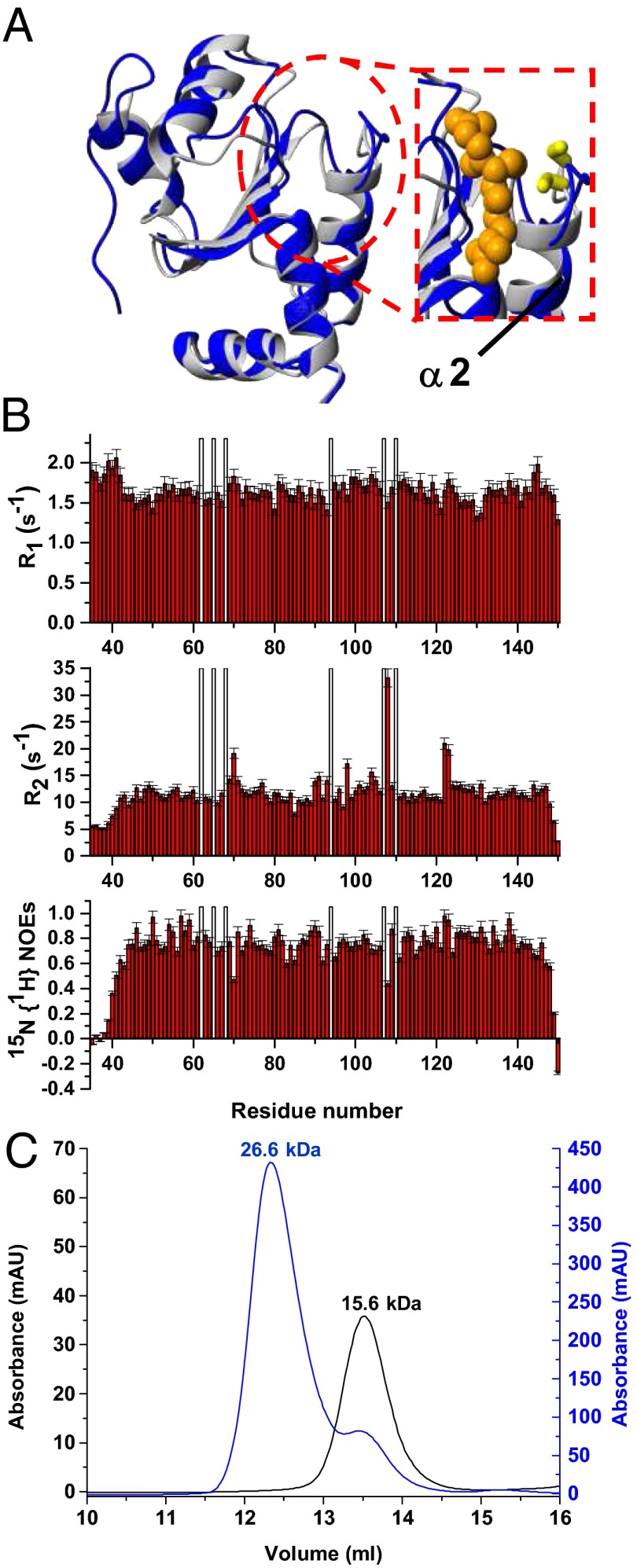
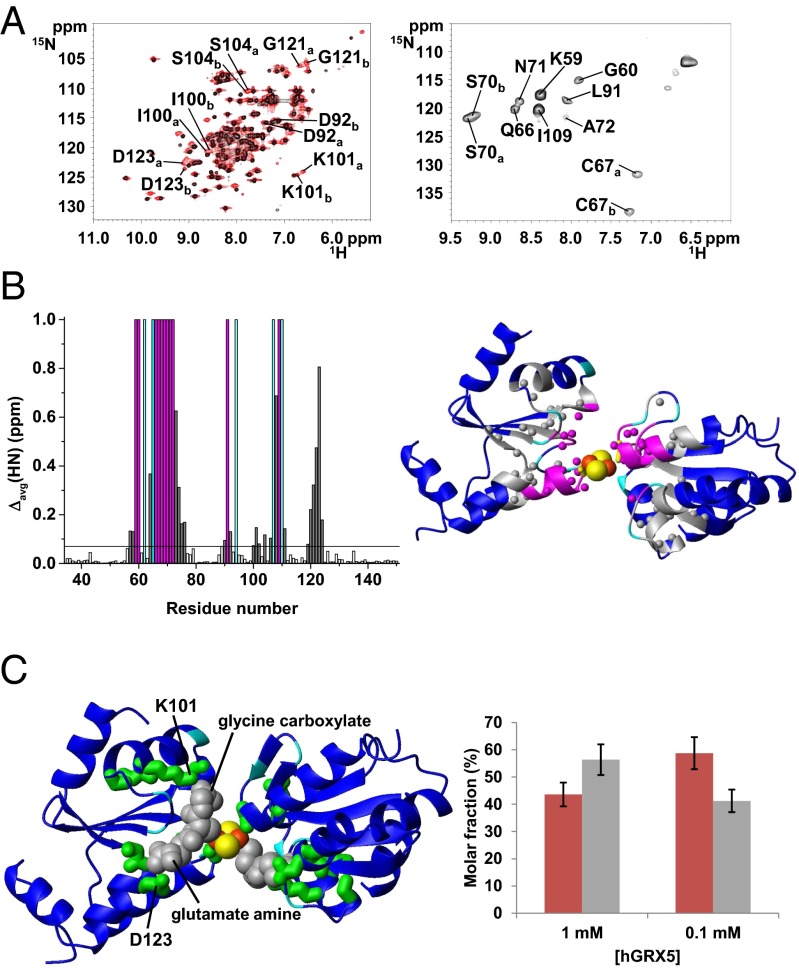
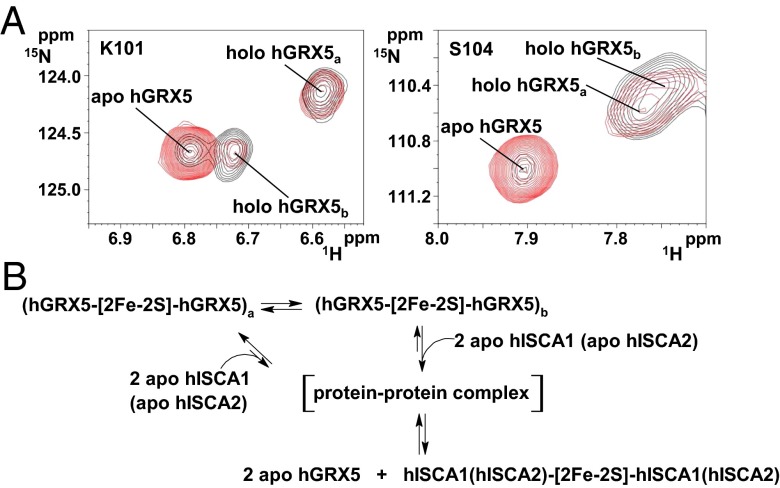
The tetrameric structural organization found in the crystal structure of holo hGRX5 shows that two [2Fe-2S] clusters are completely buried by four protein subunits and four iron-bound GSH molecules (15). The cluster-binding sites are not accessible to a [2Fe-2S]–receiving partner protein, and therefore are not suitable for the cluster transfer. We, therefore, characterized the structures of apo and holo hGRX5 in solution by NMR. Apo hGRX5 has the same backbone conformation as each monomer of the crystal structure of holo hGRX5 (PDB ID code 2WUL; rmsd 1.45 Å), with the typical thioredoxin fold (Fig. 4A). However, the absence of the cluster makes the backbone conformation in the region around the cluster-binding site less structured. Indeed, in apo hGRX5 helix α2, which follows the iron binding Cys-67, is shortened by two residues (Gly-68 and Phe-69) with respect to the holo form (Fig. 4A). Furthermore, in the apo protein some residues (Gly-68, Ser-70, Thr-108, Cys-122, and Asp-123) located in the cluster-/GSH-binding region experience backbone conformational motions (Fig. 4B; for details see SI Text). Gel filtration (Fig. 4C and SI Text) and 15N NMR relaxation data (for details see SI Text) on the holo/apo states of hGRX5 showed that cluster binding induces a change of the quaternary structure, passing in solution from an apo monomer to a holo dimer (τm = 13.80 ± 1.89 ns of holo hGRX5 consistent with a dimeric state). The tetrameric state of holo hGRX5 reported in the previous structural study (15) could be the result of different experimental conditions, in particular a much higher protein concentration, and/or crystal packing interactions. Resonance assignment of the 1H-15N HSQC spectrum of holo hGRX5 shows that 22 residues have chemical shifts different from those of the apo protein (Fig. 5A). These chemical shift changes include residues 57–58, 64, 73–76, 90, 92, 100–102, 104, 106, 108, 111, and 119–124 (shown as gray bars in Fig. 5B), which surround a sphere of about 10-Å radius centered on the cluster, once they are mapped on the two subunits coordinating the cluster in the crystal structure of holo hGRX5 (shown as gray spheres in Fig. 5B). The NH signals of the 11 residues located inside this sphere (shown as magenta bars and as magenta spheres in Fig. 5B) are not detected in the standard, diamagnetic 1H-15N HSQC spectrum. A 1H-15N inversion recovery filtered HSQC experiment, with antiphase detection (1H-15N IR-HSQC-AP), specifically designed to detect fast-relaxing signals (25, 26), allowed us to recover 9 out of the 11 missing NH signals (Fig. 5A), which have been assigned as described in SI Text (Table S1). The 1H T1 values of these NH signals (Table S1) increase with increasing the distance of the NH from the cluster with the expected r−6 dependence, being the distances taken from the two cluster-coordinating subunits in the crystal structure of holo hGRX5. Deviations from the expected T1 values are observed only for Gly-60 and Gln-66, which are facing each other on the extended loop containing the iron binding Cys-67, suggesting that the loop region undergoes some local reorientation in solution with respect to the crystal structure. In conclusion, both diamagnetic and paramagnetic NMR data consistently indicate that holo hGRX5 in solution has the two subunits of the dimer in the same relative orientation as found in the crystal structure of holo hGRX5. Therefore, holo hGRX5 in solution is a symmetric dimer bridged by a [2Fe-2S]2+ cluster, which is coordinated by Cys-67 and a GSH molecule per subunit. However, in the 1H-15N IR-HSQC-AP experiment, two sets of signals for Cys-67 and Ser-70 were identified (Fig. 5A) and two CO signals for Cys-67 were also observed in the paramagnetic 1D 13C NMR spectrum (Fig. S2), thus indicating that the symmetric dimer of holo hGRX5 exists in solution as a mixture of two forms (holo hGRX5a and holo hGRX5b, hereafter). These two dimeric species are in equilibrium, as indicated indeed by the change of their relative populations upon varying protein concentration (Fig. 5C). The paramagnetic NMR data are consistent with a fully saturated cluster binding site in both holo hGRX5a and holo hGRX5b, according to the holo hGRX5-GSH titration data described before (Fig. 1B). Indeed, the 1H-15N IR-HSQC-AP experiment shows only two cross-peaks for the NHs of the Cys ligands, whereas three cross-peaks for the NHs of the Cys ligands would be expected if two species, one with an incompletely saturated binding site (i.e., an asymmetric coordinated-cluster species providing two Cys signals) and one with a completely saturated binding site (i.e., a symmetric coordinated-cluster species providing a Cys signal), would be present in solution. This case is never observed in the paramagnetic NMR data for any sample preparation. Consistent with the presence of two states in solution for holo hGRX5, in the diamagnetic 1H-15N HSQC spectrum of holo hGRX5, 6 out of the 22 residues surrounding the “paramagnetic sphere” have pairs of NH signals with their chemical shifts both different from those of the apo protein (Fig. 5A). Once these residues (six detected in the diamagnetic 1H-15N HSQC spectrum and two in the paramagnetic 1H-15N HSQC spectrum) are mapped on the dimeric structure of holo hGRX5, they are all surrounding the iron-bound GSH molecule (Fig. 5C). In particular, among these eight residues, the two charged residues, Lys-101 and Asp-123, are particularly important for engaging electrostatic interactions with the glycine carboxylate and the glutamate amine group of GSH, respectively (Fig. 5C). These data therefore suggest that holo hGRX5a and holo hGRX5b differ in the way the cluster-bound GSH molecule interacts with the protein.
Fig. 4.
Solution structure and dynamics of apo hGRX5 and effects on the aggregation state of hGRX5 induced by [2Fe-2S] cluster binding. (A) Overlay of the solution NMR structure of apo hGRX5 (blue) with a subunit of the tetrameric crystal structure of holo hGRX5 (gray). A zoom of the GSH binding site is shown on the right, where the side chain of the iron binding cysteine and the atoms of glutathione molecule (obtained from the crystal structure) are shown as yellow sticks and orange spheres, respectively. (B) 15N relaxation parameters R1, R2, 15N{1H} NOE versus residue number of apo hGRX5 obtained at 600 MHz and 298 K. The protein was in 50 mM phosphate buffer (pH 7.0) containing 5 mM DTT, 5 mM GSH, and 10% (vol/vol) D2O. White bars indicate Pro residues and the unassigned backbone NH of Gly-68. (C) [2Fe-2S] cluster binding results in a change of the quaternary structure of hGRX5. Analytical gel filtration of apo hGRX5 (black, left y axis) and of holo hGRX5 (blue, right y axis) in degassed 50 mM potassium phosphate buffer (pH 7.0) containing 5 mM DTT and 5 mM GSH. All prepared holo hGRX5 samples contained a residual amount of apo form (ranging from 10 to 30%) that was not possible to remove by gel filtration column in the last purification step (see SI Text for details). Apparent molecular weights are reported on the top of the peaks according to the markers run under the same conditions. The molecular weight of apo hGRX5, as obtained by MALDI-MS and electrospray ionization-MS data, is 13,158 Da, in agreement with the theoretical value (13,157.9 Da) for a monomeric protein state.
Fig. 5.
Structural characterization of holo hGRX5 by solution NMR. (A) (Left) An overlay of 1H-15N HSQC spectra of 15N-labeled 1 mM apo hGRX5 (black) and holo hGRX5 (red) at 900 MHz. (Right) The 1H-15N IR-HSQC-AP spectrum of 1 mM holo hGRX5 at 800 MHz is displayed reporting the paramagnetic NH signals and their assignment. The residues having two sets of signals in holo hGRX5 are indicated with a and b labels. (B) (Left) Backbone weighted average chemical shift differences Δavg(HN) between apo and holo hGRX5. Meaningful chemical shift differences (threshold of 0.07 ppm) are shown in gray. The NH signals of 11 residues of holo hGRX5 (in magenta) are not detected in the diamagnetic 1H-15N HSQC experiments. (Right) Mapping the meaningful chemical shift variations (in gray) on the two [2Fe-2S]-bridged subunits of the crystal structure of holo hGRX5. Magenta and gray spheres represent backbone NHs. The side chain of the iron ligand Cys-67 is shown in yellow. Pro residues, iron, and sulfur atoms are colored in cyan, orange, and yellow, respectively. For clarity the GSH molecules are not depicted. (C) (Left) Mapping the eight residues with two sets of signals (in green) on two dimeric subunits coordinating the cluster in the crystal structure of holo hGRX5. GSH molecules are colored in gray. (Right) Molar fractions of holo hGRX5a (red) and holo hGRX5b (gray) are reported at 1 mM and 0.1 mM holo hGRX5 solution in 50 mM phosphate buffer, 5 mM DTT, and 5 mM GSH.
The [2Fe-2S]2+ Cluster Transfer Mechanism from hGRX5 to hISCA1.
To define the [2Fe-2S]2+ cluster transfer mechanism at the molecular level, the cluster transfer from dimeric holo hGRX5 to apo hISCA1 was investigated in solution by NMR. Upon addition of unlabeled apo hISCA1 to 15N-labeled holo hGRX5, the intensity of the signals of both hGRX5a and hGRX5b holo forms decreases in the 1H-15N HSQC spectra and the intensity of corresponding signals of the apo form increases. Specifically, the NMR data indicate that the holo hGRX5b form is largely consumed at substoichiometric hGRX5:hISCA1 molar ratio, whereas only ∼10% of the holo hGRX5a form is consumed (Fig. 6A). Holo hGRX5b is therefore more reactive than holo hGRX5a with respect to transferring the [2Fe-2S] cluster to apo hISCA1. 15N NMR relaxation and gel filtration data on hISCA1 showed that cluster binding by hISCA1 is accomplished by a stabilization of its quaternary structure from a monomer/dimer equilibrium in the apo state to a fully dimeric state in the holo protein (Fig. S3; for details see SI Text). A model for the transfer of the [2Fe-2S] cluster from hGRX5 to hISCA1 can therefore be proposed on the basis of this evidence (Fig. 6B): (i) Dimeric holo hGRX5 has two states in equilibrium with each other, holo hGRX5a and holo hGRX5b; (ii) dimeric holo hGRX5 specifically recognizes apo hISCA1 to transfer the [2Fe-2S]2+ cluster through a transient protein–protein intermediate; and (iii) holo hGRX5b is more reactive than holo hGRX5a to donate the cluster to apo hISCA1.
Fig. 6.
[2Fe-2S] cluster transfer mechanism from holo hGRX5 to apo hISCA1 and apo hISCA2. (A) 1H-15N HSQC spectra of two residues (Lys-101 and Ser-104) representative for holo hGRX5a and holo hGRX5b are shown in the absence (black) or in the presence of 0.8 equivalents (red) of apo hISCA1 in 50 mM phosphate buffer (pH 7), 5 mM DTT, and 5 mM GSH containing 10% D2O at 298 K. (B) The here proposed [2Fe-2S]2+ cluster transfer mechanism from holo hGRX5 to apo hISCA1 and apo hISCA2 is shown.
General Applicability of the [2Fe-2S]2+ Cluster Transfer Mechanism.
To generalize our [2Fe-2S]2+ cluster transfer model, a further physiological partner of hGRX5 [human ISCA2 (hISCA2) hereafter; SI Text and Fig. S1] was selected and the cluster transfer with hGRX5 and their protein–protein recognition were investigated by performing the following NMR experiments: 15N-labeled holo hGRX5/apo hISCA2 titration and 15N-labeled apo hGRX5/[2Fe-2S]2+-hISCA2 titration. From these experiments (Fig. S4) it results that (i) the [2Fe-2S]2+ cluster is transferred from hGRX5 to hISCA2, as observed for hISCA1, and (ii) the region of hGRX5 recognizing hISCA2 involves the same residues found in the hISCA1/hGRX5 interaction. Therefore, hGRX5 transfers the [2Fe-2S]2+ cluster following the same mechanism proposed for the hISCA1/hGRX5 interaction (Fig. 6B). Moreover, these data show that the hGRX5 interface identified here is specifically recognized by both partner proteins, thus suggesting that the observed cluster transfer mechanism can be of general applicability toward other physiological partners (Fig. S5). Many other mitochondrial proteins indeed need [2Fe-2S] clusters to be functional. Most of them are part of the respiratory chain complexes (27), and a few others are involved in Fe/S protein biogenesis or heme biosynthesis [i.e., mitochondrial ferredoxin (27) and ferrochelatase (28)]. In addition to hISCA1 and hISCA2, the mechanism of the [2Fe-2S]2+ cluster transfer might be operative also for these [2Fe-2S] mitochondrial target proteins. Because hISCA1 and hISCA2, together with IBA57, are required in vivo for the synthesis of [4Fe-4S] clusters (14, 19) and they can bind a [2Fe-2S] cluster in vitro (18), the [2Fe-2S]2+ cluster transfer process described here might represent an intermediate step in the formation of a [4Fe-4S] cluster (Fig. S5). Indeed, a possible scenario is that once hISCA1 and hISCA2 have independently received the [2Fe-2S] cluster from hGRX5 they can assemble together and with IBA57 to form an in vivo-detected hetero-complex (19), coupling the two hGRX5-donated [2Fe-2S] clusters to generate a [4Fe-4S] cluster in the complex.
Further NMR experiments were performed to investigate the interaction between the apo forms of hGRX5 and hISCA1 or hISCA2. Specifically, possible protein–protein interactions were analyzed by titrating 15N-labeled apo hGRX5 with unlabeled apo hISCA1 or apo hISCA2. In both cases very low chemical shift variations [Δavg(HN) = 0.02 ppm maximal variation, Fig. S6] were observed in the regions sizably affected by apo–holo interactions, indicating that the apo proteins interact negligibly, whereas no other significant chemical shift changes are detected on the other parts of the proteins. These data provide evidence for a cluster-mediated protein–protein interaction in which the [2Fe-2S] cluster is essential to promote the formation of a transient protein–protein intermediate between the partners. This behavior is typical of metallochaperones and is a general behavior already observed in copper transfer processes (29, 30).
The physiological relevance of the interface of hGRX5 interacting with hISCA1 and hISCA2 was verified by investigating the interaction of hGRX5 with the nonphysiological partner anamorsin, which is a human [2Fe-2S] protein involved in the cytosolic iron–sulfur protein assembly machinery (25, 31, 32). Specifically, when titrating 15N-labeled apo hGRX5 with unlabeled [2Fe-2S]–anamorsin no significant chemical shift changes are observed (Fig. S7), indicating that (i) no interaction occurs between the two proteins, at variance with what was observed in the corresponding experiments performed with hGRX5 and the physiological partner proteins hISCA1 (Fig. 3) or hISCA2 (Fig. S4), and (ii) cluster transfer from [2Fe-2S]–anamorsin to apo hGRX5 does not take place. These data are therefore consistent with a well-defined specificity of the protein–protein recognition for physiological partners and strongly argue for a physiologically relevant role of the protein–protein-driven cluster transfer mechanism described here between hGRX5 and hISCAs.
Conclusions
The structural plasticity of the dimeric state of holo hGRX5 observed in solution is the crucial factor that allows an efficient cluster transfer to the partner proteins hISCA1 and hISCA2 through a specific and cluster-mediated protein–protein recognition mechanism. The dimeric state of holo hGRX5 also prevents the [2Fe-2S] cluster from being released in the presence of physiological concentrations of GSH, suggesting that holo hGRX5 works as a metallochaperone that specifically transfers the [2Fe-2S] cluster to partner proteins. In such a way, the [2Fe-2S] cluster, once de novo-synthesized on ISCU (33, 34), can be safely transferred from one protein to another, up to its final target protein. The mode of cluster transfer based on a switch between two conformational states of holo hGRX5 described here could be a general mechanism used by monothiol glutaredoxins in Fe/S protein assembly pathways. In this way, monothiol glutaredoxins indeed have the possibility of efficiently releasing the cluster to multiple partner proteins. Drawing a parallel with copper trafficking pathways (30, 35), hGRX5 behaves similarly to copper chaperones (36), being characterized by a structural plasticity around the metal or cofactor binding site, which plays a role in modulating the protein recognition and in the formation of metal-mediated protein complexes.
Materials and Methods
Interaction Between Glutathione and Apo/Holo hGRX5.
The interaction between GSH and apo/holo hGRX5 was followed by NMR titrating 15N-labeled apo/holo hGRX5 (0.5–1 mM) with increasing amounts of glutathione (from 5 mM to 20 mM), in 50 mM phosphate buffer (pH 7.0), 5 mM GSH, and 5 mM DTT containing 10% (vol/vol) D2O at 298 K. Chemical shift changes were followed by 1H-15N HSQC spectra after addition of increasing amounts of GSH.
[2Fe-2S]2+ Cluster Transfer and Interaction Between hGRX5 and hISCA1 or hISCA2 or Anamorsin.
To monitor cluster transfer, holo hGRX5 was incubated with the His6-tagged apo form of hISCA1 (protein ratio ∼1:1) for 1 h at 4 °C in 50 mM phosphate buffer (pH 7.0), 5 mM GSH, and 5 mM DTT and, after separating the two proteins through affinity chromatography, UV/visible and EPR spectra were recorded as described in SI Text. The same experiment was performed for His6-tagged holo hISCA1 and apo hGRX5.
Cluster transfer and protein–protein interaction between hGRX5 and hISCA1 or hISCA2 was followed by NMR titrating 15N-labeled holo hGRX5 with unlabeled apo hISCA1 or apo hISCA2, and 15N-labeled apo hGRX5 with unlabeled holo hISCA1 or holo hISCA2, respectively, in 50 mM phosphate buffer (pH 7.0), 5 mM GSH, and 5 mM DTT containing 10% (vol/vol) D2O at 298 K. Protein–protein interaction between anamorsin and hGRX5 was followed by NMR titrating 15N-labeled apo hGRX5 with unlabeled [2Fe-2S]–anamorsin. Chemical shift changes were followed by 1H-15N HSQC spectra after addition of increasing amounts of the unlabeled partner. Protein–protein interaction between 15N-labeled apo hGRX5 and unlabeled apo hISCA1 or hISCA2 was investigated by NMR monitoring chemical shift changes in the 1H-15N HSQC maps of hGRX5 upon addition of the unlabeled partner.
Structural Characterization of Apo hGRX5 by Solution NMR.
Standard 1H-detected triple-resonance NMR experiments for backbone resonance assignment were recorded on 0.5–1 mM 13C, 15N-labeled samples of apo hGRX5 at 298K. The side chain assignment of apo hGRX5 was performed using total correlation spectroscopy- and NOESY-based NMR experiments following standard procedures. All NMR data were processed using the Topspin software package and were analyzed with the program CARA (http://cara.nmr-software.org/portal/). Secondary structure analysis has been performed by TALOS+ (37). Structure calculations of apo hGRX5 were performed with the software package UNIO (ATNOS/CANDID/CYANA) (38). The 20 conformers with the lowest residual target function values were subjected to restrained energy minimization in explicit water with the program AMBER 12 (39, 40). The quality of the structures was evaluated by the programs PSVS and iCing. The conformational and energetic analysis of the final restrained energy minimized family of 20 conformers of apo hGRX5 are reported in Table S2.
Structural Characterization of Holo hGRX5 by Solution NMR.
One-dimensional 1H paramagnetic NMR spectra of 1 mM holo hGRX5 [in D2O, 50 mM phosphate buffer (pH 7.0), 5 mM GSH, and 5 mM DTT] were recorded at 298 K on a Bruker Avance 600-MHz spectrometer, equipped with a selective 1H probe head (SEL 1H 5 mm), to detect Hα and Hβ signals of the iron ligands. These spectra were acquired by means of the super-WEFT sequence with a recycle time of 50 ms. One-dimensional 13C paramagnetic NMR experiments were performed to detect CO, Cα, and Cβ signals of the iron ligands (41) on a Bruker Avance 700-MHz spectrometer equipped with a triple resonance cryoprobe with an inner coil optimized to detect 13C resonance (TXO), operating at 175 MHz 13C Larmor frequency; 49,152 scans were acquired over a 100-kHz spectral window (carrier frequency at 100 ppm), using 20 ms and 250 ms as acquisition and recycle delays, respectively. Four fast-relaxing signals, with similar signal intensities, were observed at 215, 212, 145, and 105 ppm, whose T1 values were obtained from inversion recovery 13C experiments (Fig. S2). Six 1D experiments were performed, using the experimental conditions described above, with 2, 5, 10, 20, 40, and 60 ms as variable delay.
To detect resonances in the proximity of the [2Fe-2S] cluster, 1H-15N HSQC experiments tailored to the detection of fast-relaxing signals and 1H-15N inversion recovery HSQC antiphase experiments (1H-15N IR-HSQC-AP) were performed as previously described (25, 26). The assignment procedure of the resonances observed in paramagnetic-optimized 1H-15N HSQC experiments of holo hGRX5 is described in SI Text and their resonance assignment is summarized in Table S1.
Supplementary Material
Acknowledgments
This work was supported by the WeNMR project (European FP7 e-Infrastructure Grant 261572), Bio-NMR (European FP7 Grant 261863), Research Projects of National Interest Grant 2009FAKHZT_001, and Ente Cassa di Risparmio. This work was also supported by the European Integrated Structural Biology Infrastructure (INSTRUCT), which is part of the European Strategy Forum on Research Infrastructures and supported by national member subscriptions. Specifically, we thank the INSTRUCT Core Centre Magnetic Resonance Center (Italy).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates, chemical shifts, structural restraints, and resonance assignments of the apo form of human GRX5 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2MMZ) and BioMagResBank (Entry ID 103808), respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400102111/-/DCSupplemental.
References
- 1.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460(7257):831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 2.Rouhier N, Couturier J, Johnson MK, Jacquot JP. Glutaredoxins: Roles in iron homeostasis. Trends Biochem Sci. 2010;35(1):43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hider RC, Kong XL. Glutathione: A key component of the cytoplasmic labile iron pool. Biometals. 2011;24(6):1179–1187. doi: 10.1007/s10534-011-9476-8. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Outten CE. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry. 2012;51(22):4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780(11):1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Mapolelo DT, et al. Monothiol glutaredoxins and A-type proteins: Partners in Fe-S cluster trafficking. Dalton Trans. 2013;42(9):3107–3115. doi: 10.1039/c2dt32263c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shakamuri P, Zhang B, Johnson MK. Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins. J Am Chem Soc. 2012;134(37):15213–15216. doi: 10.1021/ja306061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi W, et al. Glutathione complexed Fe-S centers. J Am Chem Soc. 2012;134(26):10745–10748. doi: 10.1021/ja302186j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, et al. Glutathione regulates the transfer of iron-sulfur cluster from monothiol and dithiol glutaredoxins to apo ferredoxin. Protein Cell. 2012;3(9):714–721. doi: 10.1007/s13238-012-2051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lill R, et al. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823(9):1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Mühlenhoff U. The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol Biol Cell. 2013;24(12):1830–1841. doi: 10.1091/mbc.E12-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mühlenhoff U, Gerber J, Richhardt N, Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22(18):4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Manzaneque MT, Tamarit J, Bellí G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13(4):1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelling C, Dawes IW, Richhardt N, Lill R, Mühlenhoff U. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol Cell Biol. 2008;28(5):1851–1861. doi: 10.1128/MCB.01963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson C, et al. The crystal structure of human GLRX5: Iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J. 2011;433(2):303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 16.Vilella F, et al. Evolution and cellular function of monothiol glutaredoxins: Involvement in iron-sulphur cluster assembly. Comp Funct Genomics. 2004;5(4):328–341. doi: 10.1002/cfg.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KD, Chung WH, Kim HJ, Lee KC, Roe JH. Monothiol glutaredoxin Grx5 interacts with Fe-S scaffold proteins Isa1 and Isa2 and supports Fe-S assembly and DNA integrity in mitochondria of fission yeast. Biochem Biophys Res Commun. 2010;392(3):467–472. doi: 10.1016/j.bbrc.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 18.Wu G, et al. Iron-sulfur cluster biosynthesis: characterization of Schizosaccharomyces pombe Isa1. J Biol Inorg Chem. 2002;7(4-5):526–532. doi: 10.1007/s00775-001-0330-2. [DOI] [PubMed] [Google Scholar]
- 19.Mühlenhoff U, Richter N, Pines O, Pierik AJ, Lill R. Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J Biol Chem. 2011;286(48):41205–41216. doi: 10.1074/jbc.M111.296152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen LT, Culotta VC. Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol Cell Biol. 2000;20(11):3918–3927. doi: 10.1128/mcb.20.11.3918-3927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheftel AD, et al. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol Biol Cell. 2012;23(7):1157–1166. doi: 10.1091/mbc.E11-09-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couturier J, et al. Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins. J Biol Chem. 2011;286(31):27515–27527. doi: 10.1074/jbc.M111.228726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwema T, et al. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48(26):6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 24.Rouhier N, et al. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci USA. 2007;104(18):7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banci L, et al. Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis. Proc Natl Acad Sci USA. 2013;110(18):7136–7141. doi: 10.1073/pnas.1302378110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciofi-Baffoni S, Gallo A, Muzzioli R, Piccioli M. The IR-(15)N-HSQC-AP experiment: A new tool for NMR spectroscopy of paramagnetic molecules. J Biomol NMR. 2014;58(2):123–128. doi: 10.1007/s10858-013-9810-2. [DOI] [PubMed] [Google Scholar]
- 27.Lill R, Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 28.Shepherd M, Dailey TA, Dailey HA. A new class of [2Fe-2S]-cluster-containing protoporphyrin (IX) ferrochelatases. Biochem J. 2006;397(1):47–52. doi: 10.1042/BJ20051967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banci L, et al. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat Chem Biol. 2006;2(7):367–368. doi: 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- 30.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Cellular copper distribution: A mechanistic systems biology approach. Cell Mol Life Sci. 2010;67(15):2563–2589. doi: 10.1007/s00018-010-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banci L, et al. Anamorsin is a [2Fe-2S] cluster-containing substrate of the Mia40-dependent mitochondrial protein trapping machinery. Chem Biol. 2011;18(6):794–804. doi: 10.1016/j.chembiol.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Banci L, et al. Human anamorsin binds [2Fe-2S] clusters with unique electronic properties. J Biol Inorg Chem. 2013;18(8):883–893. doi: 10.1007/s00775-013-1033-1. [DOI] [PubMed] [Google Scholar]
- 33.Markley JL, et al. Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery. FEBS Lett. 2013;587(8):1172–1179. doi: 10.1016/j.febslet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansy SS, Cowan JA. Iron-sulfur cluster biosynthesis: Toward an understanding of cellular machinery and molecular mechanism. Acc Chem Res. 2004;37(9):719–725. doi: 10.1021/ar0301781. [DOI] [PubMed] [Google Scholar]
- 35.O’Halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem. 2000;275(33):25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 36.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319(1):209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 39.Case DA, et al. AMBER 12. San Francisco: Univ of California; 2012. [Google Scholar]
- 40.Bertini I, Case DA, Ferella L, Giachetti A, Rosato A. A Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics. 2011;27(17):2384–2390. doi: 10.1093/bioinformatics/btr415. [DOI] [PubMed] [Google Scholar]
- 41.Machonkin TE, Westler WM, Markley JL. Paramagnetic NMR spectroscopy and density functional calculations in the analysis of the geometric and electronic structures of iron-sulfur proteins. Inorg Chem. 2005;44(4):779–797. doi: 10.1021/ic048624j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.