Significance
Recent archaeological studies of crop domestication have suggested a relatively slow spread and fixation of some key domestication traits, such as the loss of seed shattering. In contrast, genetic studies often indicate that domestication traits have a fairly simple genetic basis, which should facilitate their rapid evolution under selection. Here we examine previously underexplored factors that could account for this apparent disconnect: the roles of gene-by-gene interactions (epistasis) and gene-by-environment effects in shaping the rate of phenotypic evolution during domestication. Analysis of a Setaria mapping population, together with a review of evidence from the literature, suggests that these genetic factors, although important, are unlikely to have played a major role in constraining the rate of phenotypic evolution during domestication.
Keywords: QTL, genotype-by-environment interactions, G × E, Setaria, domestication syndrome
Abstract
Domestication is a multifaceted evolutionary process, involving changes in individual genes, genetic interactions, and emergent phenotypes. There has been extensive discussion of the phenotypic characteristics of plant domestication, and recent research has started to identify the specific genes and mutational mechanisms that control domestication traits. However, there is an apparent disconnect between the simple genetic architecture described for many crop domestication traits, which should facilitate rapid phenotypic change under selection, and the slow rate of change reported from the archeobotanical record. A possible explanation involves the middle ground between individual genetic changes and their expression during development, where gene-by-gene (epistatic) and gene-by-environment interactions can modify the expression of phenotypes and opportunities for selection. These aspects of genetic architecture have the potential to significantly slow the speed of phenotypic evolution during crop domestication and improvement. Here we examine whether epistatic and gene-by-environment interactions have shaped how domestication traits have evolved. We review available evidence from the literature, and we analyze two domestication-related traits, shattering and flowering time, in a mapping population derived from a cross between domesticated foxtail millet and its wild progenitor. We find that compared with wild progenitor alleles, those favored during domestication often have large phenotypic effects and are relatively insensitive to genetic background and environmental effects. Consistent selection should thus be able to rapidly change traits during domestication. We conclude that if phenotypic evolution was slow during crop domestication, this is more likely due to cultural or historical factors than epistatic or environmental constraints.
Domestication is a process of evolutionary change that leads to increased dependence and associated phenotypic modifications in both domesticator and domesticated. Much is known about the phenotypes that change during plant domestication, including the so-called domestication syndrome described from annual crops, which emphasizes retention of seed on the seed head, reduction in lateral branching, reduction in seed dormancy, and increase in seed size (1). Other phenotypic changes occurring during domestication and improvement, such as shifts in flowering time and grain composition, have also been extensively studied (2–4). Recent molecular research has begun to identify and functionally characterize a growing number of the major-effect domestication and crop improvement genes (5–8). However, there is disagreement over the speed of phenotypic evolution during domestication, with an apparent disconnect between the relatively simple genetic architecture of many domestication-related traits, which should facilitate rapid change, and the protracted length of time seen in the archeobotanical record for some domestication phenotypes to become widespread (9–12).
The genetics of domestication traits generally suggest that phenotypic changes during crop domestication could potentially occur rapidly. Many domestication alleles segregate as Mendelian loci with large effects (7), and the rate of self-pollination in many crops is high, allowing for the expression of both recessive and dominant domestication alleles even when initially at low frequencies in populations. However, archeobotanical evidence from the major cereal crops rice and wheat (10, 13) and some population genetic simulations (9, 14) suggest that genetic modifications underlying phenotypic change may only slowly be translated into geographically widespread domestication phenotypes. Scenarios to explain this apparent discrepancy have focused on the strength of selection during domestication, mediated by the ability of early farmers to recognize and use favorable mutations, as well as population genetic processes such as introgression from wild populations (15). It is also possible that the vagaries of human history, including wars, epidemics, and other interruptions to food production (16), may have led to protracted domestication rates. There has been much debate as to whether the slow increase in domesticated phenotypes is real or a preservation artifact and what assumptions can be made about the botanical knowledge of early farmers (9–12, 17). Here we explore whether background effects such as gene-by-gene interactions (epistasis) and gene-by-environment (G × E) effects can affect the efficacy of selection on domestication alleles. In theory, this middle ground between molecular genetic change and its expression during development could have significantly slowed the process of phenotypic evolution during crop domestication.
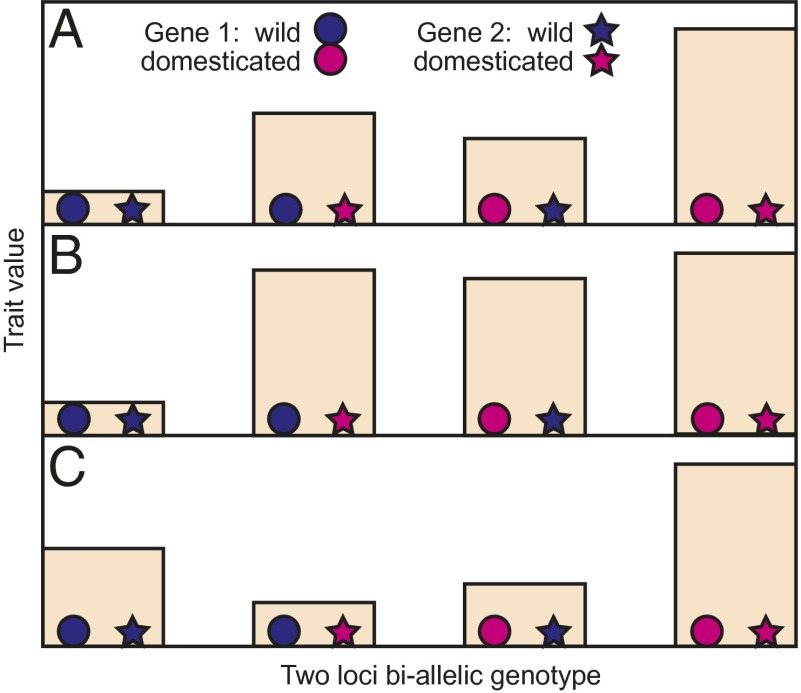
Epistasis refers to genetic interactions between loci, which may be either biallelic or higher order in nature. The term has been defined in a number of different ways [reviewed by Phillips (18)]; here we restrict our use to describe the phenomenon whereby the phenotypic effects of a given allele at a gene can vary depending on the allelic composition of the other loci that make up an individual’s genotype. This definition has been described as “compositional epistasis” (18) and is synonymous with “physiological epistasis” as used by Cheverud and Routman (19) and “functional epistasis” as used by Carter et al. (20). Epistasis has been shown to contribute to additive genetic variances and covariances, thereby affecting the response to selection; depending on the direction of these effects in morphospace, it may dramatically change the response to selection in only a few generations (20). In the context of domestication genetics, various outcomes are possible for any pair of loci, from complete lack of gene interaction, with only additive effects explaining the phenotype (Fig. 1A), to the effects of wild and domestication alleles differing across genetic backgrounds (Fig. 1 B and C).
Fig. 1.
Three sets of phenotypic outcomes that could result from biallelic epistatic interactions between two loci. Circles and stars represent genotypes homozygous for alleles that affect a domestication trait, where higher trait values are selectively favored during domestication. Pink genotypes have alleles from the domesticated population. Blue genotypes have alleles drawn from a population of close wild relatives. (A) No interaction. Additive effects of alleles explain all of the phenotypic variation. (B) Interaction such that domestication alleles at one or the other locus give trait values close to the domesticate double homozygote. (C) Less than additive interaction such that the wild–domesticate gene pair combinations have genotypic values lower than wild or domesticate double homozygotes.
The geographical distribution and population structure of many crops and their wild relatives may predispose these species to epistatic effects. Both crop species and their wild progenitors are often characterized by substantial population genetic structure, with distinct genetic subpopulations that are geographically separated and evolutionarily diverged (21, 22). This is especially true for inbreeding species, where local adaptation is not easily homogenized by introgression from other populations and where local variants can readily become established and persist through time. If adaptive differentiation among populations has led to evolutionary divergence in gene regulatory networks, a domestication allele that was initially selected upon in one population could, in theory, have very different phenotypic effects when introgressed into a population growing in a different region.
G × E interactions refer to situations in which the relative effects of alleles change across environments. The visible morphological traits selected during domestication, such as branching, inflorescence development, and seed size, are strongly affected by the environment (23), and the phenotypic effects of favorable domestication alleles may have been difficult for early farmers to consistently detect. If we assume that early agriculturalists, like those today, sought to reduce the impacts of both predictable and unpredictable environmental variation on crops, a resulting expectation is that domestication alleles would be less sensitive to environmental changes than predomestication, ancestral alleles. Such phenotypic stability would facilitate selection, enabling the spread of domesticated crops away from their native environments into other climates and latitudes (24).
Here we investigate whether epistasis and/or G × E interactions could help explain the apparent contradiction between the genetic simplicity of many domestication-related traits (both domestication and improvement traits) and the slow rate of phenotypic progress observed for some crops in the archeobotanical record. We review studies of epistasis and show that a number of genes affecting domestication-related traits are epistatic in segregating populations containing wild and domestication alleles. In many cases, domestication alleles have more stable effects in diverse genetic backgrounds than do ancestral alleles, suggesting that selection for these “robust” alleles would be effective in creating rapid phenotypic change. Similarly, the effects of domestication alleles differ across environments but are often less sensitive to environmental variation than wild (predomestication) alleles. As a complement to the literature review, we also examine the effects of epistasis on shattering and the effects of epistasis and G × E on flowering time in a cross between domesticated foxtail millet (Setaria italica) and its wild progenitor, green millet (Setaria viridis). We find loci of major-effect and epistatic and G × E interactions that significantly explain phenotypic variation. Major-effect loci appear to be less influenced by genetic background than are minor-effect loci. We suggest that for many domestication-related traits, wild populations segregate for alleles that are sensitive to genetic background and environment but that the process of domestication has favored relatively insensitive loci that have large effects on traits and are highly responsive to selection. Thus, the simple genetic architecture observed for many domestication-related traits may be the evolutionary outcome of successful selection for alleles that are relatively insensitive to background effects.
Epistasis Affects Domestication and Crop Improvement Traits
Both domestication traits (i.e., traits that distinguish a crop from its wild ancestor) and crop improvement traits (i.e., agronomic traits that vary among landraces or varieties of a crop) can be affected by epistatic interactions, and epistasis has long been recognized as an important component in the genetic architecture of crop plants. An extreme example is demonstrated by the maize opaque7 mutant, which produces opaque, soft, and floury kernels. In the inbred dent corn line in which it was discovered (W22), the recessive allele o7 segregates in a simple Mendelian fashion (25). However, in testcrosses with other maize varieties grown in similar environments, F2 and backcross populations segregate for the opaque7 phenotype with greatly reduced frequencies (25). Among 139 genotypes tested, 118 showed significant deviations from the expected 3:1 segregation of normal to opaque kernels. A single F2 population could have as few as 3.1% opaque kernels. This wide phenotypic variation thus suggests that the visibility of the opaque7 phenotype for selection during crop breeding would strongly depend on the genetic background in which it arose.
A number of quantitative trait loci (QTL) analyses of domestication traits from biparental populations derived from wild × domesticated plant crosses have reported that the combined effect of two (or more) alleles significantly differs from the sum of the separate allelic effects. In a study of vegetative architecture in an F2:3 mapping population derived from a cross between domesticated foxtail millet and its wild ancestor green millet, 10 highly significant epistatic interactions between loci were found, with the proportion of variance explained ranging from 16% to 41% (26). Three interactions were between QTLs that had significant additive effects; two were between a QTL and a marker in another part of the genome, and the remaining five involved loci that showed no significant additive effects (26). Other studies have also identified epistatic interactions between loci that have no significant main effects and between QTL and other loci. A study on epistatic interactions controlling domestication and improvement traits in a backcrossed doubled haploid population of wild barley (Hordeum vulgare ssp. spontaneum) and domesticated barley (H. vulgare L.) revealed that a significant proportion of phenotypic variation was explained by interactions between loci that did not show significant main effects (27). Such findings are disconcerting given that many studies have failed to consider epistatic effects altogether.
The impact that genetic interactions have on domestication traits can be most clearly observed in lines where wild alleles have been individually introgressed into the genetic background of a modern cultivar, followed by crossing of different introgressed lines. A particularly well characterized example of epistasis is that involving the teosinte branched1 (tb1) locus in maize, which affects apical dominance and lateral inflorescence (ear) development (28, 29). These are key domestication traits that distinguish maize (Zea mays L. ssp. mays) from its wild ancestor, teosinte (Z. mays L. subsp. parviglumis), and there has been strong selection at this locus during maize domestication (30). The tb1 locus, on chromosome 1, interacts with another locus on chromosome 3 to affect the sexuality of the inflorescences (31), and the allelic state of the chromosome 3 gene has a dramatic impact on tb1 effect size. In teosinte, the lateral branches terminate in male inflorescences (tassels), whereas in maize they terminate in female inflorescences (ears). When maize is introgressed with teosinte alleles at either locus, both teosinte alleles have little effect on inflorescence sex, with either allele only changing ∼20% of the spikelets from female to male. However, when both loci are homozygous for the teosinte alleles, the inflorescence produces up to 90% male spikelets (31). The maize tb1 allele is known to segregate in teosinte populations (30), although these alleles are likely currently under strong negative selection (32). One would expect that rare teosinte plants homozygous for maize alleles would exhibit strongly feminized inflorescences.
The maize–teosinte tb1 example illustrates a common theme in wild–domesticate systems, where the ancestral allele is sensitive to other modifying ancestral alleles but the domestication allele is insensitive. This pattern is well documented in other crop species such as barley, where there are two genes that control shattering, Btr1 and Btr2. Wild alleles of both genes are required to confer shattering, but domestication alleles at either locus convert the inflorescence to nonshattering, regardless of the allelic state of the other locus (33–35) (Fig. 1B). Similarly, investigation of domestication-related traits in a wild × domesticated barley population found that for several traits controlled by interacting loci (heading date, plant height, and yield), the wild phenotype was only generated with the introgression of both wild alleles into an introgression line (27). Independence of a major domestication locus from its genetic background was also found by Gu et al. (36) for the qSD12 locus that controls ∼50% of the variation in seed dormancy in rice, whereas minor loci showed multiple epistatic interactions. Similar genetic architectures exist for vegetative architecture in maize and foxtail millet (26, 37) and shattering in sorghum, wheat, and rice (35, 38, 39).
When introduced into domesticated germplasm, epistatic interactions may mean that the effects of interacting pairs of wild alleles are less than the sum of the effects of single wild alleles (Fig. 1C). For example, working in tomato, Eshed and Zamir (40) reported frequent less-than-additive epistasis in which the effect of double heterozygotes of wild and domestication alleles was smaller than the sum of the effects of single heterozygotes. They suggest that such effects are indicative of interacting loci in the same genetic pathway. Von Korff et al. (27), in their investigation of heading date, plant height, and yield in a wild × domesticated barley population, also found less-than-additive epistatic effects.
Alleles that are relatively insensitive to genetic backgrounds within cultivated populations can regain sensitivity within wild populations. For example, the rice sh4 locus accounts for a large proportion of shattering variation in populations derived from crosses between domesticated Oryza sativa and its wild progenitors, the annual species O. nivara (41) and perennial species O. rufipogon (42). However, in some genetic backgrounds, wild rice plants can be homozygous for the sh4 domestication allele and yet still show a shattering phenotype (43). Similarly, weedy rice strains in the United States are almost entirely fixed for the sh4 domestication allele, yet are highly shattering (44). It is possible that novel modifiers of sh4 have evolved within weedy rice.
Not all studies report significant epistatic interactions for domestication-related genes. In some cases, mapping studies do not explicitly examine epistatic interactions (e.g., ref. 42), or they only compare them between significant QTL (e.g., ref. 45). In QTL mapping approaches, first-order effects are commonly fitted before second- and higher-order (epistatic) effects, making it difficult to statistically detect epistasis (46). Even when epistasis is tested for among all possible pairs of loci (47), statistical and genetic factors can interfere with successful detection. For example, recent simulation studies have suggested that incomplete linkage disequilibrium (LD) between causal variants and observed SNPs will erode estimates of epistatic variance, leading to inflation of the size and relative importance of additive effects (48). This phenomenon may explain the scarcity of epistatic interactions found in association and multiparental mapping studies such as the nested association mapping (NAM) maize population studies on flowering time, inflorescence architecture, plant height, leaf architecture, and disease resistance (2, 49–52). In general, epistatic interactions have been reported more often in inbreeding species such as rice and Arabidopsis than in outcrossing species such as maize, consistent with the larger LD exhibited by inbreeding species (46). Another potential reason for the lack of significant epistatic interactions detected in the NAM maize studies may be the very high statistical stringency required to control for false positives, given the large number of pairwise marker comparisons required.
Environmental Interactions with Domestication Genes
As with alleles with stable effects across genetic backgrounds, alleles with consistent effects across environments are easier and more stable selection targets than those whose effects are sensitive to environmental variation. Domestication may thus have been marked by the selection of alleles with a lack of sensitivity to environmental variation. For example, teosinte is strongly affected by crowding, so that when a wild tb1 allele from teosinte is introgressed into maize, it produces a highly branched phenotype in uncrowded growing conditions but results in repressed lateral branches resembling domesticated maize in high-density growing conditions (28). In contrast, a tb1 domestication allele in maize produces an unbranched phenotype regardless of whether plants are grown in high- or low-density environments. The effect of a teosinte tb1 allele on the percentage of staminate flowers also varies between densities, with plants at low densities having ∼20% staminate flowers and those at high densities having only ∼5% staminate flowers (31). This is in contrast to the complete insensitivity of the maize tb1 allele, where plants grown under both densities produce no staminate flowers. The environmental sensitivity of the wild allele may have weakened selection by early agriculturalists when plants with either wild or domesticated alleles were grown together as a crop because the two alleles would have produced similar phenotypes when grown at high densities (31).
In maize plants homozygous with teosinte alleles of both tb1 and the locus on chromosome 3 (see above), the percentage of staminate flowers rises to over 90% at low densities but only to ∼50% at high densities (31). Thus, the phenotypic outcome of the epistatic interaction between these two loci is also significantly affected by differences in planting density.
G × E effects are also important for crop improvement traits, especially as cultivation of early-domesticated crops spreads them into new habitats. For example, the genetic control of flowering time must be modified to reconcile changing photoperiod cues with the plant’s reproductive cycle as plants are moved north and south from their center of origin (53). The spread of domesticated crops can lead to selection for insensitive alleles at flowering time genes. For example, teosinte is a tropical short-day plant and will not flower in the longer days of higher latitudes. The major gene affecting photoperiod response in Z. mays, ZmCCT, has teosinte alleles highly sensitive to day length but less sensitive or insensitive maize alleles (24). The reduction in sensitivity allows many temperate maize varieties to flower in longer-day photoperiod regimes than can tropical maize or teosinte.
Genetic Control of Shattering and Flowering Time in Setaria
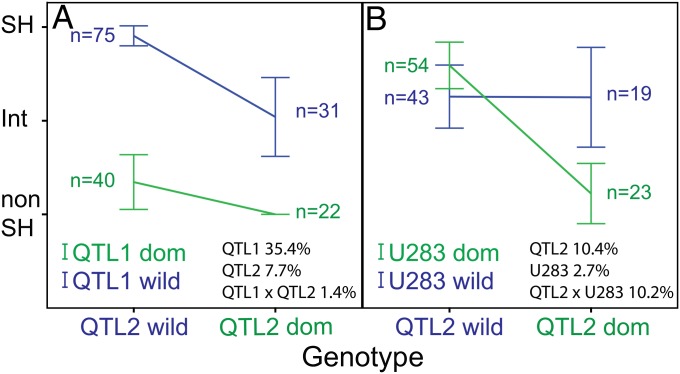
Previous work on shattering by Devos and coworkers (38, 54) established that there are two QTL that control shattering in foxtail millet. We have confirmed this using an F7 recombinant inbred line (RIL) mapping population derived from a cross between domesticated foxtail millet (S. italica) and its wild ancestor, green millet (S. viridis) (55). We find two significant QTL: QTL1 (closest marker U30) that contributes over 35% of the phenotypic variation and QTL2 (closest marker U372) that contributes ∼8%. There are no significant interactions at the Bonferroni-corrected genome-wide P < 0.05 level (P < 10−7 for each interaction). However, when we used the more liberal significance level recommended by Holland (47) (P < 0.0014 for each interaction), there were 23 significant interactions, of which three were between the minor QTL (QTL2) and other loci without main effects. Notably, the other 20 interactions were between loci where neither had a main effect. There was no significant interaction between the major locus QTL1 and any other locus, congruent with the theme that domestication alleles at major-effect QTL are often insensitive to genetic interactions (Fig. 1B). The combined effect of the wild and domestication alleles of the two QTL loci is demonstrated in Fig. 2A, where the degree of shattering in plants with a wild green millet allele of QTL1 is in part a function of the allele at the minor-effect locus, QTL2, whereas shattering in plants with the domesticated foxtail millet allele at QTL1 is unaffected by QTL2. In contrast, Fig. 2B shows the domesticated allele of QTL2 to be more sensitive to the state of an interacting locus without a significant main effect (U283) than is the wild allele.
Fig. 2.
Epistatic interactions for shattering in a cross between foxtail millet and green millet. The x axis represents the genotypes of homozygous RILs at one locus, and the color of the lines represents the different genotypes of a second locus. The y axis represents values of the shattering trait (SH, shattering; Int, intermediate shattering; NonSH, nonshattering). (A) Interaction between QTL2 (chromosome V) and QTL1 (chromosome IX) (P < 0.05 for single test; nonsignificant at the multiple test corrected significance level of P < 0.0014). (B) Significant interaction (P < 0.0014) between QTL2 and U283, a locus without a significant main effect. Percentages after each locus name and after interaction term reflect percent variance explained.
Flowering time in Setaria is strongly affected by environmental variation, including both photoperiod and temperature (56, 57). We searched for QTL and epistatic interactions for flowering time across eight trials that differed in environmental conditions, including photoperiod differences from short day (12 h light and dark) to long day (16 h light and 8 h dark) (58). We had previously reported stable QTL associations across trials, together with evidence of QTL × environment interactions (58) (Figs. S1 and S2). Of the eight QTL shared across at least two trials, four had significant epistatic interactions with other non-QTL loci. We used analysis of variance to explore the effects on flowering time of wild and domesticated alleles of QTL and their interacting loci across trials (Table 1 and Tables S1 and S2). For all QTL analyses, variation among trials was significant. QTL main effects were significant in all but one case, that of QTL III-1, where only the interaction between QTL III-1 and another locus was significant. In the other three epistatic interactions, both the QTL main effect and the interaction effect accounted for significant proportions of the flowering time variation.
Table 1.
ANOVA of flowering time in a foxtail by green millet cross, using QTL, interacting loci (if any), and trial
| QTL | QTL marker (locus 1) | Interacting marker (locus 2) | Trial | Locus 1 × locus 2 | Locus 1 × trial | Locus 1 × trial | Locus 1 × locus 2 × trial | QTL allele variance [epistasis] | QTL allele variance [trial] | QTL allele variance [overall] |
| II-1 | 66.00*** | 12.07** | 23.05*** | 71.5*** | 3.48** | 0.80 | 2.11* | n.s. | n.s. | n.s. |
| III-1 | 0.65 n.s. | 0.55 n.s. | 63.3*** | 36.69*** | 0.16 | 0.71 | 0.33 | <* | n.s. | n.s. |
| IV-1 | 297.27*** | – | 104.35*** | – | 42.54*** | – | – | n.s. | <** | n.s. |
| V-1 | 114.52*** | – | 76.59*** | – | 1.53 | – | – | <*** | >*** | <*** |
| V-2 | 62.88*** | – | 71.33*** | – | 0.97 | – | – | <* | n.s. | <* |
| V-3 | 72.53*** | – | 62.66*** | – | 0.62 | – | – | n.s. | n.s. | n.s. |
| VII-1 | 131.89*** | 104.58*** | 65.89*** | 114.50*** | 6.33*** | 3.65** | 3.50** | <*** | >*** | <*** |
| VIII-1 | 73.06*** | 0.02 | 81.11*** | 70.83*** | 1.28 | 1.08 | 2.35* | <** | <* | n.s. |
Each QTL is represented by the map marker closest to the peak of that QTL (Table S1). Locus 2 represents the interacting locus as identified from the Epistacy analysis (Materials and Methods). Columns 2–8 give ANOVA results for each QTL, interacting locus (if any), trial, and relevant interaction terms. Values in these columns are F values, and asterisks denote significance level: *P < 0.05, **P < 0.01, and ***P < 0.001. The last three columns give the results from Levene’s test for homogeneity of variance for each QTL. Terms in square brackets in column headings denote variation included in analysis: [epistasis], variation after removing trial effect; [trial], variation after removing genotypic effect; [overall], variation with both trial and genotypic effect included. Significant results signify that one allele has significantly more or less variation in flowering time than the other. The direction of the angle bracket signifies whether the domestication allele had less (<) or more (>) variation than the wild allele. n.s., not significant.
Significant G × E effects were observed for three QTL and for one locus interacting with QTL VII-1. Three QTL also showed significant interactions between pairs of epistatic loci and environment. When compared across trials, 15 interactions were found in common across two trials, 5 across three trials, and 2 across four trials (Table S2). Resampling analyses suggest that those interactions found in at least three trials are unlikely to have occurred by chance (P < 0.001). Each shared interaction displayed the same direction of effects in the trials in which they occurred (e.g., examples in Fig. S3).
We assessed whether the flowering time data support the theme that domestication alleles of major-effect loci are more robust to genomic or environmental context than wild alleles. Using Levene’s test for homogeneity of variance (59), we found that flowering time varied significantly more among genotypes with QTL alleles derived from the wild relative than among genotypes with QTL alleles derived from the domesticate, supporting the view that major-effect loci are robust to genetic background (Table 1). This was true when assessed across genotypes or across both genotypes and trials. However, when assessed across just trials, there was no consistent trend in whether flowering time variances differed between the genotypes with wild or domestication alleles. This is in contrast to the effect of density reported above on maize and teosinte branching and staminate flower production, perhaps because consistent timing of flowering has been under selection not only in the crop, foxtail millet, but also in the wild species green millet, which is now a weed of cultivation.
Conclusions
One of the main findings of our review, supported by our analysis of shattering and flowering time in foxtail millet, is that the phenotypic effects of major domestication alleles are notably less affected by genetic background than are wild alleles, whereas for minor-effect loci, other segregating alleles affect expression. The relative independence of major-effect loci indicates that selection is likely to have favored alleles with large and stable effects. The control of shattering in foxtail millet follows this trend, with a robust major-effect locus and a minor-effect locus that is involved in interactions with several other loci. Similarly, the alleles that control flowering time in domesticated millet often have more stable effects than alleles in the wild relative, across different genetic backgrounds. As with genetic background, the literature review suggests that the effect of the domestication allele is generally more stable than the wild allele across diverse environments, although it is clear from the study of flowering time in Setaria that this phenomenon depends on the trait under selection.
Our principal finding, that major domestication alleles are less sensitive to environmental changes and different genetic backgrounds than are wild alleles, suggests that selection on those alleles should be effective and that phenotypic change could therefore be rapid during domestication. This does not accord with archeobotanical observations of a protracted period of phenotypic change for characters such as shattering. How can we explain this apparent paradox? Patterns of crop use, such as the harvesting of grain while still green or the continued harvesting of wild species, may partially explain this paradox (10). For some traits, an additional genetic possibility may involve the limited availability of robust domestication alleles early in the domestication process. Phenotypic change would initially be slow if selection acted on domestication alleles with low phenotypic stability; the rate of evolution would subsequently increase if the early selected alleles were later displaced by more favorable robust alleles. This reasoning has been followed by Zhang et al. (60), who suggested that the sh4 locus controlling reduced shattering in rice arose relatively late in the domestication process, with initial selection acting on other loci with less favorable phenotypic effects. This explanation could reconcile the differences between the strong genetic signature for selection on sh4 and the protracted time for the domesticated phenotype to predominate in the archeobotanical record. However, this explanation needs to be tested, for example, by estimating the relative time of appearance during the domestication process of the various loci that control a domestication trait. Such investigations will require more complete genome information to accurately assess epistatic interactions. As advances in population and quantitative genetics continue to expand our knowledge of major- and minor-effect genes, we will have a greater ability to assess the pervasiveness of epistatic and G × E effects on the process of crop domestication and the history of domestication alleles.
Materials and Methods
Plant Materials and Phenotyping.
F7 RILs were produced by single-seed descent from an F2 mapping population derived from a cross between foxtail millet (S. italica variety B100) and green millet (S. viridis A10) (55). A set of 182 RILs were grown for phenotyping in eight field, greenhouse, and growth chamber trials. Two field, two greenhouse, and one growth chamber trial were performed at Oklahoma State University; two field trials took place at the University of Georgia (Athens, GA); and one growth chamber trial was at the Boyce Thompson Institute (Ithaca, NY). Trials varied in photoperiod, seed pretreatment, light intensity, time of year, and experimental design (58). All eight trials were phenotyped for flowering time (58); one field trial (F2_OK, field, OK) was phenotyped for seed shattering. Flowering time was measured as the time from germination to first emergence of the culm inflorescence (58). Shattering was measured on a qualitative scale, with 1 being nonshattering, 2 being intermediate, and 3 being shattering. Shattering was assessed by measuring the ease with which seeds were detached from the rachis by either pulling or bending with tweezers under a dissecting microscope, so that the nonshattering outcomes of breaking of the rhachilla above the glumes or along the length of the rachilla could be contrasted with shattering where the grain and glumes separate cleanly from the rhachilla along an abscission zone under the glumes. Intermediate shattering referred to those situations where some seeds separated at the abscission zone and some did not. The distinction between shattering and nonshattering closely parallels the method for analyzing shattering reported for the related panicoid grass, sorghum (38). Twenty seeds were assessed for each of four replicates of each RIL, and measurements were repeated by a second observer.
Map Construction and QTL Analysis.
Map construction used 684 SNP, simple sequence repeat, and sequence-tagged site markers (58). QTL analyses were performed in QTLCartographer, using composite interval mapping (CIM) and joint CIM with significance levels established by permutation (58).
Epistasis.
Pairwise epistatic interactions were calculated using the program Epistacy (47), initially with a pairwise significance level of P < 2.3 × 10−7 (creating a Bonferroni-adjusted genome-wide significance level of P < 0.05) (58) and subsequently with a pairwise significance level of P < 0.0014, following the adjustment for multiple testing advocated by Holland (47). Partial R2 terms for locus main effects and their interaction were calculated in Statistical Package for the Social Sciences (SPSS) version 19 by dividing the partial (type III) sum of squares by the corrected total sum of squares (47).
Statistical Tests and Data Visualization.
SPSS version 19 (SPSS Inc.) was used for statistical analysis and to create graphs of phenotypic means and 95% confidence intervals for epistatic interactions. Tests for effect of genetic background on QTL allele variation for flowering time were analyzed using Levene’s test for homogeneity of variances (59), after removing trial variation in an initial ANOVA analysis. Tests for effect of environment on QTL allele variation for flowering time were similarly analyzed after removing genotype (RIL) variation in an initial ANOVA analysis. Heat maps of epistatic interactions between all marker pairs were created in R version 2.13.2 (61). To more easily visualize colocalized interactions between trials in the heat maps, markers were grouped into threes along each chromosome, and the most significant P value for each group of three was chosen to represent that group. MATLAB version 8.1 (The MathWorks, Inc.) was used to write a resampling program to calculate the probability of trials sharing epistatic interactions between the same pairs of loci, given the number of significant interactions observed in each trial. Each trial was run for 1,000,000 replicates, and the 2.5 and 97.5 percentiles were calculated.
Supplementary Material
Acknowledgments
A.N.D. acknowledges Jessica Stromski and the staff of the Cimarron Valley Research Station for their help in field work and funding from Department of Energy (DE-FG02-08ER64636) and Oklahoma Center for the Advancement of Science and Technology (PSB08-007 and PS11-035B). L.L. acknowledges the support of the Natural Sciences and Engineering Research Council and the Ontario Research Fund. K.M.O. acknowledges funding on domestication research by the National Science Foundation Plant Genome Research Program (IOS-1032023).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308940110/-/DCSupplemental.
References
- 1.Harlan JR. Comparative evolution of cereals. Evolution. 1973;27(2):311–325. doi: 10.1111/j.1558-5646.1973.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 2.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325(5941):714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 3.Xue WY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 4.Kovach MJ, Calingacion MN, Fitzgerald MA, McCouch SR. The origin and evolution of fragrance in rice (Oryza sativa L.) Proc Natl Acad Sci USA. 2009;106(34):14444–14449. doi: 10.1073/pnas.0904077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger JC, Chapman MA, Burke JM. Molecular insights into the evolution of crop plants. Am J Bot. 2008;95(2):113–122. doi: 10.3732/ajb.95.2.113. [DOI] [PubMed] [Google Scholar]
- 6.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127(7):1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Gross BL, Olsen KM. Genetic perspectives on crop domestication. Trends Plant Sci. 2010;15(9):529–537. doi: 10.1016/j.tplants.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen KM, Wendel JF. A bountiful harvest: Genomic insights into crop domestication phenotypes. Annu Rev Plant Biol. 2013;64:47–70. doi: 10.1146/annurev-arplant-050312-120048. [DOI] [PubMed] [Google Scholar]
- 9.Allaby RG, Brown TA, Fuller DQ. A simulation of the effect of inbreeding on crop domestication genetics with comments on the integration of archaeobotany and genetics: A reply to Honne and Heun. Vegetation History and Archaeobotany. 2010;19(2):151–158. [Google Scholar]
- 10.Fuller DQ. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann Bot (Lond) 2007;100(5):903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heun M, Abbo S, Lev-Yadun S, Gopher A. A critical review of the protracted domestication model for Near-Eastern founder crops: Linear regression, long-distance gene flow, archaeological, and archaeobotanical evidence. J Exp Bot. 2012;63(12):4333–4341. doi: 10.1093/jxb/ers162. [DOI] [PubMed] [Google Scholar]
- 12.Purugganan MD, Fuller DQ. Archaeological data reveal slow rates of evolution during plant domestication. Evolution. 2011;65(1):171–183. doi: 10.1111/j.1558-5646.2010.01093.x. [DOI] [PubMed] [Google Scholar]
- 13.Tanno K, Willcox G. How fast was wild wheat domesticated? Science. 2006;311(5769):1886. doi: 10.1126/science.1124635. [DOI] [PubMed] [Google Scholar]
- 14.Allaby RG, Fuller DQ, Brown TA. The genetic expectations of a protracted model for the origins of domesticated crops. Proc Natl Acad Sci USA. 2008;105(37):13982–13986. doi: 10.1073/pnas.0803780105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allaby R. Integrating the processes in the evolutionary system of domestication. J Exp Bot. 2010;61(4):935–944. doi: 10.1093/jxb/erp382. [DOI] [PubMed] [Google Scholar]
- 16. Tucker RP (2012) War and the environment. A Companion to Global Environmental History, eds McNeill JR, Mauldin, ES (John Wiley, Chichester, UK), pp 319–339.
- 17.Abbo S, Lev-Yadun S, Gopher A. Plant domestication and crop evolution in the near east: On events and processes. Crit Rev Plant Sci. 2012;31(3):241–257. [Google Scholar]
- 18.Phillips PC. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9(11):855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheverud JM, Routman EJ. Epistasis and its contribution to genetic variance components. Genetics. 1995;139(3):1455–1461. doi: 10.1093/genetics/139.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter AJR, Hermisson J, Hansen TF. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor Popul Biol. 2005;68(3):179–196. doi: 10.1016/j.tpb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Huang P, et al. Phylogeography of Asian wild rice, Oryza rufipogon: A genome-wide view. Mol Ecol. 2012;21(18):4593–4604. doi: 10.1111/j.1365-294X.2012.05625.x. [DOI] [PubMed] [Google Scholar]
- 22.Morrell PL, Clegg MT. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci USA. 2007;104(9):3289–3294. doi: 10.1073/pnas.0611377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadras VO, Slafer GA. Environmental modulation of yield components in cereals: Heritabilities reveal a hierarchy of phenotypic plasticities. Field Crops Res. 2012;127:215–224. [Google Scholar]
- 24.Hung HY, et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA. 2012;109(28):E1913–E1921. doi: 10.1073/pnas.1203189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McWhirter KS, Brink RA. Canalization of endosperm development on opaque-7 maize. In: Walden D, editor. Maize Breeding and Genetics. New York: John Wiley; 1978. pp. 373–385. [Google Scholar]
- 26.Doust AN, Kellogg EA. Effect of genotype and environment on branching in weedy green millet (Setaria viridis) and domesticated foxtail millet (Setaria italica) (Poaceae) Mol Ecol. 2006;15(5):1335–1349. doi: 10.1111/j.1365-294X.2005.02791.x. [DOI] [PubMed] [Google Scholar]
- 27.von Korff M, Léon J, Pillen K. Detection of epistatic interactions between exotic alleles introgressed from wild barley (H. vulgare ssp. spontaneum) Theor Appl Genet. 2010;121(8):1455–1464. doi: 10.1007/s00122-010-1401-y. [DOI] [PubMed] [Google Scholar]
- 28.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386(6624):485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 29.Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162(4):1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43(11):1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukens LN, Doebley J. Epistatic and environmental interactions for quantitative trait loci involved in maize evolution. Genet Res. 1999;74(3):291–302. [Google Scholar]
- 32.Hufford MB, et al. The genomic signature of crop-wild introgression in maize. PLoS Genet. 2013;9(5):e1003477. doi: 10.1371/journal.pgen.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azhacuvel P, Vidya-Saraswathi D, Komatsuda T. High-resolution linkage mapping for the non-brittle rachis locus btr1 in cultivated x wild barley (Hordeum vulgare) Plant Sci. 2006;170(6):1087–1094. [Google Scholar]
- 34.Azhaguvel P, Komatsuda T. A phylogenetic analysis based on nucleotide sequence of a marker linked to the brittle rachis locus indicates a diphyletic origin of barley. Ann Bot (Lond) 2007;100(5):1009–1015. doi: 10.1093/aob/mcm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakuma S, Salomon B, Komatsuda T. The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol. 2011;52(5):738–749. doi: 10.1093/pcp/pcr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu XY, Kianian SF, Hareland GA, Hoffer BL, Foley ME. Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa) Theor Appl Genet. 2005;110(6):1108–1118. doi: 10.1007/s00122-005-1939-2. [DOI] [PubMed] [Google Scholar]
- 37.Doebley J, Stec A, Gustus C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics. 1995;141(1):333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin ZW, et al. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 2012;44(6):720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onishi K, Takagi K, Kontani M, Tanaka T, Sano Y. Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome. 2007;50(8):757–766. doi: 10.1139/g07-051. [DOI] [PubMed] [Google Scholar]
- 40.Eshed Y, Zamir D. Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics. 1996;143(4):1807–1817. doi: 10.1093/genetics/143.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li CB, Zhou AL, Sang T. Rice domestication by reducing shattering. Science. 2006;311(5769):1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 42.Cai HW, Morishima H. Genomic regions affecting seed shattering and seed dormancy in rice. Theor Appl Genet. 2000;100(6):840–846. [Google Scholar]
- 43.Ishikawa R, et al. Allelic interaction at seed-shattering loci in the genetic backgrounds of wild and cultivated rice species. Genes Genet Syst. 2010;85(4):265–271. doi: 10.1266/ggs.85.265. [DOI] [PubMed] [Google Scholar]
- 44.Thurber CS, et al. Molecular evolution of shattering loci in U.S. weedy rice. Mol Ecol. 2010;19(16):3271–3284. doi: 10.1111/j.1365-294X.2010.04708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagai K, et al. Two novel QTLs regulate internode elongation in deepwater rice during the early vegetative stage. Breed Sci. 2012;62(2):178–185. doi: 10.1270/jsbbs.62.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holland J. Epistasis and plant breeding. Plant Breed Rev. 2001;21:27–92. [Google Scholar]
- 47.Holland J. EPISTACY: A SAS program for detecting two-locus epistatic interactions using genetic marker information. J Hered. 1998;89(4):374–375. [Google Scholar]
- 48.Hemani G, Knott S, Haley C. An evolutionary perspective on epistasis and the missing heritability. PLoS Genet. 2013;9(2):e1003295. doi: 10.1371/journal.pgen.1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coles ND, McMullen MD, Balint-Kurti PJ, Pratt RC, Holland JB. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics. 2010;184(3):799–812. doi: 10.1534/genetics.109.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coles ND, Zila CT, Holland JB. Allelic effect variation at key photoperiod response quantitative trait loci in maize. Crop Sci. 2011;51(3):1036–1049. [Google Scholar]
- 51.Kump KL, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet. 2011;43(2):163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- 52.Tian F, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet. 2011;43(2):159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 53.Olsen KM, Wendel JF. Crop plants as models for understanding plant adaptation and diversification. Front Plant Sci. 2013;4:290. doi: 10.3389/fpls.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devos KM, Gale MD. Genome relationships: The grass model in current research. Plant Cell. 2000;12(5):637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bennetzen JL, et al. Reference genome sequence of the model plant Setaria. Nat Biotechnol. 2012;30(6):555–561. doi: 10.1038/nbt.2196. [DOI] [PubMed] [Google Scholar]
- 56. Kokubu T, Ishimine Y, Miyaji Y (1977) Variations of growth-period of Italian millet strains, Setaria italica BEAUV. and their responses to day-length and temperature: II. Changes of growth-period strains gathered from different districts, both native and foreign, due to the different seeding dates. Mem Fac Agric Kagoshima Univ 13(22):55–75.
- 57.Schreiber MM, Oliver LR. Two new varieties of Setaria viridis. Weed Sci. 1971;19(4):424–427. [Google Scholar]
- 58. Mauro-Herrera M, et al. (2013) Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3 3(2):283–295. [DOI] [PMC free article] [PubMed]
- 59.Levene H. Robust tests for equality of variances. In: Olkin I, Ghurye SG, Hoeffding W, Madow WG, Mann HB, editors. Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling. Stanford, CA: Stanford University Press; 1960. pp. 278–292. [Google Scholar]
- 60.Zhang LB, et al. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009;184(3):708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 61. Team RDC (2011) R: A Language and Environment for Statistical Computing (Team RDC, Vienna)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.