Significance
This study provides detailed insights into the workings of a protein that is a key determinant of drug resistance in the malaria parasite. We found that two main lineages of mutational routes lead to chloroquine transport via the chloroquine resistance transporter (PfCRT) and that a low level of chloroquine transport is conferred by as few as two mutations. However, the attainment of full transport activity is a rigid process that requires the mutations be added in a specific order to avoid decreases in chloroquine transport. Our finding that diverse forms of mutant PfCRT are all limited in their capacity to transport chloroquine indicates that resistance should be overcome by reoptimizing the chloroquine dosage.
Keywords: drug resistance, evolutionary biochemistry, Xenopus oocytes
Abstract
Mutations in the chloroquine resistance transporter (PfCRT) are the primary determinant of chloroquine (CQ) resistance in the malaria parasite Plasmodium falciparum. A number of distinct PfCRT haplotypes, containing between 4 and 10 mutations, have given rise to CQ resistance in different parts of the world. Here we present a detailed molecular analysis of the number of mutations (and the order of addition) required to confer CQ transport activity upon the PfCRT as well as a kinetic characterization of diverse forms of PfCRT. We measured the ability of more than 100 variants of PfCRT to transport CQ when expressed at the surface of Xenopus laevis oocytes. Multiple mutational pathways led to saturable CQ transport via PfCRT, but these could be separated into two main lineages. Moreover, the attainment of full activity followed a rigid process in which mutations had to be added in a specific order to avoid reductions in CQ transport activity. A minimum of two mutations sufficed for (low) CQ transport activity, and as few as four conferred full activity. The finding that diverse PfCRT variants are all limited in their capacity to transport CQ suggests that resistance could be overcome by reoptimizing the CQ dosage.
Chloroquine (CQ) was the mainstay of malaria treatment for several decades before the emergence and spread of CQ-resistant (CQR) parasites rendered it ineffective in many malarious regions. The loss of CQ efficacy was disastrous for world health, and another disaster will ensue if resistance to the current first-line treatments (the artemisinin-based therapies) continues to develop in Southeast Asia (1). Hence, there is a dire need for a readily deployable, cost-effective strategy that does not rely on artemisinin derivatives. Recent clinical trials conducted in the Republic of Guinea-Bissau have suggested that CQ resistance can be overcome simply by reoptimizing the CQ dose regimen (2–4), highlighting the potential continuing utility of CQ as an antimalarial. However, different CQR strains vary in the degree of CQ resistance they exhibit, and it is not known whether CQR malaria from other regions is susceptible to treatment by a revised dosage of CQ.
CQ is a diprotic weak base that accumulates to very high levels within the parasite’s acidic (pH ∼5) digestive vacuole (DV) (5–7) via weak-base trapping (8). Here CQ is thought to exert its antimalarial effect by preventing the detoxification of the heme released from digested host hemoglobin. Resistance to CQ is attributed primarily to mutations in the Plasmodium falciparum chloroquine resistance transporter (PfCRT), an integral membrane protein localized to the DV, and is associated with a significant reduction in CQ accumulation within the DV (9–12). Mutations in PfCRT can also modulate the parasite’s susceptibility to other current antimalarial drugs (13, 14). We recently established a robust system for the functional characterization of PfCRT in Xenopus laevis oocytes (15). The oocyte system allows interactions with PfCRT to be studied directly and in isolation, without confounding effects such as the binding of drug to heme, other targets, or transporters within the parasite. Using this system, we provided a mechanistic explanation for the phenomenon of CQ resistance by showing that mutant PfCRT from the CQR strain Dd2 (PfCRTDd2) (Fig. 1A) mediates saturable CQ transport, whereas the wild-type form of the protein from CQ-sensitive (CQS) strains does not (Fig. S1 and ref. 15). CQR parasites that have arisen independently in Columbia, Peru, Papua New Guinea, the Philippines, and Southeast Asia (Fig. 1A) have distinct PfCRT haplotypes. These haplotypes all contain a mutation at position 76—the replacement of the positively-charged lysine (K) with an uncharged residue, usually threonine (T)—but are otherwise diverse, containing at least 4 and up to 10 mutations (13). Here, we used the Xenopus oocyte system to determine whether these PfCRT proteins also mediate the saturable transport of CQ and, if so, to ascertain the number of mutations (and the order of addition) required to confer CQ transport activity upon PfCRT.
Fig. 1.
Saturable CQ transport by CQR PfCRT variants expressed in Xenopus oocytes. (A) The haplotype and origin of seven variants of CQR PfCRT. Residues that differ from the wild-type amino acid sequence (HB3) are shaded gray. (B) Immunolocalization of PfCRT in the Xenopus oocyte. In each case, the expression of the PfCRT variant resulted in a fluorescent band external to the pigment layer, indicating that the protein was expressed in the oocyte plasma membrane. The band was not present in noninjected oocytes. (C) Saturability of CQ transport via CQR variants of PfCRT. The concentration-dependence of CQ uptake was calculated by subtracting the accumulation measured in oocytes expressing PfCRTHB3 from that in oocytes expressing CQR PfCRT at each CQ concentration. The Km and Vmax values derived from these data are presented in Table 1. Uptake is shown as mean ± SEM from at least three separate experiments (in each experiment, measurements were made from 10 oocytes per treatment).
Results
Saturability of CQ Transport via a Diverse Range of PfCRT Haplotypes.
The haplotypes of PfCRT harbored by seven distinct CQR strains (Dd2, K1, GB4, Ecu1110, Ph1, Ph2, and 7G8) (Fig. 1A) as well as the CQS form of the protein (PfCRTHB3) were expressed in oocytes, with the localization of the expressed protein to the oocyte plasma membrane confirmed by immunofluorescence microscopy assays (Fig. 1B and Fig. S2). Semiquantitative Western blot analyses showed that the PfCRT proteins were present at similar levels in the oocyte membrane in each case (Figs. S3 and S4). The ability of the oocytes to transport [3H]CQ was measured in an acidic medium (pH = 6), in which the majority of CQ was protonated. All the CQR forms of PfCRT mediated CQ transport, albeit to different extents (Figs. 1C and 2). Kinetic analyses revealed that the CQR PfCRT proteins can be separated into two main groups according to their apparent Michaelis–Menten constants (Km) and maximum velocities (Vmax) for CQ transport; the GB4, K1, and Dd2 variants of PfCRT have a lower affinity for CQ and higher capacity for transport than the Ecu1110, Ph1, Ph2, and 7G8 proteins (Table 1). A similar trend was observed for the PfCRT-mediated transport of monodesethyl-CQ (the main metabolite of CQ) (Fig. S5A). The finding that the CQ Km values range from 117 to 293 µM is of physiological significance given that a clinical course of CQ is estimated to result in 200–600 µM of the drug in the DV of CQR parasites (Table S1). Under these conditions, the transporter would already be operating near or at maximum capacity. Hence, a relatively modest increase in the CQ concentration could lead to lethal levels of the drug accumulating within the DV of CQR parasites.
Fig. 2.
CQ transport activity of variants of the Dd2, Ecu1110, or Ph1 forms of PfCRT in Xenopus oocytes. One or more of the mutations present in the (A) Dd2 and (B) Ecu1110 or Ph1 forms of PfCRT were replaced with the wild-type amino acid residue, and the resulting variants were tested for the ability to transport [3H]CQ into oocytes. Twenty chimeras of PfCRTDd2 (representing the ET lineage) and PfCRT7G8 or PfCRTEcu1110 (representing the TD lineage) were also evaluated for CQ transport activity. Uptake is shown as the mean ± SEM from 3–15 separate experiments (in each experiment measurements were made from 10 oocytes per treatment). The China-e and 783 haplotypes of PfCRT were identified in CQR isolates from China and Cambodia, respectively (57, 58). ns, no significant difference (P > 0.05) in CQ accumulation between oocytes expressing a CQR PfCRT variant and the control (noninjected and PfCRTHB3-expressing) oocytes.
Table 1.
Apparent Km and Vmax values for CQ transport via a range of CQR PfCRT variants
| PfCRT variant | Apparent Km µM | Apparent Vmax pmol per oocyte/h | n |
| HB3 | – | – | – |
| Dd2 | 232 ± 11 | 61 ± 6 | 15 |
| GB4 | 275 ± 16 | 57 ± 5 | 7 |
| K1 | 293 ± 10 | 79 ± 17 | 6 |
| Ecu1110 | 191 ± 17 | 36 ± 4 | 5 |
| 7G8 | 117 ± 6 | 9 ± 1 | 6 |
| Ph1 | 186 ± 12 | 22 ± 3 | 6 |
| Ph2 | 127 ± 12 | 12 ± 1 | 7 |
| Dd2T76I | 209 ± 10 | 58 ± 9 | 3 |
| Dd2T76D | 242 ± 26 | 53 ± 4 | 3 |
| Dd2T76S | 235 ± 13 | 41 ± 8 | 3 |
| Dd2T76N | 232 ± 26 | 22 ± 4 | 3 |
| Dd2T76W | 221 ± 23 | 26 ± 6 | 3 |
| D4 | 282 ± 11 | 30 ± 4 | 3 |
| D17 | 247 ± 49 | 61 ± 11 | 4 |
| D26 | 356 ± 38 | 48 ± 9 | 4 |
| D27 | 483 ± 52 | 49 ± 8 | 4 |
| D32 | 839 ± 69 | 40 ± 10 | 4 |
CQR PfCRT-mediated transport was calculated by subtracting the uptake of CQ measured in oocytes expressing PfCRTHB3 from that in oocytes expressing CQR PfCRT. The data are shown in Fig. 1C and Fig. S5 B and C. All values are mean ± SEM from at least three separate experiments (in each experiment measurements were made from 10 oocytes per treatment).
The Minimal Requirement for CQ Transport via PfCRT.
The roles of the different mutations in the transport of CQ via PfCRT were examined by generating ∼60 variants of PfCRTDd2 and PfCRTEcu110 in which one or more of the eight (PfCRTDd2) or four (PfCRTEcu110) resistance-associated mutations was replaced with the wild-type amino acid residue (Fig. 2). The mutant proteins were confirmed to be located at the oocyte surface and/or were shown to be present at similar levels in the oocyte membrane (Figs. S4 and S6 A and B). We have reported previously that reversal of the K76T mutation abolishes CQ transport via PfCRTDd2 (ref. 15 and PfCRT variant D3 in Fig. 2A), and here the same change abrogated the CQ transport activity of PfCRTEcu1110 (PfCRT variant E1 in Fig. 2B). Reversal of either the N75E mutation in PfCRTDd2 (D2) or the N326D mutation in either PfCRTEcu1110 (E3) or PfCRTPh1 (P2) also substantially reduced the protein’s ability to transport CQ (by ∼80%, 98%, and 92%, respectively) (Fig. 2). Reversal of the other mutations affected CQ transport activity to a lesser extent. Reinstatement of the wild-type residue at position 220 of PfCRTDd2 (D4) reduced the PfCRT-mediated uptake of CQ by ∼54% (Fig. 2A), and modest decreases in CQ transport activity were observed when M74I (D1), N326S (PfCRT783), or R371I (D6) were reversed in PfCRTDd2 (∼18%, 21%, and 14%, respectively). Reversal of A220S (E2) or I356T (E5) in PfCRTEcu1110 or A144T in PfCRTPh1 (P1) also resulted in minor decreases in CQ uptake. In contrast, reinstatement of the wild-type residue at position 271 (D5) or 356 (PfCRTK1) of PfCRTDd2 resulted in a small increase in the uptake of CQ.
These observations indicated that K76T, N75E (PfCRTDd2), and N326D (PfCRTEcu1110 and PfCRTPh1) play pivotal roles in enabling PfCRT to mediate CQ transport. Nevertheless, introduction of K76T or N75E into PfCRTHB3 (D38 and D39, respectively) did not result in significant uptake of CQ. However, the addition of both mutations to PfCRTHB3 produced modest but significant CQ transport activity (D32; ∼19% of PfCRTDd2). None of the other Dd2 mutations imparted CQ transport activity when combined with K76T (D31, D33–D37). Likewise, the introduction of K76T and N326D into PfCRTHB3 (E6) was sufficient to confer a low level of CQ transport activity (∼45% of PfCRTEcu1110), whereas neither of the other two Ecu1110 mutations imparted CQ transport activity when combined with K76T (D33 and E7). Hence, the minimal requirement for (low) CQ transport activity was N75E and K76T in PfCRTDd2 and K76T and N326D in PfCRTEcu1110. Given that all known PfCRT haplotypes contain either N75E/D or N326D (13), these results indicate that PfCRT acquires the ability to transport CQ via one of two main mutational routes, both of which entail the introduction of K76T plus the replacement of an asparagine (N75 or N326) with an acidic residue. These two mutational routes are referred to henceforth as “ET” (referring to 75E and 76T) and “TD” (referring to 76T and 326D). Somewhat surprisingly, the combination of N75E and N326D resulted in a decrease, rather than an increase, in CQ transport activity; the addition of N75E to PfCRTEcu1110 (C9) or S326D to PfCRTDd2 (C14) significantly reduced CQ uptake.
The Roles of Other Resistance-Associated Mutations in the Transport of CQ via PfCRT.
The finding that the introduction of 76T and 75E (D32) or 76T and 326D (E6) into PfCRTHB3 resulted in only low levels of CQ uptake indicated that at least one of the other CQ resistance-associated mutations is required for full CQ transport activity. The addition of A220S or Q271E to the D32 variant of PfCRTDd2 (giving rise to D26 and D27, respectively) resulted in a marked increase in CQ transport activity, whereas all other mutations either were without effect (D25, D28, D30; P > 0.05) or reduced CQ uptake via PfCRT (D29; P < 0.05). Introduction of A220S and Q271E into D32 (to generate D17; N75E-K76T-A220S-Q271E) produced CQ transport activity on par with that measured for PfCRTDd2 (P > 0.05). Indeed, further investigation of the transport properties of these minimal mutants of PfCRTDd2 revealed that the kinetic parameters for CQ transport did not differ between D17 and PfCRTDd2, whereas variants possessing only two or three of these mutations (D26, D27, and D32) mediated CQ transport with lower affinity and/or capacity (Table 1 and Fig. S5B). However, in several cases the addition of one of the remaining mutations to D17 abolished CQ transport or caused significant decreases in activity (Fig. 2A). Introduction of M74I or R371I into D17 caused significant reductions in CQ transport, and N326S abolished uptake altogether, whereas the addition of I356T to D17 (to generate D16; N75E-K76T-A220S-Q271E-I356T) had no effect on the ability of the protein to mediate CQ uptake. Likewise, the addition of M74I to D16 did not alter CQ transport activity, but incorporation of N326S or R371I into D16 reduced the PfCRT-mediated uptake of CQ by ∼20%. Indeed, regardless of the order in which the remaining mutations were added to D16, all pathways resulted in a transient ∼20% reduction in CQ transport activity before the full complement of Dd2 mutations was attained.
In the case of PfCRTEcu1110, full CQ transport activity was attained by the introduction of I356L into E6 (E2; ∼83% of PfCRTEcu1110), followed by the addition of the one remaining Ecu1110 mutation (A220S) to E2, which increased CQ transport activity by ∼17%. However, when this order was switched, such that A220S was introduced into E6 (resulting in E5; K76T-A220S-N326D), there was a small but significant reduction in the protein’s ability to transport CQ (P < 0.05). The introduction of C72S into PfCRTEcu1110 (to generate PfCRT7G8) also resulted in a significant decrease in the PfCRT-mediated uptake of CQ (∼65%; P < 0.01). Likewise, the addition of C72S to PfCRTPh1 (to generate PfCRTPh2) or to PfCRTDd2 (resulting in C16) also reduced CQ transport (P < 0.01). Hence, for both PfCRTDd2 and PfCRTEcu1110, the pathway to the full complement of mutations followed a constrained progression in which mutations had to be added in a particular order to avoid significant reductions in CQ transport activity (Fig. 3 and Fig. S7).
Fig. 3.
Key mutational routes to CQR PfCRT. These schematics were constructed from the data presented in Fig. 2. The effect of a given mutation on the ability of PfCRT to transport [3H]CQ into oocytes is represented as an increase (green arrow; P < 0.05), a decrease (red arrow; P < 0.05), or no change (blue arrow; P > 0.05). The PfCRT haplotypes of the field isolates (underlined) and mutants are listed in Fig. 2. (A) The main mutational routes to PfCRTDd2. The introduction of both 76T and 75E (D32) results in a modest but significant level of CQ uptake and is the foundation of the routes leading to the Dd2, GB4, K1, China-e, and 783 haplotypes. The complete summary of the mutational pathways leading to PfCRTDd2 is presented in Fig. S7. (B) Mutational routes to PfCRTEcu1110. This pathway begins with the introduction of both K76T and N326D and leads to haplotypes such as Ecu1110, 7G8, and Ph1.
Transport of CQ via Chimeras of the ET and TD Lineages of PfCRT.
A chimera in which the N-terminal half of PfCRTDd2 was joined to the C-terminal half of PfCRT7G8 (C10; M74I-N75E-K76T-A220S-S326D-I356L) retained CQ transport activity, albeit at a level only slightly above that measured for PfCRT7G8. This result is consistent with the findings of a previous study in which a transgenic parasite line expressing this Dd2-7G8 hybrid of PfCRT was shown to possess the same level of CQ resistance as an isogenic parasite line expressing PfCRT7G8 (16). In a number of cases, the addition of one or more of the remaining Dd2 mutations to C10 decreased the PfCRT-mediated uptake of CQ (e.g., C11–13). In contrast, introduction of three of the four remaining Dd2 mutations (Q271E, N326S, and R371I) into C10 resulted in CQ transport activity greater than that measured for PfCRTDd2 (C15; 121% of PfCRTDd2; P < 0.05). It is interesting that both C15 and PfCRTK1 differ from PfCRTDd2 only at position 356, where they contain a hydrophobic residue (L or I, respectively) instead of a hydroxyl residue (T), and that both proteins possess ∼20% more CQ transport activity than PfCRTDd2. Hence, depending on which other mutations were already present, the ability of PfCRT to transport CQ increased (D9), decreased (PfCRTK1), or remained the same (D16) following the addition of I356T (Fig. 2A).
The two chimeras constructed from the C-terminal half of PfCRTDd2 and the N-terminal half of either PfCRTEcu1110 (C1; K76T-A220S-Q271E-N326S-I356T-R371I) or PfCRT7G8 (C2; C72S-K76T-A220S-Q271E-N326S-I356T-R371I) did not mediate CQ transport. The addition of T356L to C2 restored a low level of CQ transport activity (C5; 2.6% of PfCRTDd2), but the introduction of other residues of the 7G8 haplotype (271Q, 326D, or 371R) into C2 did not.
None of the ET-TD hybrids tested here possessed CQ transport activities intermediate between the two parent proteins, suggesting that, for the most part, the mutational routes taken by these two lineages are mutually exclusive solutions for attaining PfCRT-mediated CQ transport.
The Role of Position 76 in the Transport of CQ via PfCRT.
The only residues other than T to be detected at position 76 in CQR parasites are A (identified in the CQR field isolate J9) (17) and I and N (present in laboratory-generated CQR lines) (18). The influence of the residue at position 76 on the ability of PfCRTDd2 to transport CQ was investigated by replacing T with each of the other 19 amino acids (Fig. 4). The introduction of proline or a positively charged residue (R, H, or K) resulted in the loss of CQ transport activity. All the remaining mutants transported CQ, albeit at levels significantly below that measured in oocytes expressing PfCRTDd2 (P < 0.001). For at least five of these PfCRT variants, the reduction in CQ transport activity was associated with a decrease in the Vmax for CQ transport rather than with a change in the affinity for CQ or in the level of the protein at the oocyte surface (Fig. S5C and Table 1). These findings indicate that the predominance of 76T among CQR field isolates results at least partly from its ability to confer high-capacity CQ transport.
Fig. 4.
CQ transport activity of T76X mutants of PfCRTDd2 in Xenopus oocytes. The threonine at position 76 of PfCRTDd2 was replaced by each of the other 19 standard α-amino acids, and the resulting T76X mutants were tested for the ability to transport [3H]CQ into oocytes. Uptake is shown as the mean ± SEM from 3–10 separate experiments (in each experiment, measurements were made from 10 oocytes per treatment). ns, no significant difference (P > 0.05) in CQ accumulation between oocytes expressing a T76X mutant and the control (noninjected and PfCRTHB3-expressing) oocytes. See Figs. S4C, S5C, and S6C for additional data.
CQ Responses of CQS Parasites Transfected with a Minimal Mutant of PfCRTDd2 or PfCRTEcu1110.
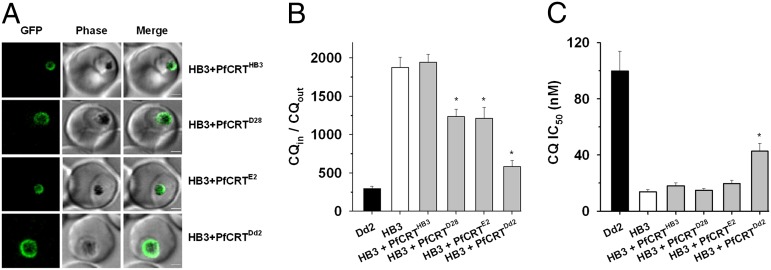
Two minimal mutants of PfCRT—D28 (N75E-K76T-N326S) and E2 (K76T-N326D-I356L)—that possessed significant (but not full) CQ transport activity in the oocyte system were expressed in P. falciparum-infected erythrocytes to assess their ability to alter the parasite’s response to CQ. The CQS strain HB3 was transfected with the native coding sequence (encoding the D28, E2, Dd2, or HB3 versions of PfCRT) flanked by the PfCRT promoter (5′) and a GFP sequence (3′). The GFP-tagged proteins localized to the DV membrane (Fig. 5A), and Western blot analysis confirmed that significant amounts of both the native and GFP-tagged forms of the protein were expressed in the transfectant lines (Fig. S8). Expression of PfCRTD28 or PfCRTE2 decreased CQ accumulation (P < 0.01) but did not reduce it to the level measured in the HB3 + PfCRTDd2 line (Fig. 5 B and C). Moreover, the fact that PfCRTDd2 (but not PfCRTD28 or PfCRTE2) increased the CQ IC50 in HB3 parasites indicates that the extent to which mutant PfCRT alters the parasite’s sensitivity to CQ is dependent on its capacity for CQ transport.
Fig. 5.
CQ responses of CQS parasites (strain HB3) transfected with a minimal mutant of PfCRTDd2 or PfCRTEcu1110 (D28 and E2, respectively). (A) The HB3, Dd2, D28, and E2 versions of PfCRT were expressed episomally as GFP-tagged proteins in the CQS P. falciparum strain HB3. In each case, live-cell imaging revealed that fluorescence was restricted to the region surrounding the hemozoin crystals, consistent with PfCRT-GFP localizing to the DV [note that PfCRT has been shown previously to localize to the parasite’s DV (9, 18)]. (Scale bars: 2 µm.) (B) Episomal expression of PfCRTD28 or PfCRTE2 reduced CQ accumulation, albeit to a lesser extent than PfCRTDd2. (C) The CQ transport activity of PfCRTDd2 (but not of PfCRTD28 or PfCRTE2) was sufficient to increase the CQ IC50 in HB3 parasites. Asterisks indicate a significant difference (P < 0.01) from the HB3 strain. The data are shown as mean ± SEM from at least six separate experiments performed on different days and with cells from different donors.
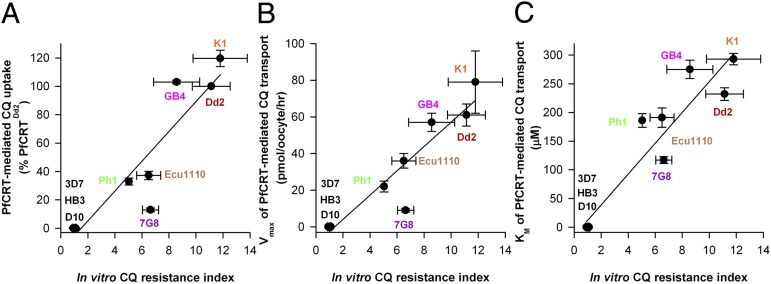
Indeed, there was also a strong positive correlation between the capacities of the K1, Dd2, GB4, Ecu1110, Ph1, and 7G8 forms of PfCRT to transport CQ in the oocyte system and the level of CQ resistance exhibited by the respective parasite strain (Fig. 6 and Table S2). For each of these CQR variants of PfCRT, the in vitro CQ resistance index of the relevant parasite strain (listed in Table S2) was plotted either against the CQ uptake values presented in Fig. 2 (Fig. 6A) or against the Vmax and Km values presented in Table 1 (Fig. 6 B and C, respectively). The CQ resistance index is the CQ IC50 in a CQR strain divided by the CQ IC50 in a CQS strain. The resulting R2 values were 0.858 (CQ uptake), 0.889 (Vmax), and 0.891 (Km). Exclusion of 7G8 from the analyses improved the correlation between the CQ resistance index and CQ uptake or Vmax (R2 values of 0.944 and 0.980, respectively) but had less effect on the relationship with Km (0.909).
Fig. 6.
Correlation between the CQ transport activity of PfCRT in oocytes and the in vitro CQ resistance index. For the Dd2, K1, GB4, Ecu1110, Ph1, and 7G8 variants of PfCRT, the in vitro CQ resistance index of the relevant parasite strain (listed in Table S2) was plotted against (A) the CQ uptake values presented in Fig. 2 or the (B) Vmax and (C) Km values presented in Table 1. The CQ resistance index is the CQ IC50 in a CQR strain divided by the CQ IC50 in a CQS strain. In each case, there was a strong positive correlation between the level of CQ resistance exhibited by parasite strains and the CQ transport properties of the corresponding forms of CQR PfCRT (A: R2 = 0.858; B: R2 = 0.889; C: R2 = 0.891). All values are shown as mean ± SEM.
Discussion
The Core CQ Resistance-Conferring Mutations in PfCRT Involve the Loss of a Positive Charge and the Gain of Negative Charges.
The findings presented here reveal that the minimum requirement for (low) CQ transport activity in both the ET and TD lineages of CQR PfCRT is two mutations. One of these changes is the loss of the positively charged K at position 76 [a mutation that has been detected in all CQR parasites characterized to date (13)]. The second is the introduction of a negative charge at either position 75 (in the ET lineage) or position 326 (in the TD lineage). When introduced together, the loss of the positive charge (K76T) and the gain of the negative charge (N75E or N326D) are sufficient to confer a low level of CQ transport activity. Moreover, only four of the eight mutations carried by the Dd2 version of PfCRT (N75E, K76T, A220S, and Q271E) are required to confer CQ transport activity approximately equivalent to that measured for PfCRTDd2. It is worth noting that one of these mutations (K76T) causes the loss of a positive charge and the gain of a hydroxyl group, two (N75E and Q271E) result in the gain of a negative charge, and the fourth (A220S) entails the introduction of a second hydroxyl group. Likewise, three of the four mutations harbored by the Ecu1110 and Ph1 forms of PfCRT involve the loss of a positive charge and the gain of a hydroxyl group (K76T), the introduction of a negative charge (N326D), or the gain of a second hydroxyl group (A220S or A144T in PfCRTEcu1110 and PfCRTPh1, respectively). Together, these changes may provide a Circe effect (19) that attracts the protonated forms of CQ toward the binding site via electrostatic forces, thereby increasing the rate of CQ transport.
Multiple Mutational Pathways Lead to CQR PfCRT, but Many Incur Transient Decreases in CQ Transport Activity en Route to the Full Complement of Mutations.
Although variant D17 (N75E-K76T-A220S-Q27IE) represents the minimum number of mutations required to achieve the level of CQ transport activity displayed by PfCRTDd2, several of the routes that lead from D17 entail small (∼20%) but significant transient reductions in CQ transport activity before the full complement of Dd2 mutations is attained. This observation, together with the fact that D17 and several of its derivatives have not been identified in field isolates, raises the question of whether the route involving D17, D16 (N75E-K76T-A220S-Q271E-I356T), and D7 (M74I-N75E-K76T-A220S-Q271E-I356T) is the optimal solution for the development of PfCRT-mediated CQ resistance in the ET lineage. One possibility is that the D17 protein underwent a 50–60% decrease in CQ transport activity through the acquisition of either M74I or R371I, the former of which results in the China-e haplotype of CQR PfCRT (M74I-N75E-K76T-A220S-Q271E). Addition of R371I to PfCRTChina-e restores full CQ transport activity and yields PfCRTGB4 (M74I-N75E-K76T-A220S-Q271E-R371I). The introduction of one of the remaining Dd2 mutations, N326S or I356T, into PfCRTGB4 leads to either a small increase (PfCRTK1; M74I-N75E-K76T-A220S-Q271E-N326S-R371I) or a decrease (PfCRT783; M74I-N75E-K76T-A220S-Q271E-I356T-R371I) in CQ transport activity. These two haplotypes converge on PfCRTDd2 with the addition of the final mutation (N326S or I356T). Given that, with the exception of D17, all these PfCRT variants have been identified within the parasite populations of Southeast Asia and China, this path may represent one of the mutational routes PfCRT navigated to produce its GB4, K1, and Dd2 haplotypes.
Although all the routes originating from D17 (N75E-K76T-A220S-Q27IE) entail one or more transient decreases in the ability of PfCRT to transport CQ, some of the intermediate haplotypes with reduced CQ transport activities may in fact represent favorable tradeoffs between conferring a moderate level of CQ resistance and maintaining the normal physiological role of the protein, which is essential but as yet unknown (16, 20, 21). Indeed, PfCRT haplotypes of the ET lineage, particularly those containing M74I-N75E-K76T, are associated with a significant decrease in the fitness of the parasite, to the extent that these CQR strains are out-competed by wild-type CQS parasites when CQ is withdrawn (22, 23). Hence, it is possible that one or more of the intermediate haplotypes of the ET lineage possesses sufficient CQ transport activity to ensure survival of the parasite when exposed to medium levels of CQ pressure and that the additional mutations found in the GB4, K1, and Dd2 haplotypes further impair the normal function of the protein and are advantageous only when CQ use is high. Furthermore, it is important to note that mutations in PfCRT are likely to have been accompanied at various points by changes in other genes, some of which may have served to maintain or even increase the parasite’s resistance to CQ when a new mutation in PfCRT decreased its CQ transport activity. One such possible modulator is the multidrug resistance transporter 1 (PfMDR1), certain mutations in which have been shown to increase the in vitro CQ resistance of some (but not all) parasites harboring mutant PfCRT (24, 25). Other proteins that have been implicated in the CQ resistance phenotype and which also may have served to offset decreases in the CQ transport activity of PfCRT include the multidrug resistance-associated proteins 1 and 2 (PfMRP1 and PfMRP2) (26, 27).
Alternatively, the ET variants of PfCRT may have evolved the ability to transport CQ via pathways that do not entail significant decreases in CQ transport activity but which require the addition of most or all of the eight mutations to achieve full CQ transport activity. One such route branches at either D32 (N75E-K76T) or D26 (N75E-K76T-A220S) with the addition of R371I (resulting in D30 and D20, respectively). In both cases, the gain of 371I did not alter the ability of the protein to transport CQ. D30 (N75E-K76T-R371I), in turn, can be converted to D20 with the introduction of A220S. The full complement of Dd2 mutations is then achieved by acquiring Q271E (D10), followed by I356T (D9), then M74I (PfCRT783), and finally N326S (PfCRTDd2). D10 also can lead to PfCRTGB4 (with the addition of M74I) and to PfCRTK1 (with the addition of N326S to PfCRTGB4). This mutational route offers two advantages. First, each mutation either maintains or increases the capacity of PfCRT for CQ transport; hence the parasite is not reliant on existing and/or simultaneous changes in other genes (e.g., those encoding PfMDR1, PfMRP1, and PfMRP2) to avoid periods of reduced resistance to CQ. Second, it describes a pathway by which one or more compensatory changes (e.g., perhaps R371I and/or M74I) could arise at an early stage to maintain the normal physiological function of the protein while it develops the ability to transport CQ.
It is worth noting that a modest level of CQ transport can be achieved in the ET lineage in the absence of N75E (D45, D46, D40, and D2). These four variants have three mutations in common: K76T (the key CQ resistance-conferring mutation), Q271E (which results in the gain of a negative charge), and N326S (which introduces a hydroxyl group). This set of three mutations conferred a very low level of CQ transport activity when introduced into PfCRTHB3 (D45; ∼5% of PfCRTDd2). Indeed, D45 mediated considerably less CQ transport than the D32 variant (N75E-K76T). Hence, it is unlikely that the K76T-Q271E-N326S route represents the main mutational pathway by which PfCRT gains the ability to confer CQ resistance.
The mutational routes to PfCRTEcu1110 are considerably fewer and much less complex than those identified for PfCRTDd2, as perhaps is expected, given that PfCRTEcu1110 contains half the number of mutations present in PfCRTDd2. The finding that C72S reduces CQ transport activity when introduced into PfCRTEcu1110 (to generate PfCRT7G8) is intriguing. This mutation also caused a decrease in CQ uptake when added to PfCRTPh1 (to generate PfCRTPh2) and to PfCRTDd2 (C16). The observation that C72S is carried by a number of PfCRT field haplotypes (13), together with the finding that it decreases CQ transport activity when introduced into three different PfCRT proteins, suggests that this mutation arose in response to pressure exerted by another antimalarial drug. Alternatively, C72S may improve the stability of the protein and/or increase its capacity to perform its normal physiological function.
PfCRT Acquires the Ability to Transport CQ via a Rigid Process.
Our results reveal that two distinct mutational pathways—the ET and TD lineages—lead to CQ transport via PfCRT. However, although there are multiple mutational routes by which PfCRT may attain full CQ transport activity, our findings indicate that this process is in fact rigid and laden with pitfalls. For example, the ability of the protein to transport CQ is reduced significantly if the CQ resistance-conferring mutations are not added in a specific order or if any amino acid other than T is introduced into position 76. Furthermore, many of the 20 chimeras generated from PfCRTDd2 (representing the ET lineage) and from PfCRT7G8 or PfCRTEcu1110 (representing the TD lineage) possess little or no CQ transport activity, indicating that several mutations from one lineage are not compatible or interchangeable with mutations from the other. Moreover, it is interesting that several of the Dd2 mutations demonstrate epistasis (28). For instance, the N326S mutation can cause a decrease (e.g., when introduced into D17, D16, or D7) or an increase (e.g., when introduced into PfCRT783 or PfCRTChina-e) in CQ transport activity; its effect depends on which other Dd2 mutations are already present. A more complex example is the role of Q271E. This mutation is necessary for establishing high levels of CQ transport activity, yet reversal of Q271E in PfCRTDd2 increases the uptake of CQ. Moreover, the addition of I356T can result in an increase, decrease, or no change in the ability of PfCRT to transport CQ. These seemingly contradictory observations suggest that repurposing the substrate-binding site and translocation pore of PfCRT for CQ transport requires the rearrangement of complex interactions between a number of amino acid residues.
At certain steps within a given mutational pathway, the reversal or addition of some mutations (e.g., M74I and R371I) has little or no effect on CQ transport activity. This finding raises the possibility that these mutations have arisen in response to other selection pressures, such as the requirement for PfCRT to maintain its normal physiological role and/or to confer cross-resistance to other antimalarial drugs.
The Capacity of Different CQR PfCRT Variants to Transport CQ Generally Correlates with the in Vitro CQ Responses of the Corresponding Parasite Strains.
We found that the relative abilities of the K1, Dd2, GB4, Ecu1110, Ph1, and 7G8 haplotypes of PfCRT to transport CQ in the oocyte system generally correlate with the level of CQ resistance exhibited by the respective parasite strain (the R2 values were between 0.858 and 0.980). This relationship was not altogether unexpected, given that (i) we observed a marked clustering of CQ transport activities according to the lineage of the PfCRT variant, such that the ET variants (K1, GB4, and Dd2) exhibited a greater capacity for CQ transport than the TD variants (Ph1, Ecu1110, and 7G8), and (ii) the levels of CQ resistance displayed by Old World P. falciparum strains (e.g., Dd2 and GB4) generally are greater than those measured in New World strains (e.g., Ecu1110 and 7G8) (29). This finding indicates that the capacity of PfCRT for mediating CQ transport is a significant factor in determining the level of CQ resistance displayed by a parasite. The observation that 7G8 is somewhat of an outlier is consistent with previous findings indicating that the 7G8 haplotype of PfMDR1 contributes to the CQ resistance phenotype to a greater extent than the PfMDR1 variants of other strains (29). Furthermore, although our data indicate that the CQ transport properties of PfCRT play a key role in determining the magnitude of resistance displayed by a CQR parasite, changes elsewhere in the genome are known to have the ability to increase (or decrease) resistance to CQ. Indeed, even CQR strains that carry the same version of pfcrt as well as the same copy number and haplotype of pfmdr1 can vary in their in vitro CQ responses (29), indicating that additional genetic elements contribute to the CQ resistance phenotype.
These considerations aside, the finding that the field TD haplotypes possessed significantly less CQ transport activity than the ET field haplotypes (and that this trend is reflected in the CQ responses of the respective strains) raises the question of why two different lineages of CQR PfCRT with distinct CQ transport properties have emerged and become established in the P. falciparum population. One possibility is that the ET variants arose under high levels of CQ pressure and were thus driven to evolve large capacities for CQ transport, albeit at the cost of parasite fitness (22, 23) (presumably caused, at least in part, by a reduction in the ability of the ET variants to fulfill the normal function of PfCRT). In contrast, a lower level of CQ pressure, such as might have been prevalent in areas of low malaria transmission and/or where the use of CQ as a prophylactic was largely unregulated and erratic (30, 31), may have resulted in the emergence of PfCRT variants with low to moderate CQ transport activities but with little or no associated fitness cost [noting that parasites harboring TD variants of PfCRT do not appear to be less fit than wild-type strains (32)]. Alternatively, or in addition, the TD haplotypes may be the product of selection pressures from both CQ and another antimalarial drug. For instance, in many of the regions where TD haplotypes are prevalent, amodiaquine was deployed before CQ resistance emerged (33). Moreover, parasites carrying TD variants of PfCRT typically exhibit higher levels of resistance to monodesethylamodiaquine (the active metabolite of amodiaquine) than those harboring ET versions of the protein (29). Hence, differences in the prevailing drug environments, and perhaps also in the rate of parasite transmission (34), may have led to the emergence and expansion of the ET lineage of in some parasite populations and of the TD lineage in others.
Clinical Relevance of the Saturability of CQ Transport via a Range of Different CQR PfCRT Variants.
Our study has revealed that PfCRT proteins from a broad range of CQR field isolates have the ability to transport CQ as well as its main metabolite, desethyl-CQ. However, in all cases the saturability of transport is such that the protein will already be operating near or at maximum capacity during a standard course of CQ. The demonstration that diverse CQR PfCRT proteins saturate within clinically relevant concentrations has significant implications for the redeployment of CQ against CQR P. falciparum. A recent randomized clinical trial has revealed that double-dose CQ (a twice-daily dose regimen which totals 50 mg CQ/kg over 3 d) is as effective as the current gold-standard antimalarial (artemether-lumefantrine) in treating P. falciparum infections in the Republic of Guinea-Bissau (3). The CQR PfCRT haplotypes common to this region belong to the ET lineage (35). We have shown that these proteins have a lower affinity and higher capacity for CQ transport than the haplotypes of the TD lineage. Hence, it is likely that CQR malaria from regions around the world will be treated by a reoptimized CQ dosage (e.g., a dose regimen that entails twice-daily administration or a slow-release formulation that consistently achieves levels of CQ that saturate CQR forms of PfCRT). A reexamination of the CQ dose regimen is both overdue and relevant; CQ remains a first-line treatment and/or prophylactic in 21 countries (36), and a CQ-azithromycin combination is in Phase IIb/III development for preventative treatment in pregnant women (37). Moreover, super-CQR strains have not emerged despite almost 20 y of widespread deployment of high-dose CQ regimens in Guinea-Bissau.
The finding that PfCRT-mediated resistance saturates and therefore may be overcome by a modest increase in the dosage may also influence the use of several other current and future quinolines that exhibit, or are likely to develop, cross-resistance with CQ. In the former case [e.g., amodiaquine and pyronaridine (29, 38)], resistance could be overcome by a reassessment of the optimum dosage. In the latter case [e.g., ferroquine and naphthoquine (39, 40)], the dose could be designed to overwhelm the PfCRT-based resistance mechanism and hence significantly retard the emergence of resistance. These possibilities have important ramifications, given that the quinolines currently serve as the long-acting partner in all antimalarial combination therapies and will continue to do so for the foreseeable future; with the single exception of the quinolone ELQ-300 (41), no other type of long-acting antimalarial is in the development pipeline (42). Taken together, our findings provide novel, detailed, and unexpected insights into the workings of a protein that is a key determinant of drug resistance in the malaria parasite.
Materials and Methods
Expression of PfCRT in X. laevis Oocytes and Measurements of CQ and Desethyl-CQ Transport.
A codon-harmonized version of the PfCRT sequence encoding a retention motif-free form of the protein was expressed at the plasma membrane of X. laevis oocytes using an approach described previously (15). In vitro transcription, oocyte preparation, and cRNA microinjection (20 ng per oocyte) were performed as outlined elsewhere (43). [3H]CQ (0.25 µM; 20 Ci/mmol) and [3H]desethyl-CQ (0.25 µM; 20 Ci/mmol) uptake by oocytes was measured 3–6 d postinjection as detailed previously (43). The measurements were made over 1–2 h at 27.5 °C and in medium that, unless otherwise specified, contained 96 mM NaCl, 2 mM KCl, 2 mM MgCl2, 1.8 mM CaCl2, 10 mM Mes, 10 mM Tris⋅base (pH 6.0), and 15 µM unlabeled CQ (for [3H]CQ uptake). In all cases, at least three separate experiments were performed (on oocytes from different frogs), and in each experiment measurements were made from 10 oocytes per treatment.
Site-Directed Mutagenesis of PfCRT.
Mutations were introduced into the PfCRT coding sequence via site-directed mutagenesis using the primer pairs listed in Table S3. Primers were designed according to either the QuickChange site-directed mutagenesis protocol or the partial overlap method of Zheng et al. (44). The introduction of multiple mutations into the PfCRT coding sequence within a single reaction was achieved by using the sense and anti-sense primers of the mutation closest to the 5′ end together with the sense primer of each of the remaining mutation sites. All of the resulting coding sequences were verified by sequencing.
Oocyte Membrane Preparation and Western Blot Analysis.
Membranes were isolated from PfCRT-expressing oocytes as described elsewhere (45) with the following minor modifications. Twenty oocytes were homogenized in 1 mL of 20 mM Tris⋅HCl (pH 8) supplemented with the Complete-mini EDTA–free protease inhibitor cocktail (Roche). The homogenate was centrifuged at 100 × g (10 min; 4 °C), and the supernatant was collected and mixed by inversion. A 37-µL aliquot was diluted to 400 µL with 20 mM Tris⋅HCl (pH 8.0) and centrifuged at 21,000 × g (20 min; 4 °C). The pellet was washed with 1 mL of a buffer containing 20 mM Tris⋅HCl (pH 8), 1 M NaCl, and the protease inhibitor cocktail, and was centrifuged at 21,000 × g (10 min; 4 °C). The pellet was solubilized in 25 µL of a solution comprising 1% Triton X-100, 20 mM Tris⋅HCl (pH 8), 10 mM NaCl, 150 mM DTT, 10% (vol/vol) b-mercaptoethanol, and 25% (vol/vol ) NuPage sample loading buffer (Life Technologies). The samples were separated on a 4–14% bis-Tris SDS-polyacrylamide gel (Life Technologies) and transferred to a nitrocellulose membrane. The membrane was probed with rabbit anti-PfCRT antibody (1:4,000; Genscript) followed by HRP-conjugated goat anti-rabbit antibody (1:8,000; Life Technologies). Anti-PfCRT is an affinity-purified polyclonal antibody raised against the N-terminal peptide MKFASKKNNQKNSS. This sequence is present in all the PfCRT variants included for study. The PfCRT band for each variant was detected by chemiluminescence (Pierce), quantified using the Image J software (46), and expressed as a percentage of the intensity measured for the PfCRTDd2 band. Total protein staining was used to evaluate sample loading and efficiency of transfer (47, 48). Briefly, membranes were rinsed with milliQ water, immersed in Ponceau S staining solution for 5 min, and rinsed with milliQ water until the background was white. Samples found to differ from the average in total protein content were excluded from the analysis. In all cases, at least three separate experiments were performed (on oocytes from different frogs), and in each experiment measurements were averaged from two independent replicates.
Immunofluorescence Analysis.
Oocytes expressing PfCRT were fixed and labeled with antibodies (20 ng per oocyte) 3 d postinjection using a protocol described elsewhere (43). Rabbit anti-PfCRT and Alexa Fluor 488 goat anti-rabbit antibody were used at concentrations of 1:100 and 1:500, respectively. Oocytes were embedded in an acrylic resin as described previously (49), and slices were viewed using a Leica Microsystems inverted confocal laser microscope.
Parasite Protein Preparation.
Blood cultures of P. falciparum were maintained as described previously (50) and synchronized by sorbitol treatment (51). A 10-mL aliquot (15% parasitemia; 2–3% hematocrit) was centrifuged (3,000 × g, 5 min), and the pellet was resuspended in 1 mL of PBS supplemented with Complete-mini EDTA–free protease inhibitor cocktail. Saponin was added (100 µL of a 1% solution) to lyse the red blood cells, and the sample was centrifuged (15,800 × g, 5 min). The pellet was washed twice in PBS supplemented with the protease inhibitor cocktail and then was triturated 15 times through a 25-G syringe. A 15-µL aliquot was combined with 147.5 µL of PBS supplemented with protease inhibitor cocktail, 25 µL of β-mercaptoethanol [10% (vol/vol) final concentration], and 62.5 µL NuPage sample loading buffer [25% (vol/vol) final concentration]. Western blot analysis was performed as described for the preparations of oocyte membrane protein.
Transfection of Parasites and Live-Cell Imaging.
Native coding sequences of pfcrt (encoding the D28, E2, Dd2, or HB3 versions of PfCRT), together with a GFP-coding sequence, were inserted into the pARL1a+ transfection vector (52). The resulting transfection vectors contained the PfCRT coding sequence flanked by the PfCRT promoter (5′) and a GFP sequence (3′) and contained the hDHFR selectable marker. Transfection of plasmid (100 µg) into the CQS P. falciparum strain HB3 was performed as described previously (53), and the transfectants were selected with WR99210 (5 nM). Transfectants were detected in blood smears 28–42 d following transfection, and the parasites were cultured in the absence of WR99210 for 2 d before commencement of the proliferation assays. Live-cell imaging was carried out on parasites of each transfectant line at the trophozoite stage (26–28 h postinvasion) of the intraerythrocytic life cycle according to a method outlined previously (54). Fluorescence was restricted to the DV in more than 95% of the transfectant parasites that were examined.
Measurements of Parasite Proliferation and CQ Accumulation.
The accumulation of [3H]CQ in mature trophozoite-infected erythrocytes was measured 36 h postinvasion using a protocol described in full elsewhere (55). Parasite growth was measured in 96-well plates using the [3H]hypoxanthine incorporation method (56). The IC50 values were determined by nonlinear regression using the equation y = a/[1 + (x/IC50)c], where y is the percent parasite proliferation, a is the maximal change in the percent parasite proliferation, x is the concentration of CQ, and c is a fitted constant.
Statistics.
Statistical comparisons were made using the Student t test for paired or unpaired samples or ANOVA in conjunction with Tukey’s multiple comparisons test.
Supplementary Material
Acknowledgments
We thank Eileen Baker, Rachel Slatyer, Marina Müller, and Stefan Prior for technical assistance. This work was supported by Australian National Health and Medical Research Council (NHMRC) Grant 1007035 and the European Union-funded Network of Excellence EviMalaR. R.E.M. was supported by NHMRC Fellowships 520320 and 1053082 and by the L’Oréal Australia For Women in Science program.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322965111/-/DCSupplemental.
References
- 1.Dondorp AM, et al. Artemisinin resistance: Current status and scenarios for containment. Nat Rev Microbiol. 2010;8(4):272–280. doi: 10.1038/nrmicro2331. [DOI] [PubMed] [Google Scholar]
- 2.Kofoed PE, et al. Different doses of amodiaquine and chloroquine for treatment of uncomplicated malaria in children in Guinea-Bissau: Implications for future treatment recommendations. Trans R Soc Trop Med Hyg. 2007;101(3):231–238. doi: 10.1016/j.trstmh.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Ursing J, et al. Similar efficacy and tolerability of double-dose chloroquine and artemether-lumefantrine for treatment of Plasmodium falciparum infection in Guinea-Bissau: A randomized trial. J Infect Dis. 2011;203(1):109–116. doi: 10.1093/infdis/jiq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ursing J, et al. Chloroquine resistant P. falciparum prevalence is low and unchanged between 1990 and 2005 in Guinea-Bissau: An effect of high chloroquine dosage? Infect Genet Evol. 2007;7(5):555–561. doi: 10.1016/j.meegid.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Hayward R, Saliba KJ, Kirk K. The pH of the digestive vacuole of Plasmodium falciparum is not associated with chloroquine resistance. J Cell Sci. 2006;119(Pt 6):1016–1025. doi: 10.1242/jcs.02795. [DOI] [PubMed] [Google Scholar]
- 6.Klonis N, et al. Evaluation of pH during cytostomal endocytosis and vacuolar catabolism of haemoglobin in Plasmodium falciparum. Biochem J. 2007;407(3):343–354. doi: 10.1042/BJ20070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn Y, Rohrbach P, Lanzer M. Quantitative pH measurements in Plasmodium falciparum-infected erythrocytes using pHluorin. Cell Microbiol. 2007;9(4):1004–1013. doi: 10.1111/j.1462-5822.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 8.Bray PG, et al. PfCRT and the trans-vacuolar proton electrochemical gradient: Regulating the access of chloroquine to ferriprotoporphyrin IX. Mol Microbiol. 2006;62(1):238–251. doi: 10.1111/j.1365-2958.2006.05368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6(4):861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch CD. Chloroquine resistance in malaria: A deficiency of chloroquine binding. Proc Natl Acad Sci USA. 1969;64(4):1181–1187. doi: 10.1073/pnas.64.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogstad DJ, et al. Efflux of chloroquine from Plasmodium falciparum: Mechanism of chloroquine resistance. Science. 1987;238(4831):1283–1285. doi: 10.1126/science.3317830. [DOI] [PubMed] [Google Scholar]
- 12.Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298(5591):210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers RL, Nash MN, Martin RE. Know your enemy: Understanding the role of PfCRT in drug resistance could lead to new antimalarial tactics. Cell Mol Life Sci. 2012;69(12):1967–1995. doi: 10.1007/s00018-011-0906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333(6043):724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RE, et al. Chloroquine transport via the malaria parasite’s chloroquine resistance transporter. Science. 2009;325(5948):1680–1682. doi: 10.1126/science.1175667. [DOI] [PubMed] [Google Scholar]
- 16.Lakshmanan V, et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24(13):2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaijaroenkul W, et al. Sequence and gene expression of chloroquine resistance transporter (pfcrt) in the association of in vitro drugs resistance of Plasmodium falciparum. Malar J. 2011;10(1):42. doi: 10.1186/1475-2875-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper RA, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61(1):35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 19.Jencks WP. Binding energy, specificity, and enzymic catalysis: The Circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- 20.Ecker A, Lewis RE, Ekland EH, Jayabalasingham B, Fidock DA. Tricks in Plasmodium’s molecular repertoire—escaping 3’UTR excision-based conditional silencing of the chloroquine resistance transporter gene. Int J Parasitol. 2012;42(11):969–974. doi: 10.1016/j.ijpara.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waller KL, et al. Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J Biol Chem. 2003;278(35):33593–33601. doi: 10.1074/jbc.M302215200. [DOI] [PubMed] [Google Scholar]
- 22.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187(12):1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People’s Republic of China. Am J Trop Med Hyg. 2005;72(4):410–414. [PubMed] [Google Scholar]
- 24.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403(6772):906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 25.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57(4):913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 26.Mok S, et al. Structural polymorphism in the promoter of pfmrp2 confers Plasmodium falciparum tolerance to quinoline drugs. Mol Microbiol. 2013;91(5):918–934. doi: 10.1111/mmi.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu J, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49(4):977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 28.Harms MJ, Thornton JW. Evolutionary biochemistry: Revealing the historical and physical causes of protein properties. Nat Rev Genet. 2013;14(8):559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sá JM, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106(45):18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giglioli G, Rutten FJ, Ramjattan S. Interruption of malaria transmission by chloroquinized salt in Guyana, with observations on a chloroquine-resistant strain of Plasmodium falciparum. Bull World Health Organ. 1967;36(2):283–301. [PMC free article] [PubMed] [Google Scholar]
- 31.Pinotti M. [New method for the control of malaria by the use of drugs mixed with kitchen salt in daily diet] Rev Bras Med. 1953;10(4):241–246. [PubMed] [Google Scholar]
- 32.Sa JM, Twu O. Protecting the malaria drug arsenal: Halting the rise and spread of amodiaquine resistance by monitoring the PfCRT SVMNT type. Malar J. 2010;9:374. doi: 10.1186/1475-2875-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh I, Kalyanum TS. The superiority of “camoquin” over other antimalarials. BMJ. 1952;2(4779):312–315. doi: 10.1136/bmj.2.4779.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallick PK, et al. Mutant pfcrt “SVMNT” haplotype and wild type pfmdr1 “N86” are endemic in Plasmodium vivax dominated areas of India under high chloroquine exposure. Malar J. 2012;11:16. doi: 10.1186/1475-2875-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ursing J, Kofoed PE, Rodrigues A, Rombo L, Gil JP. Plasmodium falciparum genotypes associated with chloroquine and amodiaquine resistance in Guinea-Bissau. Am J Trop Med Hyg. 2007;76(5):844–848. [PubMed] [Google Scholar]
- 36.World Health Organization 2012. World Malaria Report 2012. (WHO, Geneva), p 200.
- 37.Chandra RS, et al. Creative solutions to extraordinary challenges in clinical trials: Methodology of a phase III trial of azithromycin and chloroquine fixed-dose combination in pregnant women in Africa. Malar J. 2013;12(1):122. doi: 10.1186/1475-2875-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Issaka M, et al. Ex vivo responses of Plasmodium falciparum clinical isolates to conventional and new antimalarial drugs in Niger. Antimicrob Agents Chemother. 2013;57(7):3415–3419. doi: 10.1128/AAC.02383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubar F, et al. The antimalarial ferroquine: Role of the metal and intramolecular hydrogen bond in activity and resistance. ACS Chem Biol. 2011;6(3):275–287. doi: 10.1021/cb100322v. [DOI] [PubMed] [Google Scholar]
- 40.Wang JY, et al. Naphthoquine phosphate and its combination with artemisinine. Acta Trop. 2004;89(3):375–381. doi: 10.1016/j.actatropica.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Nilsen A, et al. Quinolone-3-diarylethers: A new class of antimalarial drug. Sci Transl Med. 2013;5(177):77ra37. doi: 10.1126/scitranslmed.3005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burrows JN, van Huijsduijnen RH, Möhrle JJ, Oeuvray C, Wells TN. Designing the next generation of medicines for malaria control and eradication. Malar J. 2013;12:187. doi: 10.1186/1475-2875-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bröer S. Xenopus laevis Oocytes. Methods Mol Biol. 2010;637:295–310. doi: 10.1007/978-1-60761-700-6_16. [DOI] [PubMed] [Google Scholar]
- 44.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32(14):e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergeron MJ, et al. Frog oocytes to unveil the structure and supramolecular organization of human transport proteins. PLoS ONE. 2011;6(7):e21901. doi: 10.1371/journal.pone.0021901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirois I, et al. Caspase-3-dependent export of TCTP: A novel pathway for antiapoptotic intercellular communication. Cell Death Differ. 2011;18(3):549–562. doi: 10.1038/cdd.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welinder C, Ekblad L. Coomassie staining as loading control in Western blot analysis. J Proteome Res. 2011;10(3):1416–1419. doi: 10.1021/pr1011476. [DOI] [PubMed] [Google Scholar]
- 49.Bröer S, et al. Comparison of lactate transport in astroglial cells and monocarboxylate transporter 1 (MCT 1) expressing Xenopus laevis oocytes. Expression of two different monocarboxylate transporters in astroglial cells and neurons. J Biol Chem. 1997;272(48):30096–30102. doi: 10.1074/jbc.272.48.30096. [DOI] [PubMed] [Google Scholar]
- 50.Allen RJ, Kirk K. Plasmodium falciparum culture: The benefits of shaking. Mol Biochem Parasitol. 2010;169(1):63–65. doi: 10.1016/j.molbiopara.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65(3):418–420. [PubMed] [Google Scholar]
- 52.Crabb BS, et al. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol Biol. 2004;270:263–276. doi: 10.1385/1-59259-793-9:263. [DOI] [PubMed] [Google Scholar]
- 53.Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA. 1997;94(20):10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhn Y, et al. Trafficking of the phosphoprotein PfCRT to the digestive vacuolar membrane in Plasmodium falciparum. Traffic. 2010;11(2):236–249. doi: 10.1111/j.1600-0854.2009.01018.x. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez CP, Stein W, Lanzer M. Trans stimulation provides evidence for a drug efflux carrier as the mechanism of chloroquine resistance in Plasmodium falciparum. Biochemistry. 2003;42(31):9383–9394. doi: 10.1021/bi034269h. [DOI] [PubMed] [Google Scholar]
- 56.Ferdig MT, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52(4):985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 57.Durrand V, et al. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol Biochem Parasitol. 2004;136(2):273–285. doi: 10.1016/j.molbiopara.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 58.Yang Z, et al. Molecular analysis of chloroquine resistance in Plasmodium falciparum in Yunnan Province. Trop Med Int Health. 2007;12(9):1051–1060. doi: 10.1111/j.1365-3156.2007.01882.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.