Significance
Symbiotic microbes are essential for the survival of many multicellular organisms, yet the factors promoting cooperative symbioses remain poorly understood. Three genera of solitary wasps cultivate antibiotic-producing Streptomyces bacteria for defense of their larvae against pathogens. Here we show that the wasp ancestor acquired the protective symbionts from the soil at least 68 million years ago. Although mother-to-offspring symbiont transmission dominates, exchange between unrelated individuals and uptake of opportunistic microorganisms from the environment occasionally occurs. However, experimental infections of female beewolves reveal that the wasps selectively block transmission of nonnative bacteria to their offspring. These findings suggest a previously unknown mechanism to maintain a specific symbiont over long evolutionary timescales and help to explain the persistence of bacterial mutualists in insects.
Keywords: protective symbiosis, cospeciation, mutualism stability, Hymenoptera, Crabronidae
Abstract
Many insects rely on symbiotic microbes for survival, growth, or reproduction. Over evolutionary timescales, the association with intracellular symbionts is stabilized by partner fidelity through strictly vertical symbiont transmission, resulting in congruent host and symbiont phylogenies. However, little is known about how symbioses with extracellular symbionts, representing the majority of insect-associated microorganisms, evolve and remain stable despite opportunities for horizontal exchange and de novo acquisition of symbionts from the environment. Here we demonstrate that host control over symbiont transmission (partner choice) reinforces partner fidelity between solitary wasps and antibiotic-producing bacteria and thereby stabilizes this Cretaceous-age defensive mutualism. Phylogenetic analyses show that three genera of beewolf wasps (Philanthus, Trachypus, and Philanthinus) cultivate a distinct clade of Streptomyces bacteria for protection against pathogenic fungi. The symbionts were acquired from a soil-dwelling ancestor at least 68 million years ago, and vertical transmission via the brood cell and the cocoon surface resulted in host–symbiont codiversification. However, the external mode of transmission also provides opportunities for horizontal transfer, and beewolf species have indeed exchanged symbiont strains, possibly through predation or nest reuse. Experimental infection with nonnative bacteria reveals that—despite successful colonization of the antennal gland reservoirs—transmission to the cocoon is selectively blocked. Thus, partner choice can play an important role even in predominantly vertically transmitted symbioses by stabilizing the cooperative association over evolutionary timescales.
Cooperation is ubiquitous in nature, yet it presents a conundrum to evolutionary biology because acts that are beneficial to the receiver but costly to the actor should not be favored by natural selection (1). In interspecific associations (i.e., symbioses), the two most important models to explain the maintenance of cooperation are partner fidelity and partner choice (2, 3). In partner-fidelity associations, host and symbiont interact repeatedly and reward cooperating individuals while punishing cheaters, thereby reinforcing mutually beneficial interactions (2, 4). In partner-choice associations, individuals may interact only once, but one member can select its partner in advance of any possible exploitation (2, 4). Partner choice appears to select for cooperative strains among environmentally acquired microbial symbionts, e.g., the bioluminescent Vibrio fischeri bacteria of squids (5), the nitrogen-fixing rhizobia of legumes (6), and mycorrhizal fungi of plants (7). By contrast, partner fidelity is generally assumed to be the major stabilizing force in the widespread and ecologically important vertically transmitted symbioses of insects (4).
However, localization and transmission routes of mutualistic bacteria in insects are diverse, and the differences across symbiotic systems have important implications for the evolutionary trajectory of the associations. Symbionts with an obligate intracellular lifestyle are usually tightly integrated into the host’s metabolism (e.g., ref. 8) and development (9), and the mutual interdependence of both partners coincides with perfect vertical symbiont transmission. Over evolutionary timescales, the high degree of partner fidelity results in host–symbiont cocladogenesis, and, concordantly, phylogenies of hosts and their intracellular symbionts are often found to be congruent (10–13). Although such a pattern is also observed for some extracellular symbioses with especially tight host–symbiont integration (14, 15), the ability of many extracellularly transmitted symbionts to spend part of their life cycle outside of the host’s body is often reflected in more or less extensive horizontal transmission or de novo acquisition of symbionts from the environment (16, 17). In these cases, partner choice mechanisms are expected to ensure specificity in the establishment and maintenance of the association (18). The nature of such control mechanisms, however, remains poorly understood.
Although many of the well-studied mutualistic associations in insects have a nutritional basis (19, 20), an increasing number of symbioses for the defense of the host against predators (21), parasitoids (22), or pathogens (23–25) have recently been discovered. Among defensive symbionts, Actinobacteria are particularly prevalent, probably due to their ubiquity in the soil and their ability to produce secondary metabolites with antibiotic properties (23). Antibiotic-producing actinobacterial symbionts have been discovered on the cuticle of leaf-cutting ants (26), in the fungal galleries of a bark beetle (27), and in the antennae and on cocoons of beewolf wasps (28). While in the former two cases the symbionts have been implicated in the defense of the hosts’ nutritional resources against competing fungi (26, 27), the beewolves’ bacteria protect the offspring in the cocoon against pathogenic microorganisms (28, 29).
Beewolves are solitary wasps in the genera Philanthus, Trachypus, and Philanthinus (Hymenoptera, Crabronidae, Philanthini). They engage in a defensive alliance with the Actinobacterium ‘Candidatus Streptomyces philanthi’ (CaSP) (28, 30, 31), which is cultivated by female beewolves in specialized antennal gland reservoirs (32). The uniqueness and complexity of the glands suggest a long history of host adaptation towards cultivating its actinobacterial symbionts (32). From the antennae, the streptomycetes are secreted into the brood cell, taken up by the larva, and incorporated into its cocoon (33), where they provide protection against pathogenic fungi and bacteria (28) by producing at least nine different antimicrobial compounds (29). Weeks or months later, eclosing adult females acquire the bacteria from the cocoon surface (33), thus completing the vertical transmission of CaSP. However, this mode of transmission provides opportunities for the horizontal transfer of symbionts among beewolf species or the de novo uptake of bacteria from the environment. Despite these opportunities, a monophyletic clade of CaSP strains has previously been found in 31 species of beewolves, suggesting an ancient and highly coevolved relationship (30, 31, 34).
Here we combine cophylogenetic analyses of beewolves and their vertically transmitted defensive symbionts with experimental manipulation of symbiont infection status and subsequent observations of transmission from female antennal gland reservoirs into the brood cell to (i) reconstruct the coevolutionary history of the symbiosis, (ii) estimate the age of the symbiosis, (iii) elucidate the ancestral lifestyle of the symbionts, and (iv) assess the importance of partner fidelity and partner choice for the long-term stability of the association.
Results and Discussion
Age of the Beewolf–Streptomyces Symbiosis.
To reconstruct the phylogenetic relationships across beewolves and closely related wasps, we determined sequences of five nuclear [28S rRNA (28S), wingless (wnt), long-wavelength rhodopsin (lwrh), arginine kinase (argK), and elongation factor 1α (ef1a)] and one mitochondrial gene [cytochrome oxidase (coxI)] for 50 Philanthini (Philanthus, Trachypus, Philanthinus) that engage in a defensive symbiosis with CaSP, as well as several outgroup taxa that lack antennal symbionts (34) (SI Appendix, Tables S1–S3). Based on the concatenated alignment of 5,521 bp, phylogenetic analyses strongly support monophyly of the three genera with antennal symbionts (Fig. 1 and SI Appendix, Fig. S1). As previously hypothesized (35), our results indicate that Trachypus renders Philanthus paraphyletic. Because we included representatives of all genera in the subfamily Philanthinae (sensu 35) except for the very rare Pseudoscolia (which is probably most closely related to Cerceris and Eucerceris; see ref. 35), we conclude that the symbiosis with CaSP in antennal gland reservoirs had a single origin in the ancestor of the tribe Philanthini.
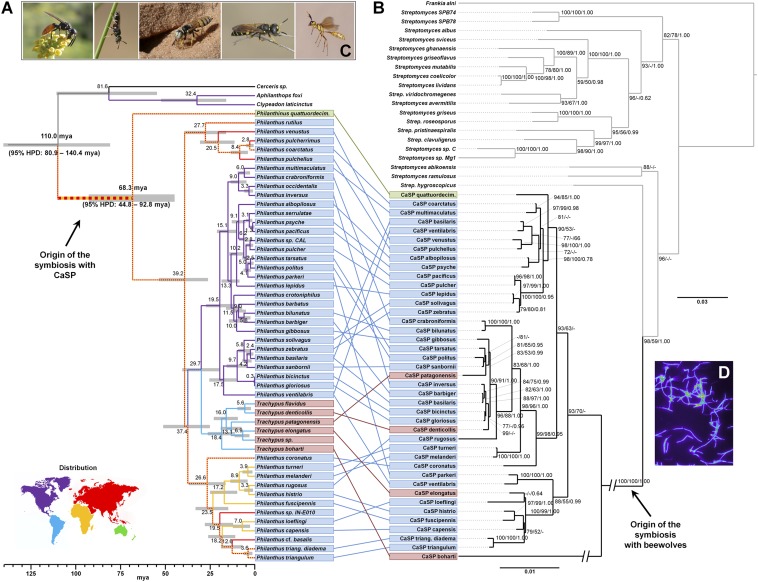
Fig. 1.
Cophylogenetic analysis of beewolves (A) and their defensive antennal symbionts (B). Node ages in the host phylogeny are shown in Mya with 95% HPD interval bars. Branches are color-coded according to the geographic distribution of the host species (see world map: hatched yellow and red branches indicate occurrence in Africa and/or Eurasia). Colored boxes around host and symbiont names denote host genera (green, Philanthinus; blue, Philanthus; red, Trachypus). Host–symbiont associations are shown by connecting lines. Values at the nodes of the symbiont phylogeny are local support values from the FastTree analysis (GTR model), bootstrap values from PHYML, and Bayesian posteriors, respectively. The origin of the symbiosis is highlighted in both phylogenies by arrows. (C) Photographs of selected Philanthini host species: Philanthus loefflingi male, Philanthus pulcherrimus male, Philanthus basilaris female at its nest entrance, Philanthus coronatus male, and Trachypus boharti female (from left to right). (D) Fluorescence micrograph of CaSP from the antennal gland secretion of a female P. triangulum (in false colors).
Three fossil calibration points were used to infer minimum ages of divergence within the beewolf phylogeny: (i) Psammaecius sepultus (Bembecinae) from Florissant beds in Colorado (36, 37), which date back to the latest Eocene (∼34.1 Mya) (38); (ii) Cerceris berlandi from late Stampian shales (∼30 Mya) in France (39); and (iii) two Philanthini fossils from Colorado (Philanthus saxigenus and Prophilanthus destructus, ∼34.1 Mya) (40, 41) and one from France (Philanthus annulatus, ∼30 Mya) (41). Due to the somewhat doubtful systematic affiliation of the Philanthus and Prophilanthus fossils, the analyses were also repeated excluding these fossil calibration points, which did not significantly affect the age estimation for the origin of the symbiosis (SI Appendix, Table S4). The age for the root was set to 140 ± 10 Mya (mean ± SD) because both the divergence of Sphecidae from other Apoidea and that of Crabronidae from bees have been estimated to have occurred in the period 130–150 Mya (42, 43), coincident with the rise of the angiosperms.
Different substitution models (GTR, GTR+I+G, HKY, HKY+G, HKY+I+G) with various parameter settings and age priors consistently dated the origin of the beewolf–Streptomyces to the late Cretaceous (SI Appendix, Table S4). The HKY+G substitution model with fixed input tree, relaxed uncorrelated log-normal clock model, and the inclusion of the Cerceris, Psammaecius, and root calibration points yielded an age estimate of 68.3 Mya [95% highest posterior density (HPD) interval: 44.8–92.8 Mya] to 110.0 Mya (95% HPD interval: 80.9–140.4 Mya) for the origin of the association with Streptomyces (Fig. 1 and SI Appendix, Figs. S2–S4 for phylogenetic trees based on other model parameters). Thus, the beewolf–Streptomyces symbiosis evolved more recently than many of the intimate nutritional mutualisms in insects, e.g., the aphid–Buchnera (160–280 Mya; see ref. 12), cockroach–Blattabacterium (135–250 Mya; see ref. 13), planthopper–Vidania (>130 Mya; see ref. 44), and Auchenorrhyncha–Sulcia (260–280 Mya; see ref. 45) associations. However, the beewolf symbiosis is probably more ancient than the functionally similar defensive association between leaf-cutter ants and antibiotic-producing Pseudonocardia bacteria because fungus farming did not evolve in ants before around 50 Mya (46). To our knowledge, the beewolf–Streptomyces mutualism represents the first defensive symbiosis in insects with a reliable age estimate.
Prevalence of Antennal Streptomyces Symbionts Across Beewolves.
To assess the prevalence of antennal symbionts across beewolf host species, we screened 338 females from 34 species and subspecies for the presence of CaSP using diagnostic 16S rRNA gene primers (34). We detected CaSP in 93% of all individuals, and prevalence ranged from 67 to 100% within species, with the exception of Philanthus cf. basalis (SI Appendix, Table S5). We tested apparently symbiont-free individuals for other eubacterial taxa and occasionally found Actinobacteria other than CaSP, Proteobacteria, or Tenericutes, in or on female beewolf antennae (SI Appendix, Fig. S5). Amycolatopsis was found in the antennae of both available individuals of P. cf. basalis and in two Philanthus triangulum individuals (of 68) from Germany. For P. cf. basalis, we verified the replacement of CaSP by Amycolatopsis and its growth in the antennal gland reservoirs by fluorescence in situ hybridization (FISH) (SI Appendix, Figs. S6 and S7). Whether these symbiont replacements represent rare individual cases or a complete lineage replacement in P. cf. basalis cannot be determined because of the small sample size (n = 2). The occurrence of Proteobacteria (Wolbachia, Serratia) and Tenericutes (Spiroplasma) probably represents systemic infections of the hosts, including the antennal hemolymph, rather than specialized colonization of the antennal gland reservoirs.
Host–Symbiont Coevolutionary History.
We reconstructed the phylogeny of CaSP symbionts from 34 Philanthus, four Trachypus, and one Philanthinus host species, using partial sequences of 16S rRNA, elongation factor-G and -Tu (fus-tuf), gyrase B (gyrB), and gyrase A (gyrA) (SI Appendix, Tables S6 and S7). The consistently clean sequencing signals indicated that each beewolf individual generally cultivates a single dominant symbiont strain in its antennae. Like previous analyses based only on 16S rRNA gene sequences of all available Streptomyces-type strains (31), both Bayesian and maximum-likelihood analyses provided strong support for the monophyly of the symbiont clade within Streptomyces (Fig. 1 and SI Appendix, Fig. S8), implying a single origin of the association. Randomization tests yielded evidence for overall cocladogenesis of beewolves and CaSP (Parafit: P = 0.001; TreeMap: P = 0.003, Jane3: P < 0.05), providing evidence for partner fidelity over evolutionary timescales and thereby corroborating earlier findings of vertical symbiont transmission (33). However, a comparison of the phylogenies also revealed numerous discrepancies between host and symbiont trees, indicating horizontal transmission of symbionts among host species (Fig. 1).
To explore the prevalence of ongoing symbiont exchange within and across beewolf populations, we sequenced gyrA from the symbionts of 109 beewolf individuals in 41 species (SI Appendix, Table S6). The topology of the gyrA tree was very similar to the multigene phylogeny, and symbiont sequences from individuals of the same host species were identical or clustered together for all but three species (SI Appendix, Fig. S9). Although for Philanthus gibbosus CaSP strains were closely related, this was not the case for Philanthus ventilabris and P. basilaris. These latter two species occur sympatrically with other beewolves, and interspecific predation among Philanthus has occasionally been observed (47), so it is conceivable that some lineages have recently acquired symbionts horizontally from congeneric beewolf females that served as larval provisions [specifically, P. ventilabris and P. basilaris may have acquired symbionts from the two smaller sympatric species Philanthus parkeri and Philanthus barbiger, respectively (Fig. 1)]. A second possible explanation for horizontal transfer of symbionts is reuse of nests and brood cells that occurs in some beewolf species (47). A third alternative is that a reservoir of CaSP spores might subsist in beewolf habitats and thereby facilitate diffuse horizontal exchange (48). Consistent with the latter two hypotheses, we detected CaSP DNA in sand from used beewolf observation cages by pyrosequencing bacterial 16S rRNA amplicons [385 of 7,123 total sequences = 5.4% (SI Appendix, Fig. S10)]. Although we cannot at present exclude the possibility of the amplification originating from dead CaSP cells, the long-term survival of the symbionts in the brood cell during beewolf hibernation indicates that CaSP can survive unfavorable environmental conditions as metabolically inactive cells (33, 48).
Partner Choice and Maintenance of Specificity in the Symbiotic Association.
Considering the ample opportunities for opportunistic Actinobacteria to be taken up by beewolf females, how is specificity maintained in the beewolf–CaSP symbiosis? Behavioral observations in field-collected P. triangulum indicate that females harboring opportunistic Actinobacteria in their antennal gland reservoirs usually do not apply visible amounts of symbiont-containing antennal gland secretion (AGS) to their brood cells [homogeneity test with Yates’s correction, χ2 = 5.49, P = 0.019 (SI Appendix, Table S8)]. Of seven beewolf females harboring opportunistic bacteria, only one was observed to secrete AGS, suggesting the possibility for partner choice during symbiont transmission.
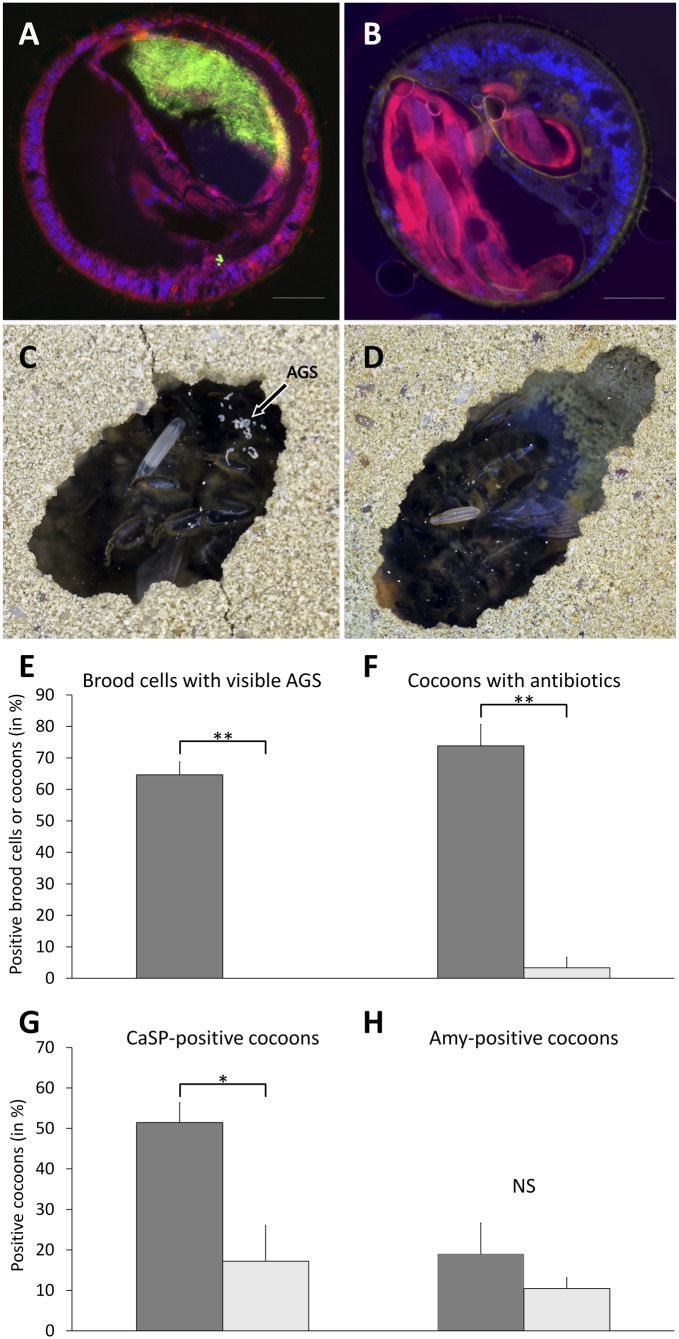
To experimentally test for partner choice, we manipulated symbiont infection status by infecting aposymbiotic P. triangulum females with either a culture of their native symbiont (CaSP) or a culture of Amycolatopsis strain alb_538-2 (Amy) isolated from a female Philanthus albopilosus antenna. Because Amycolatopsis strains were repeatedly detected in the antennae of different beewolf species (SI Appendix, Fig. S5) and could successfully colonize the antennal gland reservoirs (SI Appendix, Fig. S6), we used an Amycolatopsis isolate as a representative opportunistic Actinobacterium. Diagnostic PCRs and FISH of female antennae revealed that both CaSP and Amy can successfully colonize the antennal gland reservoirs upon experimental reinfection (Fig. 2), with 46.2% (6 of 13) and 66.7% of females (6 of 9) being successfully infected, respectively. Although AGS was visible in 64.6% of the brood cells of CaSP-infected beewolves, not a single brood cell was positive for AGS after Amy infection (Fig. 2, Wilcoxon test, Z = 2.987, P = 0.004). Concordantly, although some cocoons of Amy-infected females were positive for CaSP [probably due to some residual CaSP cells in the observation cages (SI Appendix, Fig. S10)], diagnostic PCRs and GC-MS analyses revealed significantly higher prevalence of CaSP and their antibiotics on cocoons of CaSP-infected vs. Amy-infected females (Fig. 2, Wilcoxon tests, CaSP presence: Z = 2.470, P = 0.013; antibiotic presence: Z = 2.872, P = 0.004). By contrast, Amycolatopsis was detected in equally low frequencies on cocoons of both CaSP- and Amy-infected females (Fig. 2, Wilcoxon test, Z = 0.558, P = 0.577), indicating occasional contamination from the surrounding soil. Although we experimentally infected beewolves with one opportunistic Amycolatopsis strain only, the results—taken together with the observation that field-collected beewolf females infected with opportunistic Streptomyces strains did not secrete AGS (SI Appendix, Table S8)—provide strong evidence for partner choice during symbiont transmission, most likely by blocking the AGS application to the brood cell upon infection with opportunistic bacteria (Fig. 2 and SI Appendix, Table S9).
Fig. 2.
Partner choice during symbiont transmission in the beewolf-CaSP symbiosis. (A and B) Fluorescence micrographs of female P. triangulum antennae (cross-sections) after experimental infection with CaSP (A) and Amycolatopsis (Amy) (B), respectively. Staining of bacteria was achieved with the CaSP-specific probe Cy5-SPT177 (green) and the Amy-specific probe Amy_16S (red). Host cell nuclei were counterstained with DAPI (blue). Scale bars represent 100 µm. (C and D) Examples of brood cells with (C) and without (D) visible amounts of symbiont-containing AGS after infection with CaSP and Amy, respectively. The position of the AGS on the brood cell ceiling is indicated by an arrow. (E–H) Symbiont transmission success after experimental infection with CaSP (dark gray bars) and Amy (light gray bars), assessed as the proportion of brood cells containing visible amounts of AGS (E), the proportion of cocoons containing CaSP-produced antibiotics (F), and the proportion of cocoons positive for CaSP (G) and Amy (H) in diagnostic PCRs. Significant differences between CaSP and Amy infection treatments are indicated by asterisks (Wilcoxon rank-sum tests: *P < 0.05, **P < 0.01).
In several marine and terrestrial symbioses with horizontal transmission, partner choice has been found to be important to prevent the establishment of nonnative symbionts and/or to sanction noncooperative individuals (“cheaters”) (5–7, 49). To be selectively favored, however, host punishment of cheating symbionts must either have a direct benefit for the host (49) or increase cooperation levels in future interactions with the same host individual or its offspring (50). In beewolves, three mutually nonexclusive scenarios may explain the selective advantage of partner choice during symbiont transmission: (i) Keeping opportunistic bacteria confined to the gland reservoirs may limit the spread of potentially pathogenic microbes to the cocoon and thereby reduce the risk of infection in the offspring. (ii) Because beewolves possess gland reservoirs in five antennomeres of each antenna (32), selectively blocking transmission of nonnative bacteria from individual reservoirs may enhance the chances of successfully endowing the offspring with beneficial symbionts while simultaneously limiting pathogen exposure. It is conceivable that immune effector molecules (e.g., antimicrobial peptides) differentially affect physiology or morphology of symbiotic and opportunistic bacteria in the antennal gland reservoirs (51), respectively, which could have an impact on their transmission into the brood cell. (iii) Avoiding transmission of opportunistic bacteria likely saves the host resources that would otherwise be used by the remaining bacteria to grow and fill up the gland reservoirs again.
Conclusions.
The observed pattern of diffuse codiversification between beewolves and defensive Streptomyces symbionts indicates that, despite the fact that they are localized in specialized antennal gland reservoirs, their extracellular lifestyle and external route of transmission allow for horizontal symbiont replacement and uptake of opportunistic Actinobacteria. However, in contrast to other insect symbioses that rely on partner choice rather than fidelity (17, 18, 52), only a distinct monophyletic clade of symbionts appears to be able to successfully establish a long-term association with the host. Thus, the beewolf–Streptomyces mutualism presents an interesting intermediate case between strictly vertically transmitted primary symbionts and more loosely associated secondary symbionts. Partner choice at the point of symbiont transmission apparently reinforced host–symbiont fidelity and thereby promoted the long-term stability of the mutualistic association with a specific clade of symbionts since origin of the association in the Cretaceous.
Materials and Methods
Insect Specimens.
Specimens of 43 Philanthus species and subspecies from North America, Europe, India, and South Africa, six Trachypus species from South America, and one Philanthinus species from Turkey were collected or kindly supplied by colleagues (SI Appendix, Table S1). Species were identified using published keys for the North American (53–55) and South African Philanthus (56) and for the South American Trachypus species (57), respectively. Indian specimens were identified by comparison with the original descriptions as well as the reference collection at the Natural History Museum in London. Fresh beewolf specimens were freeze-killed or placed directly into 70% or 95% ethanol and stored until DNA extraction. As outgroup taxa, crabronid species of the closely related genera Aphilanthops, Clypeadon, and Cerceris were collected, and additional sequences for the more distantly related Bembix, Bicyrtes, and Apis mellifera (Apidae) were obtained from the National Center for Biotechnology Information database (SI Appendix, Table S1).
Reconstruction of the Host Phylogeny.
DNA was extracted from insect specimens, and partial sequences of coxI (841 bp), 28S (865 bp), wnt [comprising 378 bp of coding sequence (cds)], lwrh [comprising 608 bp of cds and 156 bp of noncoding sequence (ncs)], argK (with 825 bp cds and 111 bp ncs), and ef1a (including 1,041 bp cds and 696 bp ncs) were amplified and sequenced as described previously (SI Appendix, Tables S2 and S3). Sequences were aligned using BioEdit 7.0.5.3 (58) and SeaView 4.2.6 (59), and phylogenetic trees were reconstructed using maximum parsimony, maximum likelihood, and Bayesian inference (SI Appendix).
Dating of the Host Phylogeny.
Divergence time estimations were inferred using BEAST v1.7.5 (60). Various substitution models and parameter settings were tested, and four calibration points (Psammaecius sepultus, Cerceris berlandi, the age of three Philanthini fossils, and the root age) were used for the dating analyses (see SI Appendix for details). Evaluation and comparison of model parameters were performed using Tracer v1.5 (61), and consensus trees were visualized with FigTree v1.3.1 (62), including HPD intervals (Fig. 1 and SI Appendix, Figs. S2–S4).
Reconstruction of the Symbiont Phylogeny.
Genomic DNA was extracted from whole beewolf antennae and used for amplification and sequencing of partial fus-tuf, gyrA, gyrB, and 16S rRNA genes (SI Appendix, Table S6). Reference sequences of all Streptomyces species for which fully sequenced or good draft genomes were available were retrieved from the National Center for Biotechnology Information database (SI Appendix, Table S7), and cultures of three strains that are closely related to CaSP based on 16S rRNA sequences (Streptomyces ramulosus DSM 40100, Streptomyces abikoensis DSM 40831, and Streptomyces mutabilis DSM 40169) were additionally obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) for amplification and sequencing. The concatenated alignment of 4,653 bp (1,391 bp of 16S rDNA, 639 bp of fus, 930 bp of tuf, 249 bp of fus-tuf intergenic spacer, 765 bp of gyrB, and 549 bp of gyrA) was used for phylogenetic reconstruction by approximately maximum-likelihood analysis (FastTree 2.1) (63), maximum likelihood (PHYML) analysis (64), and Bayesian inference (MrBayes 3.1.2) (65, 66, 67).
Host–Symbiont Cophylogenetic Analysis.
To test for codiversification between hosts and symbionts, three different methods were used. First, host and symbiont trees were imported into TreeMap 1.0 (68). Both trees were randomized (1,000 replicates), and the number of observed codiversification events (21) was compared with the resulting distribution of codiversification events in the randomized dataset. Second, host and symbiont distance matrices were computed in BioEdit 7.0.5.3 (58) based on the concatenated alignments, and permutation tests (1,000 replicates) were run as implemented in ParaFit (69). Third, host and symbiont trees were imported into Jane 3 (70) and tested for congruence by using both edge- and node-based cost models. In addition to an analysis using the default cost parameters, a second analysis with the cost for symbiont loss reduced to 1 was performed. The number of generations was set to 30, and the population size to 500 for both analyses, as neither parameter appeared to influence the results (several combinations tested). Statistical assessment of the observed cost of the optimal trees was achieved by randomizing the symbiont tree (β = −1) or permuting host–symbiont associations (100 resamplings, respectively). For visualization, a tanglegram was reconstructed and optimized in Dendroscope V3.0.13beta (71) and used as a template for visualization of the comparative phylogenies in Microsoft PowerPoint, including both branch lengths (both trees) and divergence time estimates (host tree only) (Fig. 1). In the symbiont tree, a reduced set of free-living Streptomyces strains was included for better visualization of the relationships among CaSP isolates. The monophyly of the symbiont clade and the within-clade relationships were identical to the full bacterial tree (SI Appendix, Fig. S8).
Detection of CaSP and Other Bacteria in Philanthus Antennae.
To determine the prevalence of CaSP and other bacteria in beewolf antennae, antennal DNA extracts were screened with CaSP-specific primers as well as primers targeting Amycolatopsis, Actinobacteria in general, and eubacteria in general (SI Appendix, Tables S2 and S3). General eubacterial PCR products were separated by temperature-gradient gel electrophoresis before sequencing as described earlier (72). Sequences of actinobacterial 16S rRNA were aligned to the SILVA small subunit (SSU) ribosomal database (73) using the SINA aligner (74) and imported into ARB (75). An alignment including reference sequences was exported, and phylogenetic reconstruction was achieved using FastTree 2.1 (63). The presence of Amycolatopsis in the antennal gland reservoirs of the two investigated individuals of P. cf. basalis was confirmed by FISH (SI Appendix, Figs. S6 and S7).
Detection of CaSP in Sand Surrounding Beewolf Nests.
To assess the possibility for horizontal uptake of CaSP from nest material, we screened sand from observation cages that had previously been occupied by beewolves for the presence of CaSP using bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) of bacterial 16S rRNA genes (SI Appendix). DNA was extracted from sand, and bacterial 16S rRNA amplicons were generated with primers Gray28F and Gray519r and sequenced commercially (76, 77). QIIME (78) was used for quality trimming, denoising, and analysis of the reads by clustering into operational taxonomic units (97% similarity cutoff). The number of CaSP amplicons was assessed using a custom-made Perl script.
Partner Choice Assays.
Because CaSP is acquired by female beewolves from the cocoon surface shortly before emergence, aposymbiotic beewolf females can be generated by carefully removing the developing beewolf from the cocoon 1–2 d before emergence. Anesthetized females were reinfected with an in vitro culture of ‘Ca. S. philanthi biovar triangulum’ strain 23Af2 or Amycolatopsis strain alb538-1 (Amy) that was isolated from the antenna of a P. albopilosus female by applying a dense culture suspension to the antennal surface and simultaneously bending the antenna carefully with forceps. Subsequently, females were reared in observation cages as described previously (79) and provided with honey and bees ad libitum. For each brood cell, the presence of the AGS was assessed by careful visual inspection. After death, each female’s antennae were subjected to diagnostic PCR and FISH, using the specific primer pairs Strep_phil_185 (fwd3)/Act-A19 and Amy_16S_1F/Amytop_16S_3R as well as probes SPT177 and Amy_16S to assess the reinfection success of CaSP and Amy, respectively (SI Appendix, Tables S2 and S3). Specificity of primers was assessed in silico and in vitro by testing CaSP and Amy DNA from pure cultures as well as several other actinobacterial strains. Offspring cocoons were removed from the cages 8–10 d after cocoon spinning and tested qualitatively for the presence of CaSP-produced antibiotics (piericidin A1, B1, and streptochlorin) by methanol extraction and GC-MS as described earlier (48, 80). Additionally, the presence of CaSP and Amy was assessed by diagnostic PCRs as described above. The percentage of brood cells containing visible amounts of AGS and of cocoons positive for symbionts or antibiotics was calculated for each female and compared between treatment groups using Wilcoxon rank-sum tests using SPSS17.0.
Supplementary Material
Acknowledgments
We thank Johannes Kroiss, Christine Michel, Michael Ohl, Fred and Sarah Gess, Kumar Ghorpadé, Thomas Schmitt, Carlo Polidori, Dirk Koedam, Toshko Ljubomirov, Erol Yildirim, Faruk Gürbüz, Kari Smith, and Eva Sonnleitner for field work or generous gifts of specimens; Thomas Datzmann for support with the phylogenetic analysis; and Benjamin Weiss for help with FISH. Permits were issued by the nature conservation boards of KwaZulu Natal (Permit 4362/2004), Eastern Cape Province (WRO44/04WR, WRO9/04WR, WRO74/06WR, WRO75/06WR, CRO135/11CR, CRO136/11CR, CRO179/10CR, and CRO180/10CR) and Western Cape Province (001-202-00026, 001-506-00001, AAA004-00053-0035, AAA004-00089-0011, AAA004-00683-0035, and 0046-AAA004-00008) of South Africa, and the Brazilian Ministry of the Environment (MMA/SISBIO/22861-1). We were financially supported by the Max Planck Society (M.K.), German Science Foundation Grants DFG-STR532/2-2 (to E.S. and M.K.) and DFG-KA2846/2-1 (to M.K.), the Evangelische Studienwerk Villigst e.V. (K.R.-M.), the Volkswagen Foundation (M.K.), the Unibund Würzburg (M.K.), the German National Academic Foundation (M.K.), and the Arthur von Gwinner Foundation (M.K. and G.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information database. For a list of accession numbers, see SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400457111/-/DCSupplemental.
References
- 1.Axelrod R, Hamilton WD. The evolution of cooperation. Science. 1981;211(4489):1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- 2.Bull JJ, Rice WR. Distinguishing mechanisms for the evolution of co-operation. J Theor Biol. 1991;149(1):63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- 3.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. The evolution of cooperation. Q Rev Biol. 2004;79(2):135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- 4.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyholm SV, McFall-Ngai MJ. The winnowing: Establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2(8):632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 6.Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425(6953):78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- 7.Kiers ET, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333(6044):880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- 8.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21(16):1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koga R, Meng X-Y, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci USA. 2012;109(20):E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumann L, Baumann P. Cospeciation between the primary endosymbionts of mealybugs and their hosts. Curr Microbiol. 2005;50(2):84–87. doi: 10.1007/s00284-004-4437-x. [DOI] [PubMed] [Google Scholar]
- 11.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 12.Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc Roy Soc B-Biol Sci. 1993;253(1337):167–171. [Google Scholar]
- 13.Bandi C, et al. The establishment of intracellular symbiosis in an ancestor of cockroaches and termites. Proc Roy Soc B-Biol Sci. 1995;259(1356):293–299. doi: 10.1098/rspb.1995.0043. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi Y, et al. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009;7(2):2. doi: 10.1186/1741-7007-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 2006;4(10):e337. doi: 10.1371/journal.pbio.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prado SS, Almeida RPP. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr Microbiol. 2009;58(1):64–69. doi: 10.1007/s00284-008-9267-9. [DOI] [PubMed] [Google Scholar]
- 17.Cafaro MJ, et al. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc Roy Soc B-Biol Sci. 2011;278(1713):1814–1822. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang MM, Poulsen M, Currie CR. Symbiont recognition of mutualistic bacteria by Acromyrmex leaf-cutting ants. ISME J. 2007;1(4):313–320. doi: 10.1038/ismej.2007.41. [DOI] [PubMed] [Google Scholar]
- 19.Feldhaar H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol. 2011;36(5):533–543. [Google Scholar]
- 20.Douglas AE. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23(1):38–47. [Google Scholar]
- 21.Kellner RLL. Molecular identification of an endosymbiotic bacterium associated with pederin biosynthesis in Paederus sabaeus (Coleoptera: Staphylinidae) Insect Biochem Mol Biol. 2002;32(4):389–395. doi: 10.1016/s0965-1748(01)00115-1. [DOI] [PubMed] [Google Scholar]
- 22.Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. 2003;100(4):1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaltenpoth M. Actinobacteria as mutualists: General healthcare for insects? Trends Microbiol. 2009;17(12):529–535. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6(12):e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brownlie JC, Johnson KN. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009;17(8):348–354. doi: 10.1016/j.tim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398(6729):701–704. [Google Scholar]
- 27.Scott JJ, et al. Bacterial protection of beetle-fungus mutualism. Science. 2008;322(5898):63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaltenpoth M, Göttler W, Herzner G, Strohm E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 2005;15(5):475–479. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 29.Kroiss J, et al. Symbiotic Streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6(4):261–263. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- 30.Kaltenpoth M, Schmitt T, Polidori C, Koedam D, Strohm E. Symbiotic streptomycetes in antennal glands of the South American digger wasp genus Trachypus (Hymenoptera, Crabronidae) Physiol Entomol. 2010;35(2):196–200. [Google Scholar]
- 31.Kaltenpoth M, Yildirim E, Gürbüz MF, Herzner G, Strohm E. Refining the roots of the beewolf-Streptomyces symbiosis: Antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae) Appl Environ Microbiol. 2012;78(3):822–827. doi: 10.1128/AEM.06809-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goettler W, Kaltenpoth M, Herzner G, Strohm E. Morphology and ultrastructure of a bacteria cultivation organ: The antennal glands of female European beewolves, Philanthus triangulum (Hymenoptera, Crabronidae) Arthropod Struct Dev. 2007;36(1):1–9. doi: 10.1016/j.asd.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Kaltenpoth M, Goettler W, Koehler S, Strohm E. Life cycle and population dynamics of a protective insect symbiont reveal severe bottlenecks during vertical transmission. Evol Ecol. 2010;24(2):463–477. [Google Scholar]
- 34.Kaltenpoth M, et al. ‘Candidatus Streptomyces philanthi’, an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int J Syst Evol Microbiol. 2006;56(6):1403–1411. doi: 10.1099/ijs.0.64117-0. [DOI] [PubMed] [Google Scholar]
- 35.Alexander BA. A cladistic analysis of the subfamily Philanthinae (Hymenoptera, Sphecidae) Syst Entomol. 1992;17(2):91–108. [Google Scholar]
- 36.Cockerell TDA. Fossil Hymenoptera from Florissant, Colorado. Bull Mus Comp Zool Harvard Coll. 1906;50:31–58. [Google Scholar]
- 37.Pulawski WJ, Rasnitsyn AP. The taxonomic position of Hoplisus sepultus from the lower oligocene of Colorado. Pol Pismo Entomol. 1980;50(3):393–396. [Google Scholar]
- 38.Evanoff E, McIntosh WC, Murphey PC. Stratigraphic summary and 40AR/39AR geochronology of the Florissant Formation, Colorado. Proc Denver Mus Nat Sci. 2001;4(1):1–16. [Google Scholar]
- 39.Timon-David J. Insectes fossiles de l'Oligocene inferieur des Camoins (Bassin de Marseille). I. Dipteres brachyceres. II. Hymenopteres [Fossil insects of the lower Oligocene in Camoins (Bassin de Marseille). I. Diptera, Brachycera. II. Hymenoptera] Bull Soc Entomol Fr. 1944;48:40–45. French. [Google Scholar]
- 40.Rohwer SA. Three new fossil insects from Florissant, Colorado. Am J Sci. 1909;28(168):533–536. [Google Scholar]
- 41.Theobald N. Les insectes fossiles des terrains oligocenes de France [The fossil insects of oligocene soils in France] Mem Soc Sci Nancy. 1937;473:1–473. French. [Google Scholar]
- 42.Cardinal S, Danforth BN. Bees diversified in the age of eudicots. Proc Roy Soc B-Biol Sci. 2013;280(1755) doi: 10.1098/rspb.2012.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimaldi D, Engel MS. Evolution of the Insects. New York: Cambridge University Press; 2005. pp. 1–755. [Google Scholar]
- 44.Urban JM, Cryan JR. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea) BMC Evol Biol. 2012;12:87. doi: 10.1186/1471-2148-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: An ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71(12):8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci USA. 2008;105(14):5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans HE, O’Neill KM. The Natural History and Behavior of North American Beewolves. Ithaca, NY: Cornell University Press; 1988. pp. 1–278. [Google Scholar]
- 48.Koehler S, Doubský J, Kaltenpoth M. Dynamics of symbiont-mediated antibiotic production reveal efficient long-term protection for beewolf offspring. Front Zool. 2013;10(1):3. doi: 10.1186/1742-9994-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jander KC, Herre EA. Host sanctions and pollinator cheating in the fig tree-fig wasp mutualism. Proc Roy Soc B-Biol Sci. 2010;277(1687):1481–1488. doi: 10.1098/rspb.2009.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bshary R, Grutter AS. Asymmetric cheating opportunities and partner control in a cleaner fish mutualism. Anim Behav. 2002;63(2):547–555. [Google Scholar]
- 51.Login FH, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334(6054):362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 52.Kikuchi Y, Hosokawa T, Fukatsu T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 2011;5(3):446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bohart RM, Grissell EE. California wasps of the subfamiliy Philanthinae (Hymenoptera: Sphecidae) Bull Calif Insect Surv. 1975;19:1–57. [Google Scholar]
- 54.Ferguson GR. Revision of the Philanthus zebratus group (Hymenoptera, Philanthidae) J NY Entomol Soc. 1983;91(4):289–303. [Google Scholar]
- 55.Ferguson GR. Two new species in the genus Philanthus and a key to the politus group (Hymenoptera, Philanthidae) Pan-Pac Entomol. 1983;59(1-4):55–63. [Google Scholar]
- 56.Arnold G. The Sphegidae of South Africa, part VI. Ann Transvaal Mus. 1925;11:137–175. [Google Scholar]
- 57.Rubio E. Revisión del género Trachypus Klug (Hymenoptera: Sphecidae) [Revision of the genus Trachypus Klug (Hymenoptera: Sphecidae)] Revista Fac Agron Univ Zulia. 1975;3(1):7–87. Spanish. [Google Scholar]
- 58.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 59.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 60.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rambaut A, Drummond AJ. 2007. Tracer v1.4. Available from http://beast.bio.ed.ac.uk/Tracer. Accessed April 17, 2013.
- 62.Rambaut A. 2010. FigTree v1.3.1: Tree figure drawing tool. Available from http://tree.bio.ed.ac.uk/software/figtree. Accessed May 19, 2011.
- 63.Price MN, Dehal PS, Arkin AP. FastTree 2: Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 65.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 66.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294(5550):2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 67.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 68.Page RDM. 1995. TreeMap for Windows version 1.0a. Available from http://taxonomy.zoology.gla.ac.uk/rod/treemap.html. Accessed March 9, 2011.
- 69.Legendre P, Desdevises Y, Bazin E. A statistical test for host-parasite coevolution. Syst Biol. 2002;51(2):217–234. doi: 10.1080/10635150252899734. [DOI] [PubMed] [Google Scholar]
- 70.Conow C, Fielder D, Ovadia Y, Libeskind-Hadas R. Jane: A new tool for the cophylogeny reconstruction problem. Algorithms Mol Biol. 2010;5:16. doi: 10.1186/1748-7188-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stoll S, Gadau J, Gross R, Feldhaar H. Bacterial microbiota associated with ants of the genus Tetraponera. Biol J Linn Soc Lond. 2007;90(3):399–412. [Google Scholar]
- 73.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pruesse E, Peplies J, Glöckner FO. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28(14):1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32(4):1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishak HD, et al. Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol. 2011;61(4):821–831. doi: 10.1007/s00248-010-9793-4. [DOI] [PubMed] [Google Scholar]
- 77.Sun Y, Wolcott RD, Dowd SE. Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. Methods Mol Biol. 2011;733:129–141. doi: 10.1007/978-1-61779-089-8_9. [DOI] [PubMed] [Google Scholar]
- 78.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strohm E, Linsenmair KE. Leaving the cradle: How beewolves (Philanthus triangulum F.) obtain the necessary spatial information for emergence. Zoology. 1995;98(3):137–146. [Google Scholar]
- 80.Koehler S, Kaltenpoth M. Maternal and environmental effects on symbiont-mediated antimicrobial defense. J Chem Ecol. 2013;39(7):978–988. doi: 10.1007/s10886-013-0304-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.