Significance
Tumor heterogeneity is a poorly understood phenomenon. We lack biomarkers for identifying aggressive primary tumor subsets that give rise to metastases and impact early cancer detection and treatment. Many solid tumors are known to accumulate hyaluronan (HA), a glycosaminoglycan, which is also produced by the tumor cells themselves. We report a quantitative approach for uncovering breast cancer heterogeneity using fluorescent HA to detect differential binding patterns to CD44 and RHAMM/HMMR receptors. This approach permits identification of tumor-cell subsets that bind high levels of HA, and may be applicable to other ligands/receptors and disease models. Despite representing the invasive/metastatic subset of parental tumors, unexpectedly, the high HA-binding subset was slow-growing, and is thus likely to be a source of dormancy and relapse.
Keywords: tumor cell heterogeneity, hyaluronan binding, heterogeneity index
Abstract
Tumor heterogeneity confounds cancer diagnosis and the outcome of therapy, necessitating analysis of tumor cell subsets within the tumor mass. Elevated expression of hyaluronan (HA) and HA receptors, receptor for HA-mediated motility (RHAMM)/HA-mediated motility receptor and cluster designation 44 (CD44), in breast tumors correlates with poor outcome. We hypothesized that a probe for detecting HA–HA receptor interactions may reveal breast cancer (BCa) cell heterogeneity relevant to tumor progression. A fluorescent HA (F-HA) probe containing a mixture of polymer sizes typical of tumor microenvironments (10–480 kDa), multiplexed profiling, and flow cytometry were used to monitor HA binding to BCa cell lines of different molecular subtypes. Formulae were developed to quantify binding heterogeneity and to measure invasion in vivo. Two subsets exhibiting differential binding (HA−/low vs. HAhigh) were isolated and characterized for morphology, growth, and invasion in culture and as xenografts in vivo. F-HA–binding amounts and degree of heterogeneity varied with BCa subtype, were highest in the malignant basal-like cell lines, and decreased upon reversion to a nonmalignant phenotype. Binding amounts correlated with CD44 and RHAMM displayed but binding heterogeneity appeared to arise from a differential ability of HA receptor-positive subpopulations to interact with F-HA. HAhigh subpopulations exhibited significantly higher local invasion and lung micrometastases but, unexpectedly, lower proliferation than either unsorted parental cells or the HA−/low subpopulation. Querying F-HA binding to aggressive tumor cells reveals a previously undetected form of heterogeneity that predicts invasive/metastatic behavior and that may aid both early identification of cancer patients susceptible to metastasis, and detection/therapy of invasive BCa subpopulations.
Breast tumors display substantial heterogeneity driven by genetic and epigenetic mechanisms (1–3). These processes select and support tumor cell subpopulations with distinct phenotypes in proliferation, metastatic/invasive proclivity, and treatment susceptibility that contribute to clinical outcomes. Currently, there is a paucity of biomarkers to identify these subpopulations (3–12). Although detection of genetic heterogeneity may itself be a breast cancer (BCa) prognostic marker (3, 13–15), the phenotypes manifested from this diversity are context-dependent. Therefore, phenotypic markers provide additional powerful tools for biological information required to design diagnostics and therapeutics. Glycomic approaches have enormous potential for revealing tumor cell phenotypic heterogeneity because glycans are themselves highly heterogeneous and their complexity reflects the nutritional, microenvironmental, and genetic dynamics of the tumors (16–18).
We used hyaluronan (HA) as a model carbohydrate ligand for probing heterogeneity in glycosaminoglycan–BCa cell receptor interactions. We reasoned this approach would reveal previously undetected cellular and functional heterogeneity linked to malignant progression because the diversity of cell glycosylation patterns, which can occur as covalent and noncovalent modifications of proteins and lipids as well as different sizes of such polysaccharides as HA, is unrivaled (16, 17, 19). In particular, tumor and wound microenvironments contain different sizes of HA polymers that bind differentially to cell receptors to activate signaling pathways regulating cell migration, invasion, survival, and proliferation (19–22).
More than other related glycosaminoglycans, HA accumulation within BCa tumor cells and peritumor stroma is a predictor of poor outcome (23) and of the conversion of the preinvasive form of BCa, ductal carcinoma in situ, to an early invasive form of BCa (24). HA is a nonantigenic and large, relatively simple, unbranched polymer, but the manner in which it is metabolized is highly complex (19, 25). There are literally thousands of different HA sizes in remodeling microenvironments, including tumors. HA polymers bind to cells via at least six known receptors (16, 19, 20, 26–32). Two of these, cluster designation 44 (CD44) and receptor for HA-mediated motility/HA-mediated motility receptor (RHAMM/HMMR), form multivalent complexes with different ranges of HA sizes (19, 29, 33), and both receptors are implicated in BCa progression (19–21, 23, 29, 30, 33–36). Elevated CD44 expression in the peritumor stroma is associated with increased relapse (37), and in primary BCa cell subsets may contribute to tumor initiation and progression (38–40). Elevated RHAMM expression in BCa tumor subsets is a prognostic indicator of poor outcome and increased metastasis (22, 33, 41). RHAMM polymorphisms may also be a factor in BCa susceptibility (42, 43).
We postulated that multivalent interactions resulting from mixture of a polydisperse population of fluorescent HA (F-HA) sizes, typical of those found in remodeling microenvironments of wounds and tumors (19, 20, 29), with cellular HA receptors would uncover a heterogeneous binding pattern useful for sorting tumor cells into distinct subsets. We interrogated the binding of F-HA to BCa lines of different molecular subtypes, and related binding/uptake patterns to CD44 and RHAMM display, and to tumor cell growth, invasion, and metastasis.
Results
CD44 and RHAMM Coexpression Is Highest in Aggressive Triple-Negative BCa.
Primary breast tumors and most BCa lines group into subtypes, with different hormone receptor levels and human epidermal growth factor receptor-2 (HER2) expression (44–47). Triple-negative BCa is estrogen receptor-negative (ER−), progesterone receptor-negative (PR−) and HER2−, and is currently one of the most difficult BCa to treat because it is commonly chemoresistant and exhibits early metastases with consequent poor 5-y survival rates (48–52). Few of these BCa subtypes have been characterized for CD44 or RHAMM expression (31, 53, 54), and none to our knowledge have been examined for HA binding. Because HA receptor display is a prerequisite for HA binding, we used mRNA microarray repositories in Oncomine (http://oncomine.com) to assess expression of known HA receptors (19) in primary BCa. Coexpression of CD44 and RHAMM mRNA occurred in all subtypes but was highest in triple-negative tumors (SI Results and Tables S1 and S2). In general agreement with Oncomine databanks, CD44 and RHAMM proteins were displayed in human BCa lines of different molecular subtypes (Table S3).
HA Binding Is Heterogeneous.
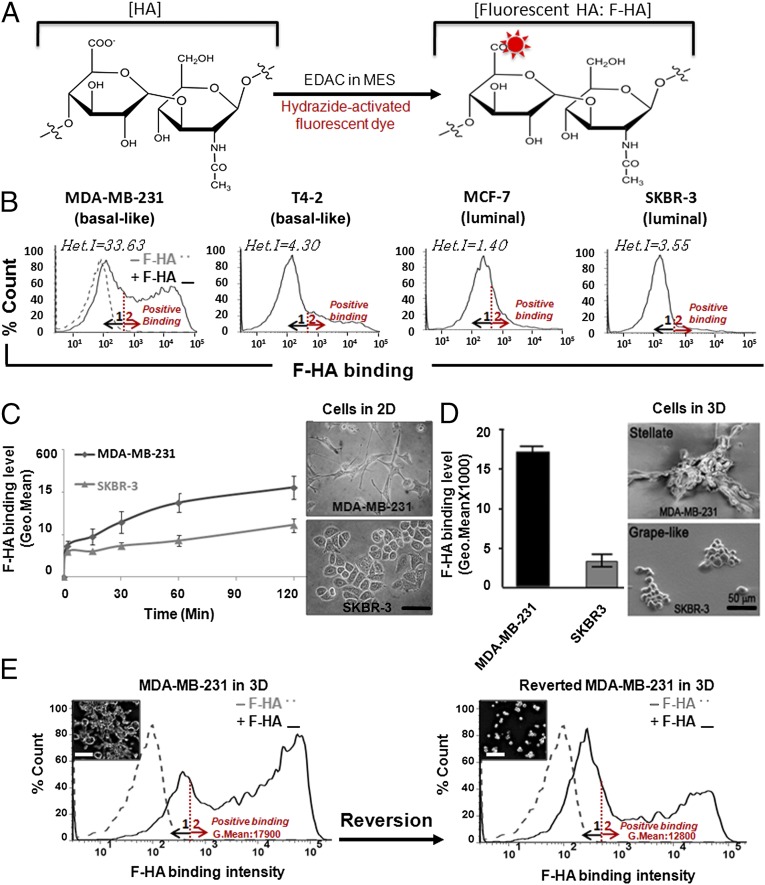
To determine whether HA binding revealed BCa cellular heterogeneity related to HA receptor display, we synthesized a F-HA probe containing a polydisperse mixture of polymer sizes typical of tumor microenvironments (Fig. 1A), and quantified F-HA binding using flow cytometry (Fig. 1B and Table 1). Each cell line bound F-HA, but the total amount that bound varied (as calculated by fluorescence geometric mean value of positive binding of peak 2) and correlated with the levels of CD44 and RHAMM protein display. Thus, MDA-MB-231 and T4-2 cells, which express high levels of CD44 and RHAMM, showed higher F-HA binding than MCF-7 or SKBR-3 cells, which display lower levels of CD44 and RHAMM (Table 1).
Fig. 1.
F-HA binding to BCa cell lines is heterogeneous and linked to malignant phenotype. (A) Schematic for fluorescent labeling of HA with hydrazide functionalized A647 or TR dyes. (B) HA-binding profiles measured by FACS after addition of A647-HA (+F-HA) to cells for 45 min at 4 °C (n = 3–5). Positive binding was determined by FACS geometric mean values of populations within peak 2, and the gray dashed line represents background fluorescent level of intact cells before addition of F-HA (−F-HA), which was similar for all cell types. All F-HA–binding profiles span from 0 to 105 fluorescent signals. The graphs indicate heterogeneity indices of the profiles as measured by Het.I = median × (AUC2/AUC1). (C and D) Comparison of MDA-MB-231 (basal-like) and SKBR-3 (luminal) HA-binding levels from 2 to 120 min after addition of F-HA to cells grown in 2D and 3D laminin-rich gels, respectively. n ≥ 3. (Scale bar: 50 µm.) Error bars represent SD and normalized SD, respectively. (Upper and Lower Left in C and D) Optical and scanning electron micrographs of MDA-MB-231 and SKBR-3 cells in 2D and 3D culture before F-HA binding. F-HA–binding profiles and morphology of MDA-MB-231 cells before (E, Left) and after reversion (E, Right) (57) to a more nonmalignant phenotype. n = 3. (Scale bar: 80 µm.)
Table 1.
Comparison of HA binding and HA receptor display by human BCa cell lines
| Basal-like | Luminal | |||
| MDA-MB-231 | T4-2 | MCF-7 | SKBR-3 | |
| HA-binding level | HA4+ | HA2+ | HA+ | HA+ |
| CD44 display | CD443+ | CD442+ | CD44low/+ | CD44− |
| RHAMM display | RHAMM+ | RHAMM+ | RHAMM2+ | RHAMM2+ |
Scores are determined by FACS geometric mean and median values of cell-surface–bound F-HA or CD44 and RHAMM displays. Degree of positivity is measured by comparing the geometric mean of sample data with that of control (intact cells before addition of F-HA or isotype-matched nonimmune IgG treated) sample data.
Unexpectedly, F-HA binding was heterogeneous in all cell lines (+F-HA in Fig. 1B). Thus, profiles followed a non-Gaussian pattern composed of either two major peaks (e.g., MDA-MB-231) or one peak and a positively skewed tail (e.g., SKBR-3). A Shapiro–Wilk normality test confirmed that F-HA–binding profiles significantly deviated from normality as measured by the value W (55) (MDA-MB-231: 0.8528; T4-2: 0.7245; MCF-7: 0.6505; and SKBR-3: 0.6265; P < 0.001). However, this measure did not allow us to compare the degree of heterogeneity among the F-HA–binding profiles. Therefore, we developed a heterogeneity index (Het.I), which measures the median of the total F-HA binding and is weighted by the differential binding:
where AUC2/AUC1 is a ratio of areas under the curves of positive binding (AUC2 in Fig. 1B; area 2 shown by the red arrow) and the first binding peak (AUC1 in Fig. 1B; area 1 shown by the black arrow). Het.I was highest for MDA-MB-231 (33.6) followed by T4, SKBR-3, and MCF-7 (4.3, 3.5, and 1.4, respectively). The rank orders for both binding heterogeneity and binding levels were independent of the type of fluorescent label used and exposure time to F-HA from 2 to 120 min (Fig. 1C).
To determine if differences in F-HA binding persisted under physiologically relevant conditions, we grew cells in 3D laminin-rich gels, which mimic aspects of tumor microenvironments (56). The rank order of F-HA–binding levels did not change (e.g., MDA-MB-231 vs. SKBR-3 in Fig. 1D), although the overall level of binding and Het.I increased due to amplification of peak 2 in 3D culture (compare MDA-MB-231 in Fig. 1 B and E). Thus, these properties were stable regardless of culture conditions (2D vs. 3D).
The most malignant cell line, the triple-negative MDA-MB-231 (48, 49), exhibited the highest F-HA binding and Het.I. To determine whether these two parameters were functionally linked to malignancy, we took advantage of our ability to revert MDA-MB-231 cells to a more nonmalignant phenotype by ablating β1 integrin and PI3K signaling in 3D culture (57). The F-HA–binding profile of reverted MDA-MB-231 cells remained heterogeneous; however, both the degree of binding heterogeneity (indicated by Het.I) and total F-HA binding were reduced by 4.8- and 1.4-fold, respectively (Fig. 1E). These results indicate that HA-binding level and binding heterogeneity are linked to a malignant phenotype.
HA-Binding Heterogeneity Arises from Distinct Abilities of HA Receptor-Positive Subpopulations to Interact with HA.
To examine whether differences in tumor cell HA receptor display accounted for quantitative differences in F-HA–binding heterogeneity (Fig. 1B), we measured cell-surface CD44 and RHAMM before F-HA exposure using flow cytometry (Fig. S1). Flow cytometry profiles lacked the second peak and positively skewed tails observed for F-HA binding, showing that neither CD44 nor RHAMM display mirrored F-HA–binding heterogeneity. Therefore, the highly heterogeneous flow cytometry profiles of F-HA binding did not appear to arise from a similarly heterogeneous display of HA receptors by tumor cell subpopulations.
Previous studies have shown that HA receptors generally do not bind constitutively to HA (58, 59). We therefore postulated that HA-binding heterogeneity arose from differences in receptor activation or binding affinity, which would be detected as a differential binding of the CD44+ and/or RHAMM+ subpopulations to F-HA. We developed a sensitive multiplexed detection assay to simultaneously monitor CD44 and RHAMM display as well as F-HA binding to live BCa cells (Fig. 2). We used MDA-MB-231 cells because these coexpress CD44 and RHAMM, exhibit the highest F-HA binding and heterogeneity, and are widely used as a malignant triple-negative BCa model (48–52).
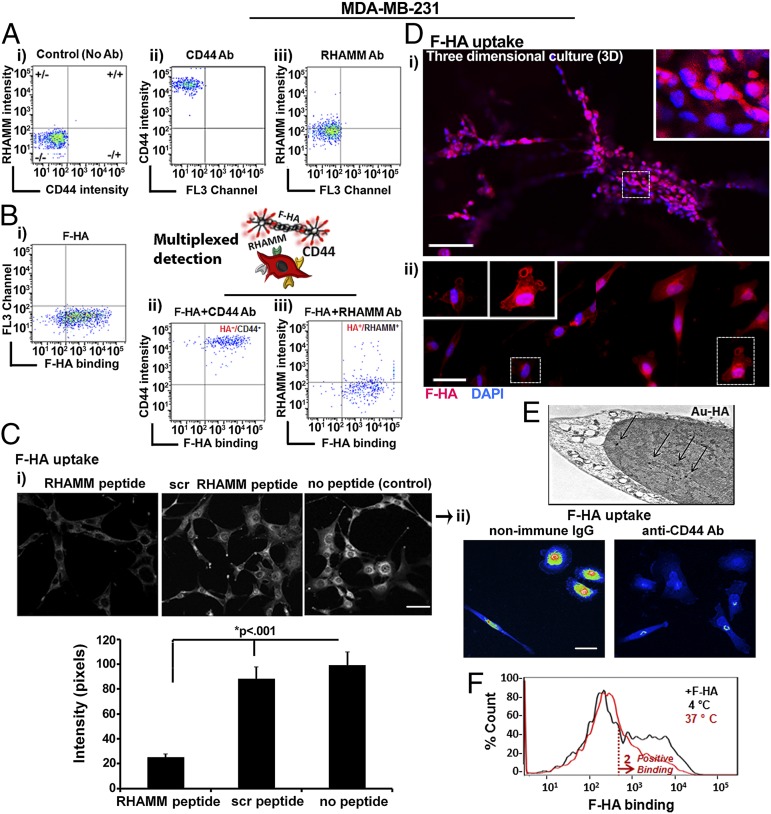
Fig. 2.
HA-binding heterogeneity arises from distinct abilities of HA receptor-positive subpopulations to interact with HA. The fluorescent characteristics of cells after incubation with CD44 and RHAMM antibodies (Ab) in the presence or absence of F-HA are shown as dot plots; (A, i) Background fluorescent level of control cells without Ab. (A, ii) CD44 Ab. (A, iii) RHAMM Ab. (B) Demonstration of F-HA binding to live cells coexpressing CD44 and RHAMM. (B, i) F-HA–binding pattern. (B, ii and iii) Multiplexed detection of F-HA interaction with RHAMM and CD44. n = 3. (C, i) Fluorescence images of MDA-MB-231 cells cultured in 2D and then exposed to F-HA in the absence of a RHAMM mimetic peptide, scrambled peptide, or no peptide (Upper), and quantification of F-HA signal in the nucleus (Lower). n = 12–63. (Scale bars: 150 µm in C, i, Upper and 50 µm in C, ii.) Error bars represent SE. (C, ii) Heat maps of F-HA uptake by MDA-MB-231 cells in the presence of antibodies to isotype matched nonimmune IgG or to CD44. (D) Fluorescent images of F-HA uptake in 3D (i), 2D (ii), and subcellular localization (Insets). n = 5. (Scale bars: 100 µm in D, i and 50 µm in D, ii.) Images in D, ii were taken from different regions of the same uptake assay and merged side by side. (E) TEM analysis of accumulated Au-HA nanoparticles (shown by arrows) in the nucleus. (F) FACS analysis of F-HA binding at 4 °C vs. 37 °C. n = 3.
As expected, controls (untreated or incubated with nonimmune IgG) exhibited background staining (e.g., grouping into CD44−/RHAMM− quadrants) (Fig. 2A, i) whereas incubation with CD44 and RHAMM antibodies resulted in staining above these control levels (Fig. 2 A, ii and iii) consistent with previous studies (31, 54). All cells displayed high CD44 levels (Fig. 2A, ii) whereas the majority of the cells exhibited low but detectable RHAMM display (Fig. 2A, iii). F-HA binding was heterogeneous and spanned from −/− to −/+ quadrants as expected from Fig. 1 (Fig. 2B, i). Multiplexed analyses showed that some CD44+ cells did not bind to F-HA (+/− quadrant in Fig. 2B, ii). However, all F-HA+ cells displayed CD44 (HA+/CD44+ in Fig. 2B, ii) and all RHAMM+ tumor cells bound F-HA (HA+/RHAMM+ in Fig. 2B, iii). Consistent with these multiplexed assays, a RHAMM mimetic peptide significantly reduced F-HA probe uptake (Fig. 2C, i), and antibodies to CD44 almost completely blocked binding and uptake of the F-HA probe into MDA-MB-231 cells (Fig. 2C, ii). These results confirmed the importance of CD44 and RHAMM in F-HA binding to MDA-MB-231 tumor cells and suggested that differential binding of F-HA to RHAMM and CD44 might generate some of the observed probe binding heterogeneity.
We next analyzed the internalization and subcellular trafficking of the F-HA probe to determine whether these might also play a role in binding heterogeneity. Fluorescence microscopy confirmed F-HA uptake was heterogeneous in 3D and in 2D cultures (Fig. 2D); F-HA accumulated at the cell membrane, particularly in cell processes resembling invadopodia (Fig. 2D, ii), and was internalized into multiple subcellular compartments including the cytoplasm and nucleus (Fig. 2C, i; no peptide control and 2D). Transmission electron microscopy (TEM) analysis confirmed the presence of Au-HA nanoparticles in the nucleus (Fig. 2E). Simultaneous exposure of cells to F-HA and the RHAMM mimetic peptide (29), which blocks HA–RHAMM interactions, prevented nuclear accumulation of F-HA and reduced its cytoplasmic and perinuclear accumulation (Fig. 2C, i). Although CD44 is the acknowledged endocytic receptor that internalizes HA in most cell types (60), these results suggested that RHAMM had a function in moving internalized F-HA into the nucleus. To validate F-HA internalization by flow cytometry, we compared F-HA binding at 4 °C vs. 37 °C. Binding heterogeneity was reduced at 4 °C (Fig. 2F) confirming F-HA binding/uptake is temperature-dependent, consistent with a role for receptor internalization and trafficking in this phenotype.
To determine whether dynamic changes in tumor subpopulations that affect their ability to bind F-HA also contribute to heterogeneity, we assessed whether F-HA–binding properties are stable or dynamic within the two subpopulations. Subpopulations that bound no or low amounts of F-HA (HA−/low in Fig. S2A, i) were therefore isolated and then reexposed to F-HA after growth in 2D culture for 7 d, which is a time when the F-HA probe had been depleted (Fig. S2A, ii). Readdition of F-HA to these cells (HA−/low+F-HA in Fig. S2B) resulted in HA−/low cells retaining the first binding peak but demonstrated a failure of these cells to generate the second peak to the extent of the parent cell population, as quantified by their nearly twofold reduced Het.I. These results suggest that the HA−/low bind F-HA stochastically, but the subpopulation of parental tumor cells that binds high levels of F-HA appears to be stable. These results also suggest that F-HA–binding heterogeneity is not due to dynamic changes per se in parental tumor cell subpopulations.
Collectively, these findings suggest that (i) use of F-HA uncovers a stable form of BCa heterogeneity not previously detected with anti-CD44 or -RHAMM antibodies, and (ii) F-HA binding likely results from a combinatorial model of stochastic and stable binding as well as differential receptor display/binding affinity and trafficking to generate heterogeneity.
Sorting of BCa Cells Based on HA Binding Reveals Subpopulations with Distinct Phenotypes.
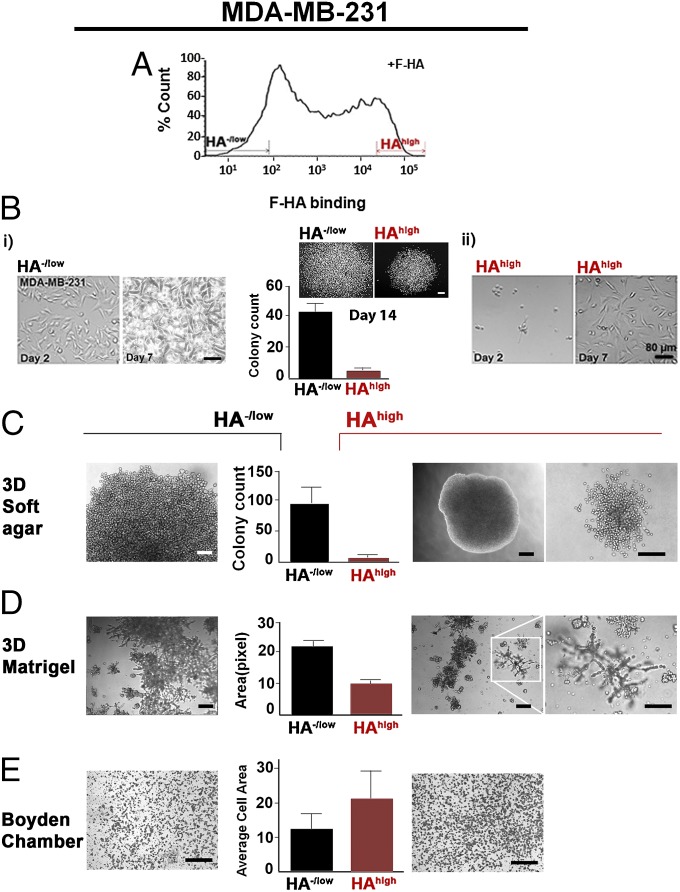
To gain insight into the functional significance of HA binding/uptake heterogeneity, we isolated HA−/low and HAhigh subpopulations using FACS (Becton Dickinson) (Fig. 3A). Sorted cells were expanded in 2D then examined for their proliferative and invasive potential. Despite the disappearance of F-HA by 7 d in tumor cells (Fig. S2A, ii), the HA−/low and HAhigh subpopulations exhibited distinct and stable phenotypes (Fig. 3 B–E). HA−/low cells uniformly adhered to the substratum and proliferated rapidly (Fig. 3B, i, Left) reaching confluence by day 7 (Fig. 3B, i, Right). HAhigh subpopulations attached more slowly (Fig. 3B, ii, Left) and failed to reach confluence by day 7 (Fig. 3B, ii, Right). In 2D colony-forming assays, HA−/low cells achieved a higher number and a 23% larger size of colonies than HAhigh cells (Fig. 3B, Center). Both subpopulations proliferated in 3D soft agar and Matrigel (BD Biosciences), but HA−/low cells formed larger colonies than HAhigh cells (Fig. 3 C and D). The morphology of HAhigh colonies in the 3D cultures suggested that they might be more invasive than HA−/low colonies (Fig. 3 C and D). Matrigel invasion assays confirmed the above (Fig. 3E).
Fig. 3.
Sorting of BCa cells based on HA binding reveals subpopulations with distinct phenotypes. (A) Identification and isolation of HA−/low and HAhigh subpopulations based on differential binding of F-HA to MDA-MB-231 cells. n ≥ 20. (B) Markers indicate 8% selection. Two-dimensional morphology of (i) HA−/low and (ii) HAhigh subpopulations grown from the same number of cells on glass culture chambers from 2 to 7 d after sorting. n = 3–5. (Scale bars: 80 µm). (B, Center) Growth status of the two subpopulations measured by clonogenic abilities up to 14 d. P < 0.001; n = 5. (Scale bar: 200 µm.) Error bars represent SD. (C) Colony-forming ability of (Left) HA−/low and (Right) HAhigh on 3D soft agar by 2.5 mo after cell seeding. (C, Center) Quantification of colonies derived from HA−/low and HAhigh subpopulations. P < 0.0001; n = 5. (Scale bars: 80 µm.) Error bars represent SD. (D) Morphology of (Left) HA−/low and (Right) HAhigh subpopulation on 3D laminin-rich gels. The white inset is an example of invaded structures through Matrigel and (Right) increased magnification of the structures. n = 3. (Scale bars: 300 µm.) (D, Center) Quantification of the occupied surface area by colonies derived from HA−/low and HAhigh subpopulations. P < 0.06; n = 3. Error bars represent SD. (E, Center) Quantification of HA−/low and HAhigh subpopulation invasion through Matrigel-coated Boyden chambers as the average area occupied by (Left) HA−/low and (Right) HAhigh cells that were passed through chambers. P < 0.0005; n = 3. (Scale bar: 300 µm.) Error bars represent SD.
Real-Time Analysis of HA Uptake and Extravasation in Chick Chorioallantoic Membranes.
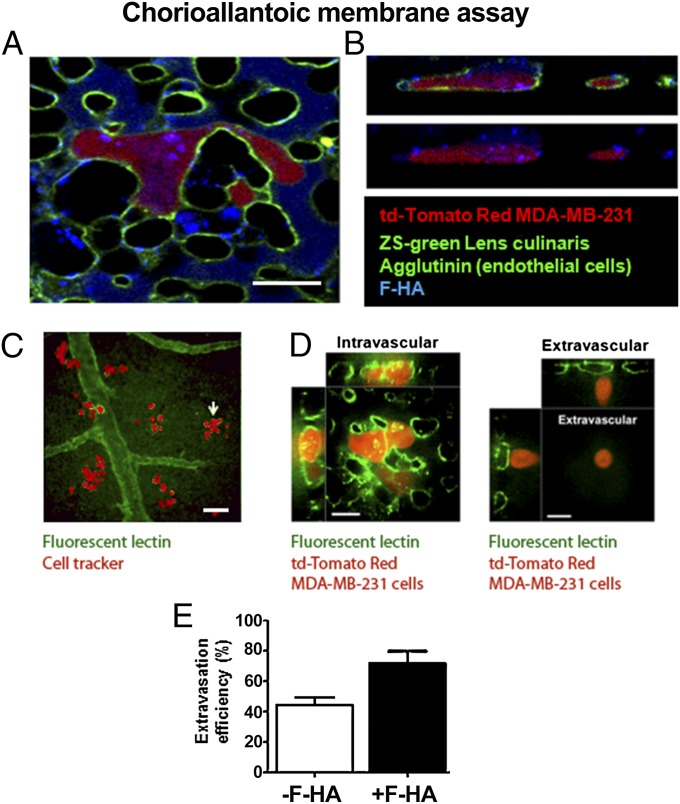
To determine if F-HA binding/uptake occurs in an in vivo-like microenvironment and is associated with invasion, we used the chick chorioallantoic membrane (CAM) model, which allows for real-time monitoring of early uptake events and tumor cell interactions within microvessels (61–64). The endothelial lumen was marked (green) before injection of fluorescent MDA-MB-231 cells (red) mixed with F-HA (blue) into a CAM vein (Fig. 4 A–E). Ten to 15 min after injection, F-HA was present within the blood vessels but had not yet been taken up into the interstitial spaces (Fig. 4A). Two hours after injection, F-HA binding/uptake occurred in the CAM vessels and F-HA vesicles were present inside tumor cells that had flattened onto the vessel endothelium, but had not yet extravasated into interstitial spaces (Fig. 4 A and B). All adherent tumor cells had taken up F-HA. To assess whether increased HA uptake identified invasive cells, the interaction of MDA-MB-231 cells with CAM microvessels was monitored before and after exposure to F-HA (Fig. 4 C–E). A subset of parental untreated MDA-MB-231 cells labeled with cell tracker (pseudocolored red) extravasated and formed viable interstitial tumor colonies surrounding and at a distance from vessels (pseudocolored green) by 72 h (Fig. 4C). MDA-MB-231 cells injected together with F-HA showed more efficient extravasation than untreated tumor cells by 24 h (Fig. 4E). Thus, tumor cells bind to and take up HA in this model, and are more efficient at attaching to and extravasating through endothelium than untreated cells.
Fig. 4.
MDA-MB-231 cells take up F-HA in an in vivo-like microenvironment, flatten onto CAM endothelia, and extravasate. The luminal surface of microvessel endothelia was marked with fluorescein-labeled Lens culinaris agglutinin before the i.v. injection of tdTomato Red MDA-MB-231 tumor cells mixed with F-HA. (A) MDA-MB-231 BCa cells took up F-HA ex ovo and rapidly flattened along the endothelium of the CAM (2 h after injection). (Scale bar: 50 µm.) (B) Confocal Z-stack images taken 2 h after injection confirmed the presence of F-HA (vessel lumen = blue) vesicles inside tumor cells. (Same magnification as A.) (C) Cell tracker-tagged MDA-MB-231 cells (pseudocolored red) that have extravasated and formed colonies 72 h after their injection into CAM vessels (pseudocolored green). (Scale bar: 100 µm.) (D) Intravital imaging of (Left) representative intravascular td-Tomato Red MDA-MB-231 cells and (Right) extravasated td-Tomato Red MDA-MB-231 24 h after injection of tumor cells (blood vessels marked by green fluorescent lectin). (Scale bars: 100 µm.) (E) Quantification of extravasation efficiency following injection of the same number of cells into CAM veins and exposure to F-HA or control buffer after 24 h. P = 0.0178; n = 3. Error bars represent SD.
Xenografts of the HAhigh Subpopulation Form Slow-Growing but Invasive Tumors.
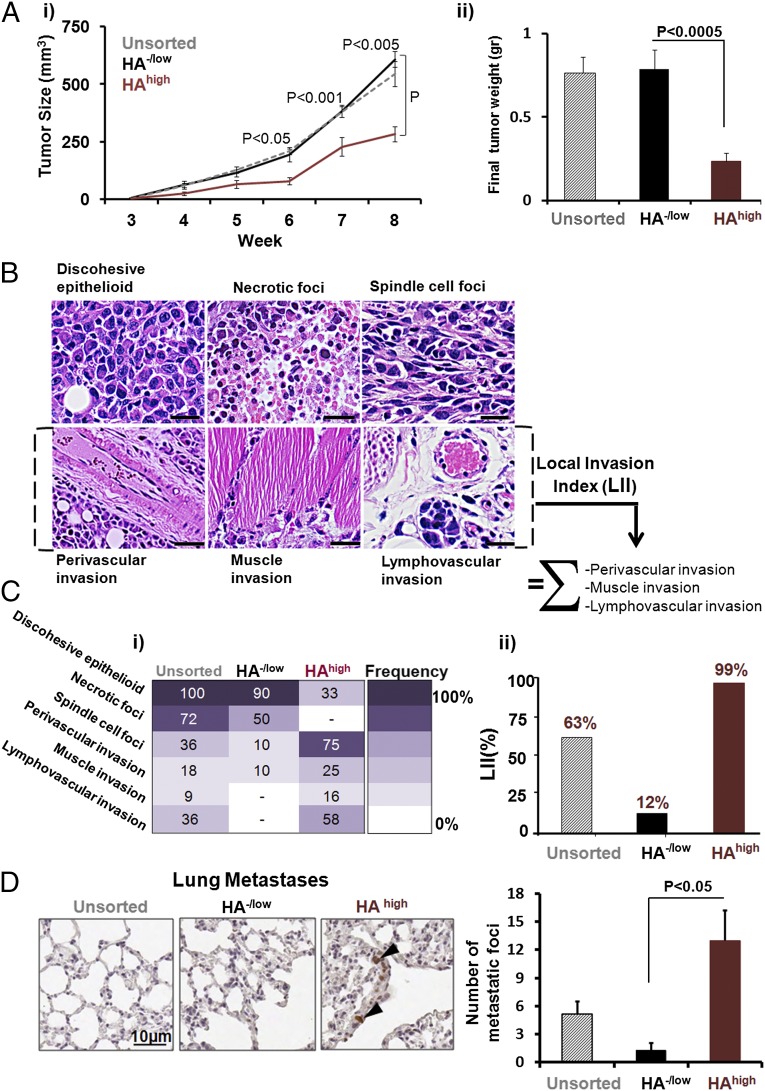
To determine whether these phenotypes resulted in detectable differences in tumor growth and invasion/metastasis, we compared the tumorigenic characteristics of parental unsorted MDA-MB-231 (with or without F-HA) with sorted HA−/low and HAhigh MDA-MB-231 tumor cells grown as xenografts in severely immunocompromised mice. Thus, all tumor cells were first embedded in Matrigel then injected into the fourth mammary fat pads of female nonobese diabetic severe combined immune-deficient (NOD-SCID) IL2 receptor (IL2R)−/− mice. Because gross tumor and histological patterns of unsorted parental cells +F-HA were similar to those of unsorted parental cells −F-HA, we restricted further analyses to unsorted parental cells (−F-HA). Parental tumors (unsorted, gray-dashed in Fig. 5A, i) grew at a rate comparable to the HA−/low tumor subpopulation (black in Fig. 5A, i), whereas the HAhigh subpopulation (red in Fig. 5A, i) grew more slowly. Wet weight measurements at week 8 mirrored this difference in tumor growth (Fig. 5A, ii).
Fig. 5.
HA−/low and HAhigh subpopulations both form tumors in immunocompromised mice, but the HAhigh subpopulation results in slow-growing and more invasive/metastatic tumors. Xenografts were formed in intact fat pads of the fourth mammary gland after injection of cells embedded in Matrigel. (A, i) Growth curve of xenograft tumors derived from unsorted parental tumor cells (n = 11, gray dashed line), HA−/low (n = 10, black line), and HAhigh subpopulations (n = 12, red line). Error bars represent SEM. (A, ii) Wet weight measurements of the tumors derived from untreated parental tumor cells (cross-hatched bar), HA−/low (black bar), and HAhigh subpopulations (red bar). (B) Identification of six distinct pathological phenotypes by H&E staining of tumors. (Scale bars: 100 µm in all panels except for perivascular invasion, which is 200 µm). (C, i) Quantification of histological signatures. The intensity bar indicates frequency of phenotype occurrence in the mice cohort. (C, ii) Definition of LII and comparison of LII among unsorted parental tumor cells (cross-hatched bar), HA−/low (black bar), and HAhigh subpopulations (red bar). (D) Microscopic evidence (Left) and quantification (Right) of distant metastases to lung tissues. n = 4. Error bars represent SEM.
H&E-stained tissue sections showed that all tumor xenografts formed high-grade adenocarcinomas. We observed six histological phenotypes of parental xenografts (Fig. 5B), and analyzed the distribution of histological “signatures” on a per tumor per mouse basis (Fig. 5C, i). Both HA−/low and HAhigh tumors exhibited part of the parental histology signatures and these occurred at different frequencies. Spindle foci, perivascular invasion, muscle invasion, and lymphovascular invasion occurred at a higher frequency in HAhigh than either HA−/low or parental tumors (Fig. 5C, i), likely because sorting based on F-HA probe binding allowed enrichment of a highly invasive subpopulation present in small amounts in the parental line. HA−/low tumors exhibited fewer spindle cell foci and less perivascular invasion than parental or HAhigh tumors, and showed no evidence of lymphatic or muscle invasion. To quantify these differences by a single value, we defined a local invasion index (LII) comprising the sum of perivascular, muscle, and lymphovascular invasion frequencies. The LII was 63% for parental tumors, 12% for HA−/low, and 99% for HAhigh tumors (Fig. 5C, ii), revealing an overall 8.2-fold higher invasive capacity in HAhigh compared with HA−/low subpopulations. The LII for unsorted parental tumors was not changed by their exposure to F-HA before injection into mice (66% vs. 63%, respectively). Lung and liver tissues were examined for evidence of metastases. Proliferating human tumor cells were identified by human-specific anti-Ki67 antibodies. No metastases were detected in liver, but micrometastases were observed in lung (Fig. 5D). Quantification of these showed a significantly greater number of micrometastases in lungs of animals injected with HAhigh vs. HA−/low subpopulations. These results show that absent or low HA binding is associated with enhanced primary tumor proliferation but low invasion and lung metastasis, whereas high HA binding is linked to enhanced invasion and metastasis but slow proliferation of primary tumor cells.
Discussion
It has long been recognized that tumors are heterogeneous and comprise multiple subpopulations that likely provide a survival and growth advantage for primary tumors and their metastases (65–68). Tumor cell heterogeneity confounds our understanding of tumor progression, relapse, and therapy (3, 7, 69). Despite advances in genotypic and phenotypic heterogeneity assessment strategies, biomarkers, in particular those identifying the presence of aggressive and/or resistant subpopulations, are urgently needed for the design of effective BCa diagnostics and therapies.
Because some phenotypic diversity can be simulated in many BCa cell lines (7, 70), we tested the possibility that patterns of exogenous ligand binding to receptors in aggressive cancer cells might offer a new method for profiling cellular and functional heterogeneity. We chose HA because the production and metabolism of this glycosaminoglycan is linked to cancer progression and poor clinical outcome (20, 21, 71, 72). We profiled multivalent interactions of F-HA and HA receptors in live cells. We discovered that subtype-specific quantitative differences among binding levels of F-HA to BCa cancer cell lines correlated with HA receptor display, but all F-HA–binding profiles were heterogeneous. To obtain a quantitative measure of binding heterogeneity, we developed Het.I. The values measured by Het.I suggested that HA binding follows a combinatorial model by which some cells bind HA stochastically, whereas other subpopulations exhibit a stable ability to bind HA (Fig. S2).
Among the different cell lines we used, the most metastatic and triple-negative basal-like MDA-MB-231 exhibited both the highest HA-binding level and HA-binding heterogeneity, suggesting that these properties correlate with the malignant state. To directly confirm this correlation without altering the oncogenic mutations of this cell line, we reverted the malignant phenotype to a more nontumorigenic and less malignant phenotype in 3D culture (57). The less malignant phenotype corresponded with a lower HA-binding level and reduced binding heterogeneity. These results suggest that F-HA probe binding to cells provides a measure for determining the degree of malignant behavior and underscores the importance of architecture and organization as crucial parameters in malignancy (73).
Because CD44/RHAMM displays did not mirror HA-binding heterogeneity (Fig. S1 and Fig. 2), we monitored multiplexed interactions of the F-HA probe with CD44 and RHAMM, and showed that CD44 and RHAMM interactions with F-HA occurred heterogeneously. Not all CD44 receptors that were present on the cells interacted with F-HA. In addition, surface-bound F-HA changed in a temperature-dependent manner and F-HA was observed in the cytoplasm and nucleus of cells at 37 °C, indicating internalization of HA as a contributing factor to uptake heterogeneity. Because the F-HA conjugate is a mixture of different sizes of HA molecules (molecular weight range: 10–480 kDa; molecular weight average: 240 kDa), it is likely that both receptor trafficking and size-related differential binding contribute to the observed heterogeneity.
Using xenografts and 3D cultures, we found a surprising link between binding heterogeneity and stable differences in the phenotypes and tumorigenicity of these subpopulations. These were either poorly invasive/metastatic and fast-growing (HA−/low) or highly invasive/metastatic but slow-growing (HAhigh). This finding is at odds with the conventional paradigm that rapidly proliferating tumors are also invasive and likely to metastasize. Our screening method has thus uncovered a type of tumor heterogeneity that may be associated with epigenetic mechanisms used by breast tumor cells for acquiring aggressive behavior and may be useful for identifying new therapeutic targets. Our results also suggest that the presence of HAhigh subpopulations in primary tumors may indicate a susceptibility for metastasis and predict tumor recurrence and therapy resistance. The probe may have revealed previously existing heterogeneity due to other as yet unknown factors, and/or it may be acting as a functional probe that signals and forces cells into a stable differentiation program to restrict proliferation but increase motility/invasion. HA has the potential to accomplish the latter effect because it is well-documented to activate signaling cascades that control such cellular functions (19–21, 71, 72); however, some of these effects have been reported as transient. Because the F-HA signal disappeared by 7 d and the LII for unsorted parental tumors was not changed by their exposure to F-HA before injection into mice, we currently favor the former possibility.
In summary, use of F-HA uncovers a stable form of BCa heterogeneity that has not been detected previously with anti-CD44 or -RHAMM antibodies. The demonstration that BCa lines, which are generally considered homogeneous phenotypically, are in fact quite heterogeneous with respect to HA binding, raises the possibility that other clinically relevant traits may stably segregate specific subpopulations in tumor cell lines, which could ultimately be applied to probing primary tumor heterogeneity. Because HA is a biocompatible natural polymer and is well-tolerated in vivo (20, 21, 74, 75), HA-based probes can be readily used as part of clinical heterogeneity profiling. The quantitative approaches and the Het.I formula for measuring HA-binding differences may facilitate incorporation of heterogeneity as a determining factor in diagnosis and therapy of metastatic cancers, and could be used to detect heterogeneity of other measurable phenotypic traits or those introduced by biophysical cues (74, 75) in 3D micro-/nanoenvironments. This type of subpopulation analysis points to the importance of taking tumor cell heterogeneity, even within tumor cell lines, into consideration, and may also provide more clinically relevant models for assessing treatment efficacy.
Materials and Methods
Two-Dimensional Cell Culture.
HMT-3522 S1, HMT-3522 T4-2 mammary epithelial cells (76–78), MCF-10A, Hs578T, MCF-7, MDA-MB-231, and SKBR-3 (American Type Culture Collection) were cultured (79) (SI Materials and Methods).
Three-Dimensional Cell Culture.
Cells were prepared for culturing on top of Matrigel (BD Biosciences), a laminin-rich gel (lrECM) (56) (SI Materials and Methods). Reversion of 3D MDA-MB-231 cells was performed using AIIB2 and LY294002 reagents (57) and cells were extracted from 3D lrECM (56) (SI Materials and Methods).
F-HA Probe Synthesis.
Medical-grade HA (Hyal Pharmaceutical), which contained a mixture of HA polymer sizes with a range from 10 kDa up to 480 kDa (molecular weight average: 240 kDa), was conjugated to Alexa Fluor or Texas Red (TR) (Invitrogen) through hydrazide coupling (SI Materials and Methods). The conjugation procedure did not detectably affect the HA polymer size range.
F-HA Probe Binding/Uptake.
Subconfluent cells were grown to maximize RHAMM surface display (31, 54), and were exposed to either Alexa Fluor 647 (A647)-HA (1 × 106 cells per mL PBS with 1 mL of A647-HA at 433 µg HA/mL) in suspension at 4 °C in the dark or to A647-HA and TR-HA at 37 °C in the dark in 2D and 3D cultures (SI Materials and Methods).
Flow Cytometry and FACS.
CD44 and RHAMM antibodies that did not block F-HA binding were used for flow cytometry. Samples were prepared for analysis (Becton Dickinson FACSCalibur and FACSVantage SE) and sorting (Becton Dickinson FACSVantage SE) into HA−/low and HAhigh subsets (SI Materials and Methods).
EM.
TEM and scanning EM (SEM) were performed on Au-HA–treated cells (80) in 2D and untreated 3D cultured cells, respectively (SI Materials and Methods).
In-Culture Functional Assays.
Phenotypes of FACS-sorted HA−/low and HAhigh subpopulations were assessed in 2D and 3D under both anchorage dependent and independent conditions (SI Materials and Methods).
Ex Ovo F-HA Uptake and Extravasation in the CAM Model.
Fertilized chicken eggs were prepared (81). A647-HA and subconfluent MDA-MB-231 cells were injected into CAM veins, and extravasated cells were quantified (82) (SI Materials and Methods).
In Vivo Tumorigenic Assessment.
All mouse experiments were performed according to E. O. Lawrence Berkeley National Laboratories Animal Welfare and Research Committee. All tumor cells were embedded in Matrigel before injection (62.5 × 103 cells) into the fourth mammary fat pads of female NOD-SCID IL2R−/− (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice (SI Materials and Methods).
Immunofluorescence Detection of Human Ki67.
Tissues were deparaffinized in two 5-min cycles of 100% xylene then rehydrated in four 2-min cycles of 100% (vol/vol) EtOH, 95% EtOH, 70% EtOH, and 50% EtOH, followed by a rinse in PBS for 2 min before staining with anti-Ki67 antibody for detection of metastases (SI Materials and Methods).
Statistical Analysis.
Oncomine data banks were investigated as described (SI Materials and Methods and SI Results). Deviation from normal distribution was analyzed by the Shapiro–Wilk normality test (55) using Prism 5.0 software (GraphPad Inc.) after log transformation. A W value of 1 indicated a normal distribution with P < 0.001 considered significant. Experimental groups were compared using Student t tests with P < 0.05 considered significant.
Supplementary Material
Acknowledgments
We thank S. Bahram Bahrami for animal work assistance and helpful discussions; Berbie Chu for animal surgery assistance; Saori Furuta, Aishwarya Jayagopal, Arya Nikjoo, Aman Kaur Mann, and Jenny Ma for laboratory assistance; and the University of Washington NanoTech User Facility, a member of the National Science Foundation National Nanotechnology at the Infrastructure Network for use of SEM. This work was supported by National Cancer Institute (NCI) of the National Institute of Health (NIH) Ruth L. Kirschstein National Research Service Award F32 CA132491 (to M.V.); US Department of Defense Breast Cancer Research Program Innovative Developmental and Exploratory Awards Grant BC044087 (to M.J.B. and E.A.T.) and Grant W81XWH0810736 (to M.J.B.); a Canadian Breast Cancer Foundation Grant (to E.A.T.); NCI of the NIH Grants R37CA064786, U54CA143836, and U01CA143233 (to M.J.B.); US Department of Energy Office of Biological and Environmental Research and Low Dose Radiation Program Contract DE-AC02-05CH1123 (to M.J.B.); in part, a grant from The Breast Cancer Research Foundation (to M.J.B.); and Canadian Cancer Society Research Institute Grant 700537 (to J.D.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402383111/-/DCSupplemental.
References
- 1.Baumgarten SC, Frasor J. Minireview: Inflammation: An instigator of more aggressive estrogen receptor (ER) positive breast cancers. Mol Endocrinol. 2012;26(3):360–371. doi: 10.1210/me.2011-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boudreau A, van’t Veer LJ, Bissell MJ. An “elite hacker”: Breast tumors exploit the normal microenvironment program to instruct their progression and biological diversity. Cell Adhes Migr. 2012;6(3):236–248. doi: 10.4161/cam.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 4.Metzger-Filho O, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30(15):1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Li S, Dunker AK, Uversky VN. Molecular profiling: An essential technology enabling personalized medicine in breast cancer. Curr Drug Targets. 2012;13(4):541–554. doi: 10.2174/138945012799499767. [DOI] [PubMed] [Google Scholar]
- 6.Petersen OW, Gudjonsson T, Villadsen R, Bissell MJ, Rønnov-Jessen L. Epithelial progenitor cell lines as models of normal breast morphogenesis and neoplasia. Cell Prolif. 2003;36(Suppl 1):33–44. doi: 10.1046/j.1365-2184.36.s.1.4.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, et al. Tumor initiating but differentiated luminal-like breast cancer cells are highly invasive in the absence of basal-like activity. Proc Natl Acad Sci USA. 2012;109(16):6124–6129. doi: 10.1073/pnas.1203203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pepin F, et al. Gene expression profiling of microdissected breast cancer microvasculature identifies distinct tumor vascular subtypes. Breast Cancer Res. 2012;14(4):R120. doi: 10.1186/bcr3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabos P, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res Treat. 2012;135(2):415–432. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeschke J, et al. Biomarkers for detection and prognosis of breast cancer identified by a functional hypermethylome screen. Epigenetics. 2012;(7):701–709. doi: 10.4161/epi.20445. [DOI] [PubMed] [Google Scholar]
- 11.Dedeurwaerder S, Fumagalli D, Fuks F. Unravelling the epigenomic dimension of breast cancers. Curr Opin Oncol. 2011;23(6):559–565. doi: 10.1097/CCO.0b013e32834bd481. [DOI] [PubMed] [Google Scholar]
- 12.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol. 2012;9(1):48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irshad S, Ellis P, Tutt A. Molecular heterogeneity of triple-negative breast cancer and its clinical implications. Curr Opin Oncol. 2011;23(6):566–577. doi: 10.1097/CCO.0b013e32834bf8ae. [DOI] [PubMed] [Google Scholar]
- 14.Burrell RA, et al. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J Cell Biochem. 2010;111(4):782–790. doi: 10.1002/jcb.22781. [DOI] [PubMed] [Google Scholar]
- 15.Rakha E, Reis-Filho JS. Basal-like breast carcinoma: From expression profiling to routine practice. Arch Pathol Lab Med. 2009;133(6):860–868. doi: 10.5858/133.6.860. [DOI] [PubMed] [Google Scholar]
- 16.Potapenko IO, et al. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol Oncol. 2010;4(2):98–118. doi: 10.1016/j.molonc.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabinovich GA, van Kooyk Y, Cobb BA. Glycobiology of immune responses. Ann N Y Acad Sci. 2012;1253:1–15. doi: 10.1111/j.1749-6632.2012.06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto S. Clinical applications of glycomic approaches for the detection of cancer and other diseases. Curr Opin Mol Ther. 2006;8(6):507–513. [PubMed] [Google Scholar]
- 19.Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol Rev. 2011;91(1):221–264. doi: 10.1152/physrev.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veiseh M, Turley EA. Hyaluronan metabolism in remodeling extracellular matrix: Probes for imaging and therapy of breast cancer. Integrative Biol. 2011;3(4):304–315. doi: 10.1039/c0ib00096e. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton SR, et al. Pharmacokinetics and pharmacodynamics of hyaluronan infused into healthy human volunteers. Open Drug Metab. 2009;3:43–55. [Google Scholar]
- 22.Maxwell CA, McCarthy J, Turley E. Cell-surface and mitotic-spindle RHAMM: Moonlighting or dual oncogenic functions? J Cell Sci. 2008;121(Pt 7):925–932. doi: 10.1242/jcs.022038. [DOI] [PubMed] [Google Scholar]
- 23.Sironen RK, et al. Hyaluronan in human malignancies. Exp Cell Res. 2011;317(4):383–391. doi: 10.1016/j.yexcr.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Corte MD, et al. Analysis of the expression of hyaluronan in intraductal and invasive carcinomas of the breast. J Cancer Res Clin Oncol. 2010;136(5):745–750. doi: 10.1007/s00432-009-0713-2. [DOI] [PubMed] [Google Scholar]
- 25.Bollyky PL, Bogdani M, Bollyky JB, Hull RL, Wight TN. The role of hyaluronan and the extracellular matrix in islet inflammation and immune regulation. Curr Diab Rep. 2012;12(5):471–480. doi: 10.1007/s11892-012-0297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson M, Stern R. Chain gangs: New aspects of hyaluronan metabolism. Biochem Res Int. 2012;2012:893947. doi: 10.1155/2012/893947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeAngelis PL. Glycosaminoglycan polysaccharide biosynthesis and production: Today and tomorrow. Appl Microbiol Biotechnol. 2012;94(2):295–305. doi: 10.1007/s00253-011-3801-6. [DOI] [PubMed] [Google Scholar]
- 28.Afratis N, et al. Glycosaminoglycans: Key players in cancer cell biology and treatment. FEBS J. 2012;279(7):1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 29.Tolg C, et al. A RHAMM mimetic peptide blocks hyaluronan signaling and reduces inflammation and fibrogenesis in excisional skin wounds. Am J Pathol. 2012;181(4):1250–1270. doi: 10.1016/j.ajpath.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao F, et al. Hyaluronan oligosaccharides promote excisional wound healing through enhanced angiogenesis. Matrix Biol. 2010;29(2):107–116. doi: 10.1016/j.matbio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Veiseh M, et al. Imaging of homeostatic, neoplastic, and injured tissues by HA-based probes. Biomacromolecules. 2012;13(1):12–22. doi: 10.1021/bm201143c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toole BP. Hyaluronan: From extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4(7):528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 33.Turley EA, Naor D. RHAMM and CD44 peptides-analytic tools and potential drugs. Front Biosci. 2012;17:1775–1794. doi: 10.2741/4018. [DOI] [PubMed] [Google Scholar]
- 34.Wang A, de la Motte C, Lauer M, Hascall V. Hyaluronan matrices in pathobiological processes. FEBS J. 2011;278(9):1412–1418. doi: 10.1111/j.1742-4658.2011.08069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neri P, Bahlis NJ. Targeting of adhesion molecules as a therapeutic strategy in multiple myeloma. Curr Cancer Drug Targets. 2012;12(7):776–796. doi: 10.2174/156800912802429337. [DOI] [PubMed] [Google Scholar]
- 36.Orian-Rousseau V. CD44, a therapeutic target for metastasising tumours. Eur J Cancer. 2010;46(7):1271–1277. doi: 10.1016/j.ejca.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 37.Auvinen P, et al. Increased hyaluronan content and stromal cell CD44 associate with HER2 positivity and poor prognosis in human breast cancer. Int J Cancer. 2013;132(3):531–539. doi: 10.1002/ijc.27707. [DOI] [PubMed] [Google Scholar]
- 38.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Idowu MO, et al. CD44(+)/CD24(-/low) cancer stem/progenitor cells are more abundant in triple-negative invasive breast carcinoma phenotype and are associated with poor outcome. Hum Pathol. 2012;43(3):364–373. doi: 10.1016/j.humpath.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Wang C, et al. The overexpression of RHAMM, a hyaluronan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin Cancer Res. 1998;4(3):567–576. [PubMed] [Google Scholar]
- 42.Kalmyrzaev B, Pharoah PD, Easton DF, Ponder BA, Dunning AM. SEARCH Team Hyaluronan-mediated motility receptor gene single nucleotide polymorphisms and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3618–3620. doi: 10.1158/1055-9965.EPI-08-0216. [DOI] [PubMed] [Google Scholar]
- 43.Maxwell CA, et al. HEBON EMBRACE SWE-BRCA BCFR GEMO Study Collaborators kConFab Interplay between BRCA1 and RHAMM regulates epithelial apicobasal polarization and may influence risk of breast cancer. PLoS Biol. 2011;9(11):e1001199. doi: 10.1371/journal.pbio.1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kristensen VN, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci USA. 2012;109(8):2802–2807. doi: 10.1073/pnas.1108781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 47.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang SJ, Saadi W, Lin F, Minh-Canh Nguyen C, Li Jeon N. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res. 2004;300(1):180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 49.Ye Q, Kantonen S, Gomez-Cambronero J. Serum deprivation confers the MDA-MB-231 breast cancer line with an EGFR/JAK3/PLD2 system that maximizes cancer cell invasion. J Mol Biol. 2013;425(4):755–766. doi: 10.1016/j.jmb.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gelmon K, et al. Targeting triple-negative breast cancer: Optimising therapeutic outcomes. Ann Oncol. 2012;23(9):2223–2234. doi: 10.1093/annonc/mds067. [DOI] [PubMed] [Google Scholar]
- 51.Tester AM, et al. Pro-matrix metalloproteinase-2 transfection increases orthotopic primary growth and experimental metastasis of MDA-MB-231 human breast cancer cells in nude mice. Cancer Res. 2004;64(2):652–658. doi: 10.1158/0008-5472.can-0384-2. [DOI] [PubMed] [Google Scholar]
- 52.Marshall JC, et al. Effect of inhibition of the lysophosphatidic acid receptor 1 on metastasis and metastatic dormancy in breast cancer. J Natl Cancer Inst. 2012;104(17):1306–1319. doi: 10.1093/jnci/djs319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheridan C, et al. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: An early step necessary for metastasis. Breast Cancer Res. 2006;8(5):R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamilton SR, et al. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282(22):16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52(3/4):591–611. [Google Scholar]
- 56.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang F, et al. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94(19):1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284(39):26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim Y, et al. CD44-epidermal growth factor receptor interaction mediates hyaluronic acid-promoted cell motility by activating protein kinase C signaling involving Akt, Rac1, Phox, reactive oxygen species, focal adhesion kinase, and MMP-2. J Biol Chem. 2008;283(33):22513–22528. doi: 10.1074/jbc.M708319200. [DOI] [PubMed] [Google Scholar]
- 60.Naor D, Nedvetzki S, Golan I, Melnik L, Faitelson Y. CD44 in cancer. Crit Rev Clin Lab Sci. 2002;39(6):527–579. doi: 10.1080/10408360290795574. [DOI] [PubMed] [Google Scholar]
- 61.Ribatti D. The chick embryo chorioallantoic membrane as an in vivo assay to study antiangiogenesis. Pharmaceuticals. 2010;3(3):482–513. doi: 10.3390/ph3030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deryugina EI, Quigley JP. Chick embryo chorioallantoic membrane model systems to study and visualize human tumor cell metastasis. Histochem Cell Biol. 2008;130(6):1119–1130. doi: 10.1007/s00418-008-0536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 2008;13(3):221–234. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leong HS, Chambers AF, Lewis JD. Assessing cancer cell migration and metastatic growth in vivo in the chick embryo using fluorescence intravital imaging. Methods Mol Biol. 2012;872:1–14. doi: 10.1007/978-1-61779-797-2_1. [DOI] [PubMed] [Google Scholar]
- 65.Heppner GH, Miller BE. Tumor heterogeneity: Biological implications and therapeutic consequences. Cancer Metastasis Rev. 1983;2(1):5–23. doi: 10.1007/BF00046903. [DOI] [PubMed] [Google Scholar]
- 66.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 67.Simpson-Herren L, Noker PE, Wagoner SD. Variability of tumor response to chemotherapy. II. Contribution of tumor heterogeneity. Cancer Chemother Pharmacol. 1988;22(2):131–136. doi: 10.1007/BF00257310. [DOI] [PubMed] [Google Scholar]
- 68.Dexter DL, Leith JT. Tumor heterogeneity and drug resistance. J Clin Oncol. 1986;4(2):244–257. doi: 10.1200/JCO.1986.4.2.244. [DOI] [PubMed] [Google Scholar]
- 69.Moore NM, Kuhn NZ, Hanlon SE, Lee JS, Nagahara LA. De-convoluting cancer’s complexity: Using a ‘physical sciences lens’ to provide a different (clearer) perspective of cancer. Preface. Phys Biol. 2011;8(1):010302. doi: 10.1088/1478-3975/8/1/010302. [DOI] [PubMed] [Google Scholar]
- 70.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 71.Toole BP, Slomiany MG. Hyaluronan: A constitutive regulator of chemoresistance and malignancy in cancer cells. Semin Cancer Biol. 2008;18(4):244–250. doi: 10.1016/j.semcancer.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Golshani R, et al. Hyaluronic acid synthase-1 expression regulates bladder cancer growth, invasion, and angiogenesis through CD44. Cancer Res. 2008;68(2):483–491. doi: 10.1158/0008-5472.CAN-07-2140. [DOI] [PubMed] [Google Scholar]
- 73.Bissell MJ, Kenny PA, Radisky DC. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: The role of extracellular matrix and its degrading enzymes. Cold Spring Harb Symp Quant Biol. 2005;70:343–356. doi: 10.1101/sqb.2005.70.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curtis MW, Sharma S, Desai TA, Russell B. Hypertrophy, gene expression, and beating of neonatal cardiac myocytes are affected by microdomain heterogeneity in 3D. Biomed Microdevices. 2010;12(6):1073–1085. doi: 10.1007/s10544-010-9461-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khaing ZZ, Schmidt CE. Advances in natural biomaterials for nerve tissue repair. Neurosci Lett. 2012;519(2):103–114. doi: 10.1016/j.neulet.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 76.Briand P, Nielsen KV, Madsen MW, Petersen OW. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res. 1996;56(9):2039–2044. [PubMed] [Google Scholar]
- 77.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89(19):9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weaver VM, Howlett AR, Langton-Webster B, Petersen OW, Bissell MJ. The development of a functionally relevant cell culture model of progressive human breast cancer. Semin Cancer Biol. 1995;6(3):175–184. doi: 10.1006/scbi.1995.0021. [DOI] [PubMed] [Google Scholar]
- 79.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 80.Kemp MM, et al. Synthesis of gold and silver nanoparticles stabilized with glycosaminoglycans having distinctive biological activities. Biomacromolecules. 2009;10(3):589–595. doi: 10.1021/bm801266t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leong HS, et al. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nat Protoc. 2010;5(8):1406–1417. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arpaia E, et al. The interaction between caveolin-1 and Rho-GTPases promotes metastasis by controlling the expression of alpha5-integrin and the activation of Src, Ras and Erk. Oncogene. 2012;31(7):884–896. doi: 10.1038/onc.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.