Abstract
Geologically and chemically distinct aquifers were screened for the presence of two genes coding for key enzymes of the reverse tricarboxylic acid (rTCA) cycle in autotrophic bacteria, 2-oxoglutarate : ferredoxin oxidoreductase (oorA) and the beta subunit of ATP citrate lyase enzymes (aclB). From 42 samples investigated, aclB genes were detected in two and oorA genes in six samples retrieved from polluted and sulfidic aquifers. aclB genes were represented by a single phylotype of almost identical sequences closely affiliated with chemolithoautotrophic Sulfurimonas species. In contrast, sequences analysis of oorA genes revealed diverse phylotypes mainly related to sequences from cultivation-independent studies.
Keywords: Groundwater, chemoautotroph, CO2 fixation, reverse tricarboxylic acid cycle
To date, six mechanisms are known by which autotrophic organisms fix carbon (7). A frequent CO2 fixation strategy in chemolithoautotrophic bacteria occurs via the Calvin-Benson-Bassham cycle (6). The enzyme responsible for the actual fixation of CO2 is ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) and genes coding for the large subunit of RubisCO are often used to analyse chemolithoautotrophs in cultivation-independent environmental studies. Several investigations have been performed in groundwater systems, which revealed the widespread occurrence of diverse forms of RubisCO and hence autotrophic bacteria assimilating CO2 via the Calvin cycle pathway in the subsurface (2, 4).
The reductive tricarboxylic acid cycle (rTCA) is, in addition to the Calvin cycle, a carbon fixation pathway which is frequently found in prokaryotes: The rTCA cycle appears to operate in phylogenetically diverse autotrophic bacteria (Aquificae, Nitrospirae, Chlorobi, Proteobacteria), including genera of anoxic phototrophic bacteria, sulfate-reducing bacteria and hyperthermophilic hydrogen-oxidizing bacteria (7, 17). Although special biochemical adaptations to oxic conditions are known, this pathway involves enzymes that are sensitive to oxygen. As a result, rTCA-based CO2 fixation is mostly found in anaerobes or microaerophiles (7). Cultivation-independent investigation based on the detection of genes coding for key enzymes in the rTCA pathway were mainly performed in hydrothermal vent systems and thermal springs (e.g. 10, 14, 18, 27). The results of these studies suggest that the rTCA cycle is a major autotrophic pathway in thermal and sulfidic environments and molecular evidence also suggests that the Calvin cycle is often less prevalent in these ecosystems. On the other hand, autotrophy based on the rTCA cycle is not restricted to hot ecosystems. Several isolates from bacterial enrichment studies and environmental investigations performed with samples from terrestrial ecosystems, activated sludge, saline lakes, sulfidic caves, nonvent marine systems and an estuary revealed the presence of taxonomically diverse bacteria potentially using the rTCA cycle for CO2 fixation (e.g. 8, 11, 21, 23, 24, 29, 31, 37 and references therein); however, their ecological significance in these habitats is often not well understood.
To our knowledge, the distribution and diversity of autotrophic bacteria assimilating CO2 via the rTCA pathway has not been analyzed in groundwater systems. In the present study, 42 groundwater samples from a variety of aquifers were screened for the presence of two genes coding for key enzymes of the rTCA cycle: the alpha subunit of 2-oxoglutarate : ferredoxin oxidoreductase enzymes (oorA genes) and the beta subunit of ATP citrate lyase enzymes (aclB genes). Amplification products, which were successfully retrieved from six samples, were used to construct clone libraries and selected positive clones were sequenced in order to evaluate their phylogenetic relationships.
Successful amplification of aclB and oorA genes was achieved with samples of two groundwater systems, both contaminated with organics (Table 1): (a) At an on-site underground facility close to the city of Bitterfeld, 50 km north of Leipzig (Germany). Samples were taken from a horizontal well (19.5 meter below surface) located in a quaternary aquifer and from a column filled with original aquifer material. The groundwater is rich in sulfate and is slightly sulfidic; oxygen, nitrate and nitrite are below the detection limit (Table 1). Chlorobenzene is the main contaminant (35); (b) In groundwater influenced by activities of a former hydrogenation plant close to the city of Zeitz (Saxonia, Germany), 40 km southwest of Leipzig, where benzene production caused massive groundwater contamination. Naturally existing sulfate was found to be the main electron acceptor for microbial oxidation reactions in the central parts of the plume. Nitrate was detected on the fringes of the plume, indicating nitrate reduction as an additional important electron-accepting process at the site. Although oxygen concentrations were not measured in situ, the presence of sulfide in the investigated sampling sites indicates anoxic, sulfidic conditions in the aquifer (Table 1).
Table 1.
Description of samples and detection of aclB and oorA genes based on PCR amplification
| Sampling site | Depths (mbs) | Oxygen mg L−1 | Sulfide mg L−1 | Sulfate mg L−1 | Ammonium mg L−1 | Nitrate mg L−1 | Nitrite mg L−1 | Main pollutant | aclB | oorA |
|---|---|---|---|---|---|---|---|---|---|---|
| Municipal groundwater Salzburg (5 samples) | ||||||||||
| Groundwater samples (S1, S2, S3, S5, S6) | 2.9–10.1 | 3.6–8.5 | b.d. | 7.8–31.3 | b.d. | 2.5–39 | b.d. | Non-polluted | − | − |
| Reactor facility Bitterfeld (5 samples) | ||||||||||
| Aquifer sediment samples (B1, B2, B3, B4) | column | b.d. | n.d. | 681–871 | 5–5.4 | b.d. | b.d. | Chlorobenzene | − | − |
| Groundwater inflow from horizontal well 1 (B5) | 23 | b.d. | b.d.–1 | 714 | 5.3 | b.d. | b.d. | Chlorobenzene | + | + |
| Pilot Plant Leuna (6 samples) | ||||||||||
| Groundwater samples (L1–L6) | channels | 0.4–1.6 | b.d.–0.04 | 473–601 | 45.6–61.4 | b.d.–34.9 | b.d.–1.7 | MTBE | − | − |
| Test field Zeitz (10 samples) | ||||||||||
| Pumice sample (Z1) | column | n.d. | 23.5 | 332 | 2.5 | n.d. | n.d. | Benzene | − | − |
| Coarse sand sample (Z2) | column | n.d. | 12.2 | 391 | 1.9 | n.d. | n.d. | Benzene | − | − |
| Groundwater sample—column inflow (Z3) | n.d. | 11.1 | 391 | 2.2 | n.d. | n.d. | Benzene | − | − | |
| Groundwater sample (Z4) | 13.9 | n.d. | 0.6 | n.d | 1.1 | n.d. | n.d. | Benzene | − | − |
| Groundwater sample (Z5) | 19.1 | n.d. | 8.1 | n.d. | 5.5 | n.d. | n.d. | Benzene | + | + |
| Groundwater sample (Z6) | 42 | n.d. | 4.6 | n.d. | 3.6 | n.d. | n.d. | Benzene | − | + |
| Groundwater sample (Z7) | 47 | n.d. | 4 | n.d. | 3.7 | n.d. | n.d. | Benzene | − | − |
| Groundwater sample (Z8) | 53 | n.d. | 0.7 | n.d. | 3.3 | n.d. | n.d. | Benzene | − | + |
| Groundwater sample (Z9) | 20 | n.d. | 0.7 | n.d. | 7.7 | n.d. | n.d. | Benzene | − | + |
| Groundwater sample (Z11) | 11.5 | n.d. | 0.3 | n.d. | b.d. | n.d. | n.d. | Benzene | − | + |
| Deep subsurface—Brenner base tunnel (16 samples) | ||||||||||
| Groundwater samples (V1–V5; P1, P2; N; A1–A3; E1–E3; M1, M2) | 48–780 | n.d. | b.d. | 14–1296 | b.d.–0.24 | b.d.–2.56 | b.d.–0.04 | Non-polluted | − | − |
b.d.: below detection limit
n.d.: not determined
+ PCR amplification product (samples underlined)
− no PCR amplification product
Groundwater systems, where rTCA cycle genes were not detected, included sampling campaigns performed in non-polluted and shallow groundwater systems (Salzburg, Austria), a methyl tert-butyl ether (MTBE), BTEX and ammonia-polluted groundwater remediation test site near Leuna (Germany) and from deep subsurface samples retrieved by exploratory drilling along the projected Brenner Base Tunnel (Tyrol, Austria) (Table 1); these sampling sites are described in more detail elsewhere (4).
For DNA extraction, volumes between 300 to 500 mL groundwater were concentrated on filters (pore size 0.22 μm; Durapore, Millipore, Bedford, MA, USA) and immediately frozen until analysis. Aquifer sediments from the underground columns in Zeitz and Bitterfeld were sampled with sample lances, immediately transferred to an anoxic jar (Anaerocult A; Merck, Germany), transported at 4°C and stored at −20°C before further processing. DNA extraction of groundwater and sediment samples was performed with the FastDNA Spin Kit for soil (Qbiogene, Carlsbad, CA, USA) according to the manufacturer’s instructions and as described by Alfreider et al.(4). Two sets of oligonucleotide primers were used for PCR amplification of aclB and oorA gene fragments as described in Campbell et al.(9) and Campbell and Cary (10).
PCR products of appropriate length were cloned into a pDrive Cloning Vector (Qiagen, Valencia, CA, USA) according to the protocols provided by the manufacturer and as described in Alfreider et al.(4). Plasmid DNA was isolated (Qiagen plasmid kit; Qiagen), and clones were screened for the presence of inserts by PCR using vector-specific primers. Full PCR fragments were sequenced with capillary genetic analyzers (ABI 3730 or ABI3700; Applied Biosystems, Foster City, CA, USA) by an external sequencing facility (Macrogen, Seoul, Korea). Closest relatives to oorA and aclB nucleotide sequences and deduced amino acid sequences were obtained using NCBI’s sequence similarity search tools BLASTN and BLASTP (5), the IMG/M data management and analysis system for metagenomes (26) and CAMERA metagenomic resources (30). Deduced amino acids were aligned using Clustal W, as provided by MEGA 4 software followed by visual inspection of the alignment (33). Neighbor-joining trees applying gamma distribution as the distance method were computed with the MEGA 4 software package. Bootstrap analysis (1,000 replicates) was used to obtain confidence estimates for tree topology. Sequence data have been submitted to GenBank databases under accession numbers JF794648–JF794655 for aclB genes and JF794656–JF794687 for oorA genes.
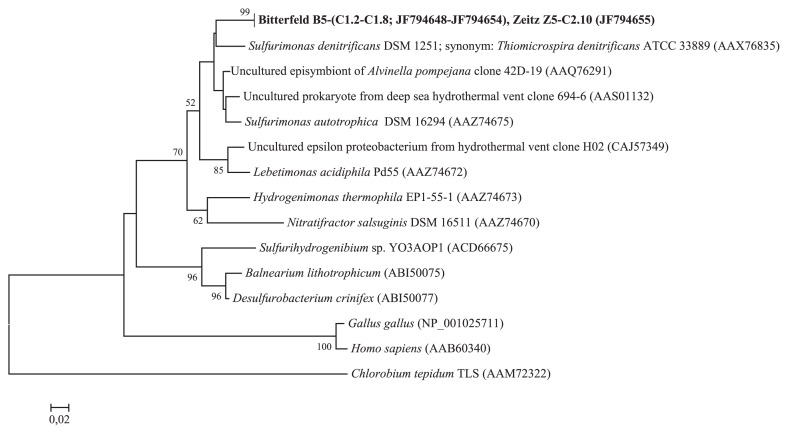
PCR screening of 42 samples revealed the presence of genes coding for oxoglutarate : ferredoxin oxidoreductase enzymes in six samples and genes coding for ATP citrate lyase enzymes were detected in two samples (Table 1). PCR products were cloned and positive clones with the proper insert length were selected for sequence analysis. ATP citrate lyase genes were detected in sampling station Bitterfeld B5 with seven identical sequences (based on deduced amino acid sequences) and one sequence was retrieved from the groundwater well Zeitz Z5 (Fig. 1). Phylogenetic analysis revealed that these sequences formed a unique cluster and showed 90 to 91% amino acid sequence similarities with ɛ-proteobacterial aclB sequences. These closest relatives include deep-sea hydrothermal Sulfurimonas isolates (32), deep-sea hydrothermal chemolithoautotrophic isolates of ɛ-Proteobacteria, epibiontic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana(9) and free-living microorganisms obtained from deep-sea hydrothermal vents (10). However, the sequence with the highest similarity to aclB genes obtained from samples B5 and Z5 was found in the non-vent bacterium Sulfurimonas denitrificans, a mesophilic sulfur oxidizer isolated from coastal marine sediments (31, 34).
Fig. 1.
Neighbor-joining phylogenetic tree calculated from deduced amino acids of aclB sequences obtained in this study (shown in bold) and public databases. GenBank accession numbers of sequences are given in parentheses. Bootstrap values are shown as percentages of 1,000 replicates and values over 50% are indicated on nodes.
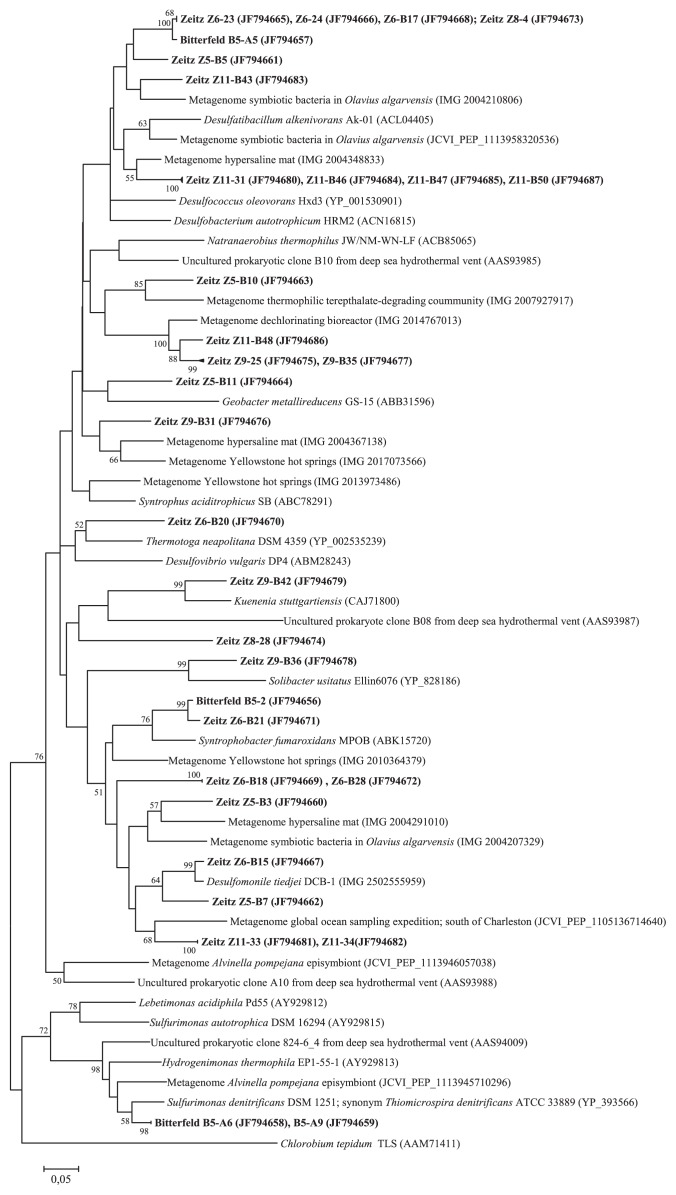
In contrast to the aclB genes, the analysis of sequences coding for 2-oxoglutarate:ferredoxin oxidoreductase enzymes showed a high degree of diversity. The amino acid sequences deduced from oorA genes sequences retrieved from horizontal well B5 (sampling station Bitterfeld) revealed three phylotypes (Fig. 2). Sequence Bitterfeld B5-A5 was affiliated with several sequences obtained from sampling stations Z5, Z6 and Z11 of the Zeitz aquifer. Sequence Bitterfeld B5-2 and the closely related sequence Zeitz Z6-B21 are affiliated with Syntrophobacter fumaroxidans str. MPOB (89% sequence similarity). Two Bitterfeld B5 clones (B5-A9 and B5-A6) were clearly separated from all other oorA genotypes retrieved in this study (Fig. 2). Both sequences clustered with oorA genes derived from deep-sea hydrothermal chemolithoautotrophic isolates and cultivation-independent studies. Sulfurimonas denitrificans was the closest relative with 94% amino acid similarity. As already mentioned above, Sulfurimonas denitrificans and other Sulfurimonas species were also the closest relatives of all aclB sequences observed at sampling station Bitterfeld B5. S. denitrificans, an obligate chemolithoautotrophic ɛ-Proteobacterium, is a metabolically versatile bacterium with the potential to use a variety of compounds as electron donors, e.g. reduced sulfur compounds, hydrogen or formate (31). S. denitrificans, which was originally isolated from coastal marine sediments (34), is also suggested to be a key player in the nitrogen cycle in the Baltic Sea, responsible for chemolithoautotrophic denitrification (8, 13). In addition to nitrate, S. denitrificans can use inorganic sulfur compounds or oxygen as electron acceptors (31).
Fig. 2.
Neighbor-joining phylogenetic tree calculated from deduced amino acids of oorA sequences obtained in this study (shown in bold) and public databases. Accession numbers of sequences are given in parentheses. Reference sequences from metagenomic libraries were obtained from the Joint Genome Institute database (IMG) and CAMERA database (JCVI_PEP_). Bootstrap values are shown as percentages of 1,000 replicates and values over 50% are indicated on nodes.
The oorA sequences derived from the sampling stations Zeitz were widespread along the phylogenetic tree. Most sequences were not closely affiliated with known oorA genotypes deposited in public databases and therefore these genotypes were also distantly related to cultivated representatives. Consequently, it would be too speculative to deduce ecophysiological characteristics from these sequences. Comparison with deduced amino acid sequences retrieved from sequences of public databases showed closest similarities ranging between 76% and 92%. For example one cluster of oorA sequences from groundwater well Zeitz Z11 (clones 31, B46, B47, B50) was affiliated (84%–88% deduced amino acid identity) with an oorA gene obtained from a metagenomic study of a hypersaline mat from Guerrero Negro, Mexico, which is characterized by steep physicochemical gradients on the millimeter scale (22). Several oorA genes retrieved in our study were affiliated with sequences of different metagenomic investigations, including a thermophilic terephthalate-degrading microbial community growing at around 55°C that was developed in a lab-scale bioreactor (IMG/M database, not published), oorA genes from a highly stable reductive dechlorinating bioreactor (IMG/M database, not published), microbial communities from Yellowstone hot springs and hot spring pools (IMG/M database, not published) and symbionts from the marine oligochaete Olavius algarvensis(38). Summarizing the phylogenetic analysis based on genes coding for 2-oxoglutarate:ferredoxin oxidoreductase enzymes revealed that the majority of sequences were affiliated with oorA sequences derived from cultivation independent studies, generally thermal ecosystems and/or environments that are characterized by distinct concentration gradients and different redox states of bio-geochemical important elements supporting reverse tricarboxylic acid cycle as the autotrophic pathway.
The presence of phylogenetically distant oorA sequences, however, suggests the occurrence of oorA phylotypes coding for 2-oxoglutarate : ferredoxin oxidoreductase enzymes involved in other (non rTCA-related) biochemical processes. A reaction that could specifically be of interest at study site Zeitz is the potential role of 2-oxoglutarate : ferredoxin oxidoreductase for aromatic ring reduction (12), since field site Zeitz is contaminated with BTEX compounds with benzene as the main pollutant. Degradation of benzene and toluene under sulfate-reducing conditions has been demonstrated at the site (36), and some of the oorA sequences are affiliated with sequences detected in sulfate-reducing or putative syntrophic bacteria (Fig. 2) that are potentially involved in anaerobic BTEX degradation.
To our knowledge, this is the first comprehensive study demonstrating the existence of bacteria with genetic potential using a pathway other than the Calvin-Benson-Bassham cycle for CO2 fixation in groundwater ecosystems. From earlier investigations based on RubisCO analysis we have learned that in samples retrieved from the same groundwater sampling sites, the Calvin cycle is widely distributed (in 85% of 48 samples) and used by diverse obligate and facultative chemolithoautotrophic Proteobacteria for CO2 fixation (4). In the present study, genes for the rTCA cycle were only found in six samples at two sampling locations, with groundwater characterized by anoxic and sulfidic conditions and contaminated with organics. One obvious reason for the limited distribution is the properties of certain enzymes in the rTCA cycle, which are sensitive to oxygen. As a result, the rTCA cycle is only found in anaerobes or microaerophiles. So far, the reductive TCA cycle has been believed to be the main strategy for chemolithoautotrophic CO2 fixation in hydrothermal vents and other hot ecosystems. The microbial communities in these habitats are usually dominated by autotrophic ɛ-Proteobacteria and Aquificales, which generally fix CO2 via the rTCA pathway (10, 17, 27). CO2 fixation based on the rTCA pathway was also shown for a small number of bacterial representatives phylogentically related to Chlorobiales, δ-Proteobacteria and magnetotactic cocci affiliated with the class of α-Proteobacteria(37), but their environmental importance has yet to be determined.
Previous studies at sampling sites Zeitz and Bitterfeld, focusing on the bacterial composition based on 16S rDNA analysis, revealed the dominance of β-Proteobacteria and a broad diversity of other phyla (1, 3). Samples from the Bitterfeld site revealed two clone sequences which were phylogentically classified into the class of ɛ-Proteobacteria. Interestingly, the 16S rDNA sequence of one of these clones (RA9C8) was affiliated with the facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium Sulfuricurvum kujiense(20) and representatives of the genus Sulfurimonas, including S. denitrificans and S. autotrophica (data not shown). A recent study also revealed 16S rDNA sequences related to ɛ-Proteobacteria at field site Zeitz (19). A phylotype affiliated to the genus Sulfurovum was found to be abundant in a column system operating under close to in situ conditions (19); this phylotype has been shown to be involved in anaerobic benzene degradation (15). Furthermore, a phylotype closely related to S. denitrificans has been recently identified in nitrate-dependent sulfide-oxidizing enrichment cultures prepared from groundwater from field site Zeitz.
The lifestyle of the versatile ɛ-Proteobacteria in organics-contaminated sulfidic sites is not well understood and should be the focus of future studies. Some members of this taxonomic group are autotrophic, using e.g. reduced sulfur compounds or hydrogen as electron donors, oxygen, nitrate or metals as electron acceptors, and fixing carbon via the rTCA cycle. In contrast, Sulfurspirillum or Arcobacter species have been described for fermentative metabolism (25). Recently, phylotypes affiliated mainly to the genus Sulfuricurvum were found to be abundant in toluene-degrading sulfate-reducing enrichment cultures prepared from tar-oil contaminated aquifer sediment samples; their function remained unclear (28). Notably, Hubert et al.(16) observed that similar phylotypes were dominant in oil formation waters from a Canadian oil sand reservoir; the authors speculate that some of these might use also sulfur-containing crude oil compounds as carbon and/or sulfur sources.
Acknowledgements
This study was funded by the Austrian Science Fund to AA (FWF P17649). We thank Jörg Ahlheim and Ralf Trabitzsch (both UFZ) for logistic support, sample collection and technical assistance in the field.
References
- 1.Alfreider A, Vogt C, Babel W. Microbial diversity in an in situ reactor system treating monochlorobenzene contaminated groundwater as revealed by 16S ribosomal DNA analysis. System Appl Microbiol. 2002;25:232–240. doi: 10.1078/0723-2020-00111. [DOI] [PubMed] [Google Scholar]
- 2.Alfreider A, Vogt C, Hoffmann D, Babel W. Diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from groundwater and aquifer microorganisms. Microb Ecol. 2003;45:317–328. doi: 10.1007/s00248-003-2004-9. [DOI] [PubMed] [Google Scholar]
- 3.Alfreider A, Vogt C. Bacterial diversity and aerobic biodegradation potential in a BTEX-contaminated aquifer. Water Air Soil Pollut. 2007;183:415–426. [Google Scholar]
- 4.Alfreider A, Vogt C, Kaiser M, Psenner R. Distribution and diversity of autotrophic bacteria in groundwater systems based on the analysis of RuBisCO genotypes. System Appl Microbiol. 2009;32:140–150. doi: 10.1016/j.syapm.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Badger MR, Bek EJ. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2acquisition by the CBB cycle. J Exp Bot. 2008;59:1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 7.Berg IA. Ecological aspects of the distribution of different autotrophic CO2fixation pathways. Appl Environ Microbiol. 2011;77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brettar I, Labrenz M, Flavier S, Botel J, Kuosa H, Christen R, Hoefle MG. Identification of a Thiomicrospira denitrificans-like epsilonproteobacterium as a catalyst for autotrophic denitrification in the central Baltic Sea. Appl Environ Microbiol. 2006;72:1364–1372. doi: 10.1128/AEM.72.2.1364-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell BJ, Stein JL, Cary SC. Evidence of chemolithoautotrophy in the bacterial community associated with Alvinella pompejana, a hydrothermal vent polychaete. Appl Environ Microbiol. 2003;69:5070–5078. doi: 10.1128/AEM.69.9.5070-5078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell BJ, Cary SC. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl Environ Microbiol. 2004;70:6282–6289. doi: 10.1128/AEM.70.10.6282-6289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell BJ, Engel AS, Porter ML, Takai K. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- 12.Doerner E, Boll M. Properties of 2-oxoglutarate: ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J Bacteriol. 2002;184:3975–3983. doi: 10.1128/JB.184.14.3975-3983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grote J, Jost G, Labrenz M, Herndl GJ, Juergens K. Epsilonproteobacteria represent the major portion of chemoautotrophic bacteria in sulfidic waters of pelagic redoxclines of the Baltic and Black Seas. Appl Environ Microbiol. 2008;74:7546–7551. doi: 10.1128/AEM.01186-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall JR, Mitchell KR, Jackson-Weaver O, Kooser AS, Cron BR, Crossey LJ, Takacs-Vesbach CD. Molecular characterization of the diversity and distribution of a thermal spring microbial community using rRNA and metabolic genes. Appl Environ Microbiol. 2008;74:4910–4922. doi: 10.1128/AEM.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrmann S, Kleinsteuber S, Chatzinotas A, Kuppardt S, Lueders T, Richnow HH, Vogt C. Functional characterization of an anaerobic benzene-degrading enrichment culture by DNA stable isotope probing. Environ Microbiol. 2010;12:401–411. doi: 10.1111/j.1462-2920.2009.02077.x. [DOI] [PubMed] [Google Scholar]
- 16.Hubert CRJ, Oldenburg TBP, Fustic M, et al. Massive dominance of Epsilonproteobacteria in formation waters from a Canadian oil sands reservoir containing severely biodegraded oil. Environ Microbiol. 2012;14:387–404. doi: 10.1111/j.1462-2920.2011.02521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huegler M, Wirsen CO, Fuchs G, Taylor CD, Sievert SM. Evidence for autotrophic CO2fixation via the reductive tricarboxylic acid cycle by members of the ɛ subdivision of proteobacteria. J Bacteriol. 2005;187:3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huegler M, Gaertner A, Imhoff JF. Functional genes as markers for sulfur cycling and CO2fixation in microbial communities of hydrothermal vents of the Logatchev field. FEMS Microbiol Ecol. 2010;73:526–537. doi: 10.1111/j.1574-6941.2010.00919.x. [DOI] [PubMed] [Google Scholar]
- 19.Kleinsteuber S, Schleinitz KM, Breitfeld J, Harms H, Richnow HH, Vogt C. Molecular characterization of bacterial communities mineralizing benzene under sulfate-reducing conditions. FEMS Microbiol Ecol. 2008;66:143–157. doi: 10.1111/j.1574-6941.2008.00536.x. [DOI] [PubMed] [Google Scholar]
- 20.Kodama Y, Watanabe K. Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int J Syst Evol Microbiol. 2004;54:2297–2300. doi: 10.1099/ijs.0.63243-0. [DOI] [PubMed] [Google Scholar]
- 21.Kovaleva OL, Touroval TP, Muyzer G, Kolganova TV, Sorokin DY. Diversity of RuBisCO and ATP citrate lyase genes in soda lake sediments. FEMS Microbiol Ecol. 2011;75:37–47. doi: 10.1111/j.1574-6941.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- 22.Kunin V, Raes J, Harris JK, et al. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4:198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Cono V, Smedile F, Bortoluzzi G, et al. Unveiling microbial life in new deep-sea hypersaline Lake Thetis. Part I: Prokaryotes and environmental settings. Environ Microbiol. 2011;13:2250–2268. doi: 10.1111/j.1462-2920.2011.02478.x. [DOI] [PubMed] [Google Scholar]
- 24.Luecker S, Wagner M, Maixner F, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luijten MLGC, de Weert J, Smidt H, Boschker HTS, de Vos WM, Schraa G, Stams AJM. Description of Sulfurospirillum halorespirans sp. nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int J Syst Evol Microbiol. 2003;53:787–793. doi: 10.1099/ijs.0.02417-0. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz VM, Ivanova NN, Szeto E, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perner M, Seifert R, Weber S, Koschinsky A, Schmidt K, Strauss H, Peters M, Haase K, Imhoff JF. Microbial CO2fixation and sulphur cycling associated with low-temperature emissions at the Lilliput hydrothermal field, southern Mid-Atlantic Ridge (9°S) Environ Microbiol. 2007;9:1186–1201. doi: 10.1111/j.1462-2920.2007.01241.x. [DOI] [PubMed] [Google Scholar]
- 28.Pilloni G, von Netzer F, Engel M, Lueders T. Electron acceptor-dependent identifcation of key anaerobic toluene degraders at a tar-oil-contaminated aquifer by Pyro-SIP. FEMS Microbiol Ecol. 2011;78:165–175. doi: 10.1111/j.1574-6941.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- 29.Porter ML, Engel AS. Diversity of uncultured epsilonproteobacteria from terrestrial sulfidic caves and springs. Appl Environ Microbiol. 2008;74:4973–4977. doi: 10.1128/AEM.02915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. CAMERA: a community resource for Metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sievert SM, Scott KM, Klotz MG, et al. The genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol. 2008;74:1145–1156. doi: 10.1128/AEM.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takai K, Campbell BJ, Cary SC, et al. Enzymatic and genetic characterization of carbon and energy metabolism by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl Environ Microbiol. 2005;71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 34.Timmer-Ten Hoor A. A new type of thiosulphate oxidizing, nitrate reducing microorganism: Thiomicrospira denitrificans sp. nov. Neth J Sea Res. 1975;9:344–350. [Google Scholar]
- 35.Vogt C, Alfreider A, Lorbeer H, Hoffmann D, Wuensche L, Babel W. Bioremediation of chlorobenzene-contaminated ground water in an in situ reactor mediated by hydrogen peroxide. J Contam Hydrol. 2004;68:121–141. doi: 10.1016/S0169-7722(03)00092-5. [DOI] [PubMed] [Google Scholar]
- 36.Vogt C, Goedeke S, Treutler HC, Weiss H, Schirmer M, Richnow HH. Benzene oxidation under sulfate-reducing conditions in columns simulating in situ conditions. Biodegradation. 2007;18:625–636. doi: 10.1007/s10532-006-9095-1. [DOI] [PubMed] [Google Scholar]
- 37.Williams TJ, Zhang CL, Scott JH, Bazylinski DA. Evidence for autotrophy via the reverse tricarboxylic acid cycle for the marine magnetotactic coccus strain MC-1. Appl Environ Microbiol. 2006;72:1322–1329. doi: 10.1128/AEM.72.2.1322-1329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woyke T, Teeling H, Ivanova NN, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]