Abstract
A number of molecular ecological studies have revealed complex and unique microbial communities in various terrestrial plant roots; however, little is known about the microbial communities of aquatic plant roots in spite of their potential use for water quality improvement in aquatic environments (e.g. floating treatment wetland system). Here, we report the microbial communities inhabiting the roots of emerged plants, reed (Phragmites australis) and Japanese loosestrife (Lythrum anceps), collected from a floating treatment wetland in a pond by both culture-independent and culture-dependent approaches. Culture-independent analysis based on 16S rRNA gene sequences revealed that the microbial compositions between the two aquatic plant roots were clearly different (e.g. the predominant microbe was Betaproteobacteria for reed and Alphaproteobacteria for Japanese loosestrife). In comparisons of microbial communities between the plant roots and pond water taken from near the plants, the microbial diversity in the plant roots (e.g. 4.40–4.26 Shannon-Weiner index) were higher than that of pond water (e.g. 3.15 Shannon-Weiner index). Furthermore, the plant roots harbored 2.5–3.5 times more phylogenetically novel clone phylotypes than pond water. The culture-dependent approach also revealed differences in the microbial composition and diversity among the two plant roots and pond water. More importantly, compared to pond water, we succeeded in isolating approximately two times more novel isolate phylotypes, including a bacterium of candidate phylum OP10 (recently named Armatimonadetes) from the plant roots. These findings suggest that aquatic plants roots are significant sources for a variety of novel organisms.
Keywords: aquatic plant, microbial community, novel microbe, Armatimonadetes
Many studies of the microbial communities associated with the roots of terrestrial plants have so far been reported (8, 10, 11, 18, 24, 32, 35, 36, 49), suggesting that the microbes might exert a beneficial, neutral or deleterious influence on plant growth. Some of these studies also revealed that the roots of different plant species interacted with different microbial communities (10, 11, 35, 36, 49), and that the growth and activity of microbial populations in the area, which is adjacent to the root system (rhizosphere) and on the external surface of the roots (rhizoplane) are, in general, greater than that root-free soil due to root exudates such as amino acids, sugars and growth factors (11, 26, 36).
Like terrestrial plants, aquatic plants living in water environments also closely interact with microbial communities on their roots. Some important microbial processes such as plant growth promotion, nitrification, denitrification, and remediation of contaminants are significantly active in aquatic plant roots as observed in terrestrial plants (1, 13, 30, 38, 45–47, 50).
Recently, a floating wetland system vegetated with aquatic plants has been used for water quality improvement and purification of various water resources such as stormwater, sewage, piggery effluent, poultry processing wastewater and water supply reservoirs (7, 14, 20, 44). In this system, water can be clarified and purified by the function of the plant roots and root microbes, while it directly passes the extensive roots hanging beneath the floating unit. To date, several previous studies have shown the important contribution of the root microbes for water quality improvement (45–47); however, there are few reports on microbes associated with the roots of aquatic plants including those grown in the floating treatment wetland system as well as natural environments.
Recently, we analyzed a root microbial community of a floating aquatic plant, Spirodela polyrrhiza, and found that it could be used to successfully isolate various novel microbes, including rarely cultivated organisms within the phylum Verrucomicrobia(28).
In the present study, we analyzed the microbial communities inhabiting the roots of two different emerged plants, reed (Phagmites australis) and Japanese loosestrife (Lythrum anceps), in the floating wetland system by culture-independent and culture-dependent approaches. The aims of this study are to obtain primary knowledge about the microbes associated with the roots of aquatic plants, and to verify whether the roots harbor unique microbial communities and are superior sources for culturing novel microbes.
Materials and Methods
Plant samples
Reed (P. australis) and Japanese loosestrife (L. anceps) were harvested from a pond (820 m2) located within Yamanashi prefectural wood park (Kanegawa-no-mori) in the summer (July 23, 2007). The pond receives water from Hirose Reservoir, which is fed by Fuefuki River located almost in Central Japan. These aquatic plants had been cultivated on a wooden floating base unit filled with fiber made from coconut husks for 5 years (from 2003 to 2007). The features of water chemistry in the pond such as nitrogen concentration, phosphorus concentration and temperature were described in our previous study (25).
Culture-independent analysis
Total nucleic acids were extracted from 0.3 g (wet weight) of plant roots by using the ISOIL kit for bead beating (Nippon Gene, Tokyo, Japan). For DNA extraction from pond water (100 mL) collected from near the plants, the Ultraclean water DNA kit (MO BIO Laboratories, Carlsbad, CA, USA) was used. PCR amplification of the 16S rRNA genes from the extracted DNA was performed using two bacterial universal primers, EUB8F (48) (5’-AGAGTTTGATCMTGGCTCAG-3’: corresponding to positions 8–27 of the Escherichia coli 16S rRNA gene) and EUB1512R (19) (5’-ACGGYTACCTTGTTACGACTT-3’; corresponding to positions 1492–1512 of the E. coli 16S rRNA gene). The reactions were conducted as previously described (28) except that the numbers of cycles was reduced to minimize the PCR bias. The cycle number was adjusted to 18–25 cycles (18, 20, and 25 cycles for reed roots, Japanese loosestrife roots, and pond water, respectively). The amplified DNA fragments were purified by an illustra GFX PCR purification kit (GE Healthcare, Little Chalfont, UK), and cloned into the E. coli strain DH5α using the pT7 Blue T-vector kit (Novagen). The clonal DNA was amplified from randomly selected recombinants by colony direct PCR using the two primers, pT7-F (5’-GATCTACTAGTCATATGGAT-3’) and pT7-R (5’-TCGGTAC CCGGGGATCCGAT-3’), which were specific to the vector sites flanking the insert. The DNA fragments obtained were subjected to restriction fragment length polymorphism (RFLP) analysis by separate digestion with two different restriction endonucleases, HhaI and HaeIII (Takara, Otsu, Japan). Coverage (C) values for each of the clone libraries were calculated by equation C=[1−(n/N)]×100 (9), where n is the number of unique clones and N is the total number of clones analyzed.
The PCR products from representative clones of each of the RFLP groups were purified with the GFX PCR DNA and Gel purification kit (GE Healthcare), and sequenced as previously described with primer EUB907R (41) (5’-CCGYCAATTCMTTT RAGTTT-3’). After checking the possible chimeric artifacts with the Bellerophon program (http://greengenes.lbl.gov/cgi-bin/nph-bel3_interface.cgi), the sequences were compared with those in the NCBI database using the BLASTn program (http://www.ncbi.nlm.nih.gov/blast/). Taxonomic classification of the clonal sequence at the level of the bacterial family was also conducted using the CLASSIFIER program (http://rdp.cme.msu.edu/classifier/classifier.jsp).
Cultivation of microbes
A low-nutrient medium, DTS (pH 7.0), containing 0.17 g Bacto tryptone (Difco), 0.03 g Bacto soytone (Difco), 0.025 g glucose, 0.05 g NaCl and 0.025 g K2HPO4 in 1 L of distilled water, was used. Approximately 0.15 g (wet weight) of plant roots were gently rinsed twice with 30 mL sterilized DTS medium in a 50 mL test tube to remove microbes which were not associated with the plants, and the roots were mechanically homogenized with 10 mL DTS medium under the conditions at 15,000 rpm for 7 min by an Ace HOMOGENIZER AM-1 (Nihonseiki, Tokyo, Japan). The homogenates or pond water samples collected from an area surrounding the plants were diluted in 10-fold steps with DTS medium. Each diluted sample (50 μL) was independently inoculated on agar (1.5%) medium plates in triplicate, and incubated at 25°C for 30 days under dark conditions.
Phylogenetic analysis of the isolates
16S rRNA genes of the isolated microbes were amplified by the colony direct PCR method using primers EUB8F and EUB1512R, and subjected to RFLP and sequencing analyses using methods similar to those described for culture-independent analysis with the exception that the restriction enzyme MspI (Takara) was used in RFLP analysis instead of HaeIII. The sequences were compared with those present in public databases using the BLASTn program. Taxonomic classification of the isolate at the family level was performed with the same method used for clone library analysis.
Diversity index
The diversity of clones and isolated microbes at the level of the RFLP phylotype were calculated by the Shannon-Weiner index [(H)=−∑(pi) (ln pi)] and Simpson’s reciprocal index, 1/D, where D equals ∑(pi)2 and where pi is the proportion of clones or isolates within each phylotype i relative to the total number of clones or isolates.
Sequencing of the 16S rRNA gene and phylogenetic analysis of strain YO-36
Phylogenetic analysis of strain YO-36 isolated was performed on the basis of 16S rRNA gene sequencing. The 16S rRNA gene of strain YO-36 was directly PCR-amplified with the universal primers 8F and 1492R (41) using AmpliTaq Gold (Applied Biosystems, Carlsbad, CA, USA). PCR was carried out in 100 μL reaction volumes in a Perkin-Elmer GeneAmp system 9700 (Perkin-Elmer Life Sciences, Boston, MA, USA) under the thermal cycle program as follows: initial denaturation at 95°C for 9 min, followed by 35 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min. The PCR product was purified with a MicroSpin S-400 HR column (GE Healthcare). Sequencing was performed with a CEQ DTCS-Quick Start kit (Beckman-Coulter, Fullerton, USA) and a CEQ-2000 automated sequence analyzer (Beckman). The sequence was compared with those from the public database using the BLASTn program. Phylogenetic analysis was performed with the ARB software package (23) using reference sequences with over 1,300 bp sequence length. After automatic and manual sequence alignments, a phylogenetic tree was constructed by the neighbor-joining (NJ) method as previously described (40). The bootstrap values were determined from 1,000 re-samplings with PAUP* 4.0 (39) for the NJ method and TREEFINDER (17) for the maximum likelihood (ML) method as previously described (40).
Nucleotide accession numbers
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of clones and isolates are AB540257–AB540432 and AB529661–AB529718, respectively.
Results and Discussion
Culture-independent analysis of microbes inhabiting roots of aquatic plants
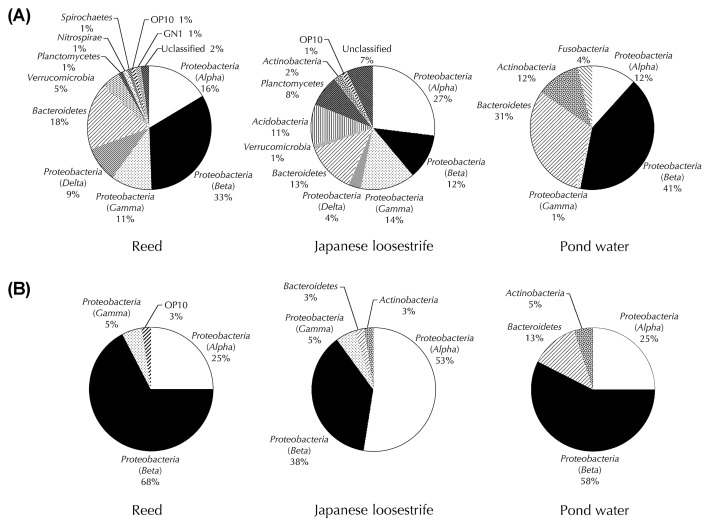
Firstly, the microbial communities inhabiting the roots of reed and Japanese loosestrife were investigated using the 16S rRNA gene libraries of these two plants. For comparison, we also analyzed the microbial community in the surrounding environment, the pond water collected from near the aquatic plants. As shown in Supplementary Table S1, the clones (85 clones from each library; plastid- and mitochondria-derived sequences were not included) grouped on the basis of RFLP analysis could be divided into 66, 74 and 36 phylotypes (further referred to as C-phylotype; clone-phylotype), from clone libraries of reed, Japanese loosestrife and pond water, respectively. The coverage values were 32.9% for reed, 25.9% for Japanese loosestrife and 72.9% for pond water. Phyloge-netic analysis based on the sequencing of a clone representing each C-phylotype showed that the clone libraries of reed, Japanese loosestrife and pond water comprised of 11, 10 and 6 bacterial divisions (phylum or class (for Proteobacteria)), respectively (Fig. 1A). Betaproteobacteria was the most abundant bacterial division in reed (33% of total clones) and pond water samples (41%). In contrast, the most predominant division in Japanese loosestrife was Alphaproteobacteria (27%). Further taxonomic classification of the clonal sequences indicated that 64 clones from reed, 45 clones from Japanese loosestrife and 73 clones from pond water were placed within 22, 15 and 11 families, respectively (Table 1). Among 27 different families detected in the two aquatic plants, only 9 families (Caulobacteraceae, Hyphomicrobiaceae, Rhodobacteraceae, Sphingomonadaceae, Comamonadaceae, Burkholderiales genera incertae, Chitinophagaceae, Saprospiraceae and Planctomycetaceae) were shared in both samples. Furthermore, the most predominant bacterial families in the roots of reed and Japanese loosestrife were different; Burkholderiales genera incertae sedis for reed (15%) and Sinobacteraceae for Japanese loosestrife (12%). It has been reported that the bacterial community inhabiting the roots of terrestrial plants such as annual plants and trees apparently varies among different species (10, 11, 35, 36, 49). Therefore, the findings shown above suggested that aquatic plants might also harbor species-specific bacterial communities on the roots as is the case for terrestrial plants. We then compared the microbial community compositions between pond water and aquatic plant roots at the level of the bacterial family. The most predominant group in pond water was Comamonadaceae (38%), unlike those of two aquatic plants. Additionally, the predominant groups of the plant samples (Burkholderiales genera incertae sedis and Sinobacteraceae) were not detected in the pond water sample. This result indicated that the aquatic plant roots formed distinct microbial communities from the pond water.
Fig. 1.
Phylogenetic distribution of the 16S rRNA gene clones (A) and isolates (B) belonging to different bacterial taxa in the roots of reed and Japanese loosestrife, and pond water.
Table 1.
Taxonomic classification of clones and isolates
| Phylum | Class | Order | Family | Number of clones | Number of isolates | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Reed | Japanese loosestrife | Pond water | Reed | Japanese loosestrife | Pond water | ||||
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | 1 | 1 | 3 | |||
| Rhizobiales | Beijerinckiaceae | 2 | |||||||
| Bradyrhizobiaceae | 1 | 1 | |||||||
| Hyphomicrobiaceae | 1 | 1 | 1 | ||||||
| Rhizobiaceae | 1 | ||||||||
| Xanthobacteraceae | 1 | ||||||||
| Unclassified | 1 | 8 | 1 | 2 | 7 | ||||
| Rhodobacterales | Rhodobacteraceae | 4 | 2 | 9 | 3 | 4 | 9 | ||
| Sphingomonadales | Erythrobacteraceae | 1 | |||||||
| Sphingomonadaceae | 1 | 5 | 4 | 4 | 1 | ||||
| Unclassified | 1 | ||||||||
| Unclassified | Unclassified | 6 | 2 | ||||||
|
| |||||||||
| Betaproteobacteria | Burkholderiales | Alcaligenaceae | 2 | 8 | |||||
| Burkholderiaceae | 2 | 1 | |||||||
| Comamonadaceae | 7 | 1 | 32 | 4 | 3 | 16 | |||
| Oxalobacteraceae | 1 | 5 | |||||||
| Burkholderiales genera incertae sedis | 13 | 2 | 19 | 4 | |||||
| Unclassified | 1 | ||||||||
| Methylophilales | Methylophilaceae | 4 | 1 | 1 | |||||
| Rhodocyclales | Rhodocyclaceae | 7 | 1 | ||||||
| Unclassified | Unclassified | 1 | 2 | ||||||
|
| |||||||||
| Deltaproteobacteria | Bdellovibrionales | Bdellovibrionaceae | 1 | ||||||
| Myxococcales | Cystobacterineae | 1 | |||||||
| Polyangiaceae | 1 | ||||||||
| Unclassified | 4 | 2 | |||||||
| Unclassified | Unclassified | 1 | |||||||
|
| |||||||||
| Gammaproteobacteria | Legionellales | Legionellaceae | 1 | ||||||
| Methylococcales | Methylococcaceae | 2 | |||||||
| Pseudomonadales | Pseudomonadaceae | 6 | 1 | ||||||
| Xanthomonadales | Sinobacteraceae | 10 | 2 | ||||||
| Unclassified | Unclassified | 1 | 2 | 1 | |||||
|
| |||||||||
| Bacteroidetes | Flavobacteria | Flavobacteriales | Cryomorphaceae | 5 | 2 | ||||
| Flavobacteriaceae | 2 | 8 | 3 | ||||||
| Sphingobacteria | Sphingobacteriales | Chitinophagaceae | 2 | 6 | 2 | 1 | 1 | ||
| Cytophagaceae | 2 | 11 | |||||||
| Saprospiraceae | 1 | 2 | |||||||
| Unclassified | 1 | 2 | |||||||
| Unclassified | Unclassified | Unclassified | 2 | 1 | 3 | 1 | |||
|
| |||||||||
| Verrucomicrobia | Opitutae | Opitutales | Opitutaceae | 3 | |||||
| Subdivision 3 | Unclassified | Unclassified | 1 | 1 | |||||
|
| |||||||||
| Acidobacteria | Acidobacteria Gp3 | Unclassified | Unclassified | 6 | |||||
| Acidobacteria Gp4 | Unclassified | Unclassified | 2 | ||||||
| Acidobacteria Gp6 | Unclassified | Unclassified | 1 | ||||||
|
| |||||||||
| Planctomycetes | Planctomycetacia | Planctomycetales | Planctomycetaceae | 1 | 5 | ||||
| Unclassified | Unclassified | Unclassified | 2 | ||||||
|
| |||||||||
| Fusobacteria | Fusobacteria | Fusobacteriales | Fusobacteriaceae | 3 | |||||
|
| |||||||||
| Nitrospira | Nitrospira | Nitrospirales | Nitrospiraceae | 1 | |||||
|
| |||||||||
| Spirochaetes | Spirochaetes | Spirochaetales | Spirochaetaceae | 1 | |||||
|
| |||||||||
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | 2 | |||||
| Unclassified | 8 | 1 | 2 | ||||||
| Unclassified | Unclassified | 1 | |||||||
| Unclassified | Unclassified | Unclassified | 1 | ||||||
|
| |||||||||
| Candidate phylum OP10 | Unclassified | Unclassified | Unclassified | 1 | 1 | 1 | |||
|
| |||||||||
| Candidate phylum GN1 | Unclassified | Unclassified | Unclassified | 1 | |||||
|
| |||||||||
| Unclassified | Unclassified | Unclassified | Unclassified | 2 | 6 | ||||
|
| |||||||||
| Total analyzed clone or isolate numbers | 85 | 85 | 85 | 40 | 40 | 40 | |||
| Total numbers of clones or isolates classified at the family level | 64 | 45 | 73 | 33 | 32 | 37 | |||
To compare the microbial diversity among the two plant roots and the pond water, the Shannon-Weiner index and Simpson’s reciprocal index were calculated based on the grouping of C-phylotype (Table 2). Both the indices scores from two aquatic plants were higher than those from pond water, suggesting that the microbial communities of aquatic plant roots were more diverse than those of their surrounding pond water.
Table 2.
Diversity indices for clones and isolates at the phylotype level
| Sample | Clone | Isolate | ||
|---|---|---|---|---|
|
|
|
|||
| Shannon-Weiner index | Simpson’s reciprocal index | Shannon-Weiner index | Simpson’s reciprocal index | |
| Reed roots | 4.04 | 45.44 | 2.82 | 12.90 |
| Japanese loosestrife roots | 4.26 | 67.52 | 2.96 | 13.56 |
| Pond water | 3.15 | 15.67 | 2.04 | 6.30 |
In the terrestrial environments, the microbial density and structures inhabiting the plant roots were distinct from those of root-free bulk soil (11, 26, 36). These phenomena are thought to be due to the distinct environments of the habitats formed by the surface structure and exudates released from the roots. The data obtained in the present study would also support this hypothesis in aquatic plants roots as well as in terrestrial plants.
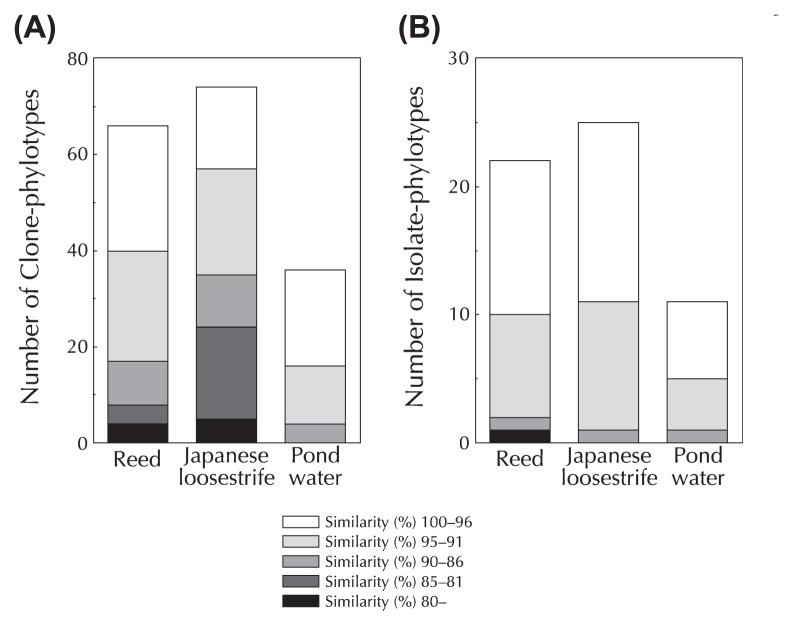
Some candidate phyla and rarely cultured groups such as the Verrucomicrobia and Acidobacteria were found in the clone libraries of roots of two aquatic plants. The Verrucomicrobia and candidate phylum OP10 (recently named the phylum Armatimonadetes(42); see below) were detected in both aquatic plants. Candidate phylum GN1 and Acidobacteria were also found in reed and Japanese loosestrife rhizospheres (Fig. 1A, Table S1); however, these phyla were not detected in the clone library of pond water (Fig. 1A, Table S1). Furthermore, unclassifiable bacterial sequences (i.e., C-phylotypes Nos. 26 and 35 from reed and Nos. 70, 78, 131 and 134 from Japanese loosestrife) were found in the clone libraries of the two aquatic plants, but not in pond water (Fig. 1A, Table S1). When clones with sequences with less than 95% similarity to the 16S rRNA gene of any known bacterial species were regarded as phylogenetically novel microbes, then more C-phylotypes from the roots of aquatic plants showed phylogenetic novelty (40 C-phylotypes for reed and 57 C-phylotypes for Japanese loosestrife), compared to pond water (16 C-phylotypes) (Fig. 2A). In addition, C-phylotypes showing less than 90% sequence similarity to any known bacterial species were detected in both reed and Japanese loosestrife rhizospheres, but not in pond water (Fig. 2A). These results clearly indicated that the roots of aquatic plants harbor far more novel organisms than pond water.
Fig. 2.
Novel clonal 16S rRNA gene sequences (A) and isolates (B) recovered from the roots of reed and Japanese loosestrife, and pond water. The similarity percentages between the clones (A) or isolates (B) and their closest species in the GenBank database are shown.
Cultivation and isolation of microbes from roots of aquatic plants
To verify whether novel organisms can be cultured from the roots of aquatic plants, microbial cultivation and isolation experiments were conducted. Homogenates of reed roots and Japanese loosestrife roots, and pond water were independently inoculated on low-nutrient medium plates and incubated at 25°C. The numbers of visible colonies continuously increased for more than 20 days. After 30 days of cultivation, maximum viable counts of 4.0×108±2.3×106 CFU g−1 (wet weight), 1.1×108±1.7×107 CFU g−1 (wet weight) and 9.7× 105±1.9×105 CFU mL−1 were obtained from reed, Japanese loosestrife, and pond water, respectively. Forty colonies were randomly selected from the medium plates with appropriate dilution (approx. 50 to 80 colonies on a plate), and their 16S rRNA genes were PCR-amplified and subjected to RFLP analysis. The isolates from the two plants were grouped into 45 phylotypes (hereafter referred to as I-phylotype (meaning isolate-phylotype) numbered 1 to 45) consisting of 22 for reed and 25 for Japanese loosestrife. By contrast, the isolates from pond water consisted of 11 I-phylotypes numbered 46 to 56 (Table 3). The phylogenetic relationships of the isolates and their closely related species were analyzed on the basis of partial 16S rRNA gene sequences (517 to 712 bp). The 16S rRNA gene sequences of the representative isolates for each I-phylotype were determined and compared with the sequences in the NCBI database. The most closely related species of the isolates are shown in Table 3. Betaproteobacteria and Alphaproteobacteria were the most abundant bacterial divisions in cultivation studies of reed roots and pond water, and in Japanese loosestrife roots, respectively, similar to the pattern in culture-independent analysis (Fig. 1B). In further taxonomic classification at the family level, the isolates from reed (33 isolates), Japanese loosestrife (32 isolates) and pond water (37 isolates) were divided into 7, 11 and 8 groups, respectively. The predominant bacterial families in reed and pond water were Burkholderiales genera incertae sedis for reed (47.5%) for reed and Comamonadaceae (40%) for pond water as found in culture-independent analysis; however, the predominant family in Japanese loosestrife was different between culture-dependent (Alcaligenaceae; 20%) and culture-independent (Sinobacteraceae; 12%) methods as described in previous studies in a wide variety of natural habitats (5, 6, 18, 21, 31).
Table 3.
Isolated microbes in this study and their related authentic species on the basis of 16S rRNA gene sequence
| Isolate- phylotype No.a | Isolate | Authentic species (Accession No.) | Identity (%) | Phylum (Class) | Length (bp) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Reed | Japanese loosestrife | Pond water | ||||||||
|
|
|
|
||||||||
| Total no. of isolates | Name of representative strain | Total no. of isolates | Name of representative strain | Total no. of isolates | Name of representative strain | |||||
| 1 | 5 | YO-23 | Rubrivivax gelatinosus strain ATCC17011 (D16213) | 97 | Proteobacteria (Beta) | 709 | ||||
| 2 | 6 | YO-32 | 1 | MI-15 | Aquincola tertiaricarbonis strain L10T (DQ656489) | 97 | Proteobacteria (Beta) | 711 | ||
| 3 | 5 | YO-28 | Methylibium fulvum strain Gsoil 328 (AB245357) | 97 | Proteobacteria (Beta) | 653 | ||||
| 4 | 1 | YO-5 | Azoarcus buckelii strain U120 (AJ315676) | 94 | Proteobacteria (Beta) | 711 | ||||
| 5 | 3 | YO-6 | Curvibacter delicatus strain LMG 4328 (AF078756) | 97 | Proteobacteria (Beta) | 696 | ||||
| 6 | 1 | YO-8 | 1 | MI-36 | Chelatovorus multitrophus strain DSM 9103T (EF457243) | 91 | Proteobacteria (Alpha) | 709 | ||
| 7 | 1 | YO-9 | Herbaspirillum seropedicae strain ATCC 35892 (Y10146) | 91 | Proteobacteria (Beta) | 710 | ||||
| 8 | 1 | YO-13 | Bosea thiooxidans strain BI-42 (AF508803) | 93 | Proteobacteria (Alpha) | 710 | ||||
| 9 | 1 | YO-14 | Rhodobacter ovatus strain JA234T (AM690348) | 98 | Proteobacteria (Alpha) | 709 | ||||
| 10 | 1 | YO-16 | Limnobacter thiooxidans strain CS-K2 (AJ289885) | 91 | Proteobacteria (Beta) | 697 | ||||
| 11 | 1 | YO-17 | Sandaracinobacter sibiricus strain RB16-17 (Y10678) | 97 | Proteobacteria (Alpha) | 710 | ||||
| 12 | 1 | YO-18 | Rhodobacter blasticus strain ATCC 33485T (DQ342322) | 98 | Proteobacteria (Alpha) | 709 | ||||
| 13 | 3 | YO-19 | Ideonella dechloratans (X72724) | 98 | Proteobacteria (Beta) | 710 | ||||
| 14 | 1 | YO-26 | Leptothrix discophora strain SS-1 (L33975) | 95 | Proteobacteria (Beta) | 517 | ||||
| 15 | 1 | YO-27 | Pseudomonas pohangensis strain H3-R18T (DQ339144) | 98 | Proteobacteria (Gamma) | 711 | ||||
| 16 | 1 | YO-29 | Paracoccus marcusii (Y12703) | 95 | Proteobacteria (Alpha) | 709 | ||||
| 17 | 1 | YO-33 | Nitrosococcus oceani strain ATCC 19707 (AY690336) | 87 | Proteobacteria (Gamma) | 708 | ||||
| 18 | 1 | YO-34 | Pelomonas aquatica strain CCUG 52575T (AM501435) | 97 | Proteobacteria (Beta) | 695 | ||||
| 19 | 1 | YO-36 | Desulfotomaculum halophilum strain SEBR 3139 (U88891; Fir-micutes) | 80 | Candidate phylum OP10 | 709 | ||||
| 20 | 1 | YO-38 | Novosphingobium hassiacum strain W-51 (AJ416411) | 97 | Proteobacteria (Alpha) | 711 | ||||
| 21 | 1 | YO-40 | Sinorhizobium adhaerens strain 5D19 (AJ420773) | 98 | Proteobacteria (Alpha) | 709 | ||||
| 22 | 2 | YO-45 | Novosphingobium resinovorum strain NCIMB 8767 (EF029110) | 94 | Proteobacteria (Alpha) | 712 | ||||
| 23 | 8 | MI-1 | Azohydromonas lata strain IAM 12665 (AB201626) | 97 | Proteobacteria (Beta) | 690 | ||||
| 24 | 3 | MI-2 | Rubrivivax gelatinosus strain ATCC 17011 (D16213) | 96 | Proteobacteria (Beta) | 696 | ||||
| 25 | 1 | MI-5 | Sphingobium japonicum strain UT26 (AF039168) | 97 | Proteobacteria (Alpha) | 712 | ||||
| 26 | 1 | MI-6 | Rhodobacter blasticus strain ATCC 33485T (DQ342322) | 98 | Proteobacteria (Alpha) | 712 | ||||
| 27 | 1 | MI-8 | Steroidobacter denitrificans strain FS (EF605262) | 93 | Proteobacteria (Gamma) | 707 | ||||
| 28 | 2 | MI-9 | Rhodobacter capsulatus strain ATCC 11166T (DQ342320) | 96 | Proteobacteria (Alpha) | 695 | ||||
| 29 | 1 | MI-10 | Niastella jeongjuensis strain GR20-13 (DQ244076) | 94 | Bacteroidetes | 710 | ||||
| 30 | 1 | MI-11 | Asticcacaulis biprosthecium strain DSM 4723T (AJ247193) | 98 | Proteobacteria (Alpha) | 710 | ||||
| 31 | 1 | MI-12 | Phenylobacterium lituiforme strain FaiI3 (AY534887) | 97 | Proteobacteria (Alpha) | 711 | ||||
| 32 | 1 | MI-13 | Novosphingobium hassiacum strain W-51 (AJ416411) | 97 | Proteobacteria (Alpha) | 711 | ||||
| 33 | 3 | MI-14 | Rhodoferax antarcticus strain Fryx1 (AY609198) | 98 | Proteobacteria (Beta) | 696 | ||||
| 34 | 3 | MI-16 | Hyphomicrobium hollandicum strain IFAM KB-677 (Y14303) | 93 | Proteobacteria (Alpha) | 660 | ||||
| 35 | 1 | MI-20 | Sphingomonas suberifaciens strain IFO 15211 (D13737) | 95 | Proteobacteria (Alpha) | 709 | ||||
| 36 | 1 | MI-26 | Caulobacter henricii strain ATCC 15253T (AJ227758) | 98 | Proteobacteria (Alpha) | 708 | ||||
| 37 | 1 | MI-30 | Methylosinus trichosporium strain NCIMB 11131 (Y18947) | 91 | Proteobacteria (Alpha) | 709 | ||||
| 38 | 1 | MI-31 | Novosphingobium resinovorum strain NCIMB 8767T (EF029110) | 96 | Proteobacteria (Alpha) | 709 | ||||
| 39 | 1 | MI-32 | Pseudodevosia insulae strain DS- 56T (EF012357) | 96 | Proteobacteria (Alpha) | 711 | ||||
| 40 | 1 | MI-33 | Acidimicrobium ferrooxidans strain DSM 10331 (AFU75647) | 90 | Actinobacteria | 712 | ||||
| 41 | 1 | MI-34 | Rhodopseudomonas cryptolactis strain DSM 9987 (AB087718) | 95 | Proteobacteria (Alpha) | 712 | ||||
| 42 | 1 | MI-35 | Bradyrhizobium elkanii strain USDA 76 (U35000) | 99 | Proteobacteria (Alpha) | 711 | ||||
| 43 | 1 | MI-37 | Steroidobacter denitrificans strain FS (EF605262) | 93 | Proteobacteria (Gamma) | 709 | ||||
| 44 | 2 | MI-40 | Devosia insulae strain DS-56T (EF012357) | 92 | Proteobacteria (Alpha) | 712 | ||||
| 45 | 1 | MI-39 | Afipia massiliensis strain CCUG 45153T (AY029562) | 93 | Proteobacteria (Alpha) | 710 | ||||
| 46 | 9 | KW-15 | Rhodobacter massiliensis strain Framboise (AF452106) | 98 | Proteobacteria (Alpha) | 709 | ||||
| 47 | 3 | KW-13 | Flavobacterium terrae strain R2A1-13T (EF117329) | 94 | Bacteroidetes | 711 | ||||
| 48 | 9 | KW-16 | Limnohabitans curvus strain MWH-C5 (AJ938026) | 98 | Proteobacteria (Beta) | 710 | ||||
| 49 | 7 | KW-29 | Limnohabitans curvus strain MWH-C5 (AJ938026) | 97 | Proteobacteria (Beta) | 710 | ||||
| 50 | 5 | KW-28 | Herbaspirillum rubrisubalbicans strain M4 (AJ238356) | 97 | Proteobacteria (Beta) | 710 | ||||
| 51 | 1 | KW-22 | Novosphingobium aromati-civorans strain SMCC B0695 (U20755) | 98 | Proteobacteria (Alpha) | 711 | ||||
| 52 | 1 | KW-39 | Pedobacter caeni strain R-21937 | 86 | Bacteroidetes | 711 | ||||
| 53 | 1 | KW-40 | Methylotenera mobila strain JLW8T (DQ287786) | 94 | Proteobacteria (Beta) | 712 | ||||
| 54 | 2 | KW-45 | Tetrasphaera nostocoidensis strain Ben 70 (DQ007320) | 91 | Actinobacteria | 709 | ||||
| 55 | 1 | KW-42 | Polynucleobacter necessarius strain STIR1 (AJ811014) | 99 | Proteobacteria (Beta) | 682 | ||||
| 56 | 1 | KW-43 | Chitinophaga ginsengisegetis strain Gsoil 040T (AB264798) | 91 | Bacteroidetes | 680 | ||||
|
| ||||||||||
| Total | 40 | 40 | ||||||||
|
| ||||||||||
| Novel microbesb | 14 (11) | 8 (5) | ||||||||
The phylotypes were defined on the basis of the results of restriction fragment length polymorphism (RFLP) analysis. The isolate-phylotypes whose sequences indicated less than 95% identity with those from authentic species are underlined.
The number of phylotypes showing phylogenetic novelty is shown in parentheses.
The scores of diversity indices based on the grouping of I-phylotypes indicated that the isolates obtained from two aquatic plant roots were more diverse than from pond water (Table 2). These results may be due to the diverse niche-associated occurrence of microbial communities on aquatic plant roots, as shown in culture-independent analysis.
Twenty I-phylotypes showing below 95% 16S rRNA gene sequence similarity (a criterion indicating novel microbes defined in culture-independent analysis; see above) to validly described species were obtained from two aquatic plants (10 I-phylotypes from reed and 11 I-phylotypes from Japanese loosestrife). By contrast, only 5 I-phylotypes showing phylogenetic novelty were recovered from pond water (Table 3, Fig. 2B). This also suggested that aquatic plants harbor more readily-cultured novel microbes in their root than their surrounding water environments.
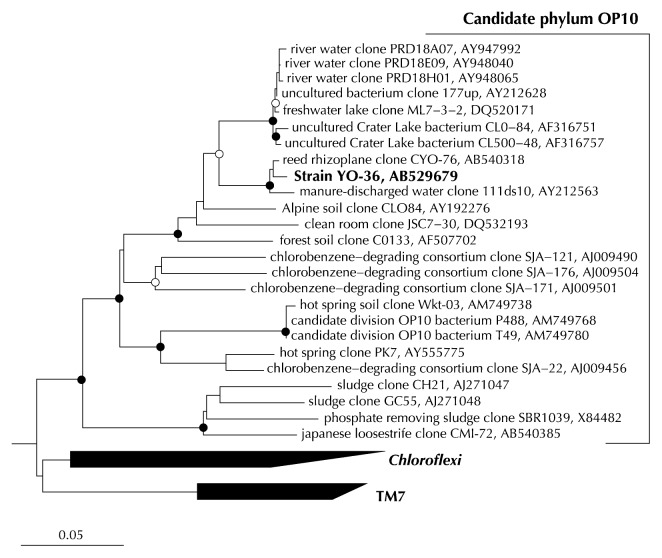
Most importantly, we succeeded in isolating a novel microorganism, strain YO-36 (I-phylotype No. 19), from the reed roots. Phylogenetic analysis based on 16S rRNA gene sequence (almost full length, >1,300 bp) clearly revealed that strain YO-36 belongs to the candidate phylum OP10 that includes clones CYO-76 (C-phylotype No. 62 from reed) and CMI-72 (C-phylotype No. 127 from Japanese loosestrife) obtained by direct molecular analysis of the samples. The highest sequence similarity of YO-36 was to clone CYO-76 (98.5%) and a previously described environmental clone (clone 111ds10 obtained from manure-discharged water) (34) (97.7%), whereas the identity with clone CMI-72 was fairly low (81.4%). Strain YO-36 showed significantly low sequence similarities (<80%) to any known validly described species but shared 90–91% similarities with some environmental clonal sequences derived from members of the candidate phylum OP10 (4, 22) (Table 3). Therefore, very recently, based on polyphasic taxonomic analyses (morphological, physiological and phylogenetic characterizations), we proposed a novel genus and species for this particular strain YO-36, Armatimonas rosea gen. nov., sp. nov., and also proposed a new phylum Armatimonadetes for the candidate phylum OP10 and a new class Armatimonadia within the phylum (42).
Concluding remarks
In this study, we clearly showed that the roots of aquatic plants harbor unique, diverse, and novel microbes. Moreover, we also succeeded in the cultivation and isolation of a diverse array of phylogenetically novel microbes (20 I-phylotypes from the roots), including an isolate within the candidate phylum OP10 by employing relatively long cultivation (30 days) using a low-nutrient agar plate medium that does not require any unique reagents, organisms and materials that have been used for the isolation of rarely cultivated groups in other studies (2, 3, 12, 15, 16, 27, 29, 33, 37, 40, 43, 51). In our previous study, a variety of novel microbes belonging to the rarely cultured phylum Verrucomirobia were successfully isolated from the roots of a floating aquatic plant, S. polyrrhiza, with no vigorous efforts, as was the case in this study. Together with the previous findings, the present study clearly demonstrated that the roots of aquatic plants can be highly intriguing sources of novel organisms. Functional analyses of these organisms would provide further insights into the interactions occurring between microbes and aquatic plants. Further study will also reveal the microbial community structure and diversity in aquatic plant roots from the natural environment as well as those from the floating treatment wetland system via advanced techniques with 16S pyrosequencing technology.
Supporting information
Fig. 3.
Phylogenetic tree showing the relationship between strain YO-36 and related sequences of the candidate phylum OP10. The tree was constructed using the neighbor-joining (NJ) method on the basis of the 16S rRNA gene sequences. Node with bootstrap values of >90% and >80%, estimated using the NJ method and maximum likelihood (ML) methods, are shown as a closed circle and an open circle, respectively. The tree was rooted against a group of 26 sequences belonging to different phyla in the domains Bacteria and Archaea. Scale bar indicates 0.05 substitutions per nucleotide position.
Acknowledgements
We are grateful to Ms. Kiyo Oishi (University of Yamanashi) for her experimental assistance. We thank Yamanashi Prefectural Wood Park at Kanegawa and the Society for the Research of Pro-Natural Civil Engineering Method of Yamanashi for their help in the cultivation of aquatic plants. We are also grateful to Dr. Cindy H. Nakatsu (Pardue University) for critically reading the manuscript. This research was partly supported by Japan Science and Technology Agency (JST), and National Natural Science Foundation of China (NSFC) project as the Japan-China Joint Research Program on “Science and Technology (S&T) for Environmental Conservation and Construction of a Society with Less Environmental Burden”.
References
- 1.Bodelier PLE, Libochant JA, Blom CWPM, Laanbroek HJ. Dynamics of nitrification and denitrification in root-oxygenated sediments and adaptation of ammonia-oxidizing bacteria to low oxygen or anoxic habitats. Appl Environ Microbiol. 1996;62:4100–4107. doi: 10.1128/aem.62.11.4100-4107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollman A, Lewis K, Epstein SS. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol. 2007;73:6386–6390. doi: 10.1128/AEM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruns A, Cypionka H, Overmann J. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the Central Baltic Sea. Appl Environ Microbiol. 2002;68:3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crump BC, Hobbie JE. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol Oceanogr. 2005;50:1718–1729. [Google Scholar]
- 5.Dunbar J, Takala S, Barns SM, Davis JA, Kuske CR. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol. 1999;65:1662–1669. doi: 10.1128/aem.65.4.1662-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis RJ, Morgan P, Weightman AJ, Fry JC. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol. 2003;69:3223–3230. doi: 10.1128/AEM.69.6.3223-3230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garbett P. An investigation into the application of floating reed bed and barley straw techniques for the remediation of eutrophic waters. Water Environ J. 2005;19:174–180. [Google Scholar]
- 8.Gilbert GS, Clayton MK, Handelsman J, Parke JL. Use of cluster and discriminant analyses to compare rhizosphere bacterial communities following biological perturbation. Microb Ecol. 1996;32:123–147. doi: 10.1007/BF00185884. [DOI] [PubMed] [Google Scholar]
- 9.Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 10.Grayston SJ, Campbell CD. Functional biodiversity of microbial communities in the rhizospheres of hybrid larch (Larix eurolepis) and Sitka spruce (Picea sitchensis) Tree Physiol. 1996;16:1031–1038. doi: 10.1093/treephys/16.11-12.1031. [DOI] [PubMed] [Google Scholar]
- 11.Grayston SJ, Wang S, Campbell CD, Edwards AC. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem. 1998;30:369–378. [Google Scholar]
- 12.Guan LL, Onuki H, Kamino K. Bacterial growth stimulation with exogenous siderophore and synthetic N-acyl homoserine lactone autoinducers under iron-limited and low-nutrient conditions. Appl Environ Microbiol. 2000;66:2797–2803. doi: 10.1128/aem.66.7.2797-2803.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann M, Saunders AM, Schramm A. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediment. Appl Environ Microbiol. 2009;75:3127–3136. doi: 10.1128/AEM.02806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard RK, Gascho GJ, Newton GL. Use of floating vegetation to remove nutrients from swine lagoon wastewater. Trans ASAE. 2004;47:1963–1972. [Google Scholar]
- 15.Isawa K, Hojo K, Yoda N, et al. Isolation and identification of a new bifidogenic growth stimulator produced by Propionibacterium freudenreichii ET-3. Biosci Biotechnol Biochem. 2002;66:679–681. doi: 10.1271/bbb.66.679. [DOI] [PubMed] [Google Scholar]
- 16.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol. 2002;68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Kaiser O, Pühler A, Selbitschka W. Phylogenetic analysis of microbial siversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb Ecol. 2001;42:136–149. doi: 10.1007/s002480000121. [DOI] [PubMed] [Google Scholar]
- 19.Kane MD, Poulsen LK, Stahl DA. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligo-nucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr-Upal M, Seasons M, Mulamoottil G. Retrofitting a stormwater management facility with a wetland component. J. Environ. Sci. Health. 2000;A35:1289–1307. [Google Scholar]
- 21.Kisand V, Wikner J. Combining culture-dependent and -independent methodologies for estimation of richness of estuarine bacterioplankton consuming riverine dissolved organic matter. Appl Environ Microbiol. 2003;69:3607–3616. doi: 10.1128/AEM.69.6.3607-3616.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipson DA, Schmidt SK. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Appl Environ Microbiol. 2004;70:2867–2879. doi: 10.1128/AEM.70.5.2867-2879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig W, Strunk O, Westram R, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch JM. Introduction: Some consequences of microbial rhizosphere complete for plant and soil. In: Lynch JM, editor. The Rhizosphere. John Wiley & Sons; Chichester, UK: 1990. pp. 1–10. [Google Scholar]
- 25.Malla R, Nagao N, Maezono K, Tanaka Y, Mori K. Formulation of a simple mathematical biomass model for selected floating and emergent macrophytes. Jpn J Water Treat Biol. 2010;46:1–15. [Google Scholar]
- 26.Manoharachary C, Mukerji K. Rhizosphere biology—an overview. In: Mukerji KG, Manoharachary C, Singh J, editors. Microbial Activity in the Rhizosphere. Springer-Verlag; Berlin Heidelberg, Germany: 2006. pp. 1–15. [Google Scholar]
- 27.Manome A, Zhang H, Tani Y, Katsuragi T, Kurane R, Tsuchida T. Application of gel microdroplet and flow cytometry techniques to selective enrichment of non-growing bacterial cells. FEMS Microbiol Lett. 2001;197:29–33. doi: 10.1111/j.1574-6968.2001.tb10578.x. [DOI] [PubMed] [Google Scholar]
- 28.Matsuzawa H, Tanaka Y, Tamaki H, Kamagata Y, Mori K. Culture-dependent and independent analyses of the microbial communities inhabiting the giant duckweed (Spirodera polyrrhiza) rhizoplane and isolation of a variety of rarely cultured organisms within the phylum Verrucomicrobia. Microbes Environ. 2010;25:302–308. doi: 10.1264/jsme2.me10144. [DOI] [PubMed] [Google Scholar]
- 29.Ohno M, Okano I, Watsuji T, Kakinuma T, Ueda K, Beppu T. Establishing the independent culture of a strictly symbiotic bacterium Symbiobacterium thermophilum from its supporting Bacillus strain. Biosci Biotechnol Biochem. 1999;63:1083–1090. doi: 10.1271/bbb.63.1083. [DOI] [PubMed] [Google Scholar]
- 30.Ottosen LDM, Risgaard-Petersen N, Nielsen LP. Direct and indirect measurements of nitrification and denitrification in the rhizosphere of aquatic macrophytes. Aquat Microb Ecol. 1999;19:81–91. [Google Scholar]
- 31.Pearce DA, Gast CJ, Lawley B, Ellis-Evans JC. Bacterioplankton community diversity in a maritime Antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS Microbiol Ecol. 2003;45:59–70. doi: 10.1016/S0168-6496(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 32.Pepper IL. Beneficial and pathogenic microbes in agriculture. In: Maier RM, Pepper IL, Gerba CP, editors. Environmental Microbiology. Academic Press; San Diego, USA: 2000. pp. 425–446. [Google Scholar]
- 33.Sangwan P, Kovac S, Davis KER, Sait M, Janssen PH. Detection and cultivation of soil verrucomicrobia. Appl Environ Microbiol. 2005;71:8402–8410. doi: 10.1128/AEM.71.12.8402-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson JM, Santo Domingo JW, Reasoner DJ. Assessment of equine fecal contamination: the search for alternative bacterial source-tracking targets. FEMS Microbiol Ecol. 2004;47:65–75. doi: 10.1016/S0168-6496(03)00250-2. [DOI] [PubMed] [Google Scholar]
- 35.Smalla K, Wieland G, Buchner A, et al. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol. 2001;67:4742–4751. doi: 10.1128/AEM.67.10.4742-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Söderberg KH, Olsson PA, Baath E. Structure and activity of the bacterial community in the rhizosphere of different plant species and the effect of arbuscular mycorrhizal colonisation. FEMS Microbiol Ecol. 2002;40:223–231. doi: 10.1111/j.1574-6941.2002.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol. 2004;70:4748–4755. doi: 10.1128/AEM.70.8.4748-4755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stout L, Nüsslein K. Biotechnological potential of aquatic-microbe interactions. Curr Opin Biotechnol. 2010;21:339–345. doi: 10.1016/j.copbio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Swofford DL. PAUP*-Phylogenetic Analysis Using Parsimony and Other Methods, Version 4. Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- 40.Tamaki H, Hanada S, Sekiguchi Y, Tanaka Y, Kamagata Y. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ Microbiol. 2009;11:1827–1834. doi: 10.1111/j.1462-2920.2009.01907.x. [DOI] [PubMed] [Google Scholar]
- 41.Tamaki H, Sekiguchi Y, Hanada S, Nakamura K, Nomura N, Matsumura M, Kamagata Y. Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl Environ Microbiol. 2005;71:2162–2169. doi: 10.1128/AEM.71.4.2162-2169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamaki H, Tanaka Y, Matsuzawa H, Muramatsu M, Meng XY, Hanada S, Mori K, Kamagata Y. Armatimonas rosea gen. nov., sp. nov., a Gram-negative, aerobic, chemoheterotrophic bacterium of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10. Int J Syst Evol Microbiol. 2011;61:1442–1447. doi: 10.1099/ijs.0.025643-0. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka Y, Hanada S, Manome A, Tsuchida T, Kurane R, Nakamura K, Kamagata Y. Catellibacterium nectariphilum gen. nov., sp. nov., which requires a diffusible compound from a strain related to the genus Sphingomonas for vigorous growth stimulation. Int J Syst Evol Microbiol. 2004;54:955–959. doi: 10.1099/ijs.0.02750-0. [DOI] [PubMed] [Google Scholar]
- 44.Todd J, Brown EJG, Wells E. Ecological design applied. Ecol Engineer. 2003;20:421–440. [Google Scholar]
- 45.Toyama T, Sato Y, Inoue D, Sei K, Chang YC, Kikuchi S, Ike M. Biodegradation of bisphenol A and bisphenol F in the rhizosphere sediment of Phragmites australis. J Biosci Bioeng. 2009;108:147–150. doi: 10.1016/j.jbiosc.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Toyama T, Yu N, Kumada H, Sei K, Ike M, Fujita M. Accelerated aromatic compounds degradation in aquatic environment by use of interaction between Spirodela polyrrhiza and bacteria in its rhizosphere. J Biosci Bioeng. 2006;101:346–353. doi: 10.1263/jbb.101.346. [DOI] [PubMed] [Google Scholar]
- 47.Uhlik O, Jecna K, Mackova M, Vlcek C, Hroudova M, Demnerova K, Paces V, Macek T. Biphenyl-metabolizing bacteria in the rhizosphere of horseradish and bulk soil contaminated by polychlorinated biphenyls as revealed by stable isotope probing. Appl Environ Microbiol. 2009;75:6471–6477. doi: 10.1128/AEM.00466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weisburg WG, Burns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wieland G, Neumann R, Backhaus H. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl Environ Microbiol. 2001;67:5849–5854. doi: 10.1128/AEM.67.12.5849-5854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaga F, Washio K, Morikawa M. Sustainable biodegradation of phenol by Acinetobacter calcoaceticus P23 isolated from the rhizosphere of duckweed Lemna aoukikusa. Environ Sci Technol. 2010;44:6470–6474. doi: 10.1021/es1007017. [DOI] [PubMed] [Google Scholar]
- 51.Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M. Cultivating uncultured. Proc. Natl. Acad. Sci USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.