Abstract
Ammonia monooxygenase subunit A gene (amoA) is frequently used as a functional gene marker for diversity analysis of ammonia-oxidizing bacteria (AOB). To select a suitable amoA primer for real-time PCR and PCR-denaturing gradient gel electrophoresis (DGGE), three reverse primers (degenerate primer amoA-2R; non-degenerate primers amoA-2R-GG and amoA-2IR) were examined. No significant differences were observed among the three primers in terms of quantitative values of amoA from environmental samples using real-time PCR. We found that PCR-DGGE analysis with the amoA-2IR primer gave the best results in this studied soil. These results indicate that amoA-2IR is a suitable primer for community analysis of AOB in the environment.
Keywords: ammonia-oxidizing bacteria (AOB), amoA primer, real-time PCR, DGGE, primer degeneracy
Chemolithoautotrophic ammonia-oxidizing bacteria(AOB) play an important role in the global cycling of nitrogen (11). AOB convert ammonia (NH3) to nitrite (NO2−) through nitrification, which consists of the following two steps: oxidation of NH3 to hydroxylamine (NH2OH) by ammonia monooxygenase (AMO) (14) and conversion of NH2OH to NO2− by hydroxylamine oxidoreductase (13, 15). During nitrification, nitrous oxide (N2O), a major greenhouse gas emitted from agricultural fields, is produced by AOB via two processes (33); chemical decomposition of intermediates such as nitrosyl hydride produced during oxidation of NH2OH to NO2−(16) and nitrifier denitrification (24, 27), in which N2O is produced during reduction of NO2−(24, 27) with NH2OH as an electron donor (25). Fertilization of agricultural fields increases N2O emissions (3); however, the extent of AOB contribution and the species involved in N2O emissions in fields are not yet known because it is difficult to isolate individual bacterial species from the environment (1). Therefore, community analysis using PCR-based methods is important to understand the ecology of AOB in the environment.
A gene for subunit A of AMO (amoA) is frequently used as the functional marker of AOB in PCR-based methods for diversity analysis of AOB (26). Rotthauwe et al.(26) constructed a primer set consisting of a single primer amoA-1F and a degenerate primer amoA-2R, having degeneracy at position 7 (K=G or T) and 9 (S=C or G) from the 5′ end (Table 1). The primer set amplified amoA fragments of Nitrosomonas and Nitrosospira belonging to the β subclass of Proteobacteria(26). Oved et al.(23) developed a PCR-denaturing gradient gel electrophoresis (DGGE) method using amoA-1F attached to the GC-clump at the 5′ end (amoA-1F-GC) (Table 1) and the amoA-2R primer set. Furthermore, they identified its usefulness for community analysis of AOB. The amoA-1F(amoA-1F-GC)/amoA-2R primer set has frequently been used for diversity analysis of AOB (4, 22, 23); however, degeneracy of the amoA-2R primer results in smears and multiple DGGE bands (7, 17). Therefore, several studies have evaluated the usefulness of non-degenerate primers that correspond to the same region as amoA-2R (7, 8, 17, 20).
Table 1.
PCR primers used in this study for the amplification of amoA gene fragments
| Primer | Sequence (5′-3′) | Tm (°C) | Reference | |
|---|---|---|---|---|
| Forward | amoA-1F | GGGGTTTCTACTGGTGGT | 46.5 | (26) |
| amoA-1F-GC* |
CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCC CGGGGGTTTCTACTGGTGGT |
84.2 | (26, 28) | |
| Reverse | amoA-2R | CCCCTCKGSAAAGCCTTCTTC (K=G or T, S=G or C) | 53.5 | (26) |
| amoA-2R-GG | CCCCTCGGGAAAGCCTTCTTC | 54.4 | (20) | |
| amoA-2IR | CCCCTCIGIAAAGCCTTCTTC | 50.5 | (7) |
GC-clump attached forward primer, amoA-1F-GC, was used in PCR-DGGE.
In amoA-2R-GG (20) and amoA-2IR (7) primers, the degenerate regions of amoA-2R are replaced by guanines and inosines, respectively (Table 1). amoA-2R-GG and amoA-2IR reportedly exhibit single-band patterns in DGGE (6, 9). Furthermore, amoA-2R-GG produces DGGE patterns similar to those of amoA-2R (7, 8). Although amoA-2IR has the potential to detect more diverse amoA sequences than normal base primers, the primer has not been used in community analysis of AOB in the environment, including soil. In this study, three primers, amoA-2R, amoA-2R-GG, and amoA-2IR, were used to analyze the quantitative capability and detectability of amoA in soil samples by real-time PCR and PCR-DGGE.
Soil samples were collected from experimental lysimeter plots, wherein N2O fluxes were monitored using an automated chamber system (2, 21), at the National Institute for Agro-Environmental Sciences, Tsukuba, Ibaraki prefecture in April, 2010. Two types of soil, gray lowland and andosol, were used in this study. In Japan, these are the most common soil types in agricultural fields. Gray lowland soil and andosol account for 22% and 18%, respectively, of all agricultural fields, including paddy fields, upland crop fields, grassland, and orchards (31). In particular, andosol, which is volcanic-ash soil, occupies 41% of the total upland crop fields in Japan (31). Gray lowland soil has the following properties: moisture, 26% (w/w); NH4-N, 1.87 mg kg−1; NO3-N, 0.55 mg kg−1; pH (H2O), 6.4; total C, 17.3 g kg−1; total N, 1.5 g kg−1; exchangeable K, 1.21 cmol(+) kg−1; and available P, 113 mg kg−1. Andosol has the following properties: moisture, 28% (w/w); NH4-N, 1.72 mg kg−1; NO3-N, 2.21 mg kg−1; pH (H2O), 6.3; total C, 31.6 g kg−1; total N, 2.6 g kg−1; exchangeable K, 1.11 cmol(+) kg−1; and available P, 211 mg kg−1. Five soil cores (diameter, 3 cm; depth, 5 cm) from each site were collected, pooled, and sieved (2 mm). Total community DNA was extracted from soil in triplicate using the FastDNA spin kit for soil (Q-Biogene/MP Biomedicals, Solon, OH, USA) as described previously (19). During extraction of DNA from andosol, 16 mg skim milk was added to prevent DNA adsorption and improve recovery (19, 30).
Real-time PCR was performed to measure the amplification efficiency of the three primer sets shown in Table 1(12). Four types of amoA clones, including the complementary sequence of amoA-2R, were constructed as templates (Fig. S1). Nitrosospira multiformis ATCC 25196 amoA (Nmul_ A2765) was used for the construction of clones with the four primer sets shown in Table S1. The PCR reaction mixture compositions and thermocycling conditions are shown in Table S2. The PCR reactions were performed using the iCycler thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). Each amplicon was purified using the QIAquick PCR Purification kit (Qiagen, Valencia, CA, USA), inserted into a pGEM-T Easy vector (Promega, Madison, WI, USA), and transformed to Escherichia coli strain DH5α (Toyobo, Tokyo, Japan). Plasmid DNA was extracted using the QIAprep MiniPrep kit (Qiagen) and sequenced using the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems/Life Technologies, Carlsbad, CA, USA) and the BigDye Terminator v3.1 DNA sequencing kit (Applied Biosystems). The plasmids were linearized using HincII restriction enzyme, purified, and diluted in distilled water. DNA concentrations were measured using a spectrophotometer (Nanodrop 1000; Thermo Scientific, Wilmington, DE, USA), and then amoA copy number in the solution was calculated based on the determined DNA concentration and the molecular weight of the linearized plasmid. Serial dilutions containing between 103 and 107amoA copies were prepared from the solution of the linearized plasmid. Real-time PCR was performed using the StepOne Real-Time PCR System (Applied Biosystems) by the SYBR Green I method. The PCR conditions are shown in Table S2. Standard curves based on serial dilutions of each constructed clone with the three primer sets (Table 1) were generated by plotting the threshold cycle for each standard calculated by StepOne software, ver. 2.1 (Applied Biosystems).
amoA copy numbers in the extracted soil DNA were measured in triplicate with each of the three primer sets shown in Table 1. The mixed clones were serially diluted as described above to obtain the standard curve. Tukey’s test was used to evaluate the significance of differences of amoA copy numbers obtained from each primer.
Primer detection sensitivity for amoA from soil DNA was examined by PCR-DGGE. PCR was performed with each of the three primer sets shown in Table 1 and the extracted soil DNA. The composition of PCR reaction mixtures and the thermal cycling conditions are shown in Table S2. To optimize the annealing temperatures, four annealing temperatures, from 52°C to 58°C in increments of 2°C, were used to examine changes in the band patterns (Table S2). All amplicons were purified and adjusted to 200 ng DNA per well and used for DGGE (n=2), which was performed using the DCode universal mutation detection system (Bio-Rad Laboratories). An 8% (w/v) polyacrylamide gel with a denaturant gradient ranging from 50% to 65% was prepared. The procedures of electrophoresis, gel staining, and gel imaging were the same as those described previously (9). The detected major DGGE bands were excised, re-amplified by PCR, and sequenced according to the methods described previously (8). All sequences obtained in this study were examined for chimeric sequences as described previously (18). The band sequences and obtained amoA sequences from the National Center for Biotechnology Information (NCBI) were translated into amino acid sequences and aligned using Clustal W (32). The neighbor-joining tree was constructed using MEGA, version 5.0 (Molecular Evolutionary Genetics Analysis [http://www.megasoftware.net/]) with the Jones–Taylor–Thornton amino acid substitution model.
The effect of primer mismatches on amplification efficiency was examined by real-time PCR using the constructed amoA clones, including the complementary sequences with amoA-2R. The standard curves obtained from each clone amplified by each of the three primer sets showed similar slope values (Table S3). The calculated amplification efficiencies of amoA-2R, amoA-2R-GG, and amoA-2IR ranged from 91.3% to 97.6% (mean: 95.0%), 93.0% to 96.7% (mean: 95.0%), and 91.1% to 94.7% (mean: 93.0%), respectively (Table S3). The mean efficiency of amoA-2IR was slightly lower than that of the amoA-2R and amoA-2R-GG primers. PCR biases derived from primer mismatches were not detected when complementary amoA sequences of amoA-2R were used as templates.
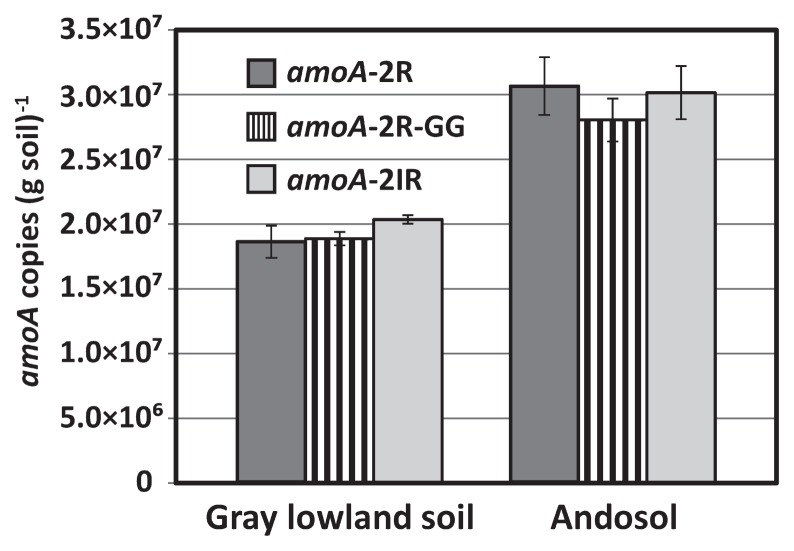
No significant differences were observed among the three primer sets in terms of quantitative values of amoA copy numbers in gray lowland soil (1.86–2.03×107 copies [g dry soil]−1) and andosol (2.80–3.06×107 copies [g soil]−1) (Fig. 1). Even though amplification efficiencies differed among the primers, each quantitative value was apparently corrected by each standard curve.
Fig. 1.
Abundance of amoA gene in gray lowland soil and andosol detected by each primer. Error bar designates SD for n=3.
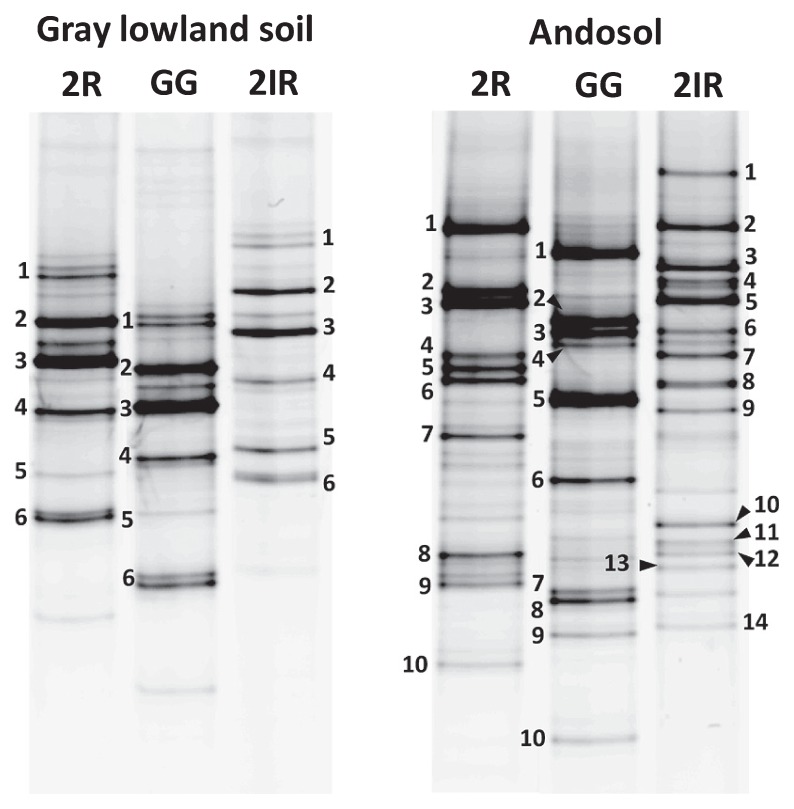
To optimize the annealing temperature for PCR-DGGE, four annealing temperatures (52, 54, 56, and 58°C) were examined. At 58°C, amoA-2R and amoA-2R-GG exhibited good amoA detectability in soil samples (Fig. S2); amoA-2IR produced more bands at 52°C than at 54°C or higher (Fig. S2). The band patterns of the soil samples observed under previously described annealing conditions are shown in Fig. 2. The patterns obtained from the three primer sets for gray lowland soil were similar, whereas the band migration distances varied with the primers. The distance variances were apparently caused by differences in the primer sequences. The band pattern obtained from andosol was more complex than that from gray lowland soil (Fig. 2). The band patterns of amoA-2R and amoA-2R-GG obtained from andosol were similar. In contrast, amoA-2IR produced a different band pattern (Fig. 2). The number of bands obtained from amoA-2IR was 14, and it was greater than 10 from amoA-2R and amoA-2R-GG.
Fig. 2.
DGGE band patterns of amoA retrieved from gray lowland soil and andosol. Band patterns were produced at an annealing temperature of 58°C (amoA-2R and amoA-2R-GG) or 52°C (amoA-2IR) (Fig. S3). Numbered bands were sequenced. 2R, amoA-2R primer; GG, amoA-2R-GG primer; 2IR, amoA-2IR primer.
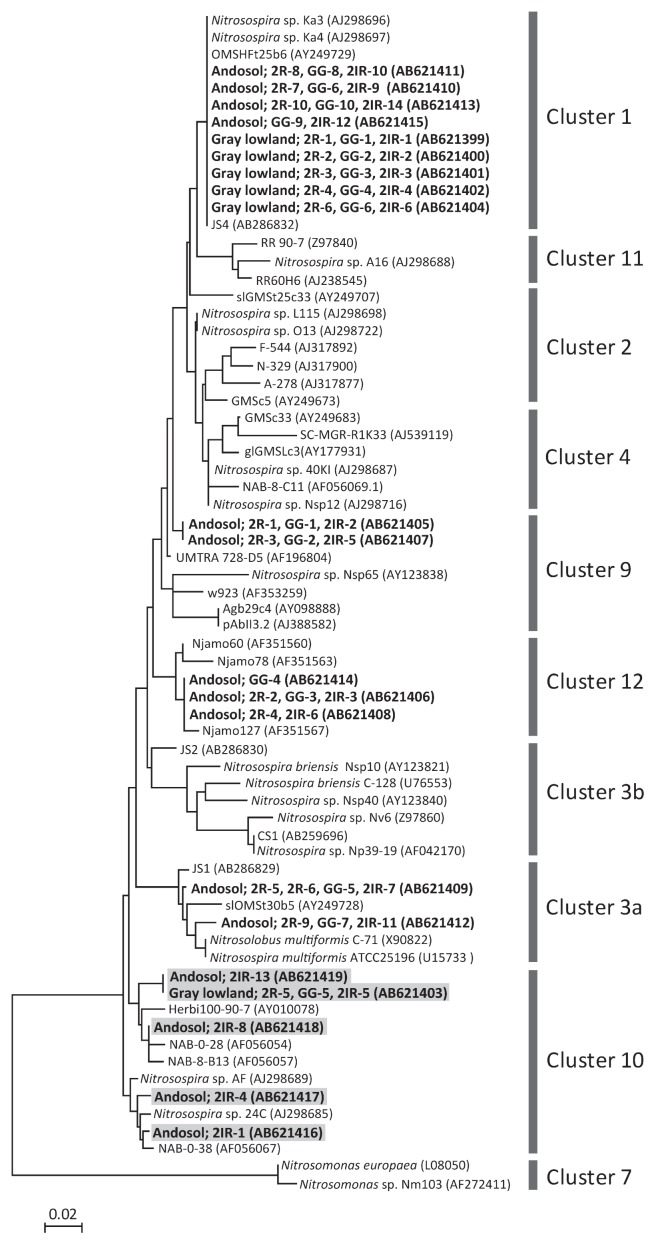
All bands to which a number was assigned in Fig. 2 were sequenced and identified as amoA fragments. Chimeric sequences were not detected. In the band pattern obtained from gray lowland soil, all bands in each lane were found to exhibit identical nucleotide sequences (Fig. 3). No difference in detectable sequences was observed among the three primers from gray lowland soil samples. In the band pattern obtained from andosol, eight bands in each lane were found to exhibit identical nucleotide sequences (Fig. 3). Sequences retrieved from amoA-2R band 5 (Andosol; 2R-5) and 6 (2R-6) had the same nucleotide sequence (Fig. 3), which seemed to be caused by amoA-2R degeneracy. amoA-2IR detected not only almost all of the amoA sequences obtained by amoA-2R and amoA-2R-GG, but also several unique sequences (Fig. 3). The unique sequences were assigned to cluster 10 on the basis of translated amino acid sequences of amoA, and were classified using the previously described (5–7) nomenclature for Nitrosospira amoA (Fig. 3). Other researchers reported that cluster 10-related amoA sequences were difficult to detect from a mixed clone library of various amoA sequences using the amoA-1F/amoA-2R primer set (5, 7). amoA-2R and amoA-2R-GG each detected only one sequence (retrieved from gray lowland bands; 2R-5 and GG-5, respectively), which was the same nucleotide sequence with 2IR-5 assigned to cluster 10; however, their bands were not as clear as those from 2IR-5 (Fig. 2). These results indicate that DGGE analysis with amoA-2IR can reproduce the pattern of the amoA sequence in this studied soil environment with higher sensitivity than amoA-2R and amoA-2R-GG. Because inosine is able to form stable pairs with both cytosine and thymine (10), the amplification bias may not be caused by amoA-2IR.
Fig. 3.
Phylogenetic tree based on partial amoA sequences (150 amino acids) retrieved from gray lowland soil and andosol in this study and obtained from NCBI. 2R, GG, and 2IR refer to the used primers: amoA-2R, amoA-2R-GG, and amoA-2IR, respectively. Subsequent numbers correspond to the DGGE bands shown in Fig. 2. Scale bar indicates two changes per 100 amino acid positions.
Avrahami et al.(7) compared the detectability of amoA genes among the three primers used in this study with 8 amoA clones. They reported that detectability with amoA-2IR was less than that with amoA-2R and amoA-2R-GG. In this study, we used total soil DNA samples to evaluate the primers and found that amoA-2IR detected several unique sequences from andosol at low annealing temperature. The low annealing temperature may increase the amoA detectability of amoA-2IR (Fig. S2) because the Tm value of amoA-2IR was 50.5°C, which was lower than that of amoA-2R (53.5°C) and amoA-2R-GG (54.4°C). Hornek et al.(17) reported that amoAr-I, which included inosine at position 13 from the 5′ end of amoA-2IR, detected Nitrosomonas-amoA sequences in environmental samples that were different from those detected by amoA-2R. In this study, Nitrosomonas-like amoA sequences were not detected from all samples. The population of Nitrosomonas species appeared to be lower than the detection limit in the soil samples. Nitrosospira-like amoA sequences have been shown to be more common and abundant than those from Nitrosomonas in soil environments (29).
In our study, amoA-2IR showed the same quantitative capability and superior detection sensitivity of amoA sequences as amoA-2R from soil samples compared with amoA-2R and amoA-2R-GG primers. Our results indicated that amoA-2IR is a suitable primer for community analysis of AOB in soil, as determined by both real-time PCR and PCR-DGGE. Our results also indicate that amoA-2IR is more useful for the analysis of AOB in andosol.
The sequences obtained in this study were deposited in the DNA Data Bank of Japan under accession numbers AB621399 to AB621419.
Supplementary Material
Acknowledgements
This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) from the Bio-oriented Technology Research Advancement Institution, Tokyo, Japan. Additional financial support was provided by the Funding Program for Next Generation World-Leading Researchers, Japan Society for the Promotion of Science.
References
- 1.Aakra Å, Utåker JB, Nes IF, Bakken LR. An evaluated improvement of the extinction dilution method for isolation of ammonia-oxidizing bacteria. J. Microbiol Methods. 1999;39:23–31. doi: 10.1016/s0167-7012(99)00094-9. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama H, Tsuruta H, Watanabe T. N2O and NO emissions from soils after the application of different chemical fertilizers. Chemosphere-Global Change Science. 2000;2:313–320. [Google Scholar]
- 3.Akiyama H, Yan XY, Yagi K. Estimations of emission factors for fertilizer-induced direct N2O emissions from agricultural soils in Japan: Summary of available data. Soil Sci Plant Nutr. 2006;52:774–787. [Google Scholar]
- 4.Ando Y, Nakagawa T, Takahashi R, Yoshihara K, Tokuyama T. Seasonal changes in abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria and their nitrification in sand of an eelgrass zone. Microbes Environ. 2009;24:21–27. doi: 10.1264/jsme2.me08536. [DOI] [PubMed] [Google Scholar]
- 5.Avrahami S, Conrad R. Cold-temperate climate: a factor for selection of ammonia oxidizers in upland soil? Can J Microbiol. 2005;51:709–714. doi: 10.1139/w05-045. [DOI] [PubMed] [Google Scholar]
- 6.Avrahami S, Conrad R, Braker G. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl Environ Microbiol. 2002;68:5685–5692. doi: 10.1128/AEM.68.11.5685-5692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avrahami S, Liesack W, Conrad R. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ Microbiol. 2003;5:691–705. doi: 10.1046/j.1462-2920.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- 8.Chu H, Fujii T, Morimoto S, Lin X, Yagi K, Hu J, Zhang J. Community structure of ammonia-oxidizing bacteria under long-term application of mineral fertilizer and organic manure in a sandy loam soil. Appl Environ Microbiol. 2007;73:485–491. doi: 10.1128/AEM.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu HY, Fujii T, Morimoto S, Lin XG, Yagi K. Population size and specific nitrification potential of soil ammonia-oxidizing bacteria under long-term fertilizer management. Soil Biol Biochem. 2008;40:1960–1963. [Google Scholar]
- 10.Crick FH. Codon–anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 11.Hayatsu M, Tago K, Saito M. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nutr. 2008;54:33–45. [Google Scholar]
- 12.Henry S, Bru D, Stres B, Hallet S, Philippot L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol. 2006;72:5181–5189. doi: 10.1128/AEM.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofman T. The biochemistry of the nitrifying organisms. 3. Composition of Nitrosomonas. Biochem J. 1953;54:293–295. doi: 10.1042/bj0540293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollocher TC, Tate ME, Nicholas DJ. Oxidation of ammonia by Nitrosomonas europaea: definite 18O-tracer evidence that hydroxylamine formation involves a monooxygenase. J Biol Chem. 1981;256:10834–10836. [PubMed] [Google Scholar]
- 15.Hooper AB, Maxwell PC, Terry KR. Hydroxylamine oxidoreductase from Nitrosomonas: absorption spectra and content of heme and metal. Biochemistry. 1978;17:2984–2989. doi: 10.1021/bi00608a007. [DOI] [PubMed] [Google Scholar]
- 16.Hooper AB, Terry KR. Hydroxylamine oxidoreductase of Nitrosomonas: Production of nitric oxide from hydroxylamine. Biochim. Biophys Acta (BBA)-Enzymology. 1979;571:12–20. doi: 10.1016/0005-2744(79)90220-1. [DOI] [PubMed] [Google Scholar]
- 17.Hornek R, Pommerening-Röser A, Koops HP, Farnleitner AH, Kreuzinger N, Kirschner A, Mach RL. Primers containing universal bases reduce multiple amoA gene specific DGGE band patterns when analysing the diversity of beta-ammonia oxidizers in the environment. J. Microbiol Methods. 2006;66:147–155. doi: 10.1016/j.mimet.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino YT, Morimoto S. Soil clone library analyses to evaluate specificity and selectivity of PCR primers targeting fungal 18S rDNA for denaturing-gradient gel electrophoresis (DGGE) Microbes Environ. 2010;25:281–287. doi: 10.1264/jsme2.me10136. [DOI] [PubMed] [Google Scholar]
- 19.Morimoto S, Ogawa N, Hasebe A, Fuji T. Isolation of effective 3-chlorobenzoate-degraders in soil using community analyses by PCR-DGGE. Microbes Environ. 2008;23:285–292. doi: 10.1264/jsme2.me08526. [DOI] [PubMed] [Google Scholar]
- 20.Nicolaisen MH, Ramsing NB. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol Methods. 2002;50:189–203. doi: 10.1016/s0167-7012(02)00026-x. [DOI] [PubMed] [Google Scholar]
- 21.Nishimura S, Sudo S, Akiyama H, Yonemura S, Yagi K, Tsuruta H. Development of a system for simultaneous and continuous measurement of carbon dioxide, methane and nitrous oxide fluxes from croplands based on the automated closed chamber method. Soil Sci Plant Nutr. 2005;51:557–564. [Google Scholar]
- 22.Onodera Y, Nakagawa T, Takahashi R, Tokuyama T. Seasonal change in vertical distribution of ammonia-oxidizing archaea and bacteria and their nitrification in temperate forest soil. Microbes Environ. 2010;25:28–35. doi: 10.1264/jsme2.me09179. [DOI] [PubMed] [Google Scholar]
- 23.Oved T, Shaviv A, Goldrath T, Mandelbaum RT, Minz D. Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl Environ Microbiol. 2001;67:3426–3433. doi: 10.1128/AEM.67.8.3426-3433.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poth M, Focht DD. 15N Kinetic analysis of N2O production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl Environ Microbiol. 1985;49:1134–1141. doi: 10.1128/aem.49.5.1134-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie GAF, Nicholas DJ. Identification of sources of nitrous-oxide produced by oxidative and reductive processes in Nitrosomonas europaea. Biochem J. 1972;126:1181–1191. doi: 10.1042/bj1261181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaw LJ, Nicol GW, Smith Z, Fear J, Prosser JI, Baggs EM. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ Microbiol. 2006;8:214–222. doi: 10.1111/j.1462-2920.2005.00882.x. [DOI] [PubMed] [Google Scholar]
- 28.Sheffield VC, Cox DR, Lerman LS, Myers RM. Attachment of a 40-base-pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephen JR, McCaig AE, Smith Z, Prosser JI, Embley TM. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takada-Hoshino Y, Matsumoto N. An improved DNA extraction method using skim milk from soils that strongly adsorb DNA. Microbes Environ. 2004;19:13–19. [Google Scholar]
- 31.Takata Y, Obara O, Nakai M, Kohyama K. Process of the decline in the cultivated soil area with land use changes in Japan. Jpn J Soil Sci Plant Nutr. 2011;82:15–24. (in Japanese) [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrage N, Velthof GL, van Beusichem ML, Oenema O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem. 2001;33:1723–1732. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.