Abstract
The diversity of purple phototrophic bacteria in algae-dominated biofilm of a streambed in Tama River, Japan was investigated. Clone library analysis of the pufM gene encoding a subunit of the photochemical reaction center of purple bacteria detected 18 operational taxonomic units (OTUs) in several classes of Proteobacteria. Most of the OTUs showed less than 85% identity to the PufM amino acid sequences of known phototrophic bacteria. These results suggest that phylogenetically divergent and unknown purple phototrophic bacteria are present in the epilithic biofilm of the river.
Keywords: Phototrophic bacteria, river, epilithic biofilm, pufM, purple bacteria
Purple phototrophic bacteria have been isolated from a variety of environments, such as sediments, soils and waters in ponds, lakes, lagoons and oceans (10, 11). As purple bacteria are metabolically versatile, e.g., photosynthesis, degradation of organic compounds, nitrogen fixation or sulfide oxidation, they play important roles in ecosystems in the light.
Microbial ecological studies on freshwater environments have found several groups of purple phototrophic bacteria belonging to Alpha- or Betaproteobacteria from lakes (12, 17) and rivers (8, 9, 20, 23). Some of these purple bacteria are known to be anaerobic anoxygenic phototrophs. The presence of aerobic anoxygenic phototrophs (AAP), which carry out photosynthetic reactions only under aerobic conditions, has been also indicated by phylogenetic or physiological studies; however, the distribution and ecophysiology of purple phototrophic bacteria in freshwater environments has not been documented.
Epilithic biofilm is important to sustain the ecosystem in freshwater environments in terms of producing organic substrates, feeding animals and degrading organic matter. The streambed biofilm is known to be mainly composed of oxygenic phototrophs, i.e., cyanobacteria and algae (2); however, no study has targeted purple phototrophic bacterial diversity in river biofilm. Dense assemblages of bacterial cells and their active respiration in biofilm possibly develop some anaerobic niches (6) even when phototrophs emit oxygen, and consequently both aerobic and anaerobic anoxygernic phototrophs may find their niches within river biofilm.
In this study, we applied a culture-dependent method and a molecular method based upon the pufM gene encoding a subunit of the photochemical reaction center to investigate the diversity of purple phototrophic bacteria in epilithic biofilm in an upstream region of a river where the amount of dissolved organic matter is limited. Phylogenies of pufM gene sequences are mostly consistent with those of the 16S rRNA gene (14), and thus the pufM gene is frequently utilized for genetic surveys of anoxygenic phototrophs (1, 4, 12, 16, 24).
Submerged cobbles of about 15 to 25 cm in longest length were collected from a streambed in riffle located in the upstream region of Tama River in Ohme City (35°47′13″N, 139°15′15″E), in the western suburbs of Tokyo, Japan in August 2009. The riffle width at the sampling site was 40 m. The water depth of the sampling site was about 20 cm. Water temperature, pH, biochemical oxygen demand (BOD) and flow velocity of the river water at the sampling time were 18°C, 7.6, 0.5 mg L−1 and 0.4 m s−1, respectively. Average values of dissolved oxygen, total nitrogen and total phosphorus in this region in July to September 2009 were 8.9±0.3 mg L−1, 0.79±0.10 mg L−1 and 0.017±0.005 mg L−1, respectively (monthly report by Bureau of Environment, Tokyo Metropolitan Government, http://www.kan-kyo.metro.tokyo.jp). A brownish biofilm of about 1 mm thickness was present on the cobbles. A total 150 cm2 area of epilithic biofilm was scraped off from the top surface of each cobble using a sterile toothbrush and suspended into 10 mL sterile distilled water.
For bacterial culture, 0.1 mL of the biofilm suspension was transferred into a 30 mL volume screw cap tube filled with PE medium (7), a semisynthetic medium containing organic compounds. The tubes were incubated at 30°C under filtered incandescent light (ca. 2,000 lux) of wavelength over 700 nm for 7 to 14 days. The cultures which showed spectral properties of purple bacteria were streaked on agar plates of PE medium. The plates were incubated anaerobically under incandescent light and red colonies were transferred and streaked onto new plates. These operations were repeated more than two times to obtain pure cultures.
Total genomic DNAs were directly extracted from the collected biofilm according to Noll et al.(15). DNA fragments of the pufM gene coding for the M subunit of the photochemical reaction center were amplified. Nested PCR was conducted to amplify pufM gene fragments from environmental DNA using primer sets pufLM-F/pufLM-R (1st PCR) (14) and M150f/M572r (2nd PCR) (16). PCR products (approximately 380 bp) were cloned with the pTAC-1 Vector (DynaExpress TA cloning kit, BioDynamics Laboratory, Tokyo, Japan). Escherichia coli JM109 competent cells (Nippon Gene, Tokyo, Japan) were transformed according to the manufacurer’s instructions. DNA sequences were determined with the BigDye v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and a DNA sequencer ABI3130xl (Applied Biosystems). Chimeric clones were checked manually and excluded from further analyses. The phylogenetic tree based on the amino acid sequences of the partial PufM was constructed using the neighbor-joining and maximum-likelihood methods with the MEGA version 5 program (19, 22).
Cultivation of the river biofilm on PE medium found red-brown colonies, all of which were similar in morphology. Among them, 6 strains were isolated to determine the pufM sequences. Sequence analysis indicated that the isolates were classified into two, one of which was designated Tisolate 25 and the other Tisolate 231.
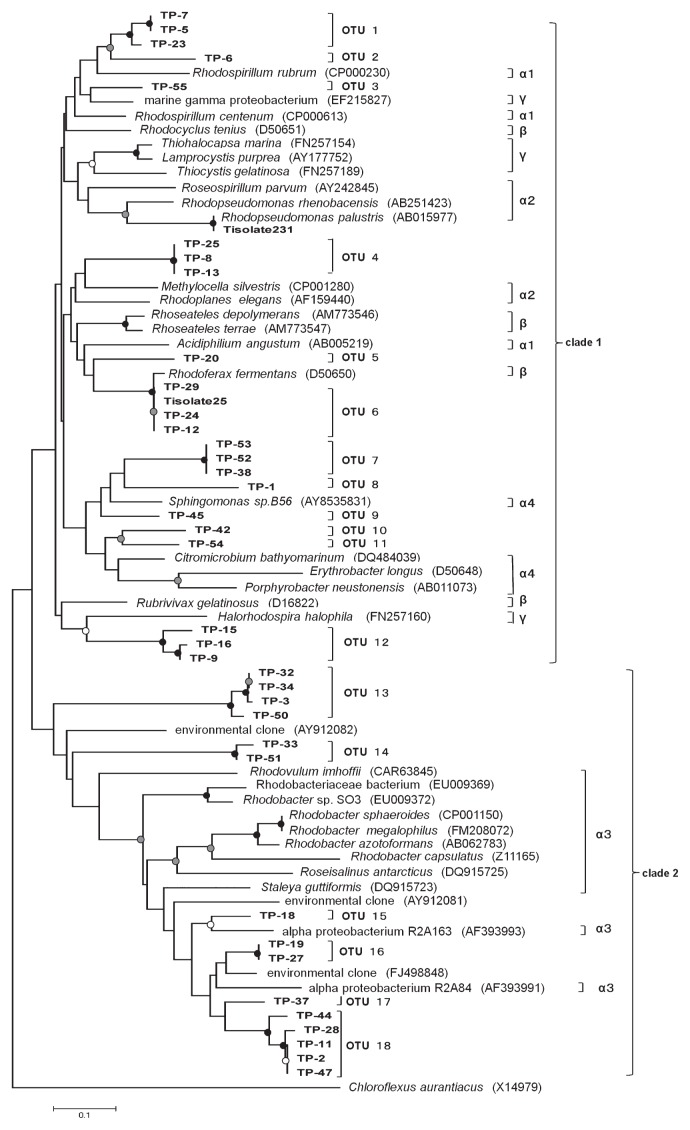
Figure 1 shows a neighbor-joining tree based on the amino acid sequences suspected from the partial pufM gene sequences from 37 clones and two isolates obtained in this study together with those from the database. A phylogenetic tree using the maximum-likelihood method showed tree topology roughly consistent with that in Fig. 1 (data not shown). Two major clades were recognized; one containing alpha-1, alpha-2, alpha-4 subclasses, beta and gamma classes of Proteobacteria and the other containing the alpha-3 subclass of Proteobacteria. This was roughly in agreement with earlier studies concerning pufLM or pufM phylogeny (4, 14). Obtained clones were grouped into 18 operational taxonomic units (OTUs). Each OTU was defined as a group having amino acid sequence identities above 90%. These OTUs were widely distributed in the phylogenetic tree and many were distantly related. No dominant OTUs in terms of numbers of clones were found, since every OTU consisted of less than 5 clones.
Fig. 1.
Phylogenetic tree of partial PufM amino acid sequences inferred from gene sequences. Chloroflexus aurantiacus was used as an outgroup. PufM amino acid sequences from environmental DNA in this study are indicated by TP-1–TP-55, and those from isolates in this study are indicated by the ‘Tisolate’ prefix. Sequences from the database are represented with their respective accession numbers after bacterial names in parentheses. OTUs are indicated to the right of the tree. Alpha-1, alpha-2, alpha-3, alpha-4 subclass, beta and gamma class of Proteobacteria are also indicated by α1, α2, α3, α4, β and γ to the right of the tree. Bootstrap values >90, 70–89 and 50–69% are indicated by black, gray and open circles, respectively. Scale bar represents the number of substitutions per site.
OTUs except OTU 6 showed less than 85% identity to the PufM sequences of the cultivated bacteria in the database. Sequences of OTU 6, which has the same sequence as that of Tisolate 25, was closely related to that of Rhodoferax fermentans (accession no. D50650, 98.4% identity). The other isolate, Tisolate 231, had 100% PufM sequence identity to Rhodopseudomonas palustris (accession no. AB015977). R. fermentans and R. palustris are known to be anaerobic anoxygenic phototrophs (11). Tisolate 25 and Tisolate 231 were grown photoheterotrophically under anaerobic conditions.
The alpha-3 clade contains 6 OTUs (OTUs 13 to 18). OTU 13 is distantly related to other members in this clade. Sequences included in OTU 14 showed very low identities to those of the database; the highest identity was 65.7% to that of an environmental clone (accession no. AY912082) (23) collected from river water. OTUs 15 to 18 formed a clade with Staleya guttiformis (now known as Sulfitobacter guttiformis) and alpha proteobacterium R2A163 and R2A84 (21), reported as aerobic anoxygenic phototrophic bacteria isolated from saline environments.
OTUs 1 and 2 were related to Rhodospirillum rubrum, belonging to alpha-1 subclass of Proteobacteria, with 75.4% and 71.4% sequence identities, respectively. OTU 3 was found to be similar to a marine gamma proteobacterium (5) with 84.1% sequence identity. OTU 4 was related to Methylocella sp. and Rhodoplanes sp. belonging to alpha-2 subclass of Proteobacteria. OTU 5 was similar to R. fermentans with 73.0% identity. It is indicated that OTUs 7 to 11 were grouped with the genus Sphingomonas, Citromicrobium, Erythorobacter and Porphyrobacter belonging to alpha-4 subclass. Phototrophic bacteria in this subclass have been known to be aerobic anoxygenic phototrophs. OTU 12 showed low identities to known sequences, and a close relative was Halorhodospira halophila, belonging to Gammaproteobacteria (69.0% identity).
In this study, we investigated the diversity of purple phototrophic bacteria in a streambed biofilm. Co-occurrence of possibly aerobic (e.g., OTUs 15 to 18) and anaerobic (e.g., OTU 6 and Tisolate 231) anoxygenic phototrophs was observed within the river biofilm as expected. Most OTUs detected by pufM clone library analysis had low identities to the sequences of cultured bacteria. Studies on bacterial communities in river biofilms using 16S rRNA gene analyses have also detected many clones of uncultured bacteria (3, 8, 13).
Phylogenetic analysis of the PufM sequences indicated that purple phototrophic bacteria in the river biofilm are widely distributed to alpha subclasses, beta and gamma classes of Proteobacteria. Such high diversity of purple phototrophic bacteria has not been reported in other environments. In French Mediterranean coast lagoon sediments, pufM clones in alpha-3 subclass of Proteobacteria accounted for 94.9% of total pufM clones (18). In the case of antarctic lake water, no pufM clones were related to alpha-3 subclass of Proteobacteria but 80% of the pufM clones were related to Betaproteobacteria(12). Microenvironments of algae-dominated river biofilm under the shallow and rapid flow of water are highly heterogeneous, differing in concentrations of dissolved oxygen, organic and inorganic compounds. The population of purple phototrophic bacteria within the epilithic biofilm microflora may be low because a nested approach was required to amplify their DNAs. The niche for purple phototrophic bacteria in the biofilm may be restricted, but seem to have largely diverged.
As purple phototrophic bacteria have bacteriochlorophylls, which have absorption bands at different wavelengths from those of chlorophylls in oxygenic phototrophs, purple photorophic bacteria can capture light energy even in algae-dominated biofilms of rivers. In the biofilm community, in addition to the primary production and degradation of organic matter, some purple phototrophic bacteria possibly contribute to oxidize sulfide produced by sulfate-reducing bacteria, since we observed sulfide production from anaerobic culture of the epilithic biofilm used in this study when illumination was stopped (data not shown).
This study demonstrated the unexpected diversity of purple photophototrophic bacteria in river biofilm. Physiological studies of yet-to-be cultured epilithic purple phototrophic bacteria will clarify the roles of these bacteria in the river ecosystem.
The nucleotide sequences determined in this study have been deposited in the GenBank/EMBL/DDBJ database under accession numbers AB670200 to AB670233.
Acknowledgements
We wish to thank Dr. K. Shimada, Tokyo Metropolitan University, for his helpful advice on this manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research (20370013) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K. M.).
References
- 1.Achenbach LA, Carey J, Madigan MT. Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments. Appl Environ Microbiol. 2001;67:2922–2926. doi: 10.1128/AEM.67.7.2922-2926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan JD. Stream ecology: Structure and Function of Running Waters. Chapman and Hall; London: 1995. [Google Scholar]
- 3.Anderson-Glenna MJ, Bakkestuen V, Clipson NJW. Spatial and temporal variability in epilithic biofilm bacterial communities along an upland river gradient. FEMS Microbiol Ecol. 2008;64:407–418. doi: 10.1111/j.1574-6941.2008.00480.x. [DOI] [PubMed] [Google Scholar]
- 4.Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 5.Cho J.-C, Stapels MD, Morris RM, Vergin KL, Schwalbach MS, Givan SA, Barofsky DF, Giovannoni SJ. Polyphyletic photosynthetic reaction centre genes in oligotrophic marine gammaproteobacteria. Environ Microbiol. 2007;9:1456–1463. doi: 10.1111/j.1462-2920.2007.01264.x. [DOI] [PubMed] [Google Scholar]
- 6.Costerton JW, de Beer D, Caldwell D, Korber DR, James GA. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanada S, Hiraishi A, Shimada K, Matsuura K. Isolation of Chloroflexus aurantiacus and related thermophilic phototrophic bacteria from Japanese hot springs using an improved isolation procedure. J Gen Appl Microbiol. 1995;41:119–130. [Google Scholar]
- 8.Honma H, Asano R, Obara M, Otawa K, Suyama Y, Nakai Y. Bacterial populations in epilithic biofilms along two oligotrophic rivers in the tohoku region in Japan. J Gen Appl Microbiol. 2009;55:359–371. doi: 10.2323/jgam.55.359. [DOI] [PubMed] [Google Scholar]
- 9.Hullar MAJ, Kaplan LA, Stahl DA. Recurring seasonal dynamics of microbial communities in stream habitats. Appl Environ Microbiol. 2006;72:713–722. doi: 10.1128/AEM.72.1.713-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imhoff JF, Trüper HG. The genus Rhodospirillum and related genera. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H, editors. The Prokaryotes. 2nd ed. Springer; New York: 1992. pp. 2141–2155. [Google Scholar]
- 11.Imhoff JF, Hiraishi A, Süling J. Anoxygenic phototrophic purple bacteria. In: Boone DR, Castenholz RW, Garity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. 2A. Springer; New York: 2005. pp. 119–153. [Google Scholar]
- 12.Karr EA, Sattley WM, Jung DO, Madigan MT, Achenbach LA. Remarkable diversity of phototrophic purple bacteria in a permanently frozen antarctic lake. Appl Environ Microbiol. 2003;69:4910–4914. doi: 10.1128/AEM.69.8.4910-4914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi Y, Kim C, Yoshimizu C, Kohzu A, Tayasu I, Nagata T. Longitudinal changes in bacterial community composition in river epilithic biofilms: Influence of nutrients and organic matter. Aquat Microb Ecol. 2009;54:135–152. [Google Scholar]
- 14.Nagashima KVP, Hiraishi A, Shimada K, Matsuura K. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J Mol Evol. 1997;45:131–136. doi: 10.1007/pl00006212. [DOI] [PubMed] [Google Scholar]
- 15.Noll M, Matthies D, Frenzel P, Derakshani M, Liesack W. Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ Microbiol. 2005;7:382–395. doi: 10.1111/j.1462-2920.2005.00700.x. [DOI] [PubMed] [Google Scholar]
- 16.Okubo Y, Futamata H, Hiraishi A. Characterization of phototrophic purple nonsulfur bacteria forming colored microbial mats in a swine wastewater ditch. Appl Environ Microbiol. 2006;72:6225–6233. doi: 10.1128/AEM.00796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page KA, Connon SA, Giovannoni SJ. Representative freshwater bacterioplankton isolated from Crater Lake, Oregon. Appl Environ Microbiol. 2004;70:6542–6550. doi: 10.1128/AEM.70.11.6542-6550.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranchou-Peyruse A, Herbert R, Caumette P, Guyoneaud R. Comparison of cultivation-dependent and molecular methods for studying the diversity of anoxygenic purple phototrophs in sediments of an eutrophic brackish lagoon. Environ Microbiol. 2006;8:1590–1599. doi: 10.1111/j.1462-2920.2006.01050.x. [DOI] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Suyama T, Shigematsu T, Takaichi S, Nodasaka Y, Fujikawa S, Hosoya H, Tokiwa Y, Kanagawa T, Hanada S. Roseateles depolymerans gen. nov., sp. nov., a new bacteriochlorophyll a-containing obligate aerobe belonging to the β-subclass of the Proteobacteria. Int J Syst Bacteriol. 1999;49:449–457. doi: 10.1099/00207713-49-2-449. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki MT, Rappé MS, Haimberger ZW, Winfied H, Adair N, Ströbel J, Giovannoni SJ. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. App Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waidner LA, Kirchman DL. Aerobic anoxygenic photosynthesis genes and operons in uncultured bacteria in the Delaware River. Environ Microbiol. 2005;7:1896–1908. doi: 10.1111/j.1462-2920.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- 24.Yutin N, Suzuki MT, Beja O. Novel primers reveal wider diversity among marine aerobic anoxygenic phototrophs. Appl Environ Microbiol. 2005;71:8958–8962. doi: 10.1128/AEM.71.12.8958-8962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]