Abstract
In this study, a microcolony technique was combined with direct fluorescent antibody staining for the specific detection and enumeration of Legionella pneumophila in freshwater samples with growth activity. This method allowed the detection of active L. pneumophila (within 48 h) in 91 bath water samples collected from 30 bathing facilities, with similar sensitivity of a conventional plate-counting method. These results suggest that the microcolony method combined with fluorescent antibody staining could be useful as a monitoring technique for the prevention of Legionnaires’ disease through the early detection of L. pneumophila in freshwater.
Keywords: Legionella pneumophila, rapid enumeration, microcolony, growth activity, freshwater
Members of the genus Legionella are ubiquitous in freshwater environments and the vast majority of Legionnaires’ disease cases are due to L. pneumophila(5). Outbreaks of Legionnaires’ disease have been associated with a wide variety of freshwater sources, including cooling towers, fountains, hot tubs, spas and so on (3, 9, 15). In Japan, the reported incidences of Legionellosis have been steadily increasing and the main source of contamination is considered to be water from bathing facilities and spas (8, 10, 11).
The rapid monitoring of Legionella species in environmental water is essential for the prevention of Legionnaires’ disease outbreaks. When an increase of Legionella species is detected in environmental water, such as a water circulation system, rapid disinfection of the water leads to efficient control of Legionellosis outbreaks. The detection and enumeration of Legionella species have been typically performed using conventional culture methods; however, this approach is time-consuming, as an incubation period of up to 10 days is required for determining the number of Legionella spp.
In recent years, PCR-based methods have been used for the detection and quantification of Legionella spp. For example, a quantitative real-time PCR technique targeting the 16S rRNA gene and mip (macrophage infectivity potentiator) genes of L. pneumophila has been used to detect targeted cells in water samples (12). On the other hands, detection methods discriminating between live and dead cells are of major interest because viable L. pneumophila with growth activity are able to increase by the proliferation and cause infection outbreaks. Therefore, the development of a rapid enumeration method for the quantification of active L. pneumophila with growth activity is required for efficient water hygiene management and control of Legionellosis.
One such approach is the microcolony method, which is based on microscopic observation of the early stages of colony formation on selective culture medium and allows the enumeration of viable target cells by growth activity (6). Several studies have reported that most bacteria in the natural environment grow to the microcolony stage, while they hardly form visible colonies (1, 7, 14). In this study, we combined the microcolony method with fluorescent antibody staining for the identification and enumeration of active L. pneumophila in environmental water samples collected from 30 bathing facilities and spas (91 samples).
L. pneumophila (JCM7571) was used to optimize the microcolony-fluorescent antibody staining (MC-FA) method. L. pneumophila was grown on buffered charcoal yeast extract agar supplemented with α-ketoglutarate (BCYEα) agar media (Eiken Chemical Co. Ltd., Tokyo, Japan) and suspended in PBS. Approximately 106 cells of L. pneumophila were trapped by vacuum on membrane filters (pore size: 0.2 μm, ANODISC 25; Whatman International, Ltd., Kent, UK), which were then transferred onto BCYEα agar medium and incubated at 37°C for 48 h. These filters were placed on filter paper (No. 2; Whatman International Ltd.) soaked with 4% formaldehyde to fix microcolonies at room temperature for 30 min. Membrane filters were then transferred onto filter paper saturated with sterile water, allowed to stand for 10 min, and then air-dried. For the enumeration of L. pneumophila microcolonies, membrane filters were stained with fluorescein isothiocyanate (FITC)-labeled anti-L. pneumophila antibodies (Monoclonal Technologies Inc., Alpharetta, GA, USA) as described below. The specificity of this fluorescent antibody was reported (4). Membrane filters were treated with 10 μl fluorescent antibodies diluted with 3% bovine serum albumin (BSA; Wako Pure Chemical Industries, Osaka, Japan) in PBS at 30°C for 30 min. Membrane filters were placed on filter paper soaked with PBS to rinse for 10 min and allowed to air dry. Microcolonies of L. pneumophila were counted by epifluorescent microscopy (E-400; Nikon, Tokyo, Japan) with 200×magnification.
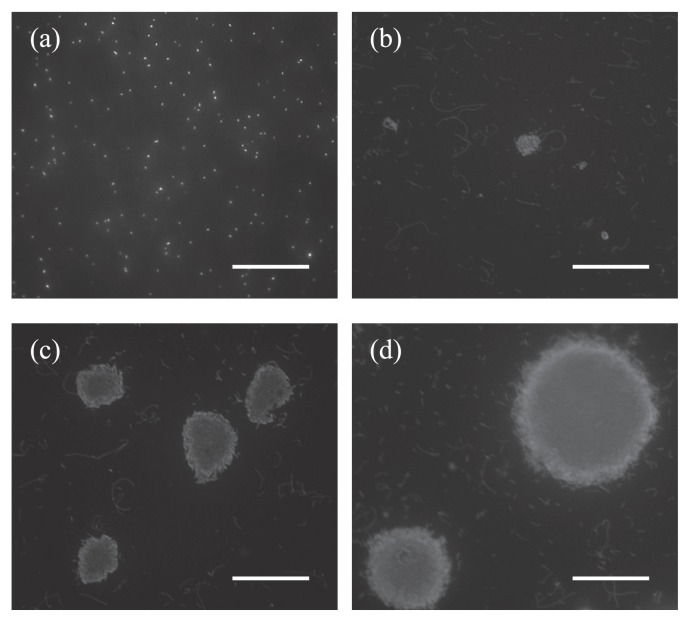
Microcolony formation of L. pneumophila was monitored for a 48-h period (Fig. 1). After 32 h, the formed microcolonies were approximately 20 μm in diameter and reached approximately 100 μm in diameter after 48 h. The number of active L. pneumophila by the MC-FA method was (1.4±0.9)×106 microcolony-forming units (mCFU) (100 mL)−1, while it was (1.8±0.7)×106 CFU (100 mL)−1 by the conventional plate-counting method. There was no significant difference in the results obtained by the two methods. Fluorescent antibody staining allowed the simple and specific detection of L. pneumophila microcolonies. With the MC-FA method, only 48 h or less was required for the detection of microcolony formation of L. pneumophila, which is markedly shorter than the ten days required for the conventional plate-counting method. An additional advantage of this method is the counting accuracy; the target bacteria can be easily distinguished from contaminants (non-biological particles) included in environmental samples, because the formed microcolonies are markedly larger (>20 μm) than single cells or contaminants in the microscopic field.
Fig. 1.
Epifluorescent micrographs of Legionella pneumophila microcolonies by the MC-FA method. L. pneumophila serogroup 1 (JCM7571) cells on membrane filters were cultured on BCYEα medium for the indicated time periods and subjected to staining with FITC-labeled anti-L. pneumophila antibodies. Before incubation (a), incubated for 24 hours (b), 32 hours (c), 48 hours (d). Scale bars are 50 μm.
After optimization of the MC-FA method, bath water samples were collected from 30 bathing facilities and spas in Japan from July 2004 to November 2006. Ninety-one samples were collected in sterilized plastic bottles and used immediately after sampling. In environmental samples, detection of Legionella cells in bath water samples by the conventional plate-counting method was performed according to JIS K 0350-50-10:2006. Samples of water (500 mL) were concentrated by filtration through a poly-carbonate filter (pore size: 0.2μm; Toyo-Roshi, Tokyo, Japan). After filtration, bacteria collected on membrane filters were resuspended in 5 mL sterilized water and sonicated for 1 min. The suspensions were heat treated at 50°C for 30 min to suppress the growth of non-Legionella bacteria and then serially diluted in sterile water. Subsequently, 100 μl of each sample was spread on Wadowsky-Yee-Okuda glycine-vancomycin-polymyxin B-amphotericin B supplemented with α-ketoglutarate (WYOα) medium (Nikken Bio Medical Laboratory, Kyoto, Japan) and incubated at 37°C for 7 days. Legionella-like colonies, which were grayish-white in color, were subcultured on both sheep blood and BCYEα agar media (Nikken Bio Medical Laboratory) and incubated at 37°C for 2 days for further verification. Colonies that grew on only BCYEα medium were subjected to slide agglutination tests using Legionella antisera kit (Denka Seiken, Tokyo, Japan) to determine the Legionella serogroup.
The number of L. pneumophila in each sample was also determined by the MC-FA method. Filtering, concentration, and heat treatment were performed by identical procedures to those described for the plate-counting method. Then, 1 mL of each sample was filtrated through a membrane filter and the filters were transferred onto WYOα medium and incubated at 37°C for 48 h. Fixation and staining were performed by identical procedures to those described previously. The whole area of each filter was scanned under an epifluorescent microscope with 200× magnification, and the lower detection limit was 1 mCFU (100 mL)−1.
In 91 bath water samples collected from bathing facilities and spas, 25 samples (28%) were positive by the MC-FA method, and 16 (17%) were positive by the plate-counting method. The number of colony-forming and microcolony-forming units in the positive samples, as determined by the plate-counting and the MC-FA method, respectively, are shown in Table 1. Nine samples were positive by the MC-FA method but negative by the plate-counting method, whereas the opposite was never found. In most bathing facilities, bath water was treated by detergents and chlorination, and L. pneumophila cells may have been damaged by treatment and impaired in their ability to form visible colonies on the selective medium. Furthermore, in environmental microbiology, it is recognized that many human pathogenic bacteria do not form visible colonies on conventional culture media when in the natural environments (2, 16).
Table 1.
Quantification of L. pneumophila in bath water samples by the plate-counting and the microcolony-fluorescent antibody staining (MC-FA) method
| Sample No. | Temperature (°C) | Cl (mg L−1) | Plate-counting method (CFU 100 mL−1) | MC-FA methoda (mCFU 100 mL−1) |
|---|---|---|---|---|
| 11 | 38.0 | <0.1 | NDb | 2 |
| 80 | 42.0 | <0.1 | ND | 2 |
| 91 | 42.0 | <0.1 | ND | 3 |
| 86 | 40.0 | <0.1 | ND | 5 |
| 12 | 37.0 | <0.1 | ND | 11 |
| 35 | 40.3 | <0.1 | ND | 12 |
| 70 | 42.0 | 1.0 | ND | 13 |
| 64 | 40.0 | <0.1 | ND | 18 |
| 36 | 41.3 | <0.1 | ND | 114 |
| 22 | 38.0 | <0.1 | 10 | 2 |
| 66 | 42.0 | <0.1 | 10 | 3 |
| 31 | 41.4 | <0.1 | 10 | 66 |
| 45 | 39.8 | <0.1 | 20 | 7 |
| 44 | 40.7 | <0.1 | 20 | 18 |
| 30 | 41.3 | <0.1 | 20 | 57 |
| 46 | 40.9 | <0.1 | 40 | 3 |
| 43 | 42.0 | <0.1 | 40 | 12 |
| 29 | 40.2 | <0.1 | 40 | 49 |
| 28 | 39.9 | <0.1 | 50 | 41 |
| 32 | 42.5 | <0.1 | 70 | 39 |
| 38 | 40.2 | <0.1 | 180 | 171 |
| 34 | 40.0 | <0.1 | 230 | 140 |
| 37 | 40.4 | <0.1 | 670 | 980 |
| 68 | 42.0 | <0.1 | 820 | 1010 |
| 39 | 28.6 | <0.1 | 1200 | 1869 |
Means of two replicates.
Under detection limit (10 CFU 100 mL−1)
On the other hand, in all positive samples by both methods (16 samples), a positive relationship was observed (r=0.791, p<0.01); in particular, for five samples containing a high concentration of active L. pneumophila (>100 CFU [100 mL]−1), the bacterial number by the MC-FA and plate-counting methods was close. We concluded that the MC-FA method is capable of rapidly identifying active L. pneumophila in water samples. These findings suggest that this MC-FA method could be useful as a rapid monitoring technique of L. pneumophila for the prevention of Legionnaires’ disease outbreaks.
It is expected that the MC-FA method described here can be applied for the detection of any target bacterium for which selective media and antibodies have been developed. Furthermore, this method with the Microcolony-Auto-Counting-System (13, 14) enables the routine bacterial monitoring of environmental samples.
Acknowledgements
This study was supported in part by JSPS Grants-in aid for Scientific Research (A) (20249007 and 21256002).
References
- 1.Baba T, Matsumoto R, Yamaguchi N, Nasu M. Bacterial population dynamics in a reverse-osmosis water purification system determined by fluorescent staining and PCR-denaturing gradient gel electrophoresis. Microbes Environ. 2009;24:163–167. doi: 10.1264/jsme2.me09120. [DOI] [PubMed] [Google Scholar]
- 2.Bej A, Mahbubani MH, Atlas RM. Detection of viable Legionella pneumophila in water by polymerase chain reaction and gene probe methods. Appl Environ Microbiol. 1991;57:597–600. doi: 10.1128/aem.57.2.597-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borella P, Montagna T, Stampi S, et al. Legionella contamination in hot water of Italian hotels. Appl Environ Microbiol. 2005;71:5805–5813. doi: 10.1128/AEM.71.10.5805-5813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delgado-Viscogliosi P, Simonart T, Parent V, et al. Rapid method for enumeration of viable Legionella pneumophila and other Legionella spp. in water. Appl Envion Microbiol. 2005;71:4086–4096. doi: 10.1128/AEM.71.7.4086-4096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai M, Yamaguchi N, Nasu M. Rapid enumeration of physiologically active bacteria in purified water used in the pharmaceutical manufacturing process. J Appl Microbiol. 1999;86:496–504. doi: 10.1046/j.1365-2672.1999.00689.x. [DOI] [PubMed] [Google Scholar]
- 7.Kenzaka T, Yamaguchi N, Utrarachkij F, Suthienkul O, Nasu M. Rapid identification and enumeration of antibiotic resistant bacteria in urban canals by microcolony-fluorescence in situ hybridization. J Health Sci. 2006;52:703–710. [Google Scholar]
- 8.Kura F, Amemura-Maegawa J, Yagita K, Endo T, Ikeno M, Tsuji H, Taguchi M, Kobayashi K, Ishii E, Watanabe H. Outbreak of Legionnaires’ disease on cruise ship linked to spa-bath filter stones contaminated with Legionella pneumophila serogroup 5. Epidemiol Infect. 2006;134:385–391. doi: 10.1017/S095026880500508X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leoni E, De Luca G, Legnani PP, Sacchetti R, Stampi S, Zanetti F. Legionella waterline colonization: detection of Legionella species in domestic, hotel and hospital hot water systems. J Appl Microbiol. 2005;98:373–379. doi: 10.1111/j.1365-2672.2004.02458.x. [DOI] [PubMed] [Google Scholar]
- 10.Nakadate T, Yamaguchi K, Inoue H. An outbreak of Legionnaires’ disease associated with a Japanese spa. Nihon kokyuki Gakkai Zasshi. 1999;37:601–607. (In Japanese) [PubMed] [Google Scholar]
- 11.Okada M, Kawano K, Kura F, Amemura-Maegawa J, Watanabe H, Yagita K, Endo T, Suzuki S. The largest outbreak of Legionellosis in Japan associated with spa baths: epidemic curve and environmental investigation. J Jpn Assoc Infect Dis. 2005;79:365–374. doi: 10.11150/kansenshogakuzasshi1970.79.365. (In Japanese) [DOI] [PubMed] [Google Scholar]
- 12.Solhang A, Bergh K. Identification and differentiation of Legionella pneumophila and Legionella spp. with real-time PCR targeting the 16S rRNA gene and species identification by mip sequencing. Appl Environ Microbiol. 2006;72:6394–6398. doi: 10.1128/AEM.02839-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, Yamaguchi N, Baba T, Amano N, Nasu M. Rapid enumeration of low numbers of moulds in tea based drinks using an automated system. Int J Food Microbiol. 2011;145:365–369. doi: 10.1016/j.ijfoodmicro.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Yamaguchi N, Someya T, Nasu M. Rapid and automated enumeration of viable bacteria in compost using a micro-colony auto counting system. J. Microbiol. Methods. 2007;71:1–6. doi: 10.1016/j.mimet.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Wery N, Bru-Adan V, Minervini C, Delgénes JP, Garrelly L, Godon JJ. Dynamics of Legionella spp. and bacterial populations during the proliferation of L. pneumophila in a cooling tower facility. Appl Environ Microbiol. 2008;74:3030–3037. doi: 10.1128/AEM.02760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Ueno D, Inoue K, Someya T. Direct viable count combined with fluorescence in situ hybridization (DVC-FISH) for specific enumeration of viable Escherichia coli in cow manure. Microbes Environ. 2009;24:33–38. doi: 10.1264/jsme2.me08543. [DOI] [PubMed] [Google Scholar]