Abstract
Cyclic-di-AMP (c-di-AMP) is an essential second messenger in Bacillus subtilis, and depletion leads to defects in the integrity of the cell wall. Levels of c-di-AMP are regulated by both the rates of synthesis (by diadenylate cyclases) and the rates of degradation (by the GdpP phosphodiesterase, formerly YybT). Little is known about the regulation of gdpP expression or GdpP activity, but mutations that inactivate GdpP lead to high-level resistance to β-lactam antibiotics. Here we demonstrate that expression of gdpP is regulated by a cis-acting antisense RNA (gdpPas) in vivo. Transcription of this antisense RNA is initiated in the middle of the gdp gene and is dependent on an alternative sigma factor, σD, previously associated with the expression of late flagellar genes, chemotaxis proteins and cell wall autolytic enzymes. Changes in σD activity can modulate GdpP protein levels by ~2.5-fold, which may provide a mechanism for the cell to upregulate c-di-AMP levels in coordination with the activation of autolytic enzymes.
Introduction
Cyclic-di-AMP (c-di-AMP) is a recently recognized second messenger molecule in bacteria. It was first identified as an endogenous metabolite in the crystal structure of DisA, which catalyses its synthesis at its DAC domain (diadenylate cyclase domain; previously DUF147 domain) (Witte et al., 2008). DAC domain-containing proteins can be found in many other (predominantly Gram-positive) bacteria and archaea (Römling, 2008). Many species only harbour one DAC domain protein, and null mutation of the sole DAC appears to be lethal in Listeria monocytogenes, Staphyloccocus (Staph.) aureus, Streptoccocus pneumoniae, Mycoplasma pulmonis and Mycoplasma genitalium (Chaudhuri et al., 2009; Corrigan et al., 2011; French et al., 2008; Glass et al., 2006; Song et al., 2005; Woodward et al., 2010). The genome of Bacillus subtilis encodes three DAC-containing proteins, DisA, YbbP and YojJ. Although single mutants of these DAC proteins are viable, the double mutant lacking both DisA and YbbP is non-viable (Luo & Helmann, 2012). The essential roles of c-di-AMP are not well understood, but recent results suggest that it is, directly or indirectly, involved in peptidoglycan (PG) homeostasis (Corrigan et al., 2011; Luo & Helmann, 2012).
Cellular levels of c-di-AMP are regulated by rates of synthesis and degradation. The DAC domain in the cyclase catalyses the synthesis of c-di-AMP from two ATP molecules, while the DHH domain in the hydrolase catalyses cleavage of c-di-AMP to linear 5′-pApA. In B. subtilis, the three c-di-AMP cyclases (DisA, YbbP and YojJ) and one hydrolase (GdpP) are subject to both transcriptional and post-transcriptional regulation. Expression of disA is regulated by both σA and σM (Eiamphungporn & Helmann, 2008), and the cyclase activity of DisA is influenced by DNA integrity. DisA forms a large octamer that moves along intact chromosomal DNA. Upon encountering a DNA double-strand break, the DisA complex pauses at the lesion site and ceases c-di-AMP synthesis, thus delaying sporulation (Bejerano-Sagie et al., 2006; Oppenheimer-Shaanan et al., 2011; Witte et al., 2008). The second cyclase, YbbP, is orthologous to the essential DAC proteins of L. monocytogenes and Staph. aureus. Its transcript is mainly dependent on σA, with possible read through from an upstream σW promoter (Cao et al., 2002; Luo & Helmann, 2012). YbbP is a membrane-localized protein that responds to cell envelope stress. Mutation of ybbP results in increased susceptibility to β-lactam antibiotics such as cefuroxime (Luo & Helmann, 2012). The third enzyme, YojJ, is a cytosolic protein. In vegetatively growing cells, YojJ is able to restore growth to a normally lethal double disA ybbP mutant only if artificially overexpressed (Luo & Helmann, 2012).
GdpP (formerly YybT) is the only known c-di-AMP phosphodiesterase (PDE) in B. subtilis. This transmembrane protein contains three functional domains: a haem-binding PAS domain, a degenerate GGDEF domain and a DHH/DHHA1 PDE domain (Rao et al., 2010, 2011). The PDE activity of GdpP can be inhibited by the alarmone ppGpp and by haem in vitro. The haem-dependent PAS inhibition can be partially relieved by nitric oxide (NO). However, the biological relevance of these haem, NO and ppGpp effects on PDE activity has not been studied in vivo. In this work, we characterize an antisense RNA transcribed from within gdpP. This cis-acting RNA is dependent on σD and it can modulate the cellular levels of GdpP.
Methods
Bacterial strains and growth conditions.
Strains used in this study are listed in Table 1. B. subtilis strains are derivatives of strain 168 or NCIB 3610. Escherichia coli strain DH5α was used for standard cloning procedures. Unless noted otherwise, all cultures were grown in Luria–Bertani (LB) broth at 37 °C with vigorous shaking. Antibiotics were added to the growth medium when appropriate: 100 µg ampicillin ml−1 for E. coli, and 1 µg erythromycin ml−1 plus 25 µg lincomycin ml−1 (MLS; macrolide-lincomycin-streptogramin B resistance), 10 µg chloramphenicol ml−1, 100 µg spectinomycin ml−1, 5 µg tetracycline ml−1 and 10 µg kanamycin ml−1 for B. subtilis. OD600 readings were taken on a Spectronic 21 spectrophotometer.
Table 1. Strains used in this study.
| Strain | Genotype | Source or reference |
| 168 | trpC2 | Laboratory stock |
| NCIB3610 | Prototrophic, undomesticated parent of 168 | Laboratory stock |
| HB15826 | 168 amyE : : PyybS-lacZ cat | This study |
| HB15827 | 168 amyE : : PgdpPas-lacZ cat | This study |
| HB15829 | 168 flgM : : spc amyE : : PyybS-lacZ cat | This study |
| HB15830 | 168 flgM : : spc amyE : : PgdpPas-lacZ cat | This study |
| HB15833 | 168 sigD : : kan amyE : : PyybS-lacZ cat | This study |
| HB15834 | 168 sigD : : kan amyE : : PgdpPas-lacZ cat | This study |
| HB15835 | 168 sigD : : kan flgM : : spc amyE : : PgdpPas-lacZ cat | This study |
| HB15885 | 168 amyE : : PD*-lacZ cat | This study |
| HB15886 | 168 sigD : : kan amyE : : PD*-lacZ cat | This study |
| HB15887 | 168 flgM : : spc amyE : : PD*-lacZ cat | This study |
| HB13056 | 168 thrC : : PfabHaF-fabHa-fabF-FLAG mls | Kingston et al. (2011) |
| HB15843 | 168 gdpP : : pMUTIN4-gdpP-FLAG mls | This study |
| HB15845 | 168 gdpP-FLAG | This study |
| HB15857 | 168 gdpP-FLAG thrC : : PfabHaF-fabHa-fabF-FLAG mls | This study |
| HB15858 | 168 sigD:kan gdpP-FLAG thrC : : PfabHaF-fabHa-fabF-FLAG mls | This study |
| HB15859 | 168 flgM : : spc gdpP-FLAG thrC : : PfabHaF-fabHa-fabF-FLAG mls | This study |
| HB15860 | 168 sigD:kan flgM : : spc gdpP-FLAG thrC : : PfabHaF-fabHa-fabF-FLAG mls | This study |
| HB15836 | 168 gdpP : : pMUTIN4-PD* mls | This study |
| HB15837 | 168 PD* | This study |
| HB15901 | 3610 PD* | This study |
| HB15902 | 3610 flgM : : cat | This study |
| HB15903 | 3610 flgM : : cat PD* | This study |
| HB15904 | 3610 flgM : : cat amyE : : Physpank-sigD kan | This study |
| HB15905 | 3610 flgM : : cat amyE : : Physpank-sigD kan PD* | This study |
| HB15906 | 3610 flgM : : cat amyE : : Physpank-sigD kan lytABC : : spc lytD : : mls lytF : : tet | This study |
| HB15907 | 3610 flgM : : cat amyE : : Physpank-sigD kan lytABC : : spc lytD : : mls lytF : : tet PD* | This study |
| HB15908 | 3610 disA : : spc flgM : : cat | This study |
| HB15909 | 3610 disA : : spc flgM : : cat PD* | This study |
| HB15910 | 3610 ybbP : : tet flgM : : spc amy : : Phag-cat-lacZ | This study |
| HB15911 | 3610 ybbP : : tet flgM : : spc PD* amy : : Phag-cat-lacZ | This study |
| HB15912 | 3610 sigM : : kan ybbP : : tet flgM : : spc amy : : Phag-cat-lacZ | This study |
| HB15913 | 3610 sigM : : kan ybbP : : tet flgM : : spc PD* amy : : Phag-cat-lacZ | This study |
| HB15914 | 3610 disA : : spc amyE : : Pspac(hy)-gdpP cat | This study |
| HB15915 | 3610 ybbP : : tet amyE : : Pspac(hy)-gdpP cat | This study |
| HB15916 | 3610 yojJ : : kan amyE : : Pspac(hy)-gdpP cat | This study |
| HB15917 | 3610 gdpP : : mls amyE : : Pspac(hy)-gdpP cat | This study |
| HB15918 | 3610 amy : : Pspac(hy)-gdpP cat | This study |
Strain construction.
All strains were generated by transformation of strain 168 to antibiotic resistance using chromosomal DNA, PCR products or plasmids as described elsewhere (Harwood & Cutting, 1990). When required, mutations in the strain 168 background were transferred to the strain NCIB 3610 background by SPP1-mediated generalized transduction, as described elsewhere (Kearns & Losick, 2005). Unless stated otherwise, all PCR products were generated using chromosomal DNA from strain 168 as a template and all constructs were verified by sequence analysis (Cornell University Life Sciences Core Laboratories Center). The oligonucleotides used in this study are listed in Table 2.
Table 2. Oligonucleotides used in this study.
| No. | Oligonucleotide | Sequence (5′–3′)* |
| 5586 | yybTanti-GSP1 | CGATGAGTGAGGTTGAGGCT |
| 5587 | yybTanti-GSP2 | GGAATACTTCGATCAAGTGCTGA |
| 5588 | yybTanti-GSP3 | ATCAATATGATTGAAGCAACAGC |
| 5589 | yybTanti-GSP4 | ACAGCGGAATTGGTGACAGA |
| 5590 | yybTanti-GSP5 | AGCCGTCACTCGTCATGGA |
| 5591 | yybTanti-GSP6 | CGCCTGAAGAAGCAATGGA |
| 5592 | yybTanti-GSP7 | GTCACCGAAAGCAGCAATGT |
| 5593 | yybTanti-GSP8 | AACCCAATGGAGAAACGAACA |
| 5594 | yybTanti-GSP9 | CGCAATCCAGCTTGGACTT |
| 5595 | yybTanti-GSP10 | CTGACGTTAAGCGTCGGTGT |
| 4549 | AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG |
| 5628 | sigD-up-for | CTGATGGAGCTCAGTCAGGT |
| 5629 | sigD-up-rev (kan) | CCTATCACCTCAAATGGTTCGCTGCCTGATCTTCATAATTCAAGGA |
| 5630 | sigD-do-for (kan) | CGAGCGCCTACGAGGAATTTGTATCGTCAGATCCATTCAAAGGCATT |
| 5631 | sigD-do-rev | GCTTCATAGAAATGACTGACATGT |
| 5632 | flgM-up-for | TTCAGCAGAAGGTATGAATATCA |
| 5633 | flgM-up-rev(spc) | CGTTACGTTATTAGCGAGCCAGTCGATATGGATTAACGGATTGTGT |
| 5634 | flgM-do-for(spc) | CAATAAACCCTTGCCCTCGCTACGTCATACAAAGTAGACGCAAATCA |
| 5635 | flgM-do-rev | CCGCTGTCTTGTATAACCATCA |
| 5879 | flgM-up-rev(cat) | CTTGATAATAAGGGTAACTATTGCCGATATGGATTAACGGATTGTGT |
| 5880 | flgM-do-for(cat) | GGGTAACTAGCCTCGCCGGTCCACGTCATACAAAGTAGACGCAAATCA |
| 5565 | PyybS-for (EcoRI) | GAGGAATTCACAAAGAGGTGAAACACAATG |
| 5566 | PyybS-rev (BamHI) | GAGGGATCCCAATCACAGGAACATAAACGA |
| 5598 | PyybTanti-for (EcoRI) | GAGGAATTCACGAGTGACGGCTTATGTGT |
| 5619 | PyybTanti-rev2 (BamHI) | GAGGGATCCGTCACCGAAAGCAGCAATGT |
| 5620 | yybT-int-for (EcoRI) | GAGGAATTCATCAAACGGGATGAGCGTCTCT |
| 5621 | yybT-PDmut-up-rev | TCGCTGCGCCTATGGAATCCATGTCGGGGAATTTATG |
| 5622 | yybT-PDmut-do-for | CATAAATTCCCCGACATGGATTCCATAGGCGCAGCGATCGGGATTTTAAAG |
| 5623 | yybT-rev (BamHI) | GAAGGATCCTCATCTCTGTACGCCTCCCT |
| 5684 | yybS-int-up-for-(EcoRI) | GAGGAATTCTTGATATTGTAGAAACTGTAGCGA |
| 5685 | yybS-up-rev-(FLAG) | CATCCGCGGTTTATCATCATCATCTTTATAATCCATTTCTATCACTCCCCACCATGT |
| 5246 | yybT-for N-flag 1 | ATGGATTATAAAGATGATGATGATAAACCGCGGATGCCAAGCTTTTATGAAAAAC |
| 5686 | yybT-int-do-rev (BamHI) | GAGGGATCCATCCAATCCTTGTGTCACATCA |
| 5298 | yybT-for | AGTGATAGAAATGCCAAGCT |
Restriction sites are underlined.
To generate promoter–lacZ fusions, a DNA fragment containing PyybS or PgdpPas was PCR-amplified with primer pairs 5565/5566 or 5598/5619, respectively, and cloned into vector pDG1661 (Guérout-Fleury et al., 1996). The resulting plasmids were linearized by digestion with ScaI and integrated into strain 168 at the amyE locus. To create the PD*-lacZ fusion, the same protocol was used except that the DNA fragment was amplified using strain HB15837 (168 PD*) as template.
To generate the at-locus marker-less mutation PD* (HB15837), we utilized an unstable integrative plasmid pMUTIN4, which harbours MLS resistance and lacZ genes (Vagner et al., 1998). A DNA fragment containing PD* was first generated using overlap-extension PCR as described previously (Gaballa et al., 1998; Ho et al., 1989) with some modifications. Briefly, the up-fragment (including 620 bp upstream of the mutation site) and down-fragment (including 900 bp downstream of the mutation) were amplified using PCR with primer pairs 5620/5621 and 5622/5623, respectively. Primers 5621 and 5622 introduce the desired PD* mutation and are complementary to each other. These up- and down-fragments were then joined using PCR with primers 5620/5623. The resulting PCR product was cloned into pMUTIN4, which was transformed into strain 168. The transformants were plated on LB agar plates supplemented with MLS and X-Gal (50 µg ml−1). The resulting strain with plasmid pMUTIN4 integrated at the gdpP locus (strain HB15836) was resistant to MLS and blue on X-Gal plates. This at-locus integration of pMUTIN4 is not stable, and is capable of looping out from the chromosome, leaving behind either the wild-type (WT) or mutant sequence in the chromosome. To loop out pMUTIN4, cells of strain HB15836 were grown overnight in LB broth (without antibiotic selection), reinoculated into LB diluted to 1 : 100, grown to OD600 0.4 and diluted to 1 : 10 000, and 100 µl of cells was plated on LB agar supplemented with X-Gal. Cells that had lost the pMUTIN4 plasmid appeared as white colonies on X-Gal plates, and were sensitive to MLS. The strain harbouring the PD* mutation was verified by PCR amplification using primers 5298/5598 and DNA sequencing. To generate PD* in the strain NCIB 3610 background, the gdpP : : pMUTIN4-PD* construct in strain HB15836 was transferred to 3610 by SPP1 transduction, followed by the pMUTIN4 loop-out assay as above.
To generate an at-locus, marker-less N-terminal FLAG-tagged gdpP strain (HB15857), a similar protocol was used as for PD* construction. A DNA fragment containing gdpP-flag was constructed by overlap extension using up-fragment primers 5684/5685 and down-fragment primers 5246/5686. The flag sequence harbours a PsiI restriction site. After looping out plasmid pMUTIN4, colonies that were white on X-Gal plates and sensitive to MLS were screened by PCR using primers 5684/5686 followed by PsiI digestion. The correct construct was verified by DNA sequencing.
Gene deletions (including constructs of sigD : : kan, flgM : : spc, flgM : : cat) were generated by replacing the coding region with an antibiotic resistance cassette using long flanking homology PCR (LFH-PCR) followed by DNA transformation as described previously (Mascher et al., 2003).
β-Galactosidase activity measurements.
Strains harbouring promoter–lacZ fusions were grown overnight in 5 ml LB broth at 30 °C with vigorous shaking. Cells from 0.5 ml culture were harvested and β-galactosidase assays were performed as described by Miller (1972). Each strain was tested in biological triplicates and repeated three times. Data are reported as the mean and sem.
5′ Rapid amplification of cDNA ends (5′-RACE).
The transcriptional start site of antisense gdpP was determined using 5′-RACE. Five pairs of primers (5586/5587, 5588/5589, 5590/5591, 5592/5593, 5594/5595) were used to map antisense transcripts initiated at the +800 to 1980 bp region relative to the gdpP start codon. For each 5′-RACE, 5 µl total RNA from a mid-exponential-phase LB culture of strain 168 cells was reverse-transcribed to cDNA using TaqMan reverse transcription reagents (Roche) and the first primer (5586, 5588, 5590, 5592 or 5594). The 3′ end of cDNA was tailed with poly-dCTP using terminal deoxynucleotidyltransferase (New England Biolabs). The tailed cDNAs were then amplified by PCR with primer AAP (4549) and the second primer (5587, 5589, 5591, 5593 or 5595, respectively). The PCR products were subject to DNA sequencing.
Immunoprecipitation and Western blotting.
The FLAG-tagged GdpP and FabF were purified using immunoprecipitation with Anti-FLAG M2 Affinity Gel (Sigma-Aldrich) and quantified by Western blotting analysis. An overnight LB culture of B. subtilis cells harbouring both gdpP-flag and/or fabF-flag constructs was inoculated into 300 ml LB broth at an initial OD600 of 0.01, and grown to OD600 0.5–0.6 at 37 °C with vigorous shaking. Cells were harvested by centrifugation and resuspended in 4 ml lysis buffer [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1 % Triton X-100, 1 mM EDTA and 2 mg lysozyme ml−1]. The cell suspension was incubated at 37 °C for 20 min, followed by sonication and centrifugation at 9000 g for 5 min at 4 °C. Anti-FLAG resins were added to the supernatant, and incubated at 4 °C overnight with gentle agitation. The anti-FLAG resins were washed three times with TBS [50 mM Tris/HCl (pH 7.4), 150 mM NaCl], and proteins bound to the resins were eluted by resuspending in 50 µl SDS-sample buffer [62.5 mM Tris/HCl (pH 6.8), 2 % SDS, 10 % (v/v) glycerol, 50 mM DTT and 0.002 % bromophenol blue] and boiled for 5 min. The samples were briefly centrifuged, and proteins in the supernatant were resolved on a 10 % SDS-PAGE gel and electrophoretically transferred to an Immunblot PVDF membrane (Bio-Rad). Membranes were blocked in TBS with 0.5 % Tween 20 (TBST) supplemented with 5 % non-fat milk for 0.5 h at room temperature, followed by incubation with anti-FLAG rabbit antibody (Sigma-Aldrich) in TBST supplemented with 0.5 % non-fat milk overnight at room temperature. The membrane was washed three times for 10 min each in TBST, and incubated with alkaline phosphatase-coupled secondary goat anti-rabbit antibody for 1 h (Sigma-Aldrich). After being washed three times for 10 min each in TBST, the membrane was cut into two pieces to separate the GdpP-FLAG- and FabF-FLAG-containing sections, and developed for 10 and 1 min, respectively. The developing reagent contained 100 mM Tris/HCl (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 300 µg BCIP (5-bromo-4-chloro-3-indolylphosphate) ml−1 and 300 µg ml−1 Nitro Blue Tetrazolium substrate (Bio-Rad). The relative level of GdpP-FLAG in each strain was normalized to the internal control FabF-FLAG using densitometry analysis with ImageJ (Girish & Vijayalakshmi, 2004).
Disc diffusion and MIC assays.
Disc diffusion assays were performed as described previously (Luo et al., 2010), with minor modifications. Mueller–Hinton (MH) broth (Sigma-Aldrich) and a MOPS-based glucose minimal medium (MM) (Bsat et al., 1996) were used for these assays. The bottom agar was 15 ml MH or MM broth supplemented with 1.5 % agar, and the top agar was 4 ml MH or MM broth supplemented with 0.75 % agar. Chemical discs were prepared using Whatman filter paper discs (7 mm diameter) and freshly made chemical stocks. For each chemical test, three different amounts were used: aztreonam 6, 30 and 60 µg; cefuroxime 3, 6 and 12 µg; cefixime 5, 10 and 15 µg; and sodium azide 5, 10 and 30 mmol. The zone of growth inhibition was measured after overnight growth at 37 °C. MIC tests were performed as described previously (Luo & Helmann, 2012), except that the media used were MH and MM broths.
Cell lysis assay.
Fresh colonies were first grown in LB, MH or MM broth to OD600 0.4, diluted 1 : 100 in fresh LB, MH or MM, respectively, and inoculated in Bioscreen microtitre plates with a total volume of 200 µl. Growth was measured spectrophotometrically (OD600) using a Bioscreen incubator (Growth Curves USA) at 37 °C with vigorous shaking. For cefuroxime-induced lysis, five concentrations of cefuroxime (0.5, 1, 2, 4 or 8 µg ml−1, final) were added to the cell culture at two growth phases (exponential phase at OD600 0.3–0.4 and transition phase at OD600 0.6–0.7).
Motility and biofilm formation.
Swimming and swarming motility tests were performed as described by Kearns & Losick (2005) and Patrick & Kearns (2009), with some modification. Freshly made LB agar was cooled to 55 °C, poured into Petri dishes (25 ml per plate) and dried in a laminar flow hood for 20 min before use. Cells were grown to mid-exponential phase (OD600 0.4) or transition phase (OD600 0.7), harvested and resuspended in phosphate buffer to OD600 10. Ten microlitres of cells was spotted on the swimming plate (LB supplemented with 0.3 % agar) or swarming plate (LB supplemented with 0.7 % agar and 5 µg tetrazolium violet ml−1) and dried in a laminar flow hood for another 10 min. For swimming tests, the plates were incubated at 37 °C, and the swimming zones were measured every 30 min until they reached the edge of the plate (~5 h). For swarming tests, plates were incubated at room temperature (22 °C) overnight. Tetrazolium violet is a redox dye that is imported into the cell and reduced to a purple-coloured formazan. It does not affect cell growth or motility, but helps to mark the swarming zone. Biofilm formation tests were performed as described by Branda et al. (2006) and Romero et al. (2010). We tested pellicle formation in Msgg liquid medium and colony morphology on Msgg-supplemented 1.5 % agar plates.
Results and Discussion
An antisense transcript is encoded within gdpP
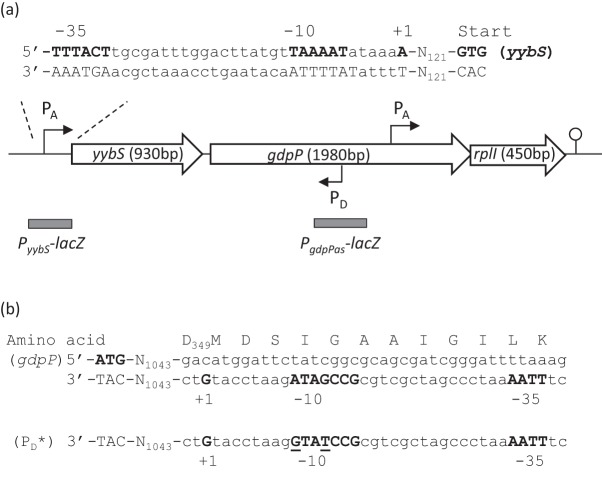
The gdpP gene (formerly yybT) is the second gene in a three-gene operon (yybS-gdpP-rplI) (Fig. 1a). Recent transcriptomic surveys using high-density cDNA tiling arrays and RNA-Seq have suggested that there are at least three transcripts emanating from this locus (Irnov et al., 2010; Nicolas et al., 2012; Rasmussen et al., 2009). The major transcript is initiated from a σA-dependent promoter located 121 bp upstream of yybS. Within the gdpP coding region, there is a putative σA-dependent promoter located upstream of the essential gene rplI (Nicolas et al., 2012). The third transcript is a putative antisense RNA encoded within gdpP. As this antisense transcript could potentially affect the expression of GdpP we investigated its expression and function.
Fig. 1.
A σD-dependent promoter in gdpP. (a) Schematic map of the yybS-gdpP-rplI operon. Promoter sites are indicated by bent arrows with a subscript to indicate the relevant holoenzyme. The −35, −10 and +1 elements of PyybS and the start codon of yybS are in bold type. The balloon indicates a transcriptional terminator. The DNA regions included in the PyybS-lacZ and PgdpPas-lacZ fusions are also illustrated. (b) Promoter sequence of gdpPas. The sense gdpP start codon and the −35, −10 and +1 elements of the antisense PgdpPas are shown in bold type. PD* mutations are underlined.
The antisense gdpP (gdpPas) was first annotated as transcript shd124 by Rasmussen et al. (2009), and more recently as S1559 by Nicolas et al. (2012). However, assignment of the start site and length for this antisense RNA differed in those two studies. While Rasmussen and co-workers suggested a 1429 nt transcript starting from genome nucleotide position 4164574 (GenBank accession no. AL009126.3), Nicolas and co-workers assigned its start site to position 4164565 and predicted its length as 667 nt. To verify expression of this antisense transcript and define its transcriptional start site, we performed 5′-RACE with total RNA isolated from exponential phase cells grown in LB broth. Five pairs of primers were used to map any antisense transcripts initiated within a region of 800–1980 bp downstream of the gdpP start codon. One transcript, designated gdpPas, was identified and its start site was mapped to 1048 bp downstream of the start codon, corresponding to the genome nucleotide location of 4164576. The start site of gdpPas is proximal to a candidate σD promoter (Fig. 1) that is positionally conserved within the genomes of closely related bacilli (Fig. 2).
Fig. 2.
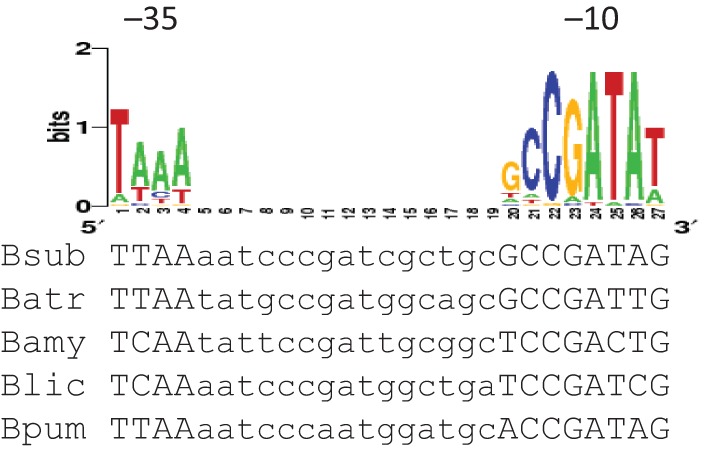
Alignment of PgdpPas elements from various bacilli species. The gdpP sequence of B. subtilis 168 (Bsub) was aligned with the corresponding sequences from Bacillus atrophaeus 1942 (Batr), Bacillus amyloliquefaciens FZB42 (Bam), Bacillus licheniformis ATCC 14580 (Blic) and Bacillus pumilus SAFR-032 (Bpum) using clustalw2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The WebLogo of the σD promoter consensus from B. subtilis is shown above the alignment (Sierro et al., 2008). The −35 and −10 elements are indicated.
We next tried to determine the size and abundance of the sense gdpP transcript and antisense gdpPas transcript using Northern blotting and radioactively labelled oligonucleotide probes. However, we were unable to detect signals corresponding to either transcript (data not shown). It is possible that the expression levels of these transcripts are below the detection threshold, or perhaps more likely that the transcripts are unstable due to the formation of an RNA–RNA duplex. By examining the tiling data in detail, we noticed the transcript signals fading away after about 600 nt (Nicolas et al., 2012; Rasmussen et al., 2009). No terminators were found in the vicinity of this region, nor was an extended RNA transcript detected in RNA prepared from a rho mutant as monitored using tiling arrays (Nicolas et al., 2012). The termination mechanism for the antisense transcript therefore remains unknown. For simplicity reasons, here we adopt the annotation of 667 nt as suggested by Nicolas et al. (2012), which is a conservative estimate.
The gdpPas transcript is dependent on σD
σD is an alternative σ factor that mainly regulates the expression of autolysins and genes involved in flagella biosynthesis and chemotaxis. The activity of σD is mediated primarily by the anti-σ factor FlgM, which sequesters σD and inhibits its activity. An flgM mutant displays increased sigD activity and hence increased expression of genes within the σD regulon (Caramori et al., 1996; Fredrick & Helmann, 1996). To determine whether the predicted gdpPas promoter is active, we constructed an ectopic promoter–lacZ reporter (amyE : : PgdpPas-lacZ) and introduced it into a WT strain (strain 168) and into isogenic strains possessing sigD and/or flgM mutations. This promoter fusion includes sequences from −230 to +60 bp relative to the transcription start site. The sense yybS promoter–lacZ fusion was also constructed and used as a control (Fig. 1a). Expression of PgdpPas-lacZ was completely eliminated in a sigD mutant or a sigD flgM double mutant, but was highly induced in an flgM mutant (Fig. 3). In contrast, transcription of PyybS-lacZ was not affected by mutations in either sigD or flgM. This result suggests that the antisense promoter is active and that it is σD-dependent.
Fig. 3.
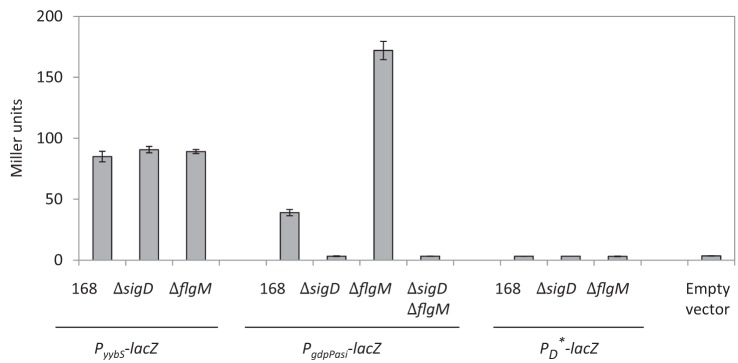
β-Galactosidase activity of PyybS-lacZ, PgdpPas-lacZ and PD*-lacZ in different strain backgrounds in LB overnight cultures. This experiment was performed in biological triplicate and repeated three times. Error bars, sem.
To further verify that the gdpPas promoter activity is due to σD, we constructed a PD*-lacZ fusion in which the −10 element GCCGACA sequence was changed to GCCTATG (mutated nucleotides underlined). These two nucleotide mutations alter two of the most conserved residues in the σD promoter consensus, yet maintain the sense GdpP amino acid sequence (Figs 1b and 2). As expected, PD*-lacZ expressed no β-galactosidase in the WT strain or the sigD/flgM mutant backgrounds (Fig. 3).
gdpPas transcription reduces the level of GdpP protein in the cell
As we could not detect any ORFs within the gdpPas transcript, and as gdpPas shares perfect complementarity with the sense gdpP mRNA, we hypothesized that gdpPas functions as a cis-acting regulatory RNA. Base-pairing between a cis-acting RNA and its target mRNA typically alters target RNA stability and/or modulates sense translation (Georg & Hess, 2011), both of which alter expression levels of the sense-encoded protein. To measure the expression level of GdpP and the effects of gdpPas transcription, we constructed an at-locus gdpP allele encoding an N-terminal FLAG-tag for immunodetection. This tagged allele of gdpP was introduced into WT and isogenic mutants of sigD, flgM and sigD flgM. Direct attempts to monitor GdpP-FLAG accumulation by Western blotting using crude cell lysates and anti-FLAG antibodies were unsuccessful, probably due to the low abundance of GdpP in all strain backgrounds. We therefore utilized immunoprecipitation to enrich GdpP-FLAG and then used Western blotting to quantify the recovered protein. As an internal control we used strains also expressing a FabF-FLAG protein (Kingston et al., 2011). Using this strategy, we determined that GdpP-FLAG accumulation increased in a sigD or a sigD flgM mutant, but decreased in an flgM mutant (relative to levels in the WT strain background) (Fig. 4). As σD activity is known to be quite heterogeneous in growing cell populations (Kearns & Losick, 2005), perhaps the most biologically relevant comparison of GfpP-FLAG accumulation is between the flgM mutant (wherein gdpas is highly expressed) and the sigD mutant (wherein gdpas is not expressed). We estimate an overall change in GfpP-FLAG accumulation of about 2.5- to threefold when comparing the sigD and flgM mutant strains. Thus, the gdpPas transcript can serve as a negative regulator of GdpP expression.
Fig. 4.
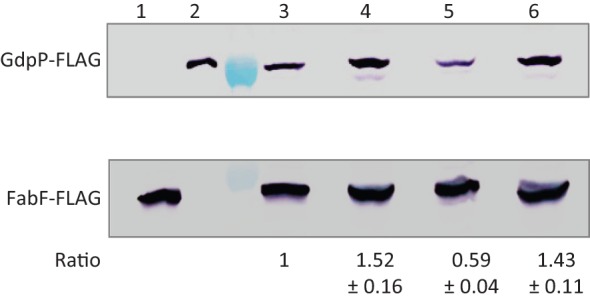
Detection of GdpP-FLAG and FabF-FLAG by immunoprecipitation followed by SDS-PAGE and Western blotting with anti-FLAG antibodies. The sections of the membrane containing GdpP-FLAG and FabF-FLAG were separated after antibody hybridization, and developed separately using a chromogenic assay. Strains used were: lane 1, fabF-FLAG (HB13056); 2, gdpP-FLAG (HB15845); 3, gdpP-FLAG fabF-FLAG (HB15857); 4, sigD gdpP-FLAG fabF-FLAG (HB15858); 5, flgM gdpP-FLAG fabF-FLAG (HB15859); 6, sigD flgM gdpP-FLAG fabF-FLAG (HB15860). The lane between lanes 2 and 3 is a protein ladder, where the 75 and 37 kDa markers are visible at the sections of GdpP-FLAG and FabF-FLAG proteins, respectively. This experiment was repeated three times, and one representative experiment is shown. The intensities of GdpP-FLAG bands were normalized with the internal protein control FabF-FLAG. The numbers below each band represent the fold change (mean±sem) of normalized GdpP-FLAG relative to strain HB15857 (lane 3).
Lack of gdpPas transcription does not lead to obvious cell lysis or motility phenotypes
We next investigated the possible biological effects of gdpPas transcription by monitoring functions known or postulated to be related to GdpP function. Previously, we showed that c-di-AMP is involved in PG homeostasis (Luo & Helmann, 2012). A gdpP mutant (exhibiting increased c-di-AMP levels) is more resistant to PG-targeting antibiotics such as the β-lactam drugs and, conversely, overexpression of gdpP confers β-lactam sensitivity. As demonstrated above, transcription of gdpPas is dependent on σD, and σD is known to regulate the expression of the major vegetative autolysins (LytC, LytD and LytF) which degrade PG (Blackman et al., 1998; Helmann et al., 1988; Lazarevic et al., 1992; Margot et al., 1994, 1999; Serizawa et al., 2004). We therefore hypothesized that σD downregulates GdpP expression (thereby leading to elevated c-di-AMP levels) as a mechanism to upregulate PG biosynthesis concomitant with the upregulation of autolysins under σD control. To test this idea, we mutated the gdpPas promoter and generated an at-locus marker-less PD* mutant, which eliminates σD-dependent gdpPas transcription without affecting the expression of other σD regulon genes (Figs 1b and 3). We predicted that cells carrying PD* would be unable to downregulate GdpP synthesis when σD is activated, and that this might lead to an imbalance between PG biosynthetic and autolytic functions. In an attempt to detect such an imbalance we treated both a WT strain (PDwt, strain 168) and the strain carrying the mutant promoter (PD*, HB15837) with the cell autolysis inducers sodium azide and three β-lactam antibiotics (cefuroxime, aztreonam and cefixime) and monitored cell lysis rates by OD600 and c.f.u. counts. We also examined chemical susceptibility by disc diffusion and MIC assays. However, the response differences between PDwt and PD* strains were either not significant or not reproducible with large standard errors (data not shown). We reasoned that this is probably due to the temporary and heterogeneous nature of σD activity in the our laboratory strain (strain 168) background.
The heterogeneity of σD is due to multiple factors, including the magnitude of processive transcription through the long fla/che (sigD) operon, expression of the anti-σ factor FlgM, and transcriptional regulators SwrA, SwrB, DegU and SlrA/SinR/SlrR (Amati et al., 2004; Calvio et al., 2008; Cozy & Kearns, 2010; Cozy et al., 2012; Fredrick & Helmann, 1996; Hsueh et al., 2011; Kearns & Losick, 2005; Tsukahara & Ogura, 2008). As a result, a growing B. subtilis population contains both σD active (ON) and inactive (OFF) cells. The fraction of σD ON cells is influenced by growth phase, nutrient levels and the parental strain background: exponential phase, the presence of amino acids in the growth medium and domestic strains (e.g. strain 168) are associated with low σD activity, whereas transition phase, the absence of amino acids and the undomesticated strain (strain NCIB 3610) appear to have a relatively high fraction of σD ON cells. There is no known specific growth condition that can universally induce σD activity in a population. To test the effect of the PD* mutation in a population with relatively high levels of σD activity, we conducted the same experiments using strain NCIB 3610 in a variety of different growth media (LB, MH and minimal medium) and growth phases (exponential, transition, early stationary and late stationary phases). However, no significant differences were observed between the PDwt and PD* strains, at least with respect to cell autolysis and β-lactam susceptibility. It has been reported that a full σD ON population can be achieved by simultaneously mutating flgM and overexpressing sigD (Cozy & Kearns, 2010). In this case, however, the highly expressed autolysins in this strain background caused rapid cell lysis and may have masked the effect of gdpPas, as no phenotypes were attributable to the PD* mutation (data not shown). To separate the effects of loss of gdpPas transcription from autolysin activity, we mutated all three major autolysins, lytC, lytD and lytF, in this σD ON strain background (flgM Pspank-sigD). Again, we did not observe differences between PD* and PDwt strains in terms of cell lysis and β-lactam susceptibility. It is important to note that this autolysin-defective strain grows mainly as chains of un-separated cells, which may obscure the analysis of cell lysis using OD600 as an indicator. We conclude that the detection of a gdpPas phenotype may require other factors besides high cellular σD activity.
We next considered the possibility that modulation of GdpP levels by gdpPas is most important under conditions where c-di-AMP synthesis is reduced. Therefore, we also introduced disA, sigM (encoding a disA activator) and ybbP mutations into NCIB 3610 strains carrying flgM and flgM PD* mutations. These three mutations are known to reduce c-di-AMP levels and result in increased sensitivity to cefuroxime (Luo & Helmann, 2012). However, we could not detect any differences between PD* and PDwt alleles in these strains in terms of either cell lysis rates or β-lactam susceptibility. Despite our various efforts, we were not able to associate gdpPas with an observable phenotype. Nevertheless, the presence of a conserved, σD-dependent gdpPas among closely related Bacillus species (Fig. 2) suggests that it likely plays a (still elusive) biological role.
Besides autolysis and β-lactam susceptibility, we also tested whether c-di-AMP affects cell motility for two reasons: (1) σD directs expression of genes involved in flagella biosynthesis and chemotaxis, and is therefore essential for cell motility; and (2) c-di-GMP, a related yet distinct secondary messenger molecule in bacteria, is known to regulate the transition between a motile and a sessile lifestyle, where cells with high c-di-GMP are non-motile and form robust biofilms (Hengge, 2009). c-di-GMP has not been detected in B. subtilis. However, a gdpP mutant (with increased c-di-AMP levels) in Staph. aureus has been reported to form about threefold more biofilm than the WT, which is reminiscent of the effect of c-di-GMP (Corrigan et al., 2011). To test cell motility and biofilm formation, strains harbouring mutations in the c-di-AMP cyclase genes (disA, ybbP and yojJ) or the hydrolase gene gdpP were constructed. An IPTG-inducible gdpP construct (amyE : : Pspac(hy)-gdpP) was introduced into these strains to reduce the level of c-di-AMP. We did not observe significant changes under these genetic conditions when compared with the WT strain NCIB 3610, suggesting that c-di-AMP may not be involved in cell motility (swimming and swarming) or biofilm formation, at least under our test conditions.
Concluding remarks
c-di-AMP is an essential second messenger in B. subtilis. Here, we show that the c-di-AMP hydrolase GdpP is subject to post-transcriptional regulation via an antisense RNA, gdpPas, which thereby represents a novel means of regulating c-di-AMP levels in the cell. Antisense RNAs are a widespread regulatory mechanism and include both trans- and cis-acting RNAs. In both cases, the regulatory RNA typically acts by annealing to its target and affecting either translation or mRNA stability (Thomason & Storz, 2010). Recent genome-wide studies have revealed over 100 potential regulatory RNA transcripts in B. subtilis (Irnov et al., 2010; Nicolas et al., 2012; Rasmussen et al., 2009). Only a few antisense RNA molecules have been studied in detail, including three cis-acting (ratA, SR4 and antisense of yabE) and two trans-acting (fsrA and bsrF) sRNAs (Eiamphungporn & Helmann, 2009; Gaballa et al., 2008; Jahn et al., 2012; Preis et al., 2009; Silvaggi et al., 2005). Antisense RNA typically base-pairs with its target RNA, forming an RNA–RNA duplex, which alters the half-life of the target RNA, and therefore influences protein product accumulation. Transcription of gdpPas is initiated in the middle of the gdpP gene and is dependent on σD. As demonstrated here, this cis-acting regulatory RNA can modulate the cellular levels of GdpP, although the biological role of this regulation remains elusive. Further studies are needed to better understand the regulatory network that links σD, the gdpPas regulatory transcript, gdpP expression and c-di-AMP levels with effects on PG homeostasis.
Acknowledgements
We thank Dr Win Chai (Harvard University) for strain NCIB 3610, and Dr Daniel Kearns (Indiana University) for strains DS793 (3610 amyE : : Phag-lacZ cat), DS2447 (NCIB 3610 lytABC : : spc lytD : : mls lytF : : tet amyE : : Phag-GFP cat) and DS3988 (NCIB 3610 ΔflgM amyE : : Physpank-sigD kan thrC : : PsigD-lacZ mls). This work was supported by NIH grant GM-047446 to J. D. H.
Abbreviations:
- c-di-AMP
cyclic-di-AMP
- DAC
diadenylate cyclase
- MLS
macrolide-lincomycin-streptogramin
- PG
peptidoglycan
- 5′-RACE
5′ rapid amplification of cDNA ends
- WT
wild-type
References
- Amati G., Bisicchia P., Galizzi A. (2004). DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol 186, 6003–6014. 10.1128/JB.186.18.6003-6014.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M., Oppenheimer-Shaanan Y., Berlatzky I., Rouvinski A., Meyerovich M., Ben-Yehuda S. (2006). A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125, 679–690. 10.1016/j.cell.2006.03.039 [DOI] [PubMed] [Google Scholar]
- Blackman S. A., Smith T. J., Foster S. J. (1998). The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144, 73–82. 10.1099/00221287-144-1-73 [DOI] [PubMed] [Google Scholar]
- Branda S. S., Chu F., Kearns D. B., Losick R., Kolter R. (2006). A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59, 1229–1238. 10.1111/j.1365-2958.2005.05020.x [DOI] [PubMed] [Google Scholar]
- Bsat N., Chen L., Helmann J. D. (1996). Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol 178, 6579–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvio C., Osera C., Amati G., Galizzi A. (2008). Autoregulation of swrAA and motility in Bacillus subtilis. J Bacteriol 190, 5720–5728. 10.1128/JB.00455-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Kobel P. A., Morshedi M. M., Wu M. F., Paddon C., Helmann J. D. (2002). Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol 316, 443–457. 10.1006/jmbi.2001.5372 [DOI] [PubMed] [Google Scholar]
- Caramori T., Barilla D., Nessi C., Sacchi L., Galizzi A. (1996). Role of FlgM in σD-dependent gene expression in Bacillus subtilis. J Bacteriol 178, 3113–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R., Allen A. G., Owen P. J., Shalom G., Stone K., Harrison M., Burgis T. A., Lockyer M., Garcia-Lara J. & other authors (2009). Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics 10, 291. 10.1186/1471-2164-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M., Abbott J. C., Burhenne H., Kaever V., Gründling A. (2011). c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7, e1002217. 10.1371/journal.ppat.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozy L. M., Kearns D. B. (2010). Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol 76, 273–285. 10.1111/j.1365-2958.2010.07112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozy L. M., Phillips A. M., Calvo R. A., Bate A. R., Hsueh Y. H., Bonneau R., Eichenberger P., Kearns D. B. (2012). SlrA/SinR/SlrR inhibits motility gene expression upstream of a hypersensitive and hysteretic switch at the level of σD in Bacillus subtilis. Mol Microbiol 83, 1210–1228. 10.1111/j.1365-2958.2012.08003.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W., Helmann J. D. (2008). The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol Microbiol 67, 830–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W., Helmann J. D. (2009). Extracytoplasmic function sigma factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA. J Bacteriol 191, 1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K., Helmann J. D. (1996). FlgM is a primary regulator of σD activity, and its absence restores motility to a sinR mutant. J Bacteriol 178, 7010–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. T., Lao P., Loraine A. E., Matthews B. T., Yu H., Dybvig K. (2008). Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol Microbiol 69, 67–76. 10.1111/j.1365-2958.2008.06262.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A., Baysse C., Koedam N., Muyldermans S., Cornelis P. (1998). Different residues in periplasmic domains of the CcmC inner membrane protein of Pseudomonas fluorescens ATCC 17400 are critical for cytochrome c biogenesis and pyoverdine-mediated iron uptake. Mol Microbiol 30, 547–555. 10.1046/j.1365-2958.1998.01085.x [DOI] [PubMed] [Google Scholar]
- Gaballa A., Antelmann H., Aguilar C., Khakh S. K., Song K. B., Smaldone G. T., Helmann J. D. (2008). The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A 105, 11927–11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J., Hess W. R. (2011). cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75, 286–300. 10.1128/MMBR.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girish V., Vijayalakshmi A. (2004). Affordable image analysis using NIH Image/ImageJ. Indian J Cancer 41, 47. [PubMed] [Google Scholar]
- Glass J. I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M. R., Maruf M., Hutchison C. A., III, Smith H. O., Venter J. C. (2006). Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A 103, 425–430. 10.1073/pnas.0510013103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury A. M., Frandsen N., Stragier P. (1996). Plasmids for ectopic integration in Bacillus subtilis. Gene 180, 57–61. 10.1016/S0378-1119(96)00404-0 [DOI] [PubMed] [Google Scholar]
- Harwood C. R., Cutting S. M. (1990). Molecular Biological Methods for Bacillus. New York: Wiley. [Google Scholar]
- Helmann J. D., Márquez L. M., Chamberlin M. J. (1988). Cloning, sequencing, and disruption of the Bacillus subtilis σ28 gene. J Bacteriol 170, 1568–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- Hsueh Y. H., Cozy L. M., Sham L. T., Calvo R. A., Gutu A. D., Winkler M. E., Kearns D. B. (2011). DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis. Mol Microbiol 81, 1092–1108. 10.1111/j.1365-2958.2011.07755.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irnov I., Sharma C. M., Vogel J., Winkler W. C. (2010). Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38, 6637–6651. 10.1093/nar/gkq454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn N., Preis H., Wiedemann C., Brantl S. (2012). BsrG/SR4 from Bacillus subtilis – the first temperature-dependent type I toxin–antitoxin system. Mol Microbiol 83, 579–598. [DOI] [PubMed] [Google Scholar]
- Kearns D. B., Losick R. (2005). Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19, 3083–3094. 10.1101/gad.1373905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston A. W., Subramanian C., Rock C. O., Helmann J. D. (2011). A σW-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Mol Microbiol 81, 69–79. 10.1111/j.1365-2958.2011.07679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V., Margot P., Soldo B., Karamata D. (1992). Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol 138, 1949–1961. 10.1099/00221287-138-9-1949 [DOI] [PubMed] [Google Scholar]
- Luo Y., Helmann J. D. (2012). Analysis of the role of Bacillus subtilis σM in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83, 623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Asai K., Sadaie Y., Helmann J. D. (2010). Transcriptomic and phenotypic characterization of a Bacillus subtilis strain without extracytoplasmic function σ factors. J Bacteriol 192, 5736–5745. 10.1128/JB.00826-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margot P., Mauël C., Karamata D. (1994). The gene of the N-acetylglucosaminidase, a Bacillus subtilis 168 cell wall hydrolase not involved in vegetative cell autolysis. Mol Microbiol 12, 535–545. 10.1111/j.1365-2958.1994.tb01040.x [DOI] [PubMed] [Google Scholar]
- Margot P., Pagni M., Karamata D. (1999). Bacillus subtilis 168 gene lytF encodes a γ-d-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative σ factor, σD. Microbiology 145, 57–65. 10.1099/13500872-145-1-57 [DOI] [PubMed] [Google Scholar]
- Mascher T., Margulis N. G., Wang T., Ye R. W., Helmann J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50, 1591–1604. [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M. & other authors (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106. 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y., Wexselblatt E., Katzhendler J., Yavin E., Ben-Yehuda S. (2011). c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep 12, 594–601. 10.1038/embor.2011.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick J. E., Kearns D. B. (2009). Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J Bacteriol 191, 7129–7133. 10.1128/JB.00905-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis H., Eckart R. A., Gudipati R. K., Heidrich N., Brantl S. (2009). CodY activates transcription of a small RNA in Bacillus subtilis. J Bacteriol 191, 5446–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F., See R. Y., Zhang D., Toh D. C., Ji Q., Liang Z. X. (2010). YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem 285, 473–482. 10.1074/jbc.M109.040238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F., Ji Q., Soehano I., Liang Z. X. (2011). Unusual heme-binding PAS domain from YybT family proteins. J Bacteriol 193, 1543–1551. 10.1128/JB.01364-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S., Nielsen H. B., Jarmer H. (2009). The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol 73, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D., Aguilar C., Losick R., Kolter R. (2010). Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107, 2230–2234. 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U. (2008). Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal 1, pe39. 10.1126/scisignal.133pe39 [DOI] [PubMed] [Google Scholar]
- Serizawa M., Yamamoto H., Yamaguchi H., Fujita Y., Kobayashi K., Ogasawara N., Sekiguchi J. (2004). Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses. Gene 329, 125–136. 10.1016/j.gene.2003.12.024 [DOI] [PubMed] [Google Scholar]
- Sierro N., Makita Y., de Hoon M., Nakai K. (2008). DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res 36 (Database issue), D93–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvaggi J. M., Perkins J. B., Losick R. (2005). Small untranslated RNA antitoxin in Bacillus subtilis. J Bacteriol 187, 6641–6650. 10.1128/JB.187.19.6641-6650.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. H., Ko K. S., Lee J. Y., Baek J. Y., Oh W. S., Yoon H. S., Jeong J. Y., Chun J. (2005). Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol Cells 19, 365–374. [PubMed] [Google Scholar]
- Thomason M. K., Storz G. (2010). Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 44, 167–188. 10.1146/annurev-genet-102209-163523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara K., Ogura M. (2008). Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol 8, 8. 10.1186/1471-2180-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V., Dervyn E., Ehrlich S. D. (1998). A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144, 3097–3104. [DOI] [PubMed] [Google Scholar]
- Witte G., Hartung S., Büttner K., Hopfner K. P. (2008). Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell 30, 167–178. [DOI] [PubMed] [Google Scholar]
- Woodward J. J., Iavarone A. T., Portnoy D. A. (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. 10.1126/science.1189801 [DOI] [PMC free article] [PubMed] [Google Scholar]