Background: Vfa1 was recently identified as a Vps4-binding protein, yet its function was unknown.
Results: Vfa1 potently stimulates Vps4 ATPase activity, and Vps4-Vfa1 interaction consists of a binding mechanism previously seen in other Vps4 regulator structures.
Conclusion: Biochemical and biophysical studies show that Vfa1 is a novel regulator of Vps4 in the multivesicular body pathway.
Significance: Vps4-Vfa1 interaction further expands the complexity of Vps4 regulation.
Keywords: ATPases, Crystal Structure, Endosomes, Membrane Trafficking, Protein-Protein Interactions, ESCRT, MVB, Vfa1, Vps4
Abstract
The endosomal sorting complexes required for transport (ESCRT) are responsible for multivesicular body biogenesis, membrane abscission during cytokinesis, and retroviral budding. They function as transiently assembled molecular complexes on the membrane, and their disassembly requires the action of the AAA-ATPase Vps4. Vps4 is regulated by a multitude of ESCRT and ESCRT-related proteins. Binding of these proteins to Vps4 is often mediated via the microtubule-interacting and trafficking (MIT) domain of Vps4. Recently, a new Vps4-binding protein Vfa1 was identified in a yeast genetic screen, where overexpression of Vfa1 caused defects in vacuolar morphology. However, the function of Vfa1 and its role in vacuolar biology were largely unknown. Here, we provide the first detailed biochemical and biophysical study of Vps4-Vfa1 interaction. The MIT domain of Vps4 binds to the C-terminal 17 residues of Vfa1. This interaction is of high affinity and greatly stimulates the ATPase activity of Vps4. The crystal structure of the Vps4-Vfa1 complex shows that Vfa1 adopts a canonical MIT-interacting motif 2 structure that has been observed previously in other Vps4-ESCRT interactions. These findings suggest that Vfa1 is a novel positive regulator of Vps4 function.
Introduction
Controlling and remodeling membrane structure is a vital process for cellular homeostasis. Topologically related events, such as multivesicular body (MVB)2 biogenesis, retroviral budding, and abscission between daughter cells during cytokinesis, require the action of a class of proteins collectively known as the endosomal sorting complexes required for transport (ESCRT) (1–9). The ESCRT proteins are unique in that they deform cellular membranes to generate curvature and bud away from the cytosol (7, 10). The molecular mechanisms underlying the function of these proteins are similar but distinct in different cellular processes, and they are best studied in the context of MVB biogenesis.
The ESCRT proteins can be grouped into five distinct multimeric protein complexes, ESCRT-0, -I, -II, -III, and Vps4 (11). It is believed that they work in a sequential manner to target cell surface receptors into MVB for eventual vacuolar/lysosomal degradation (12). Cargo molecules are first ubiquitinated and endocytosed into early endosome, where they are recognized by ESCRT-0 (13). Next, ESCRT-I and -II target and concentrate cargo molecules to the site of endosomal membrane deformation and may also directly cause change in membrane curvature (14). To complete the process of budding and vesicle formation, ESCRT-III oligomerizes and forms a filament structure on the endosomal membrane, which ultimately leads to scission of a forming vesicle (15, 16). Ubiquitin tags are removed before cargo molecules are included in the vesicle. Finally, the ATPase Vps4 is recruited to the site of vesicle formation. Through ATP hydrolysis, Vps4 is likely to drive the completion of vesicle formation in an irreversible direction and ultimately provide energy for the entire process (17, 18). Biochemically, Vps4 catalyzes the removal of ESCRT-III oligomers from the endosomal membrane (19). This serves two critical functions: first, to provide proper temporal and spatial control of the ESCRT-III oligomerization, thus generating appropriately sized vesicles (20), and second, to recycle components of ESCRT-III to allow for continuous cargo trafficking (21). The role of the ESCRT proteins in MVB biogenesis is evolutionarily conserved in eukaryotic cells (6).
Vps4 is a member of the AAA-ATPase (ATPase associated with diverse cellular activities) family. The crystal structure of Vps4 shows that it contains an N-terminal microtubule-interacting and trafficking (MIT) domain and a C-terminal AAA-ATPase domain connected by a flexible linker region (22–24). Vps4 exists in two quaternary structures, as a monomer in the absence of ATP binding and as a hexameric ring upon ATP binding (22, 25). Oligomerization of Vps4 is mediated through the AAA-ATPase domain with ATP binding sites located at the subunit interface. The hexameric ring structure of Vps4 is a transient species in solution, only stable with ATP bound. Vps4 binds to many proteins in the cell, most of which are mediated through the MIT domain (26). The MIT domain adopts a three-helix bundle structure that serves as a binding partner for the MIT-interacting motifs (MIMs) of ESCRT-III and ESCRT-III-related proteins. The MIM was initially defined as a small sequence motif (∼15–20 residues) at or near the C-terminal end of ESCRT-III and ESCRT-III-related proteins (27). There have been speculations that the MIM binds to the ESCRT-III protein core structure in the autoinhibited state of ESCRT-III. In the active state, it is exposed to the solvent and can be recognized by the MIT domain (28, 29). Hence, the MIM serves as a bifunctional switch of ESCRT-III function. Based on the structure and the mode of interaction with the MIT domain, several different types of MIM have been described (30–36). For example, a MIM1 forms a small helix that binds between the second and third helices of the Vps4 MIT domain (30, 31). Alternatively, a MIM2 adopts an extended structure that binds between the first and third helices (32). Binding of ESCRT-III to Vps4 can enhance the ATP hydrolysis by Vps4, although the mechanism of action is not clear (37). In addition to ESCRT-III and ESCRT-III-related proteins, there are also other regulators of Vps4 function. Vta1, a positive regulator of Vps4 ATPase, binds to a unique β-domain within the AAA-ATPase domain and presumably activates Vps4 by stabilizing the hexameric ring structure required for ATP hydrolysis (38, 39).
A novel Vps4-binding protein, called Vfa1 (Vps4-associated-1), was recently identified in an overexpression study of all putative genes in Saccharomyces cerevisiae to look for vacuole morphology defects (40). Overexpression of Vfa1 caused altered vacuole size, indicative of MVB dysfunction. Most interestingly, Vfa1 was found to interact with Vps4. Given the lack of knowledge about Vfa1 and how essential Vps4 regulation is, we used biochemical and biophysical methods to show that Vfa1 binds to the Vps4 MIT domain. We determined a high resolution crystal structure of the Vps4 MIT domain in complex with a C-terminal Vps4-binding fragment of Vfa1 and identified residues important for binding. Vfa1 binds to Vps4 with high affinity (∼0.47 μm) and can stimulate Vps4 ATPase activity by more than 10-fold. These results suggest that Vfa1 is a bona fide regulator of Vps4 ATPase activity with a possible role in MVB regulation.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
DNA fragments encoding Vfa1 and Vps4 and their various fragment constructs were amplified from the S. cerevisiae genomic DNA. These proteins were expressed in Escherichia coli Rosetta(DE3) cells using a modified pET28b vector with a SUMO protein tag inserted between a His6 tag and the respective protein coding region. The His6-SUMO-tagged protein was purified by Ni2+-nitrilotriacetic acid affinity chromatography following standard procedures. ULP1 protease was then added to remove the His6-SUMO tag, and the digested protein mixture was passed over a second Ni2+-nitrilotriacetic acid column. Proteins were further purified by ion exchange chromatography on a Source-S column (GE Healthcare) for Vfa1 proteins and a Source-Q column (GE Healthcare) for Vps4 proteins. The Vps4-Vfa1 complex was purified by mixing the two protein fragments in stoichiometric amounts followed by gel filtration chromatography using a HiLoadTM SuperdexTM 200 (GE Healthcare) column.
GST Pull-down Analysis
GST pull-down analysis was performed following standard procedures in phosphate-buffered saline (PBS) solution supplemented with 1 mm DTT and 0.1% (v/v) Tween 20 (22). Specific samples were also supplemented with 2 mm ATP as indicated. Purified proteins were incubated with either GST alone or GST-tagged proteins immobilized on glutathione-agarose beads for 1 h at 4 °C. The beads were then washed extensively with the buffer before bound proteins were analyzed on SDS-PAGE and visualized by Coomassie staining.
Isothermal Titration Calorimetry
To measure the binding affinity between Vfa1 and the Vps4 MIT domain (residues 1–82), Vfa1 or Vfa1 mutants were titrated against the Vps4 MIT domain using a Nano ITCTM (TA Instruments) or an ITC-200 microcalorimeter (GE Healthcare) at 25 °C. Data were processed using the NanoAnalyze software. All proteins were dialyzed against a buffer containing 50 mm HEPES (pH 7.5) and 50 mm NaCl, centrifuged to remove any particulates, and degassed before binding analysis. Vfa1 proteins were injected at 0.23 mm (or greater for mutants) against 0.06 mm Vps4 MIT domain. The concentrations of all proteins were determined using UV spectral analysis.
Malachite Green ATPase Assay
The procedure was adopted from previous studies with modifications as indicated (41). The reaction buffer used included 100 mm Tris-HCl (pH 7.4), 20 mm KCl, 6 mm MgCl2. Malachite green reagent was prepared as follows. Malachite green (0.081%, w/v), polyvinyl alcohol (2.3%, w/v), ammonium heptamolybdate tetrahydrate (5.7% (w/v) in 6 m HCl), and double-distilled water were mixed at a 2:1:1:2 ratio and then stored at 4 °C for at least 30 min. For Vps4 stimulation experiments, a 15-μl protein mixture of 0.125 μm Vps4 and 0.25 μm of Vfa1, Vfa1 mutant, or Vta1 was incubated with 10 μl of 1.2 mm ATP (all final concentrations) at 37 °C for 10 min in a 96-well plate. Protein concentrations were selected in order to see the stimulation of Vps4 hydrolysis and to avoid product inhibition. After the 10-min incubation, 80 μl of malachite green reagent was added to each well. Immediately following this step, 10 μl of 32% sodium citrate was used to inhibit the non-enzymatic hydrolysis of ATP. The samples were mixed thoroughly and incubated at 37 °C for 15 min before A620 was measured on a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA). To account for intrinsic hydrolysis, the signal from ATP in identically treated buffer lacking Vps4 was subtracted.
Structural Study
Vps4-Vfa1 complex crystals were grown using the sitting drop vapor diffusion method at 25 °C. The protein complex (17 mg/ml) was mixed in a 1:1 ratio with a reservoir solution of 26% (w/v) polyethylene glycol 4000, 10% (v/v) isopropyl alcohol, and 0.1 m HEPES, pH 6.5, in a final volume of 4 μl and equilibrated against 1 ml of reservoir solution. Clusters of sword-shaped crystals appeared after 1 day and grew to full size after 3 days. Single crystals were isolated from the clusters and cryoprotected using the reservoir solution.
Diffraction data were collected at Advanced Photon Source Beamline 21-ID-D. The Vps4-Vfa1 complex crystal diffracted to 2.3 Å. It belongs to the space group of P212121 and contains two molecular complexes in the asymmetric unit. Data were indexed, integrated, and scaled using HKL2000 (HKL Research). The PHENIX software suite was used for structure determination and refinement. The structure was solved by molecular replacement as implemented in the AutoMR module (42), using the Vps4 MIT domain (Protein Data Bank accession code 2V6X) as an initial search model. Non-crystallographic symmetry restraint was applied throughout refinement. Model building was done with COOT (43).
RESULTS
Vfa1 Binds to Vps4 in an ATP-independent Manner
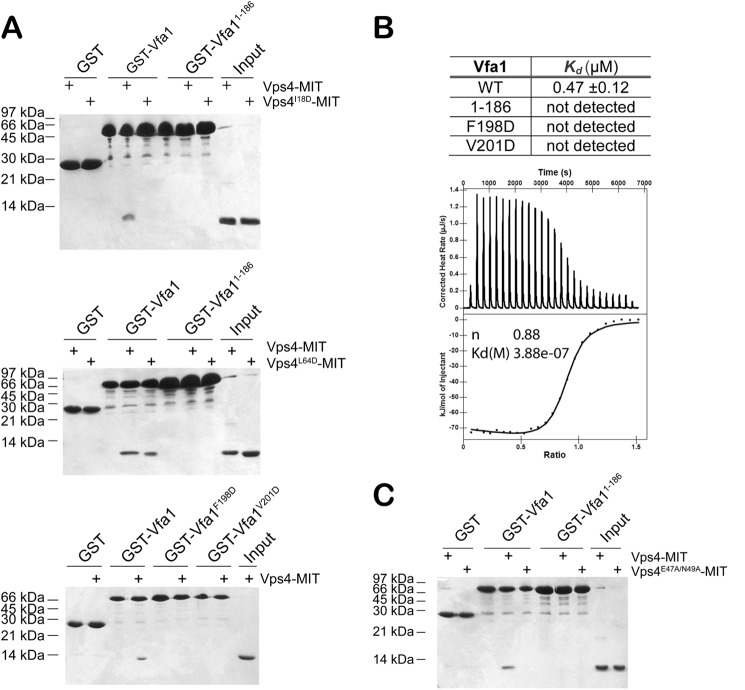
We first sought to confirm the interaction between Vps4 and Vfa1 as reported by Arlt et al. (40) using glutathione S-transferase (GST) pull-down analysis. Vps4 has been shown to exist in two oligomeric states, as a monomer and, upon binding ATP, as a hexameric ring (2, 24). Many binding partners of Vps4, including ESCRT-III and ESCRT-III-related proteins, show an ATP dependence for Vps4 binding, suggesting that they only bind to the hexameric form of Vps4 (22, 39). To maintain Vps4 in a stable hexameric structure, we used an ATPase-deficient mutant (E233Q) of Vps4 (44). We saw that Vfa1 could bind to Vps4 in both the absence and presence of ATP (Fig. 1A). We further used size exclusion chromatography to validate the interaction between Vps4 and Vfa1. Different proteins or protein mixtures (Vfa1, Vfa1 with 2 mm ATP, Vps4, Vps4 with 2 mm ATP, Vps4 plus Vfa1, and Vps4 plus Vfa1 with 2 mm ATP) were loaded onto a 120-ml Superdex-200 column, respectively. Compositions of elution peaks were examined by SDS-polyacrylamide gel analysis. Regardless of the presence of ATP, Vfa1 co-eluted with Vps4 at a position that was distinct from Vfa1 alone, suggesting that it formed a stable complex with Vps4 in solution (Fig. 1B). Therefore, Vfa1 can bind to either a monomeric or hexameric form of Vps4.
FIGURE 1.
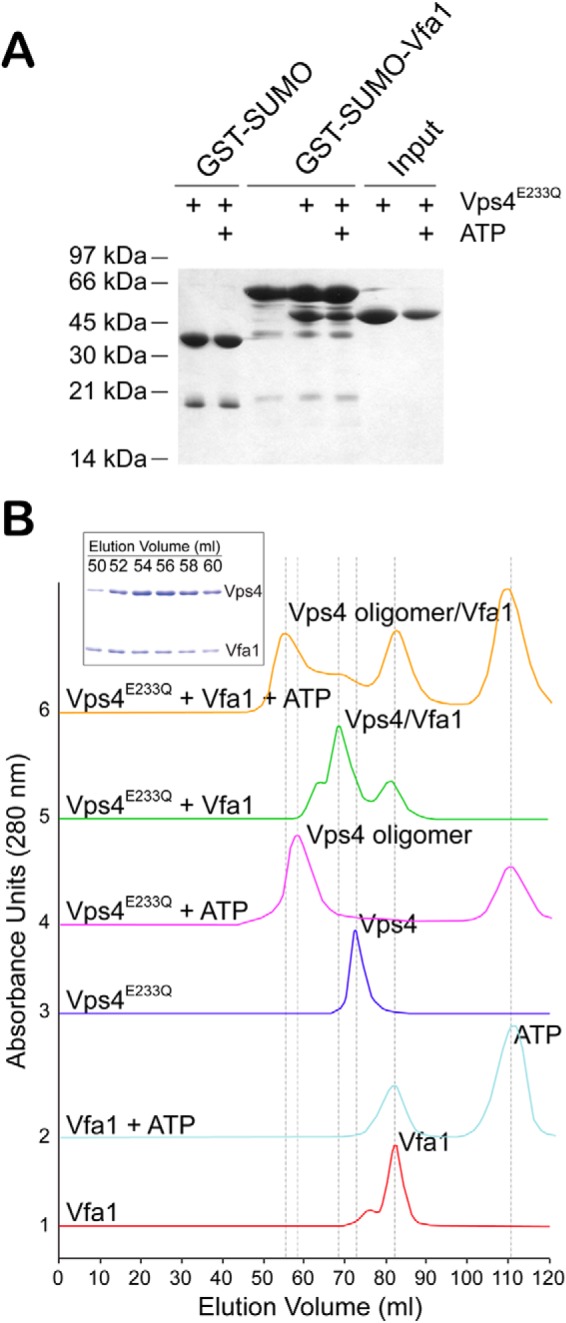
Vfa1 binds to Vps4 in an ATP-independent manner. A, GST-SUMO or GST-SUMO-Vfa1 was used to pull down Vps4E233Q. GST-SUMO was used as a tag because GST-Vfa1 runs at a position similar to that of Vps4E233Q. Binding analyses were performed in the presence or absence of ATP to determine whether Vfa1 binds to Vps4 monomer or oligomer. Proteins retained on the beads were analyzed by SDS-PAGE and visualized by Coomassie staining. B, size exclusion chromatography was used to examine compositions of molecular species in the following samples: Vfa1, Vfa1 + 2 mm ATP, Vps4E233Q, Vps4E233Q + 2 mm ATP, Vfa1 + Vps4E233Q, and Vfa1 + Vps4E233Q + 2 mm ATP. The following protein concentrations were used: Vps4, 33 μm; Vfa1, 100 μm. Peak fractions were collected, analyzed by SDS-PAGE, and visualized by Coomassie staining. The inset gel shows the compositions for the major peak in the Vfa1 + Vps4E233Q + 2 mm ATP sample run as described above.
Vps4-Vfa1 Interaction Is Mediated through the N-terminal MIT Domain of Vps4
We next identified the minimal binding domains between the two proteins. Vps4 contains an N-terminal MIT domain and a C-terminal canonical AAA-ATPase domain. Because most of the ESCRT-III and ESCRT-III-related proteins bind to the MIT domain, we reasoned that it might also be the site of binding for Vfa1. Indeed, deletion of the MIT domain (Vps4-ΔMIT) abolished the interaction between Vps4 and Vfa1. This was reminiscent of the interaction between Vps4 and Ist1, an ESCRT-III-related Vps4 regulator that binds to Vps4 via the MIT domain in an ATP-independent manner (45), but different from another regulator, Vta1, which binds to Vps4 in an ATP-dependent manner via the β-domain within the ATPase domain (Fig. 2A). ESCRT-III proteins bind to the MIT domain via C-terminal sequence motifs, known as the MIMs (46). Secondary structure prediction showed that Vfa1 was largely an α-helical protein with a C-terminal helix suggestive of a potential site for MIT interaction (data not shown). We therefore made a truncation mutant of Vfa1 [Vfa1(1–186)] with the C-terminal 17 residues deleted and tested whether it could still bind to Vps4. Removal of the C-terminal sequence abolished its interaction with Vps4 (Fig. 2B). These results show that Vps4-Vfa1 binding is mediated through the N-terminal MIT domain of Vps4.
FIGURE 2.
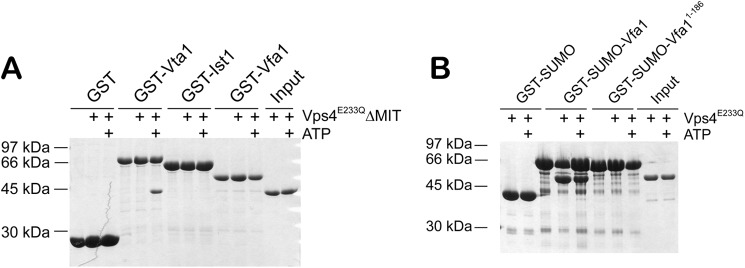
Mapping the binding interface between Vps4 and Vfa1. A, the Vps4 MIT domain is necessary for Vfa1 binding. GST, GST-Vta1, GST-Ist1, and GST-Vfa1 were used to analyze their interactions with Vps4E233QΔMIT (residues 83–437) in the presence or absence of ATP. Vta1 is known to only bind to the oligomeric form of Vps4. Ist1 is an ESCRT-III-related protein that binds to the Vps4 MIT domain. B, the C-terminal sequence of Vfa1 is necessary for Vps4 binding. GST-SUMO, GST-SUMO-Vfa1, or GST-SUMO-Vfa1(1–186) was used to analyze their interactions with Vps4E233Q in the presence or absence of ATP.
Binding of Vfa1 Stimulates the ATPase Activity of Vps4
When ESCRT-III and ESCRT-III-related proteins bind to Vps4, they often stimulate the ATPase activity of Vps4 (37, 47). To determine whether this was the case for Vfa1, we used the Malachite green ATPase assay (41) to examine how Vfa1 might affect the ATPase activity of Vps4 (Fig. 3). As a control, the addition of bovine serum albumin (BSA) did not stimulate the ATPase activity of Vps4. Vta1, a known positive regulator of Vps4, was able to increase the rate of Vps4 ATP hydrolysis 3-fold when added in a 1:2 ratio. In comparison, the addition of Vfa1 in a similar ratio increased the Vps4 ATPase activity approximately 10-fold as compared with the intrinsic activity of Vps4, although Vfa1 itself had no ATPase activity. Vfa1 therefore appeared to be a much more potent activator of Vps4 than Vta1. To examine whether this stimulation was dependent on the site of interaction that we identified, we also included Vfa1(1–186) in the Vps4 ATPase assay. No stimulation was seen upon the addition of Vfa1(1–186), demonstrating that stimulation by Vfa1 requires the specific interaction between Vps4 and Vfa1.
FIGURE 3.
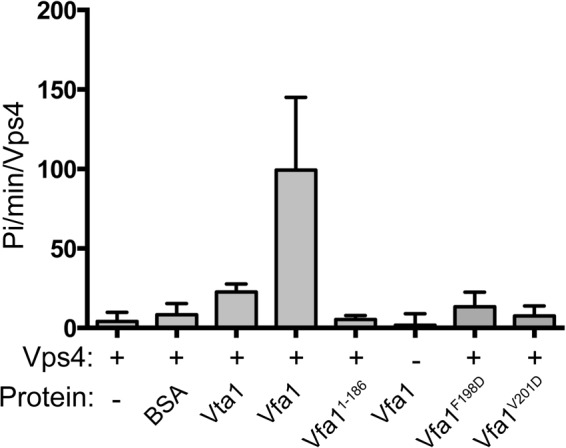
Vfa1 stimulates Vps4 ATPase activity. ATP hydrolysis by 0.125 μm Vps4 (intrinsic, first column) was assayed under conditions of 1.2 μm ATP at 37 °C (see “Experimental Procedures”). To observe stimulation of Vps4 activity, the following proteins (0.25 μm) were incubated with Vps4: bovine serum albumin (second column), Vta1 (third column), Vfa1 (fourth column), Vfa1(1–186) (fifth column), Vfa1F198D (seventh column), and Vfa1V201D (eighth column). Each experiment was done in triplicate; all experiments were repeated a minimum of three times. Intrinsic ATPase activity for Vfa1 (0.25 μm) is shown in the sixth column. Error bars, S.D.
Crystal Structure of Vps4 MIT and Vfa1 MIM
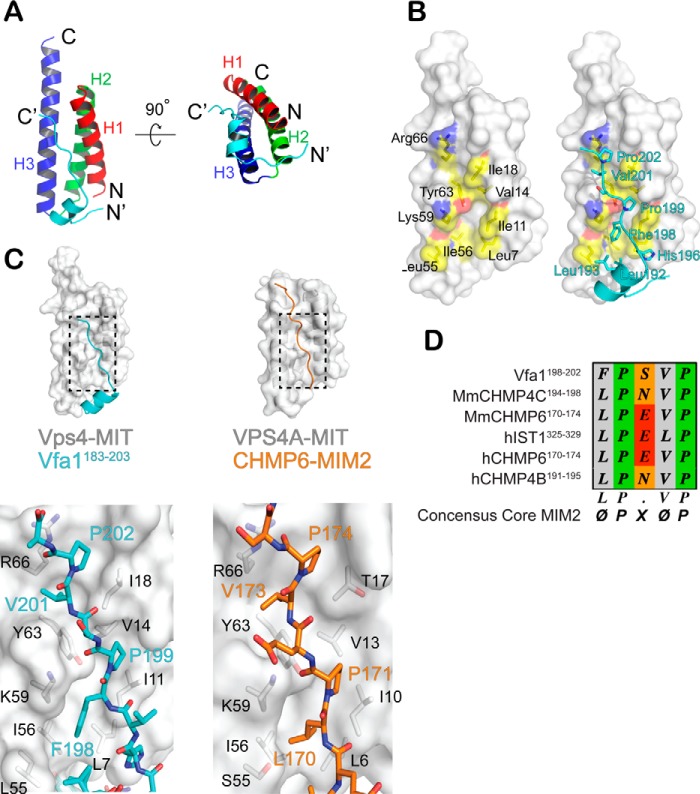
To understand the structural nature of the Vps4-Vfa1 interaction, we determined the crystal structure of the Vps4-Vfa1 complex using the Vps4 MIT domain (residues 1–82) and a C-terminal fragment of Vfa1 that encompasses the necessary Vps4-binding sequence (residues 183–203). The structure was determined by molecular replacement using the yeast Vps4 MIT domain structure as a search model (31) and refined to 2.3 Å resolution (Table 1). The refined structure has an R-factor/Rfree of 23.6%/25.7% with excellent parameters for stereochemistry. There are two molecular complexes of Vps4-Vfa1 in the asymmetric unit. The r.m.s deviations for Cα positions between the two complexes are 0.039 and 0.672 Å for Vps4 and Vfa1, respectively. The average buried surface area at the interface of Vps4 and Vfa1 is ∼900 Å2.
TABLE 1.
Crystallographic data statistics
| Data collection | |
| Space group | P212121 |
| Unit cell parameters | |
| a, b, c (Å) | 38.5, 56.7, 84.2 |
| Molecules per asymmetric unit | 2 |
| Wavelength (Å) | 0.9998 |
| Resolution (Å) | 2.3 |
| Unique reflectionsa | 8662 (840) |
| Redundancy | 7.1 (7.3) |
| Completeness (%) | 99.9 (100.0) |
| Average I/σ (I) | 34.6 (7.3) |
| Rmerge | 0.04 (0.34) |
| Refinement | |
| Resolution range (Å) | 35.00–2.30 |
| Rwork (%) | 23.6 |
| Rfree (%) | 25.7 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (degrees) | 1.40 |
| B-factor average (A2) | 43.1 |
| Ramachandran plot regions | |
| Most favored (%) | 95.9 |
| Allowed (%) | 4.1 |
| Outliers (%) | 0.0 |
| Protein Data Bank code | 4NIQ |
a Values in parentheses are for the specified high resolution bin.
As shown in Fig. 4A, the Vps4 MIT domain adopts a three-helix bundle fold (H1–H3). Binding of Vfa1 to Vps4 does not change the overall structure of the domain. The r.m.s deviations for Cα positions between the Vps4 MIT domain structure in the Vfa1 complex and other previously reported Vps4 MIT domain structures are in the range of 0.6–0.7 Å. The Vps4-binding sequence of Vfa1 can be divided into three regions: an N-terminal extended region (residues 183–188), a short two-turn α-helix in the middle (residues 189–196), and a C-terminal extended region (residues 197–203). Interactions between Vps4 and Vfa1 are mediated through all three regions. The N-terminal extended region binds to the turn between the second and third helices of the MIT domain, whereas the middle helix and the C-terminal extended region binds predominantly between the first and third helices of the MIT domain.
FIGURE 4.
The crystal structure of Vps4-Vfa1 complex. A, an overview of the Vps4-Vfa1 complex structure. Shown is a ribbon representation of Vfa1 (residues 183–203) in complex with Vps4 (1–82) in two orthogonal views. Vfa1 is colored in cyan, and the three helices of the Vps4 MIT domain are colored red, green, and blue, respectively. The N and C termini of the two proteins are labeled. B, the C-terminal extended region of Vfa1 binds in a hydrophobic pocket formed by helices 1 and 3 of the Vps4 MIT domain. Left, a surface representation of the Vps4 MIT domain and the Vfa1 binding pocket selectively colored based on the underlying atoms: carbon (yellow), oxygen (red), and nitrogen (blue). Residues that contribute to binding are labeled in black. Right, the same surface representation with Vfa1 shown in cyan. Critical hydrophobic residues utilized by Vfa1 for binding Vps4 are labeled in cyan. C, the C-terminal extended region of Vfa1 binds like a MIM2. The top panels show ribbon and surface representations of the Vps4-Vfa1 complex and the hVPS4A-CHMP6 complex (Protein Data Bank code 2K3W). The bottom panels show zoom-in views of the interactions within the two complex structures. Critical residues from the MITs are labeled in black. Residues that make close contacts from the MIM2s are in cyan for Vfa1 and orange for CHMP6. D, sequence alignment of various MIM2s.
Molecular Interactions between the C-terminal Extended Region of Vfa1 Fragment and Vps4
Vps4 presents a hydrophobic binding pocket between the first and third helices. As shown in Fig. 4B, this hydrophobic pocket includes side chains from residues Leu-7, Ile-11, Val-14, Ile-18 (helix 1) and Leu-55, Ile-56, and Tyr-63 (helix 3). In addition, the aliphatic portions of side chains from residues Lys-59 and Arg-66 also contribute to the pocket hydrophobicity, which allows for discrete interactions with the following Vfa1 residues: Leu-192, Leu-193, His-196, Phe-198, Pro-199, Val-201, and Pro-202 (Fig. 4B). A number of specific hydrogen bond interactions are also present at the interface. For example, the side chain of Vfa1 residue His-196 forms a hydrogen bond with the main chain carbonyl oxygen of Vps4 residue Gly-4. A complete list of molecular interactions observed at the interface is tabulated in Table 2.
TABLE 2.
Detailed interactions between Vps4 and Vfa1
| Vps4 | Vfa1 |
|---|---|
| Hydrogen bond interactions | |
| Gly-4 O | His-196 OE2 |
| Tyr-46 O | Ser-185 N |
| Glu-47 OE1 | Thr-187 OG1 |
| Glu-47 OE2 | Thr-187 N |
| Lys-48 N | Ser-185 O |
| Asn-49 ND2 | Thr-187 O |
| Asn-49 ND2 | Asp-188 OD1 |
| Ser-52 OG | Thr-187 O |
| Glu-62 OE2 | Val-201 N |
| Tyr-63 OH | Pro-199 O |
| Van der Waals interactions | |
| Leu-7 | Leu-192, His-196, Phe-198 |
| Ile-11 | Phe-198, Pro-199 |
| Val-14 | Pro-199 |
| Ile-18 | Pro-199, Val-201, Pro-202 |
| Leu-40 | Phe-198 |
| Lys-51 | Pro-189 |
| Leu-55 | Pro-189, Leu-192, Leu-194, Phe-199 |
| Ile-56 | Leu-192, Phe-198 |
| Lys-59 | Phe-198 |
| Tyr-63 | Phe-198, Pro-199, Val-201 |
| Arg-66 | Val-201, Pro-202 |
The interaction between Vps4 and the C-terminal extended region of the Vfa1 fragment is of particular interest. It is reminiscent of the MIT-MIM interaction as seen in the structure of human VPS4A and CHMP6, the human homolog of Vps20 (a core ESCRT-III protein) (32). The MIM involved in this type of interaction has been termed as MIM2. In both the Vps4-Vfa1 and the VPS4A-CHMP6 complexes, the MIT domains adopt similar structures. When the conformations of the bound peptides in the two complexes are compared, it is clear that the C-terminal extended region of Vfa1 fragment adopts a conformation similar to that of the CHMP6 MIM2 (Fig. 4C). Moreover, both peptides utilize hydrophobic residues, including two signature prolines, within their sequences to bind to Vps4. Specifically, CHMP6 MIM2 residues (Leu-170, Pro-171, Val-173 and Pro-174) fit into the hydrophobic pocket within the helical groove between helices 1 and 3 in a nearly identical fashion as the Vfa1 fragment.
Previous studies defined MIM2 as an MIT-binding motif that contains a conserved LP(E/D)VP sequence (32). In the Vps4-Vfa1 structure, Vfa1 utilizes a slight different 198FPSVP202 sequence at the core of the binding interface. By aligning the MIT-binding motif of Vfa1 with other MIM2 sequences, we arrived at a more general ΦPXΦP motif (where Φ represents a hydrophobic residue) that is required for a peptide sequence to bind as a MIM2 (Fig. 4D). Based on the structure shown here, the hydrophobic residues from the Vfa1 “MIM2” make contact with the hydrophobic pocket presented by Vps4 MIT domain helices 1 and 3. The variable residue X tends to be polar and always seems to point into solvent in different structures.
Additional Interaction between Vfa1 MIM and Vps4
Although the MIT-MIM2 interactions are similar in the two complex structures, CHMP6 does not contain sequence N-terminal to the MIM2 that makes additional contacts with VPS4A MIT. In Vfa1, these contacts include the N-terminal extended region and the middle helix. The middle helix is amphipathic and binds to the hydrophobic pocket formed by the first and third helices of the MIT domain via Pro-189, Leu-192, and Leu-193, as described above (Fig. 5). Interactions between the N-terminal extended region and Vps4 are largely polar. Vfa1 forms several hydrogen bond interactions with the backbone atoms from the turn between the second and third helices of the Vps4 MIT domain (Fig. 5). Specifically, Vfa1 residues 185–186 and Vps4 residues 46–48 form parallel β-sheet-like main chain hydrogen bonds. The side chain of Vps4 Glu-47 forms bidentate hydrogen bonds with the side chain and the main chain atoms of Vfa1 Thr-187. The amide nitrogen of Vps4 Asn-49 side chain forms bifurcated hydrogen bonds with the main chain carbonyl oxygen atom of Vfa1 Thr-187 and the side chain carboxyl oxygen atom of Vfa1 Asp-188.
FIGURE 5.
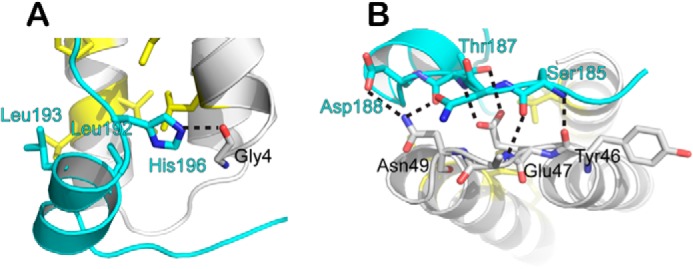
Additional interactions between Vps4 and Vfa1. Shown is a schematic representation of Vfa1 (cyan) making contacts with Vps4 (gray) beyond the MIM2 sequence. A, interactions involving the middle helix; B, interactions involving the N-terminal extended region. Residues that make distinct contacts are shown as stick models and labeled in cyan for Vfa1 and black for Vps4. Dashed lines, hydrogen bonds.
Functional Relevance of Vps4-Vfa1 Interaction
We used site-directed mutagenesis and GST pull-down analysis to evaluate the contribution of interface residues to the overall stability of the complex. Interactions between Vps4 and MIM2-containing ESCRT-III proteins have been shown previously to be disrupted by mutation of Ile-18, a residue located at the hydrophobic pocket formed by helices 1 and 3 of the Vps4 MIT domain (48). Because Ile-18 is also located at the Vps4-Vfa1 interface, we reasoned that this mutant should also prevent Vfa1 from binding. Indeed, I18D failed to bind to GST-Vfa1 in our GST pull-down analysis, suggesting that Ile-18 is essential for high affinity binding interaction (Fig. 6A). In contrast, mutation of Leu-64 of Vps4, a residue important for interactions between Vps4 and MIM1-containing ESCRT-III proteins (30), had no effect on Vfa1 binding (Fig. 6A). Moreover, Vfa1 mutants F198D and V201D also failed to bind to Vps4. As mentioned above, Phe-198 and Val-201 are part of the MIM2 sequence. Collectively, these results confirm the importance of Vfa1 MIM2 sequence to the overall stability of the complex.
FIGURE 6.
Critical residues at the Vps4-Vfa1 binding interface. A, Vfa1 binds in the first and third helical groove of Vps4 MIT as a MIM2. Top, GST, GST-Vfa1, and GST-Vfa1(1–186) were used to analyze their respective interactions with Vps4-MIT and Vps4I18D-MIT. Middle, GST, GST-Vfa1, and GST-Vfa1(1–186) were used to analyze their respective interactions with Vps4-MIT and Vps4L64D-MIT. Bottom, GST, GST-Vfa1, GST-Vfa1F198D, and GST-Vfa1V201D were used to analyze their respective interactions with Vps4-MIT. B, binding affinities of Vps4-MIT to wild-type and mutant Vfa1 as determined by an ITC assay. A representative ITC enthalpy plot for the binding of Vps4-MIT to wild-type Vfa1 is also shown. C, the region outside Vfa1 MIM2 contributes to Vps4 binding. GST, GST-Vfa1, and GST-Vfa1(1–186) were used to analyze their respective interactions with Vps4-MIT and Vps4E47A/N49A-MIT.
Using Isothermal Titration Calorimetry (ITC), we measured the binding affinity between the Vps4 MIT domain and Vfa1. The MIM2-type MIT-MIM interaction has been characterized previously as having a low micromolarity dissociation constant (KD ∼5–10 μm) (32). Here, we saw that full-length Vfa1 bound to the Vps4 MIT domain with a significantly higher affinity (KD = 0.47 μm ± 0.12) (Fig. 6B). This is ∼10-fold higher in affinity as compared with the CHMP6-hVPS4A interaction. This increase in affinity may have been due to the additional contacts between Vps4 and Vfa1 beyond the interaction that involves the C-terminal MIM2 sequence, which are not available in CHMP6. The contribution of Vfa1 sequence beyond MIM2 to the overall binding affinity of the complex was corroborated by the observation that Vps4 mutant E47A/N49A had a significantly decreased affinity for Vfa1 based on GST pull-down analysis (Fig. 6C).
To further study the functional relevance of Vps4-Vfa1 interaction, we examined how point mutations within the Vps4-Vfa1 binding interface might affect the ability of Vfa1 to stimulate Vps4 ATPase activity. Two Vfa1 mutants, F198D and V201D, which had drastically reduced affinities for Vps4 by both GST pull-down analysis and ITC, were unable to stimulate the ATPase activity of Vps4 (Fig. 3). This again suggests that the specific interaction between Vfa1 and Vps4 is important for its role as a potential positive regulator of Vps4.
DISCUSSION
Oligomerization of ESCRT-III proteins on the endosomal membrane is a critical step in generating intraluminal vesicles during MVB biogenesis. This step is terminated by the removal of ESCRT-III proteins from the membrane through the action of the AAA-ATPase Vps4. The activity of Vps4 is tightly regulated in the cell by a multitude of proteins that specifically bind to Vps4. A newly identified protein, Vfa1, which causes defects in vacuolar morphology upon overexpression in yeast, has been shown previously to bind to Vps4. In order to understand the molecular mechanism of Vfa1 function, we embarked on a detailed structural study of the interaction between Vps4 and Vfa1. We have confirmed that Vfa1 binds to Vps4 in an ATP-independent manner (40). In addition, the crystal structure of the Vps4-Vfa1 complex reveals that Vfa1 binds to the N-terminal MIT domain of Vps4, utilizing a conserved ΦPXΦP binding motif (MIM2) that is shared among some ESCRT-III and ESCRT-III-related proteins. Moreover, binding of Vfa1 to Vps4 significantly stimulates the ATP hydrolysis activity of Vps4. These data strongly suggest that Vfa1 is a positive regulator of Vps4 in the MVB pathway.
Vfa1 Binds to the Vps4 MIT Domain Using a Canonical MIM2
The MIT domain is a protein-protein interaction domain mostly found in proteins involved in membrane trafficking (Vps4, AMSH, UBPY, Vta1, Spastin, and Spartin) (26, 49). Protein sequence motifs that specifically bind to the MIT domain are collectively termed MIMs. They are often found at the C-terminal ends of ESCRT-III and ESCRT-III-related proteins and can bind to the MIT domain in a variety of manners, utilizing different binding surfaces on the MIT domain. The MIT domain of Vps4 contains two MIM-binding surfaces. In the crystal structure of the Vps4-Vps2 complex, the Vps2 C-terminal sequence forms a small helix that binds between helices 2 and 3 of the Vps4 MIT domain (31). In the solution structure of hVPS4A-CHMP6 complex, the C-terminal sequence of CHMP6 adopts an extended conformation and binds between helices 1 and 3 of the hVPS4A MIT (32). These two structures help define that any MIM that binds in a similar fashion (same orientation, with conserved binding interactions) would be a MIM1 (between helices 2 and 3) or a MIM2 (between helices 1 and 3). The complexity in binding illustrates the mechanism by which MIT domains discriminate specific binding partners. In the current work, we show that the C-terminal sequence of Vfa1 binds to the Vps4 MIT domain in a manner similar to that of CHMP6. Therefore, the structure should be classified as a MIM2. This finding strongly suggests that Vfa1 plays a role related to the function of Vps4 in the MVB pathway.
Beyond the MIM2 sequence, the molecular interfaces within the hVPS4A-CHMP6 complex and the Vps4-Vfa1 complex are significantly different. hVPS4A makes additional interactions with CHMP6 residues C-terminal to the MIM2. These interactions do not exist in Vfa1, because its MIM2 sequence is located at the very C-terminal end. For Vfa1, the sequence N-terminal to the ΦPXΦP motif makes additional contacts with Vps4 in the form of a short helix and an extended loop. As shown by mutagenesis and GST pull-down analysis, these interactions do contribute to the overall stability of the complex. The CHMP6 fragment used in the structural study does not include additional sequence N-terminal to the MIM2. It is possible that the small helix seen in the Vfa1 structure might also exist in CHMP6 if more residues were included in the structural study. Indeed, secondary structure prediction of CHMP6 indicated the possibility of a small helix upstream of the MIM2 sequence. Nevertheless, the binding mechanism beyond the canonical MIT-MIM2 interface is likely to be distinct between the two complex structures. These differences suggest that interactions outside the consensus MIM2 sequence may provide a mechanism for MIT domains to distinguish among their various binding partners. Hence, regulation of Vps4 (and other ESCRT proteins) via MIT-MIM interaction can be more finely tuned than previously thought. More MIT-MIM complex structures will be needed to properly decipher the MIT-MIM “binding code.”
Regulation of Vps4 by Vfa1
As noted previously, overexpression of Vfa1 in yeast cells leads to an alteration in vacuolar morphology (40). Because Vfa1 can bind to Vps4 in both in vivo and in vitro assays (40), these data seem to suggest that Vfa1 has an inhibitory role in Vps4 regulation. We have now shown that Vfa1 binds to the Vps4 MIT domain and contains a MIM2 sequence that has been identified previously in some ESCRT-III proteins. Binding of Vfa1 to Vps4 is ATP-independent and can greatly stimulate the ATPase activity of Vps4. Therefore, our biochemical and biophysical analysis instead suggest a positive regulatory role for Vfa1.
How does one reconcile the difference between the biochemical and biological data? The binding affinity of Vfa1 to Vps4 is about 10–20-fold higher than those of core ESCRT-III MIM2s to Vps4 (32). While in cytosol, Vfa1 can form a complex with Vps4, which may stabilize the Vps4 oligomer, and is then recruited to the endosomal membrane. Due to an avidity effect of the ESCRT-III oligomer, although individually having a weaker affinity for Vps4 than Vfa1, the ESCRT-III oligomer can displace Vfa1 from Vps4 to allow for ESCRT-III disassembly. In a Vfa1 overexpression system, however, Vfa1 would outcompete the ESCRT-III proteins and block their binding to Vps4, thus preventing the proper function of Vps4.
Deletion of Vfa1 in yeast has no observable phenotype on vacuolar morphology. It is possible that other ESCRT-III-related proteins may have overlapping function with Vfa1. Vfa1 does not seem to be essential for ESCRT-III filament disassembly but more likely plays a role in the temporal and spatial control of Vps4 function. Many known regulators of Vps4 have weak or no phenotype in their respective deletion strains (vta1, vps60, did2, ist1) (50). It is only when two or more of these genes are deleted that we see a synthetic phenotype of vacuolar morphology (47, 51). Moreover, EM tomography has often revealed more subtle effects on MVB biogenesis in a single gene deletion strain of these proteins (20). Taken together, the lack of a distinct phenotype on vacuolar morphology suggests that Vfa1 acts at a similar level as other regulators of Vps4. It will be interesting to see whether a synthetic phenotype exists when Vfa1 is deleted in conjunction with other ESCRT-III-related proteins. Yeast-two hybrid analysis showed that Vfa1 could bind to Vta1, and preliminary biochemical characterization suggested that other ESCRT-III and ESCRT-III-related proteins may also bind to Vfa1 (52) (data not shown). How Vfa1 works together with other ESCRT proteins to regulate Vps4 is a critical next step in understanding ESCRT function.
Acknowledgments
We thank the staff at the Advanced Photon Source Sector 21 (21-ID-D) for access and help with data collection. We thank J. Rauch and J. Gestwicki for assistance with Malachite green ATP assays and K. Chinnaswamy and B. Wan for access and help for ITC experiments. We thank D. Garwon for help with data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant GM095769 (to Z. X.). Use of the Advanced Photon Source, an Office of Science User Facility operated for the United States Department of Energy Office of Science by Argonne National Laboratory, was supported by the Department of Energy under Contract DE-AC02–06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
The atomic coordinates and structure factors (code 4NIQ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MVB
- multivesicular body
- ESCRT
- endosomal sorting complexes required for transport
- AAA
- ATPase associated with diverse cellular activities
- MIT
- microtubule-interacting and trafficking
- ITC
- isothermal titration calorimetry
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Hanson P. I., Cashikar A. (2012) Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28, 337–362 [DOI] [PubMed] [Google Scholar]
- 2. McCullough J., Colf L. A., Sundquist W. I. (2013) Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82, 663–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hurley J. H., Emr S. D. (2006) The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35, 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henne W. M., Buchkovich N. J., Emr S. D. (2011) The ESCRT pathway. Dev. Cell 21, 77–91 [DOI] [PubMed] [Google Scholar]
- 5. Piper R. C., Katzmann D. J. (2007) Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23, 519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winter V., Hauser M. T. (2006) Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 11, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gruenberg J., Stenmark H. (2004) The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5, 317–323 [DOI] [PubMed] [Google Scholar]
- 8. Hurley J. H., Hanson P. I. (2010) Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hill C. P., Babst M. (2012) Structure and function of the membrane deformation AAA ATPase Vps4. Biochim. Biophys. Acta 1823, 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlton J. G., Martin-Serrano J. (2007) Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912 [DOI] [PubMed] [Google Scholar]
- 11. Williams R. L., Urbé S. (2007) The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8, 355–368 [DOI] [PubMed] [Google Scholar]
- 12. Teis D., Saksena S., Emr S. D. (2008) Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589 [DOI] [PubMed] [Google Scholar]
- 13. Bilodeau P. S., Urbanowski J. L., Winistorfer S. C., Piper R. C. (2002) The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4, 534–539 [DOI] [PubMed] [Google Scholar]
- 14. Wollert T., Hurley J. H. (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elia N., Fabrikant G., Kozlov M. M., Lippincott-Schwartz J. (2012) Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys. J. 102, 2309–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Różycki B., Boura E., Hurley J. H., Hummer G. (2012) Membrane-elasticity model of Coatless vesicle budding induced by ESCRT complexes. PLoS Comput. Biol. 8, e1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elia N., Sougrat R., Spurlin T. A., Hurley J. H., Lippincott-Schwartz J. (2011) Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. U.S.A. 108, 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baumgärtel V., Ivanchenko S., Dupont A., Sergeev M., Wiseman P. W., Kräusslich H. G., Bräuchle C., Müller B., Lamb D. C. (2011) Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 13, 469–474 [DOI] [PubMed] [Google Scholar]
- 19. Lata S., Roessle M., Solomons J., Jamin M., Gottlinger H. G., Svergun D. I., Weissenhorn W. (2008) Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 378, 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nickerson D. P., West M., Henry R., Odorizzi G. (2010) Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol. Biol. Cell 21, 1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao J., Xia H., Yoshino-Koh K., Zhou J., Xu Z. (2007) Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J. Mol. Biol. 374, 655–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonciarz M. D., Whitby F. G., Eckert D. M., Kieffer C., Heroux A., Sundquist W. I., Hill C. P. (2008) Biochemical and structural studies of yeast Vps4 oligomerization. J. Mol. Biol. 384, 878–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landsberg M. J., Vajjhala P. R., Rothnagel R., Munn A. L., Hankamer B. (2009) Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure 17, 427–437 [DOI] [PubMed] [Google Scholar]
- 25. Monroe N., Han H., Gonciarz M. D., Eckert D. M., Karren M. A., Whitby F. G., Sundquist W. I., Hill C. P. (2014) The oligomeric state of the active Vps4 AAA ATPase. J. Mol. Biol. 426, 510–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hurley J. H., Yang D. (2008) MIT domainia. Dev. Cell 14, 6–8 [DOI] [PubMed] [Google Scholar]
- 27. Muzioł T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Göttlinger H., Weissenhorn W. (2006) Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10, 821–830 [DOI] [PubMed] [Google Scholar]
- 28. Shim S., Kimpler L. A., Hanson P. I. (2007) Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 29. Rózycki B., Kim Y. C., Hummer G. (2011) SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure 19, 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. (2007) ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744 [DOI] [PubMed] [Google Scholar]
- 31. Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. (2007) Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739 [DOI] [PubMed] [Google Scholar]
- 32. Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. (2008) Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 33. Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C., Hurley J. H. (2008) Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15, 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Solomons J., Sabin C., Poudevigne E., Usami Y., Hulsik D. L., Macheboeuf P., Hartlieb B., Göttlinger H., Weissenhorn W. (2011) Structural basis for ESCRT-III CHMP3 recruitment of AMSH. Structure 19, 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Z., Vild C., Ju J., Zhang X., Liu J., Shen J., Zhao B., Lan W., Gong F., Liu M., Cao C., Xu Z. (2012) Structural basis of molecular recognition between ESCRT-III-like protein Vps60 and AAA-ATPase regulator Vta1 in the multivesicular body pathway. J. Biol. Chem. 287, 43899–43908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Skalicky J. J., Arii J., Wenzel D. M., Stubblefield W. M., Katsuyama A., Uter N. T., Bajorek M., Myszka D. G., Sundquist W. I. (2012) Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport. J. Biol. Chem. 287, 43910–43926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Merrill S. A., Hanson P. I. (2010) Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem. 285, 35428–35438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. (2006) Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta 1. J. Cell Biol. 172, 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiao J., Xia H., Zhou J., Azmi I. F., Davies B. A., Katzmann D. J., Xu Z. (2008) Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell 14, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arlt H., Perz A., Ungermann C. (2011) An overexpression screen in Saccharomyces cerevisiae identifies novel genes that affect endocytic protein trafficking. Traffic 12, 1592–1603 [DOI] [PubMed] [Google Scholar]
- 41. Chang L., Bertelsen E. B., Wisén S., Larsen E. M., Zuiderweg E. R., Gestwicki J. E. (2008) High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal. Biochem. 372, 167–176 [DOI] [PubMed] [Google Scholar]
- 42. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 44. Babst M., Sato T. K., Banta L. M., Emr S. D. (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16, 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., Sundquist W. I. (2009) Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 20, 1360–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. (2005) Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. U.S.A. 102, 13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. (2008) ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell 14, 50–61 [DOI] [PubMed] [Google Scholar]
- 48. Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., Babst M. (2010) Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell 21, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Renvoisé B., Parker R. L., Yang D., Bakowska J. C., Hurley J. H., Blackstone C. (2010) SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol. Biol. Cell 21, 3293–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kotter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 51. Rue S. M., Mattei S., Saksena S., Emr S. D. (2008) Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A. L., Tavernier J., Hill D. E., Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]