Background: LolB accepts lipoproteins from LolA and anchors them to the inner leaflet of outer membranes.
Results: Membrane targeting of lipoproteins is defective in several mutants with substitutions of Leu-68 of LolB.
Conclusion: The protruding loop of LolB plays critical roles in the membrane anchoring activity.
Significance: A possible mechanism for the last step of lipoprotein sorting is proposed.
Keywords: Crystal Structure, Escherichia coli, Lipoprotein, Membrane Proteins, Protein Export, Protein Sorting, Protein Structure, LolB, Bacterial Lipoprotein, Periplasm
Abstract
The Lol system comprising five Lol proteins, LolA through LolE, sorts Escherichia coli lipoproteins to outer membranes. The LolCDE complex, an ATP binding cassette transporter in inner membranes, releases outer membrane-specific lipoproteins in an ATP-dependent manner, causing formation of the LolA-lipoprotein complex in the periplasm. LolA transports lipoproteins through the periplasm to LolB on outer membranes. LolB is itself a lipoprotein anchored to outer membranes, although the membrane anchor is functionally dispensable. LolB then localizes lipoproteins to outer membranes through largely unknown mechanisms. The crystal structure of LolB is similar to that of LolA, and it possesses a hydrophobic cavity that accommodates acyl chains of lipoproteins. To elucidate the molecular function of LolB, a periplasmic version of LolB, mLolB, was mutagenized at various conserved residues. Despite the lack of acyl chains, most defective mutants were insoluble. However, a derivative with glutamate in place of leucine 68 was soluble and unable to localize lipoproteins to outer membranes. This leucine is present in a loop protruding from mLolB into an aqueous environment, and no analogous loop is present in LolA. Thus, leucine 68 was replaced with other residues. Replacement by acidic, but not hydrophobic, residues generated for the first time mLolB derivatives that can accept but cannot localize lipoproteins to outer membranes. Moreover, deletion of the leucine with neighboring residues impaired the lipoprotein receptor activity. Based on these observations, the roles of the protruding loop of LolB in the last step of lipoprotein sorting are discussed.
Introduction
Lipoproteins are membrane proteins widely distributed in both Gram-negative and -positive bacteria (1). They are synthesized as precursors with a signal peptide and then translocated to the outer leaflet of the cytoplasmic (inner) membrane. Processing of lipoprotein precursors into mature forms takes place on the outer leaflet of the inner membrane. In Gram-negative and high GC-containing gram-positive bacteria (diderm bacteria), three enzymes, Lgt, LspA, and Lnt, sequentially process precursors into mature lipoproteins, which have three acyl chains at the conserved N-terminal Cys (2).
Mature lipoproteins in Escherichia coli and other proteobacteria are sorted to the outer membrane by the Lol system, in which an ATP binding cassette transporter in the inner membrane releases outer membrane-specific lipoproteins in an ATP-dependent manner. This causes the formation of a water-soluble complex between a lipoprotein and LolA, a periplasmic chaperone. Lipoproteins are thus able to cross the hydrophilic periplasm to the outer membrane, to which an outer membrane lipoprotein, LolB, is anchored. LolB receives a lipoprotein from LolA by connecting the entrances of their hydrophobic cavities (3) and then anchors the lipoprotein to the outer membrane.
The level of major outer membrane lipoprotein Lpp is about 106 molecules in a single cell (4, 5) and forms a lethal covalent linkage with the peptidoglycan when mislocalized in the inner membrane (6). Because of these properties, Lpp is unfavorable for the isolation of mutants defective in lipoprotein biogenesis and sorting. Indeed, deletion of the lnt gene was only possible in Lpp− cells overproducing LolCDE (7). In contrast, it has been shown that the LolB function is indispensable even in Lpp− cells (8).
Transfer of lipoproteins from LolA to LolB takes place in the direction toward higher affinity. A LolA derivative, LolA(R43L), possessing an Arg to Leu mutation in the hydrophobic cavity, strongly binds lipoproteins and, therefore, cannot transfer them to LolB (9). On the other hand, it remains largely unknown how LolB discharges and anchors a cargo to the inner leaflet of the outer membrane. Previously, five conserved Trp residues were subjected to random mutagenesis (10). Two LolB derivatives, W52P and W117A, were isolated and found to be defective in receptor activity. Only weakly defective mutants were obtained with three other Trp residues despite their strong conservation. The membrane anchor of LolB was later found to be dispensable (11), although an anchor-less LolB derivative, mLolB, is less efficient and cannot distinguish the inner and outer membranes (11). Because mLolB is expressed in the periplasm, as LolA is, the isolation and examination of mLolB mutants is expected to be easier than those of LolB, which is examined after solubilization and reconstitution into proteoliposomes (12). Moreover, the LolB functions comprising lipoprotein binding, membrane targeting, and lipoprotein incorporation into membranes can be dissected with mLolB (11). We, therefore, tried to isolate mLolB mutants by random mutagenesis of 17 conserved residues other than Trp. Most derivatives were insoluble, but mutants as to leucine 68 remained largely soluble and were useful for examining the activities after purification. We show here that the loop of mLolB protruding into an aqueous environment is critical for both the receptor and membrane targeting activities.
EXPERIMENTAL PROCEDURES
Materials
TALON Metal Affinity Resin (Clontech) and ANTI-FLAG M2 Affinity Gel (Sigma) were used to purify His- and FLAG-tagged proteins, respectively. Tran35S-label (mixture of 70% [35S]Met and 20% [35S]Cys) was purchased from MP Biochemicals. n-Dodecyl-β-d-maltopyranoside (DDM)3 was purchased from Dojindo Laboratories (Kumamoto, Japan). Anti-MalE antiserum was purchased from New England Biolabs. A Penta-His HRP Conjugate kit (Qiagen) was used to probe His-tagged Pal.
Bacterial Strains, Plasmids, and Media
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli K12 strain MC4100 (13) was used as the wild-type strain. KT50 and KT60 (8) are ΔlolB derivatives of JE5505 and JE5506 (14), respectively. DH5α (15) was used as a host for routine cloning procedures. BL21(DE3) (15) was used to prepare the Pal(Strep)-LolA(FLAG) and Pal(His)- LolA(FLAG) complexes. Cells were grown on LB broth (BD Biosciences), 2× YT medium (15), or M63 (NaCl)-maltose minimal medium (6) supplemented with ampicillin, chloramphenicol, and spectinomycin at 50, 20, and 50 μg/ml, respectively, when appropriate.
TABLE 1.
Strains, plasmids, and primers used in this study
| E. coli strains | Genotype | Reference |
|---|---|---|
| MC4100 | F− araD ΔlacU169 relA rpsL thi fibB | 13 |
| KT50 | pps his proA argE thi gal lac xyl mtl tsx lpp recA56 srl::Tn10 ΔlolB::kan | 8 |
| KT60 | pps his proA argE thi gal lac xyl mtl tsx recA56 srl::Tn10 ΔlolB::kan | 8 |
| DH5α | supE44 ΔlacU169 (ϕ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 15 |
| BL21(DE3) | F− ompT hsdS dcm gal (λDE3) | 15 |
| Plasmids | Description | Reference |
|---|---|---|
| pTTQ18 | Expression vector, bla, Ptac | Amersham |
| pMAN885EH | Expression vector, cat, PBAD | 6 |
| pCDFDuet-1 | Expression vector, aadA, PT7 | Novagen |
| pRT102 | pTTQ18 derivative encoding mLolB(His) | This study |
| pTAN21 | pTTQ18 derivative encoding Pal (no tag) | 27 |
| pYKT122 | pMAN885EH derivative encoding LolB (no tag) | 11 |
| pYKT123 | pMAN885EH derivative encoding mLolB (no tag) | 11 |
| pNAS021 | pKT021 derivative encoding LolB (no tag), aadA | This study |
| pOS141 | pCDFDuet-1 derivative encoding LolA(FLAG) and Pal(Strep) | 3 |
| pSS9 | pCDFDuet-1 derivative encoding LolA(FLAG) and Pal(His) | 18 |
| Primers | Sequence (5′ → 3′) | |
|---|---|---|
| mLolB(His)-F | cttggcactggccgtcgttt | |
| mLolB(His)-R | ttaatggtgatggtgatggtgtttcactatccagttatcca | |
| aadA-1 | agtggcggttttcatggctt | |
| aadA-2 | aatgcggatgttgcgattac | |
| Δ68 | ggcagcacggaactggag | |
| Δ67–68 | agcacggaactggagctg | |
| Δ68–69 | tgggttagtgagcagcagac | |
| Δ67–69 | gttagtgagcagcagacgg | |
Construction of Plasmids
To construct pRT102, a DNA fragment was amplified by means of PCR from pYKT102 (12) using a pair of primers, mLolB(His)-F and mLolB(His)-R (Table 1), and self-ligated after treatment with T4 polynucleotide kinase (TaKaRa Bio). To construct pNAS021, a DNA fragment was amplified by means of PCR from the chromosomal DNA of RS1104 (16) using a pair of primers, aadA-1 and aadA-2 (Table 1), and then inserted into the ScaI site of pKT021 (8). A plasmid encoding the mLolB(His) mutant with an amino acid substitution at Leu-68 was constructed with a QuikChange II Site-directed Mutagenesis kit (Stratagene) using a pair of mutagenic complementary primers. Plasmids encoding the Δ68, Δ67–68, Δ68–69, and Δ67–69 derivatives of mLolB were constructed by self-ligation of a DNA fragment that was PCR-amplified from pRT102 using a pair of primers, Δ68 plus Δ68–69, Δ68 plus Δ67–69, Δ67–68 plus Δ68–69, or Δ67–68 plus Δ67–69, respectively (Table 1), after treatment with T4 polynucleotide kinase.
Isolation of mLolB Mutants
Site-directed random mutagenesis of mLolB(His) was carried out with the QuikChange II Site-directed Mutagenesis kit with pRT102 as a template and a pair of complementary primers containing a mixture of four nucleotides at the target codon. The plasmid DNAs were amplified in DH5α and used to transform KT60 carrying pYKT122. Two hundred transformants to each target residue were selected on LB agar supplemented with 0.02% arabinose after 14 h of incubation at 37 °C and replicated on LB agar supplemented with or without 50 μm isopropyl β-d-thiogalactopyranoside (IPTG) and/or 0.001% arabinose and incubated for 14 h at 37 °C. The mutations were determined by DNA sequencing.
Subcellular Localization of mLolB Derivatives
KT60 cells harboring pYKT122 and pRT102 derivatives were grown on LB broth in the presence of 0.01% arabinose at 30 °C to the exponential growth phase, and then mLolB(His) derivatives were induced for 1 h by the addition of 50 μm IPTG. Cells were harvested, resuspended in 20 mm Tris-HCl, pH 7.5, and disrupted by sonication. After removal of unbroken cells by centrifugation at 10,000 × g for 10 min at 4 °C, membranes and aggregates were recovered by centrifugation at 100,000 × g for 1 h at 4 °C. The supernatant was saved as the soluble fraction containing the periplasm and cytoplasm.
Isolation of Pal Bound to mLolB(His) and LolA
KT50 cells harboring pRT102 derivatives were grown to the exponential growth phase on LB broth in the presence of 50 μm IPTG at 30 °C. Cells were harvested and converted into spheroplasts as previously described (17) to obtain a periplasmic fraction as a spheroplast supernatant. Periplasmic fractions were divided into two aliquots (400 μl). One aliquot was incubated with TALON resin in the presence of 2 mm MgCl2 for 30 min at 4 °C. After the resin had been washed with 20 mm Tris-HCl, pH 7.5, containing 300 mm NaCl, mLolB-His was eluted with the same buffer supplemented with 250 mm imidazole. The other aliquot was incubated with 2 μl of anti-LolA antiserum for 1 h at room temperature and then incubated with 50 μl of Protein A-agarose (Pierce) for 1 h at room temperature. After the resin had been washed with 10 mm Tris-HCl, pH 7.5, containing 150 mm NaCl, LolA was eluted with 100 mm glycine-HCl, pH 3.5. Resin-bound and -unbound Pal in the respective fractions were detected by SDS-PAGE followed by immunoblotting.
In Vitro Transfer of Pal(Strep) from LolA(FLAG) to mLolB(His) Derivatives
BL21(DE3) cells harboring pOS141 were grown in 2×YT medium at 37 °C until the culture absorbance reached 0.6. Expression of LolA(FLAG) and Pal(Strep) was then induced for 2 h by the addition of 50 μm IPTG. Cells were harvested, and the periplasmic fraction was applied to ANTI-FLAG M2 Affinity Gel, which had been equilibrated with 10 mm Tris-HCl, pH 7.5, containing 150 mm NaCl and then washed with the same buffer. LolA(FLAG) was eluted with 100 μg/ml FLAG peptide. The Pal(Strep)-LolA(FLAG) complex was purified by size exclusion chromatography as described previously (18). The purified Pal(Strep)-LolA(FLAG) complex (5 nm) was then incubated with mLolB(His) derivatives (500 nm) for 30 min at 4 or 30 °C. Each reaction mixture was applied to TALON resin pre-equilibrated with 20 mm Tris-HCl, pH 7.5, washed with 20 mm Tris-HCl, pH 7.5, containing 300 mm NaCl, and eluted with the same buffer supplemented with 250 mm imidazole. The TALON resin-bound and -unbound fractions were analyzed by SDS-PAGE followed by immunoblotting.
Purification of the Pal(His)-mLolB Complex
For technical reasons, His-tagged Pal was transferred from LolA(FLAG) to tag-less mLolB and its L68E derivative, which were also used for crystallization. The Pal(His)-LolA(FLAG) complex was purified from the periplasmic fraction of BL21(DE3) cells harboring pSS9 as described above. Tag-less mLolB and mLolB(L68E) were purified as previously described (12, 19). The Pal(His)-LolA(FLAG) complex (100 μg/ml) was incubated with 100 μg/ml mLolB or mLolB(L68E) for 1 h at 30 °C. The reaction mixtures were applied to ANTI-FLAG M2 Affinity Gel pre-equilibrated with 20 mm Tris-HCl, pH 7.5, containing 150 mm NaCl. The FLAG-unbound fraction was applied to TALON resin pre-equilibrated with the same buffer and washed with the same buffer. The Pal(His)-mLolB complex was eluted with the same buffer containing 50 mm imidazole and then further purified by size exclusion chromatography on a Superdex 75 column (GE Healthcare).
Strength of the Hydrophobic Interaction between Pal and mLolB(L68E)
MC4100 cells harboring pTAN21 (27) were grown in M63 minimal medium at 37 °C until the culture absorbance reached 0.8. Expression of Pal was then induced for 5 min by the addition of 2 mm IPTG. Cells were converted into spheroplasts, and Pal was released into the spheroplast supernatant by the addition of LolA. Wild-type mLolB(His) or its L68E derivative was added to the spheroplast supernatant fraction containing the Pal-LolA complex, and then the reaction mixture was incubated for 1 h at 30 °C to yield the Pal-mLolB(His) or Pal-mLolB(L68E)(His)complex. As a control, the Pal-LolA(His) complex was prepared by the addition of LolA(His) to spheroplasts. Pal complexed with mLolB(His), its derivative, or LolA(His) was adsorbed to TALON resin packed into a small column. To determine the intensity of the hydrophobic interaction between Pal and Lol factors, Pal bound to His-tagged Lol proteins was eluted with a linear gradient (0∼4%) of DDM at a flow rate of 0.5 ml/min. Each fraction (0.5 ml) was precipitated with 10% trichloroacetic acid and then analyzed by SDS-PAGE and immunoblotting with anti-Pal, -LolB, or -LolA antibodies.
Crystallographic Studies of mLolB Derivatives
The His tag attached to the C terminus of mLolB(L68D) (0.6 mg/ml) was removed by treatment with 6 μg/ml carboxypeptidase A (Sigma, C9268) in 20 mm Tris-HCl, pH 8.0, containing 5% (w/v) dimethylethylammonium propane sulfonate (NDSB-195; Calbiochem, 480001) and 100 mm NaCl at 4 °C for 1 day. The tag-less mLolB(L68D) was concentrated to 10 mg/ml with a Centricon YM-10 membrane (Millipore) with no further purification or desalting processes. For crystallization of mLolB(L68E), tag-less mLolB(L68E) was purified as described (12, 19). The L68D derivative was crystallized into two space groups, C2221 (a = 35.3 Å, b = 86.7 Å, c = 111.1 Å) and P6522 (a = b = 100.9 Å, c = 97.1 Å). On the other hand, the L68E variant was crystallized into space group C2221 (a = 37.1 Å, b = 87.1 Å, c = 112.4 Å). Diffraction data were collected at SPring-8 (Harima, Japan). The structures were determined by the molecular replacement method. The crystallographic and refinement statistics are listed in Table 2. The coordinates and structural factors have been deposited in the Protein Data Bank under accession numbers 3WJT (L68D in C2221), 3WJU (L68D in P6522), and 3WJV (L68E).
TABLE 2.
Crystallographic and refinement statistics
Values in parentheses refer to the highest resolution shell. r.m.s. deviations, root mean square deviations.
| Variant | L68D Orthorhombic form | L68D Hexagonal form | L68E |
|---|---|---|---|
| Crystal data | |||
| Space group | C2221 | P6522 | C2221 |
| Cell parameters | |||
| a (Å) | 35.3 | 100.9 | 37.1 |
| b (Å) | 86.7 | 100.9 | 87.1 |
| c (Å) | 111.1 | 97.1 | 112.4 |
| Resolution range (Å) | 30.0–1.55 (1.61–1.55) | 30.0–2.50 (2.59–2.50) | 30.0–2.40 (2.49–2.40) |
| Reflections (total/unique) | 242,659/24,057 | 165,029/10,408 | 41,914/7,140 |
| Redundancy | 10.1 (3.2) | 15.9 (10.2) | 5.9 (4.0) |
| Completeness (%) | 96.1 (72.4) | 98.3 (94.6) | 95.7 (85.7) |
| I/σ(I) | 52.1 (2.8) | 55.1 (4.3) | 20.5 (3.8) |
| Rsyma (%) | 5.3 (25.6) | 5.4 (30.0) | 7.0 (23.0) |
| Refinement | |||
| Protein residues | 178 (Pro-9–Lys-186) | 178 (Pro-9–Lys-186) | 176 (Ala-11–Lys-186) |
| (Double conformations) | 12 | 0 | 0 |
| Heterogen molecules | 3 × SO42 −, 1 × Cl− | 2 × SO42 − | 2 × SO42 − |
| Water molecules | 120 | 29 | 50 |
| Total atoms | 1661 | 1457 | 1464 |
| Rworkb (%) | 21.5 | 24.9 | 24.4 |
| Rfreec (%) | 23.9 | 29.1 | 28.3 |
| r.m.s. deviations | |||
| Bonds (Å) | 0.01 | 0.006 | 0.005 |
| Angle (°) | 1.4 | 1.2 | 1.3 |
a Rsym = ΣhklΣi|Ihkl,i − 〈Ihkl〉|/ΣhklΣi Ihkl,i.
b Rwork = Σhkl|Fobs − Fcalc|/Σ|Fobs|.
c Rfree was calculated with the 5% of the reflections not included for refinement as a test set.
Other Techniques
mLolB-dependent membrane incorporation of Lpp was carried out as described (11). LolB-depleted outer membranes were prepared from KT50 cells harboring pYKT123 (11). SDS-PAGE was carried out as described (20), except that Lpp was analyzed as reported (21). Immunoblotting was carried out essentially as described previously (22), and proteins were visualized using a color development substrate (5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium) or an enhanced chemiluminescence substrate (ECL Plus; GE Healthcare) followed by detection with a lumino-image analyzer (LAS-1000plus; Fujifilm).
RESULTS
Construction of mLolB Mutants
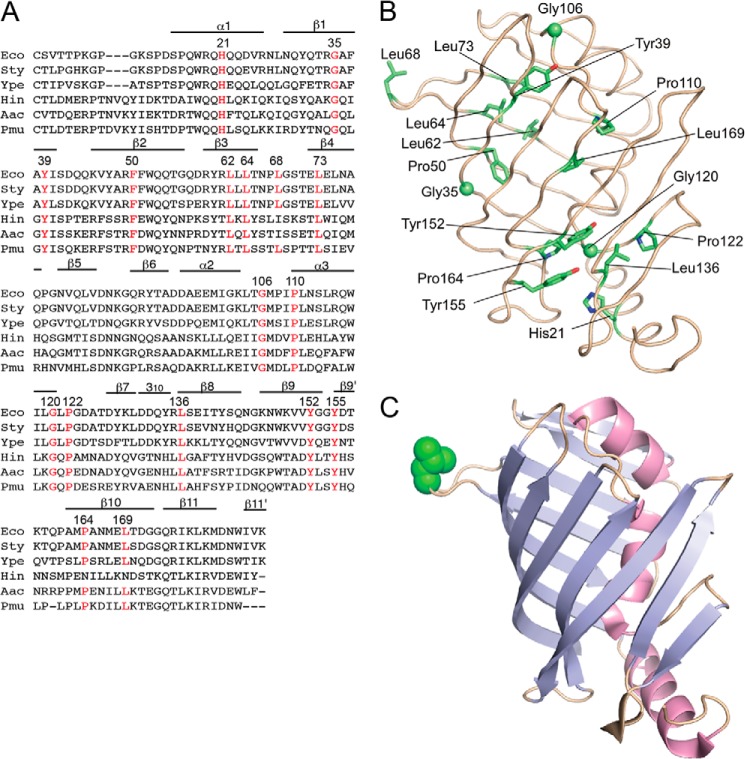
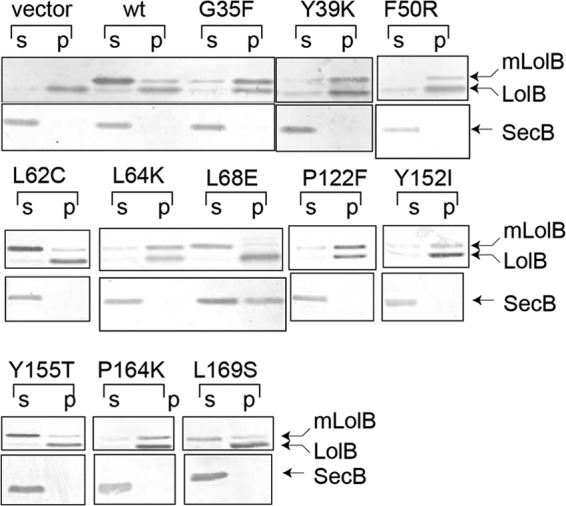
LolB sequences are highly conserved in the proteobacteria γ subdivision (Fig. 1A). E. coli KT60 carries the ΔlolB::kan mutation (8) and harbors pYKT122, which encodes LolB under the control of PBAD. Because the LolB function is essential, this strain requires arabinose for growth. Seventeen conserved residues of mLolB were subjected to random mutagenesis in pRT102 carrying mLolB(His) under the control of Ptac followed by transformation into KT60/pYKT122. The positions of the mutagenized residues are shown in the crystal structure of mLolB (23) (Fig. 1B). Among them, Leu at position 68 is unique as it is located in the protruding loop (Fig. 1C). Two hundred transformants were isolated for each mutagenized residue, and their growth was examined on LB plates with or without arabinose and/or IPTG. When KT60/pYKT122 harbored pTTQ18, an empty vector for pRT102, arabinose was essential for continuous growth. On the other hand, when the strain harbored pRT102, neither IPTG nor arabinose was required, indicating that a sufficient amount of mLolB(His) is expressed from high copy number plasmid pRT102 even in the absence of IPTG.
FIGURE 1.
Sequence alignment of LolB homologues and positions of mutated residues. A, the amino acid sequences of LolB and its homologues were aligned using ClustalW (26). Residues forming α and 310 helices and β-strands are indicated. Conserved residues that were subjected to random mutagenesis are indicated in red with their position numbers. Eco, E. coli; Sty, Salmonella enterica serovar typhimurium; Ype, Yersinia pestis; Hin, Haemophilus influenzae; Aac, Actinobacillus actinomycetemcomitans; Pmu, Pasteurella multocida. B, the positions of mutated residues are shown in the structure of mLolB. Side chains of the residues are shown as sticks, whereas Cα atoms of glycine are shown as spheres. C, mLolB is represented as a ribbon model in which strands and helices are colored cyan and red, respectively. The side chain of Leu-68 in the hydrophobic loop is shown as a CPK model in green.
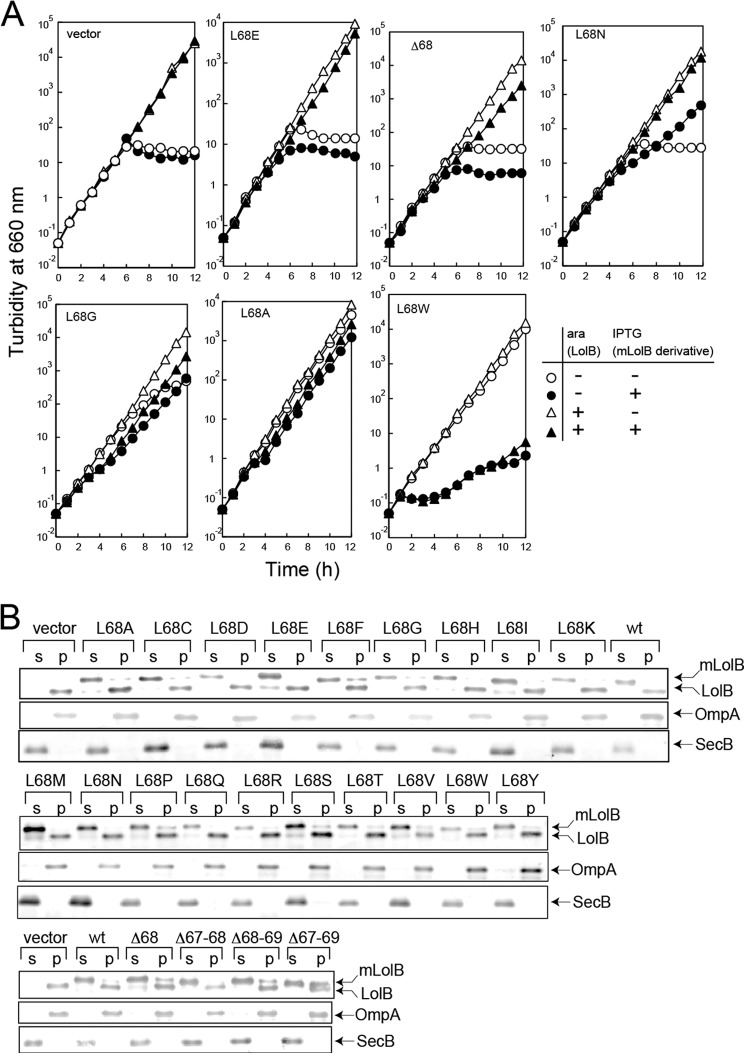
More than 40 colonies were isolated that did not grow on plates containing IPTG without arabinose (Table 3). Among them, expression of H21L, which was the sole mutant isolated on mutagenesis of His21, was not detectable. Despite repeated examinations, no mutant was isolated on mutagenesis of Leu-73 or Pro-110. Only a stop codon mutant was isolated on mutagenesis of Gly-106 and Gly-120, suggesting that the two residues can be replaced with any residue. To our surprise, the solubility of most mutants was poor (Fig. 2) despite the absence of acyl chains. The expression of some mutants was low or unstable. These properties are summarized in Table 2. Thus, outer membrane incorporation of Lpp was examined in vitro using five soluble derivatives (Fig. 3). After Lpp had been released from spheroplasts with LolA, spheroplast supernatants were isolated and incubated with the indicated mLolB derivatives in the presence of outer membranes followed by centrifugation. When no mLolB was added, Lpp remained in the supernatant fraction even after 60 min of incubation (Fig. 3A, top). Most Lpp molecules were recovered in the pellet fraction after 30 min of incubation with wild-type mLolB. On the other hand, other derivatives were variously defective in the outer membrane localization of Lpp, and their activities were quantified and are plotted in Fig. 3B. The L62C derivative was only slightly defective. In contrast, L68E and L68D exhibited little activity, whereas L68C and Y155T exhibited low but non-negligible activity. Because the replacement of Leu-68 located in the loop protruding into a hydrophilic environment caused a defective phenotype, it seemed interesting to examine the importance of Leu-68 in more detail. Thus, Leu-68 was replaced with other residues or deleted with neighboring residues. Then the growth of KT60 cells harboring pYKT122 and pRT102 derivatives on LB was examined in the presence and absence of arabinose and/or IPTG (Fig. 4A). Based on growth in the presence and absence of derivatives, their activities were classified into five groups: class 1, inactive (L68D/L68E, Δ68, Δ67–68, Δ 68–69, Δ 67–69); class 2, slightly active (L68K/L68N/L68Q/L68R); class 3, slightly defective (L68C/L68G/L68H/L68P/L68S); class 4, as active as the wild-type (L68A/L68I/L68M/L68T/L68Y); and class 5, active in low amounts but inhibitory in high amounts (L68F/L68V/L68W). Although overexpression of the class 1–4 derivatives did not inhibit the wild-type LolB function (closed triangles), class 5 derivatives such as L68W supported growth at basal expression levels (open circles) but strongly inhibited growth supported by wild-type LolB at overexpressed levels (closed triangles).
TABLE 3.
Properties of the isolated mLolB derivatives
| Target residue | Mutations | Solubilitya | Expression |
|---|---|---|---|
| His-21 | Leu | No | |
| Gly-35 | Phe (Lys, Ser, Asn, Tyr) | − | OK |
| Tyr-39 | Lys (Leu, Arg) | − | OK |
| Phe-50 | Arg (Ile) | − | OK |
| Leu-62 | Cys | + | OK |
| Leu-64 | Lys (Asp) | − | OK |
| Leu-68 | Glu (Asp, Asn, Cys, Lys) | + | OK |
| Leu-73 | No mutants | ||
| Gly-106 | Stop | OK | |
| Pro-110 | No mutant | ||
| Gly-120 | Stop | OK | |
| Pro-122 | Phe | − | Low |
| Leu-136 | Lys | − | Low |
| Tyr-152 | Ile (Lys, Ser, Thr, Asn, Gly, Asp) | − | Low |
| Tyr-155 | Thr (Asp, Lys, Asn, Pro, Phe) | + (only Thr) | Low except Thr |
| Pro-164 | Lys (Phe, Asn, Leu) | − | OK |
| Leu-169 | Thr, Ser | + | OK |
a The solubility of representative derivatives as to each target residue is shown in Fig. 2. The derivatives indicated in parentheses exhibited essentially the same solubility as the respective representative derivatives.
FIGURE 2.
Cellular levels and solubility of mLolB(His) mutants. KT60 cells harboring pYKT122 and pRT102 derivatives were grown on LB broth in the presence of 0.01% arabinose at 30 °C to their exponential growth phases. Expression of mLolB(His) derivatives was induced by the addition of 50 μm IPTG for 1 h. Cells were harvested and fractionated into soluble (s) and pellet (p) fractions containing membranes and aggregates followed by analysis by SDS-PAGE and immunoblotting with anti-LolB antibodies. As a control, SecB was examined as a soluble protein.
FIGURE 3.
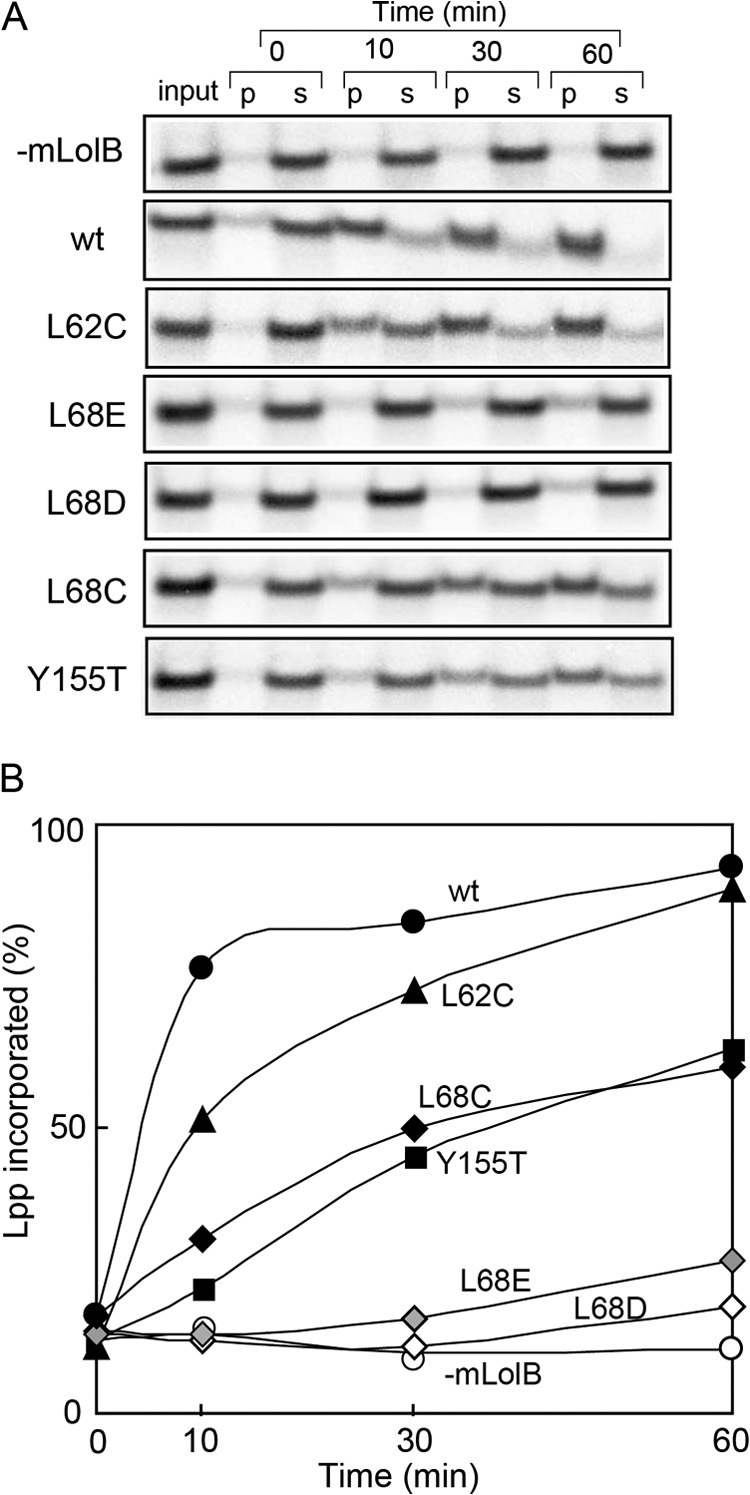
Outer membrane incorporation of Lpp by mLolB(His) mutants. A, spheroplasts prepared from MC4100 cells were labeled for 5 min with Tran35S Label in the presence of LolA(His) followed by isolation of the [35S]Lpp-LolA(His) complex with TALON resin. The complex was incubated at 30 °C for the indicated times with LolB-depleted outer membranes in the absence (−mLolB) or presence of wild-type (wt) or mutant mLolB(His). The reaction was terminated by chilling on ice. The reaction mixtures (input) were fractionated into pellets (p) and supernatants (s) followed by SDS-PAGE and autoradiography. B, the results shown in A were quantified and plotted as a function of the reaction time, taking the total amount of Lpp as 100%.
FIGURE 4.
Properties of mLolB derivatives with mutations at Leu-68. A, KT60 cells harboring both pYKT122 and the specified pRT102 derivative were grown on LB broth in the presence (+) and absence (−) of 0.02% arabinose (ara) and/or 50 μm IPTG at 30 °C for the indicated times with repeated inoculation of portions of the culture into fresh medium. Turbidity was monitored at 660 nm and plotted after correction for culture dilution. B, subcellular localization of the indicated mLolB(His) derivatives was examined as in Fig. 2. OmpA and SecB represent membrane and soluble proteins, respectively. p, pellet; s, supernatants.
All mLolB(His) derivatives of Leu-68 were obtained in the soluble fraction containing periplasmic and cytoplasmic fractions with SecB, whereas LolB expressed from pYKT122 was recovered in the pellet fraction with OmpA (Fig. 4B), indicating that these derivatives are useful for in vitro examinations.
Lpp is very inhibitory when the outer membrane sorting of lipoproteins is impaired (6). Therefore, we examined whether or not defective Leu-68 derivatives, which do not support the growth of cells expressing Lpp (Fig. 4A), exhibit any activity in cells lacking Lpp. KT50 (lpp, ΔlolB::kan) cells harboring pNAS021, which carry a temperature-sensitive replicon and lolB, were transformed with pRT102 derivatives. The transformants were incubated at 42 °C for 5 h to cure pNAS021, which confers spectinomycin-resistance, and then grown on LB plates supplemented with IPTG at 30 °C. Spectinomycin-sensitive colonies were easily isolated from KT50 cells harboring any one of the defective pRT102 derivatives and grew at 30 °C (Fig. 5), indicating that all Leu-68 derivatives including the four deletion mutants are partly functional and able to support the growth of KT50 cells lacking Lpp at 30 °C. In marked contrast, when KT50 cells were transformed with pTTQ18, no spectinomycin-sensitive cells were obtained even when >300 transformants were examined, confirming the previous observation that the LolB function is essential even in cells lacking Lpp (8). KT50 cells harboring pRT102 derivatives were variously defective at 37 °C (Fig. 5). The growth of KT50 cells expressing Δ67–68, Δ68–69, and Δ67–69 ceased shortly after the start of incubation. These deletion mutants might not be stable at 37 °C.
FIGURE 5.
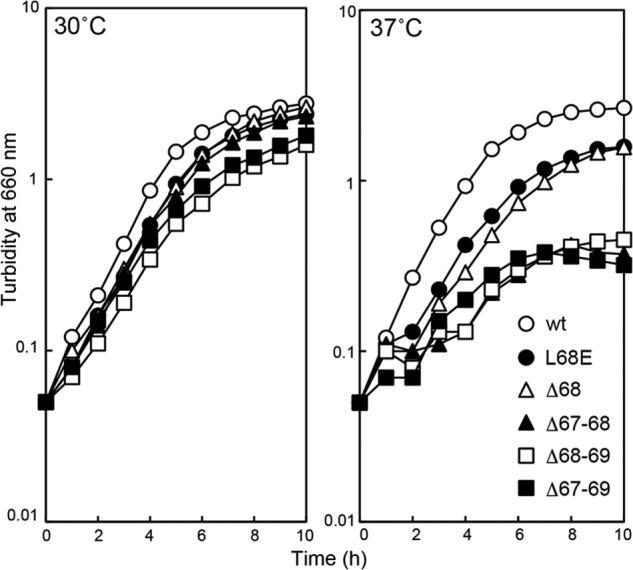
Complementation of the ΔlolB mutation by mLolB(His) derivatives. KT50 cells harboring pRT102 derivatives encoding the indicated mLolB(His) derivatives were grown on LB broth supplemented with 50 μm IPTG at 30 °C or 37 °C. Turbidity was monitored at 660 nm.
Lipoproteins accumulate in the periplasm when the function of LolB (8) is impaired. The periplasmic levels of Pal, mLolB(His), and MalE in KT50 cells grown in the presence of Leu-68 mutants of mLolB(His) were determined at 30 °C (Fig. 6A). Pal is an outer membrane-specific lipoprotein that is frequently used for in vitro examination of lipoprotein sorting. The level of mLolB(His) varied to some extent, whereas periplasmic maltose-binding protein MalE was expressed at a nearly constant level. Pal was not detected when wild-type mLolB(His) was expressed (middle of the top panel). Some derivatives caused marginal periplasmic accumulation of Pal. Significant accumulation of Pal in the periplasm took place with L68D/L68E and the four deletion mutants, indicating that the LolB function of these six derivatives is low. On the other hand, when KT50 cells harbored pNAS021 expressing LolB, no defective pRT102 derivatives caused the accumulation of Pal (Fig. 6B), indicating that the expression of LolB completely suppresses the periplasmic accumulation of Pal caused by defective mLolB(His) derivatives.
FIGURE 6.
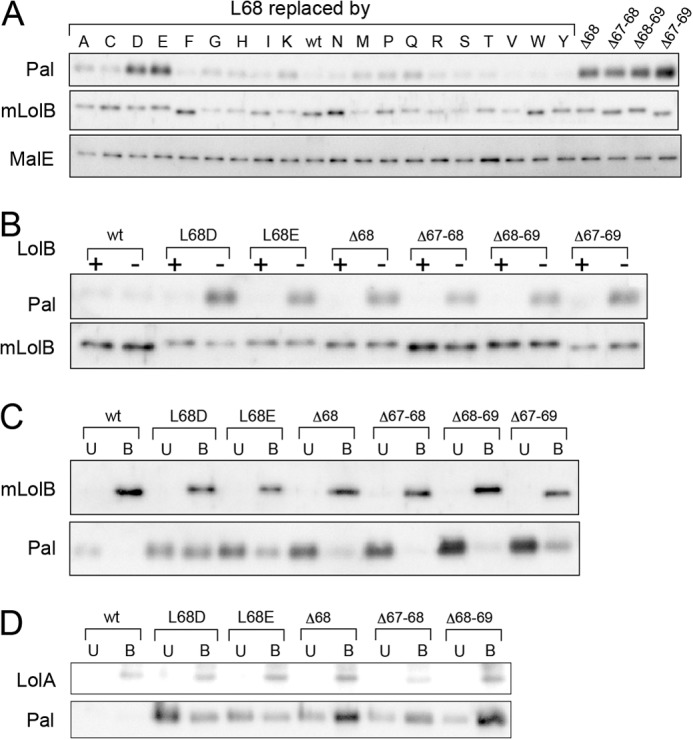
Defective mLolB(His) derivatives cause accumulation of Pal in the periplasm as a complex with mLolB or LolA. A, KT50 cells harboring pRT102 derivatives were grown on LB broth supplemented with 50 μm IPTG at 30 °C. The levels of Pal, mLolB(His), and MalE in the periplasmic fraction prepared from the cells were determined by SDS-PAGE and immunoblotting with antibodies against the respective proteins. B, KT50 cells harboring pRT102 derivatives encoding L68D/L68E and the four deletion mutants were transformed with (+) or without (−) pNAS021 encoding LolB. The levels of Pal and mLolB(His) in the periplasm were then examined as in A. wt, wild type. C, a periplasmic fraction of KT50 cells harboring pRT102 was prepared as in A and applied to TALON resin to adsorb mLolB(His) derivatives. The resin bound (B) and unbound (U) fractions were analyzed by SDS-PAGE followed by immunoblotting with anti-LolB and -Pal antibodies as described under “Experimental Procedures.” D, the periplasmic fraction mentioned in C was treated with anti-LolA antibodies followed by binding to protein A resin. The resin bound (B) and unbound (U) fractions were analyzed as in C except with anti-LolA and -Pal antibodies.
Characterization of Pal Accumulated in the Periplasm
Lipoproteins are highly hydrophobic because of their N-terminal acyl chains. Pal accumulated in the periplasm of KT50/pRT102 derivatives is, therefore, expected to form a complex with either LolA or mLolB. To identify the partner of periplasmic Pal, a defective mLolB(His) derivative in the periplasm was affinity-purified with TALON resin. The resin bound and unbound fractions were analyzed by SDS-PAGE followed by immunoblotting with anti-LolB and -Pal antibodies (Fig. 6C). All mLolB(His) molecules were recovered in the bound fraction. When L68D/L68E derivatives were expressed, Pal was recovered in not only the bound fraction but also the unbound fraction. Pal recovered in the bound fraction represents the complex formed with a His-tagged mLolB(L68D/L68E) derivative. In contrast, Pal molecules were obtained only in the unbound fraction in the presence of the three deletion mutants. These Pal molecules recovered in the unbound fraction were most likely complexed with LolA. To examine this, the periplasmic fraction was treated with anti-LolA antibodies followed by binding to protein A resin (Fig. 6D). As expected, the major fraction of Pal molecules was recovered in the bound fraction with LolA when the three deletion mutants were expressed. A portion of Pal was also obtained in the bound fraction with LolA in the presence of L68D/L68E(His), suggesting that Pal complexed with LolA after L68D/L68E(His) was saturated with Pal. Taken together, these results indicate that L68D/L68E(His) mutants can accept Pal from LolA, whereas the three deletion mutants cannot. In contrast, a small proportion of Pal molecules was recovered with the Δ67–69 mutant (Fig. 6C), suggesting that this deletion mutant accepts Pal from LolA. Detailed in vitro analysis with purified proteins is needed to determine which step(s) is impaired in this mutant.
Transfer of Pal from LolA to mLolB derivatives was examined in more detail with the purified Pal(Strep)-LolA(FLAG) complex and His-tagged mLolB Leu-68 derivatives. When the Pal(Strep)-LolA(FLAG) complex was incubated in the absence of mLolB(His), Pal remained soluble and was recovered in TALON resin-unbound fraction, whereas a significant amount of Pal was recovered in the bound fraction with mLolB(His) when the incubation was performed with mLolB(His) at both 4° and 30 °C (Fig. 7A). The Pal receptor activities of five Leu-68 derivatives purified to homogeneity were then examined by means of an in vitro assay (Fig. 7B). All derivatives accepted Pal(Strep) as did wild-type mLolB(His).
FIGURE 7.
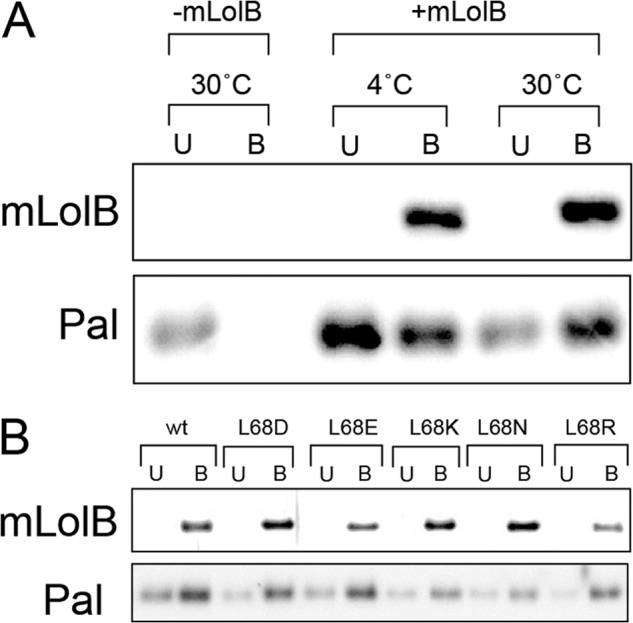
Lipoprotein receptor activity of mLolB(His) derivatives with a mutation at Leu-68. A, the Pal(Strep)-LolA(FLAG) complex was purified as described under “Experimental Procedures” and incubated with wild-type mLolB(His) for 30 min at 4 °C or 30 °C. The reaction mixtures were then applied to TALON resin to adsorb mLolB(His). The resin bound (B) and unbound (U) fractions were analyzed by SDS-PAGE followed by immunoblotting with anti-LolB and -Pal antibodies. B, the Pal-accepting activity of the indicated mLolB(His) derivatives at 30 °C was examined as in A.
Membrane Anchoring Activity of mLolB(L68E)
The results presented above suggested that the L68E derivative was defective in the membrane anchoring of Pal, which caused accumulation of Pal in the periplasm. To examine this, Pal(His)-mLolB derivatives were purified to homogeneity after in vitro transfer of Pal(His) from LolA(FLAG) to mLolB. The purified complex comprised stoichiometric amounts of Pal(His) and wild-type mLolB or mLolB(L68E) (Fig. 8A). These complexes were incubated with outer membranes and then fractionated (Fig. 8B). Wild-type mLolB caused nearly complete localization of Pal to the membrane fraction. On the other hand, most Pal molecules complexed with mLolB(L68E) remained in the supernatant, indicating that the L68E derivative is unable to catalyze membrane anchoring of Pal.
FIGURE 8.
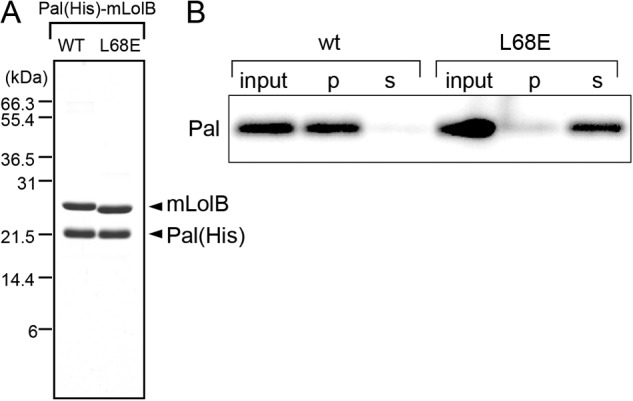
Membrane anchoring activity of mLolB(L68E). A, Pal(His)-mLolB (wt) and Pal(His)-mLolB(L68E) complexes were purified as described under “Experimental Procedures,” and aliquots (3 μg) were analyzed by SDS-PAGE followed staining with Coomassie Brilliant Blue R-250. The migration positions of molecular mass markers are shown on the left. B, the Pal(His)-mLolB (wt) and Pal(His)-mLolB(L68E) complexes were incubated with LolB-depleted outer membranes for 30 min at 30 °C. The reaction mixtures (input) were fractionated by ultracentrifugation into pellets (p) and supernatants (s) followed by analysis by SDS-PAGE and immunoblotting with anti-His tag antibodies.
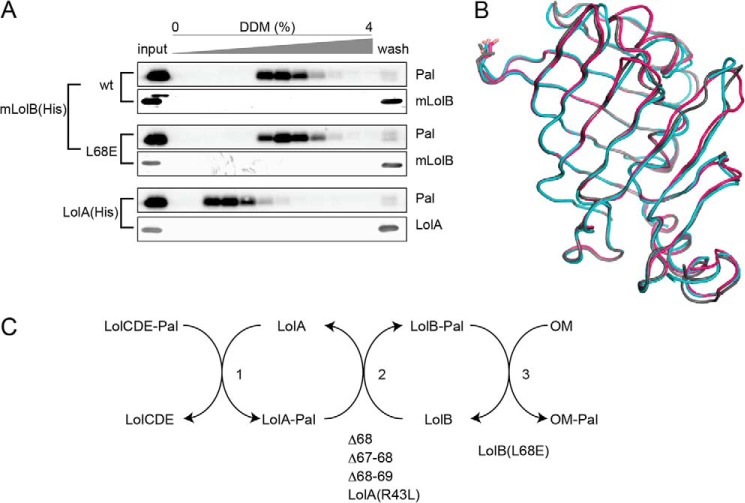
Dissociation of Pal complexed with LolA or LolB caused by a detergent, DDM, revealed that the difference in the affinity for lipoproteins is critically important for the transfer of the lipoproteins from LolA to LolB (9). The LolA(R43L) mutant cannot transfer lipoproteins to LolB because the hydrophobic interaction between lipoproteins and LolA(R43L) is as strong as that between lipoproteins and LolB (9). We examined whether or not the hydrophobic interaction between Pal and mLolB(L68E) is stronger than that between Pal and wild-type mLolB (Fig. 9A). Pal dissociated from wild-type mLolB(His) and mLolB(L68E)(His) with a similar concentration of DDM, which was significantly higher than the concentration that caused dissociation of the Pal-LolA(His) complex. These results indicate that the hydrophobic interaction with Pal is almost the same for L68E and wild-type mLolB.
FIGURE 9.
The strength of hydrophobic interaction with Pal and the crystal structure of mLolB(L68D/E). A, Pal was released from spheroplasts as a complex with LolA and then incubated with wild-type (wt) mLolB(His) or L68E(His) for 1 h at 30 °C. The reaction mixtures were applied to TALON resin and then eluted with a linear gradient of DDM. Fractions (1 ml) were taken and analyzed by SDS-PAGE and immunoblotting with anti-Pal and -LolB antibodies. The left lanes contained 50% amounts of the input samples. The hydrophobic interaction between Pal and LolA was also examined as described under “Experimental Procedures.” B, superimposition of mLolB, mLolB(L68D), and mLolB(L68E) is represented as tube models in gray, cyan, and magenta, respectively. The side chains of the residue at position 68 are shown as sticks. C, the lipoprotein transfer reaction mediated by Lol proteins and the isolated defective mutants. See “Discussion” for details.
Two derivatives of mLolB, L68D and L68E, were crystallized as described under “Experimental Procedures.” The former derivative was crystallized in two crystal forms; orthorhombic crystals belonging to space group C2221 and crystals belonging to space group P6522. The crystal structures of the two derivatives are essentially the same as that of wild-type mLolB (Fig. 9B), indicating that the property, but not structure, of the residue at position 68 is important for membrane targeting.
DISCUSSION
Although mLolB is expressed in the periplasm, as LolA is, they cannot functionally replace each other (11). Their structures are similar to each other as a whole, whereas localized structural differences are noteworthy (23). The C-terminal loop is characteristic of LolA and functions to prevent retrograde localization of lipoproteins to the inner membrane (24), whereas the loop protruding into an aqueous milieu is characteristic of LolB. Targeted mutagenesis of conserved residues revealed that Leu-68 in the protruding loop of mLolB (Fig. 1C) plays an important role in membrane targeting, as speculated from its crystal structure (23). Substitution of Leu-68 revealed that polar residues, i.e. Asp, Glu, Lys, Asn, Gln, and Arg, are not favorable, whereas hydrophobic residues, i.e. Ala, Ile, Met, Thr, and Tyr, can substitute for Leu-68 with little inactivation (Fig. 4). These results most likely indicate that the hydrophobic residue in the protruding loop initiates the membrane targeting. Replacement of Leu-68 with Trp, Phe, or Val resulted in a complex phenotype. These derivatives exhibited a dominant-negative phenotype when overexpressed but supported growth when expressed at a basal level. At present, we only speculate that these derivatives have activities but are somewhat toxic; therefore, their overexpression inhibits growth.
Deletion of Leu-68 with a neighboring residue suggested that the loop is also important for accepting lipoproteins from LolA. We previously examined the mode of interaction between LolA and mLolB by means of photosensitive cross-linking (3) and NMR (25). The two different analyses led to the same conclusion that LolA and mLolB connect through their hydrophobic cavities to form a tunnel-like structure, which allows efficient lipoprotein transfer from LolA to LolB. The NMR analyses further suggested that the convex side of mLolB is important for the LolA interaction (25). The results presented here suggest that the intact loop structure on the convex side is required for the LolA-mLolB interaction.
Lipoprotein transfer from the inner to the outer membrane through the periplasm takes place in three steps, LolCDE to LolA (step 1), LolA to LolB (step 2), and LolB to the outer membrane (step 3) (Fig. 9C). The results presented here indicate that the three deletion mutants are mainly defective in step 2, like LolA(R43L), whereas mLolB(L68D/L68E) mutants are the first examples defective in step 3. It is still unclear how the LolB-phospholipid interaction leads to anchoring of lipoproteins to the outer membrane. As proposed in this paper, Leu-68 most likely initiates membrane targeting, which then causes a conformational change of LolB to discharge lipoproteins into the lipid phase.
It was previously found that the conservation of Lol proteins such as LolCDE and LolA involved in the early step of lipoprotein processing or sorting is high compared with that of those involved in a later step. Thus, the conservation of LolB is low, and it is found only in β-, γ-, and δ-proteobacteria (2). The molecular mechanism by which mLolB mediates membrane anchoring of lipoproteins will be useful for understanding how lipoproteins are localized to the outer membrane of bacteria having an LolA homologue but no LolB homologue. Membrane targeting and anchoring of lipoproteins in these bacteria might be catalyzed by an unknown factor, which is a functional, but not structural, homologue of LolB.
Acknowledgment
We thank the beamline staff at SPring-8.
This work was supported in part by Ministry of Education, Culture, Sports, Science, and Technology of Japan grants (to H. T.) and the Target Protein Research Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K. M.).
The atomic coordinates and structure factors (codes 3WJT, 3WJU, and 3WJV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- DDM
- n-dodecyl-β-d-maltopyranoside
- IPTG
- isopropyl β-D-thiogalactopyranoside.
REFERENCES
- 1. Tokuda H., Sander P., Lee B. L., Okuda S., Grau T., Tschumi A., Brülle J. K., Kurokawa K., Nakayama H. (2014) in Bacterial Membranes: Structural and Molecular Biology, Caister Academic Press, pp. 133–177, Norfolk, UK [Google Scholar]
- 2. Okuda S., Tokuda H. (2011) Lipoprotein Sorting in Bacteria. Annu. Rev. Microbiol. 65, 239–259 [DOI] [PubMed] [Google Scholar]
- 3. Okuda S., Tokuda H. (2009) Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl. Acad. Sci. U.S.A. 106, 5877–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braun V. (1975) Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta 415, 335–377 [DOI] [PubMed] [Google Scholar]
- 5. Mizushima S. (1984) Post-translational modification and processing of outer membrane prolipoproteins in Escherichia coli. Mol. Cell. Biochem. 60, 5–15 [DOI] [PubMed] [Google Scholar]
- 6. Yakushi T., Tajima T., Matsuyama S., Tokuda H. (1997) Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J. Bacteriol. 179, 2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Narita S., Tokuda H. (2011) Overexpression of LolCDE allows deletion of the Escherichia coli gene encoding apolipoprotein N-acyltransferase. J. Bacteriol. 193, 4832–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanaka K., Matsuyama S., Tokuda H. (2001) Deletion of lolB encoding an outer membrane lipoprotein is lethal for Escherichia coli and causes the accumulation of lipoprotein localization intermediates in the periplasm. J. Bacteriol. 183, 6538–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taniguchi N., Matsuyama S., Tokuda H. (2005) Mechanisms underlying energy-independent transfer of lipoproteins from LolA to LolB, which have similar unclosed β-barrel structures. J. Biol. Chem. 280, 34481–34488 [DOI] [PubMed] [Google Scholar]
- 10. Wada R., Matsuyama S., Tokuda H. (2004) Targeted mutagenesis of five conserved tryptophan residues of LolB involved in membrane localization of Escherichia coli lipoproteins. Biochem. Biophys. Res. Commun. 323, 1069–1074 [DOI] [PubMed] [Google Scholar]
- 11. Tsukahara J., Mukaiyama K., Okuda S., Narita S., Tokuda H. (2009) Dissection of the LolB function. Lipoprotein binding, membrane targeting, and incorporation of lipoproteins into lipid bilayers. FEBS J. 276, 4496–4504 [DOI] [PubMed] [Google Scholar]
- 12. Matsuyama S., Yokota N., Tokuda H. (1997) A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16, 6947–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casadaban M. J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and μ. J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 14. Hirota Y., Suzuki H., Nishimura Y., Yasuda S. (1977) On the process of cellular division in Escherichia coli: a mutant of E. coli lacking a murein-lipoprotein. Proc. Natl. Acad. Sci. U.S.A. 74, 1417–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sambrook J., Fritsch E. F., Maniatis T. (1989) A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, Appendix A [Google Scholar]
- 16. Sugai R., Shimizu H., Nishiyama K., Tokuda H. (2001) Overexpression of yccL (gnsA) and ydfY (gnsB) increases unsaturated fatty acids, and suppresses both the temperature-sensitive fabA6 mutation and cold-sensitive secG null mutation of Escherichia coli. J. Bacteriol. 183, 5523–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsuyama S., Tajima T., Tokuda H. (1995) A novel periplasmic carrier protein involved in the sorting and transport of E. coli lipoproteins destined for the outer membrane. EMBO J. 14, 3365–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watanabe S., Oguchi Y., Yokota N., Tokuda H. (2007) Large-scale preparation of the homogeneous LolA-lipoprotein complex and efficient in vitro transfer of lipoproteins to the outer membrane in a LolB-dependent manner. Protein Sci. 16, 2741–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takeda K., Miyatake H., Yokota N., Matsuyama S., Tokuda H., Miki K. (2003) Crystallization and preliminary crystallographic study of the outer membrane lipoprotein receptor LolB, a member of the lipoprotein localization factors. Acta Crystallogr. D 59, 1224–1226 [DOI] [PubMed] [Google Scholar]
- 20. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriphage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 21. Hussain M., Ichihara S., Mizushima S. (1982) Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J. Biol. Chem. 257, 5177–5182 [PubMed] [Google Scholar]
- 22. Yamada H., Matsuyama S., Tokuda H., Mizushima S. (1989) A high concentration of SecA allows proton motive force-independent translocation of a model secretory protein into Escherichia coli membrane vesicles. J. Biol. Chem. 264, 18577–18581 [PubMed] [Google Scholar]
- 23. Takeda K., Miyatake H., Yokota N., Matsuyama S., Tokuda H., Miki K. (2003) Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22, 3199–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okuda S., Watanabe S., Tokuda H. (2008) A short helix in the C-terminal region of LolA is important for the specific membrane localization of lipoproteins. FEBS Lett. 582, 2247–2251 [DOI] [PubMed] [Google Scholar]
- 25. Nakada S., Sakakura M., Takahashi H., Okuda S., Tokuda H., Shimada I. (2009) Structural investigation of the interaction between LolA and LolB using NMR. J. Biol. Chem. 284, 24634–24643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D.G., Thompson J. D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yokota N., Kuroda T., Matsuyama S., Tokuda H. (1999) Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274, 30995–30999 [DOI] [PubMed] [Google Scholar]