Background: DOC2B promoter hypermethylation is an early and frequent event in cervical cancer.
Results: DOC2B hypermethylation induces transcriptional repression, reactivated by demethylation; ectopic expression increases Ca2+ flux and inhibits key characteristics of tumorigenesis including proliferation, motility, and invasion.
Conclusion: DOC2B gene is epigenetically regulated and inhibits cervical cancer growth.
Significance: DNA methylation regulates DOC2B gene expression in cervical cancer.
Keywords: Akt, Cell Growth, Cell Invasion, Cell Motility, ERK, DNA Promoter Methylation, DOC2B, Human Papillomavirus, Actin Remodeling, Cervical Cancer
Abstract
Double C2-like domain β (DOC2B) gene encodes for a calcium-binding protein, which is involved in neurotransmitter release, sorting, and exocytosis. We have identified the promoter region of the DOC2B gene as hypermethylated in pre-malignant, malignant cervical tissues, and cervical cancer cell lines by methylation-sensitive dimethyl sulfoxide-polymerase chain reaction and bisulfite genome sequencing; whereas, it was unmethylated in normal cervical tissues (p < 0.05). The promoter hypermethylation was inversely associated with mRNA expression in SiHa, CaSki, and HeLa cells and treatment with demethylating agent 5-aza-2-deoxycytidine restored DOC2B expression. The region −630 to +25 bp of the DOC2B gene showed robust promoter activity by a luciferase reporter assay and was inhibited by in vitro artificial methylation with Sss1 methylase prior to transient transfections. Overexpression of the DOC2B gene in SiHa cells when compared with controls showed significantly reduced colony formation, cell proliferation, induced cell cycle arrest, and repressed cell migration and invasion (p < 0.05). Ectopic expression of DOC2B resulted in anoikis-mediated cell death and repressed tumor growth in a nude mice xenograft model (p < 0.05). DOC2B expressing cells showed a significant increase in intracellular calcium level (p < 0.05), impaired AKT1 and ERK1/2 signaling, and induced actin cytoskeleton remodeling. Our results show that promoter hypermethylation and silencing of the DOC2B gene is an early and frequent event during cervical carcinogenesis and whose reduced expression due to DNA promoter methylation may lead to selective cervical tumor growth.
Introduction
Associations of genetic changes and aneuploidy with tumor growth are traditionally attributed to alterations in DNA sequence manifested as mutations, deletions, and amplifications. Inactive tumor suppressor genes cannot only serve as drivers of tumor progression due to altered or lack of protein function but may also contribute to phenotypic changes that may provide a distinct growth advantage in a hostile environment (1). Human tumors also show epigenetic alterations as heritable changes leading to abnormal gene expression; the functional consequence of which may lead to genetic changes (1). A number of key regulatory genes associated with epigenetic silencing in cervical cancer have been reported (2, 3). Elucidation of differentially methylated genes may identify new targets that could further strengthen our understanding of the molecular mechanism governing pathogenesis of cervical cancer (2).
The double C2-like domain (DOC2)3 protein family consists of two members, DOC2A and DOC2B, which are located in chromosomes 16p11.2 and 17p13.3, respectively, and share significant homology in their amino acid and nucleic acid composition. Although, DOC2A with molecular mass of 43.95 kDa is considered to be more tissue specific due to its predominant expression in brain, the 45.94-kDa DOC2B is expressed in several tissue and cell types. The domains shared by these two proteins are MUNC interacting domain and two tandem calcium binding C2 domains; however, they do not share common functions (4). DOC2B is suggested to be involved in Ca2+-dependent intracellular vesicle trafficking, ion and phospholipids binding, neurotransmitter release, and transporter activity (5, 6). It interacts with syntaxin binding protein 4 and MunC18c (7) leading to facilitation of exocytosis. Binding of calcium to DOC2B significantly increases its affinity toward phospholipids, leading to translocation of proteins from the cytosol to plasma membrane (8). Recently, DOC2B was shown as a positive SNARE regulator for GLUT4 vesicle fusion and mediates insulin-stimulated glucose transport in adipocytes as well as a regulator for delayed insulin secretion in MIN6 cells (9). It is involved in the deformation of synaptic membranes during synaptic vesicle exocytosis (10, 11). However, epigenetic regulation of the DOC2B gene and its role in tumorigenesis has not been reported.
In the present study, we have demonstrated for the first time that DOC2B gene promoter hypermethylation as an early and frequent event in cervical cancer leads to down-regulation of its expression and subsequently to altered function in cervical cancer. Our data suggests DOC2B may act as a negative growth regulator due to its impact on several tumor-associated functions in cervical cancer.
EXPERIMENTAL PROCEDURES
Cell Lines and Patient DNA Samples
MDAMB453, THP1, Jurkat, HT29, IMR32, HCT15, HepG2, PC3, CAL24, SCC4, SaoS2, WM451, MG63, WM115, SiHa, CaSki, and HeLa cells were maintained according to American Type Culture Collection guidelines; whereas, normal skin fibroblasts were grown and maintained in DMEM (HiMedia, Mumbai, India)containing 10% fetal bovine serum (FBS) (HiMedia, Mumbai, India). Cervical biopsy samples from patients who were diagnosed at the Kasturba Medical College, Manipal, India, for cervical cancer were included in the study. All participants provided informed consent in compliance with the Kasturba Hospital ethical committee approval. The clinical status of the samples was confirmed by histopathological examination. DNA was isolated from tissue biopsy, Pap smear, and cell lines by standard phenol-chloroform extraction and ethanol precipitation method.
Methylation-sensitive Arbitrarily Primed PCR (MS-AP-PCR)
For MS-AP-PCR, 2 μg of normal and tumor genomic DNA was digested with 20 units of RsaI enzyme, 20 units of RsaI and HpaII, or 20 units of RsaI and MspI (New England Biolabs) at 37 °C for 16 h. Digested DNA (100 ng) was subjected for PCR amplification using the arbitrary primers (MGCO + MGF2) in a PTC-200 Peltier thermal cycler (MJ Research) (13). The amplicons were resolved in a 8% non-denaturing polyacrylamide gel (PAGE) and visualized by silver staining. The differentially methylated bands were isolated from PAGE, reamplified, cloned into a TA vector (Promega) and sequenced in 3130 genetic analyzer (Applied Biosystems) (12, 13). The sequences were searched for similarity using the BLAT program of the University of California Southern California against HG19 release.
Methylation-sensitive Dimethyl Sulfoxide-Polymerase Chain Reaction (MS-DMSO-PCR)
The MS-DMSO-PCR was performed for the −700 to +300 bp with respect to the transcription start site of the DOC2B gene as described previously containing 0–5% of DMSO (14). The primers used for MS-DMSO-PCR are listed in Table 1.
TABLE 1.
List of primers used
The bold and underlined sequence represents the restriction sites incorporated for cloning.
| Primer name | Sequence (5′-3′) | Product size | Annealing temperature |
|---|---|---|---|
| MS-DMSO-PCR | |||
| DOC2B-MS-DMSO-F | CCCGAGGTTAGAGTGCTGTG | 1000 bp | 64 °C |
| DOC2B-MS-DMSO-F | CTCCTGGATGCTGATGGTC | ||
| BGS | |||
| DOC2B-BGS-F | GTATGTGTGTATTTGTATATTTGYGTGTG | 412 bp | 61.8 °C |
| DOC2B-BGS-R | CCCCRAACRAC CCTAACTTAACCRCTACC | ||
| RT-PCR | |||
| DOC2B-RT-PCR-F | TGGTGTGGTTCTGGGCATCCACG | 103 bp | 60 °C |
| DOC2B-RT-PCR-R | TGGGAGCTCGCTGGTGAGCGTG | ||
| GAPDH-RT-PCR-F | GGCTCCCTTGGGTATATGGT | 97 bp | 60 °C |
| GAPDH-RT-PCR-R | TTGATTTTGGAGGGATCTCG | ||
| ACTB-RT-PCR-F | GACGACATGGAGAAAATCTG | 132 bp | 60 °C |
| ACTB-RT-PCR-R | ATGATCTGGGTCATCTTCTC | ||
| Promoter construct: luciferase essay | |||
| DOC2B-Promoter-F | TTGTGGGTACCCCCAGGGTCCCCTGATCAC | 655 bp | 65 °C |
| DOC2B-Promoter-R | CCGGAAAGCTTCGGCCCTGACTTGGCCGCT | ||
Bisulfite Genomic Sequencing (BGS)
Genomic DNA (2 μg) was used for bisulfite treatment using the EZ DNA methylation kit (Zymo Research) according to the manufacturer's instructions. Primers were designed using Methyl primer express version 1 (Applied Biosystems) for −376 to +36 bp with respect to transcription start site of the DOC2B gene and is listed in Table 1. PCR products were purified and direct sequencing was performed in a 3130 Genetic analyzer according to the manufacturer's instructions using a big dye terminator kit (Applied Biosystems). The percentage of methylation for each CpG site was calculated by comparing the peak height of cytosine (C) signals with the sum of the peak heights of the cytosine and thymine (T) signals (15).
Demethylation and Reverse Transcription-PCR (RT-PCR)
The SiHa, CaSki, and HeLa cells were seeded at a density of 1 × 105 cells in a 10-cm2 culture plates and treated with 5-aza-2′-deoxycytidine (Sigma) at concentrations of 5, 10, and 20 μm daily for 3 days, total RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was synthesized using a High Capacity cDNA archive kit (Applied Biosystems) according to the manufacturer's instructions and used for RT-PCR. The nucleotide sequences used for RT-PCR are listed in Table 1.
Human Papilloma Virus (HPV) Genotyping
HPV genotyping was performed by nested PCR using PGMY09/11 and GP5+/GP6+ consensus L1 primers (16, 17). The PCR product was gel purified and subjected to direct sequencing in a 3130 Genetic analyzer using a big dye terminator kit according to the manufacturer's instructions. The HPV strains were identified by comparing using the NCBI database BLAST search.
Promoter Constructs, Artificial Methylation, Transfection, and Luciferase Assays
Luciferase reporter constructs were prepared by cloning the 655 bp (−630 to +25 bp) PCR product of the DOC2B gene into KpnI and HindIII sites of pGL3-Basic and pGL3-Enhancer vectors, respectively. The constructs were verified by DNA sequencing. All plasmid constructs were artificially methylated in vitro using SssI DNA methylase and confirmed by restriction digestion with HpaII and MspI, respectively, according to the manufacturer's instruction (New England Biolabs). The primers used are shown in Table 1.
The methylated and unmethylated constructs (1.6 μg each) were co-transfected with pRL-SV40 vector (50 ng) into 60–70% confluent cultures of SiHa cells in a 6-well tissue culture plates (Greiner bio One, Germany) with Lipotransfectamine-LTX (Invitrogen) according to the manufacturer's guidelines. The cell lysates were prepared after 48 h post-transfection and luminescence was measured in a FB12 Luminometer (Berthold, Germany) using the Dual Luciferase reporter assay kit (Promega) according to manufacturer's instructions. Reporter activity was normalized by calculating the ratio of firefly to Renilla values. All transfections were performed in duplicates and repeated twice.
Transfection and Selection of Stable DOC2B Expressing Clones
The DOC2B-pCMV6-Entry (Origene) or empty vector was transfected into SiHa cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. In brief 70–80% confluent cells were transfected with 4 μg of DOC2B cDNA construct. Stably transfected cells were established under G418 (400 μg/ml) selection for 21 days, individual clones were selected; RT-PCR was performed to confirm the gene expression.
Generation of Recombinant Retrovirus Expressing DOC2B
The pCMV-Entry-DOC2B expressing full-length DOC2B cDNA was isolated by digesting with BamHI and XhoI and subcloned into the BamHI and XhoI sites of pMX-IRES-GFP. pMX-IRES-GFP and pMX-DOC2B-IRES-GFP plasmids were transfected into the Phoenix-E packaging cells by using Lipofectamine Plus (Invitrogen) according to the manufacturer's instruction. Retroviral particles were harvested after 72 h and used to infect SiHa and HeLa cells as described previously (18). Briefly, cells were incubated with retrovirus in the presence of 8 μg/ml of hexadimethrine bromide (Sigma) for 24 h, washed, and cultured in the presence of DMEM containing 10% FBS. The cells expressing DOC2B were isolated by cell sorting using FACS Calibur (BD Biosciences) flow cytometer, confirmed by RT-PCR, and Western blot analysis. These were subsequently used for experiments.
Western Blot Analysis
Total protein was extracted and the concentration was estimated using a Bradford assay kit (Sigma). Briefly, cells were lysed in RIPA buffer and protein (30 μg) was resolved in 8% SDS-PAGE, transferred onto Nitran membrane (Sigma), blocked with 5% nonfat dry milk, and incubated separately with anti-DDK tag primary antibody for DOC2B (Origene), anti-DOC2B (Proteintech) (1:3000), phospho-AKT (Ser473), total AKT, phosphor-ERK1/2 (Thr202/Tyr204), and total ERK1/2 (Thr202/Tyr204) (1:1000) (Cell Signaling, Beverly, MA) overnight at 4 °C and subsequently by secondary antibody anti-mouse IgG-HRP and anti-rabbit IgG-HRP (Cell Signaling) (1:5000). Proteins were visualized by ECL reagent (GE Healthcare) and imaging was performed in ImageQuant LAS 4000 (GE Healthcare). Antibody against β-actin (Cell Signaling) was used to normalize the loading control.
Anoikis Assay
Cells were cultured in 6-well plates were coated with poly-2-hydroxyethyl methacrylate (poly-HEMA) as published previously. Control and DOC2B expressing cells were trypsinized, washed, and cultured on poly-HEMA-coated plates at a cell density of 1 × 105 cells/well. The number of viable cells were analyzed by FACS (FACS Calibur, BD Biosciences) after staining with propidium iodide (10 μg/ml in PBS) as published previously (19).
Anchorage-dependent and -Independent Colony Formation Assay
Colony forming assay was performed as published previously with minor modifications (20). In brief, 100 cells were plated in duplicates in 6-cm Petri plate. After 14 days, the medium was removed and the cells were washed with PBS and stained with 0.5% crystal violet in methanol for 5 min. Excessive stains were removed by washing with distilled water, photographed, and colonies were counted. The experiments were performed in duplicates and repeated three times.
In 6-well plates, 1 × 103 cells/well containing 2 ml of 0.3% Nobel agar (DMEM + 10% FBS) was overlaid on top of 3 ml of 0.6% bottom agar base (DMEM + 10% FBS) and 0.5 ml of complete medium was added every week and at the end of 4 weeks colonies were counted. The experiment was performed in triplicate and repeated 3 times. Student's t test was used for statistical analyses (21).
Cell Doubling and Growth Curve Analysis
About 3000 cells were plated in a 6-cm Petri plate for a 5-day growth curve analysis. The cells were trypsinized at each time point and counted using a hemocytometer. The experiments was performed in duplicates and repeated three times. The cell doubling time was calculated using the cell doubling time calculator.
[3H]Thymidine and [3H]Uridine Incorporation Assays
About 30,000 cells were seeded in 6-well plates. After 24 h, 1 μCi of radiolabeled thymidine and uridine (Moravek Biochemicals) were added, incubated for 6 h, trypsinized, precipitated onto GF/C glass fiber filter papers with 10% TCA, and sequentially washed with 70 and 100% alcohol using a Millipore mannifold. The membranes were dried and radioactivity was measured using liquid scintillation counter (PerkinElmer Life Sciences).
Cell Migration Assay
Transfected cells were grown to confluence in 6-well plates, starved for 24 h, and a scratch test was performed using a sterile microtip. Following the scratch, fresh medium with 10% FBS was added and migration of cells into the wound area was monitored until 72 h. The images were captured at the indicated time points using a Rolera emc2 camera and Olympus Microscope CK-41 (Olympus, Japan). The migration rate/index was calculated as published previously (22).
Actin-Phalloidin Staining
The 70–80% confluence cells grown on coverslips were fixed in 4% paraformaldehyde at 37 °C for 40 min followed by permeabilization with 0.1% Triton X-100 for 20 min at room temperature, washed with PBS, and stained overnight with 15 μg of Alexa Fluor phalloidin (Sigma). The next day, the excessive stains were removed; cells were washed with PBS, stained with Hoechst stain (1:1000), and incubated for 30 min. Images of cells were captured using a Rolera emc2 camera attached to a Olympus microscope (Olympus, Japan).
Intracellular Calcium Measurement
Intracellular Ca2+ levels were measured as published previously (23). In brief, 106 cells were collected by trypsinization and incubated in Hanks' balanced salt solution, pH 7.4, containing 4 μg/ml of Fluo-3/AM ester and 0.1% pluronic F-127 (Molecular Probes) at 37 °C for 30 min. The experiment was performed in the presence and absence of external calcium. Cells treated with ionomycin (1 mg/ml) were used as positive control. The calcium flux was monitored in 10,000 cells using FACS Calibur (BD Biosciences) and data were analyzed using CellQuest software. The median fluorescence intensity was used for subsequent analysis. The experiments were performed in duplicates and repeated three times.
Cell Cycle and Apoptosis Analysis
Cell cycle distribution was assessed by BrdU flow kit (BD Biosciences). Briefly, the transfected cells were cultured in the serum-free DMEM for 48 h followed by addition of BrdU (10 μm) for 30 min at 37 °C and cultured in complete medium for the indicated times, and cell cycle distribution was assessed. The cell cycle data were analyzed by a FACS Calibur flow cytometer using CellQuest software (BD Biosciences). The experiments were performed in duplicates and repeated three times.
Invasion Assay
In vitro cell invasion analysis was performed using the agarose spot assay as published previously (24). In brief, 0.5% agarose solution containing fibronectin (20 μg/ml) or type I collagen (100 μg/ml) was spotted onto 6-well plates (Cell Star, Germany). Stably transfected cells (1 × 106) were plated onto wells containing agarose spot in serum-free DMEM medium and incubated at 37 °C for 24–48 h. The plates were observed over a period of time to examine the movement of cells onto the agarose spot. Images of cells were captured using a Rolera emc2 camera and microscope (Olympus, Japan). The experiments were performed in duplicates and repeated three times.
In Vivo Tumorigenicity Assay
For in vivo tumorigenicity assays 5–6-week-old female BALB/c-nude mice (5 per group) were injected subcutaneously with 1 × 107 cells mixed with Matrigel (1:1) into the lower flank of the animals. The tumor growth was monitored for over 2 months and tumor volume was measured 20 days post-injection. The tumor volume (V) was determined by the length (a) and width (b) as V = ab2/2. The experimental protocols were evaluated and approved by the animal ethics committee of Manipal University. Mice were sacrificed 2 months after injection and tumor tissue and organs were removed. Tumor tissue cryosection of 5-μm thickness were taken and stained with Hematoxylin & Eosin (H&E) and Masson's trichrome and evaluated by a pathologist.
Bioinformatic and Statistical Analysis
The genomic coordinates spanning the promoter region of DOC2B was analyzed using CpG island searcher, transcriptional regulatory element database, transcription element search system (cbil.upenn.edu/cgi-bin/tess), and AliBaba2.1. All data analysis was performed using Microsoft Excel 2007 (Microsoft) and GraphPad Instat (Trial Version). Student's t test, one-way analysis of variance, and Kruskal-Wallis test were used for statistical analysis and significance. The p value less than 0.05 were considered statistically significant.
RESULTS
Identification of Differentially Methylated Regions
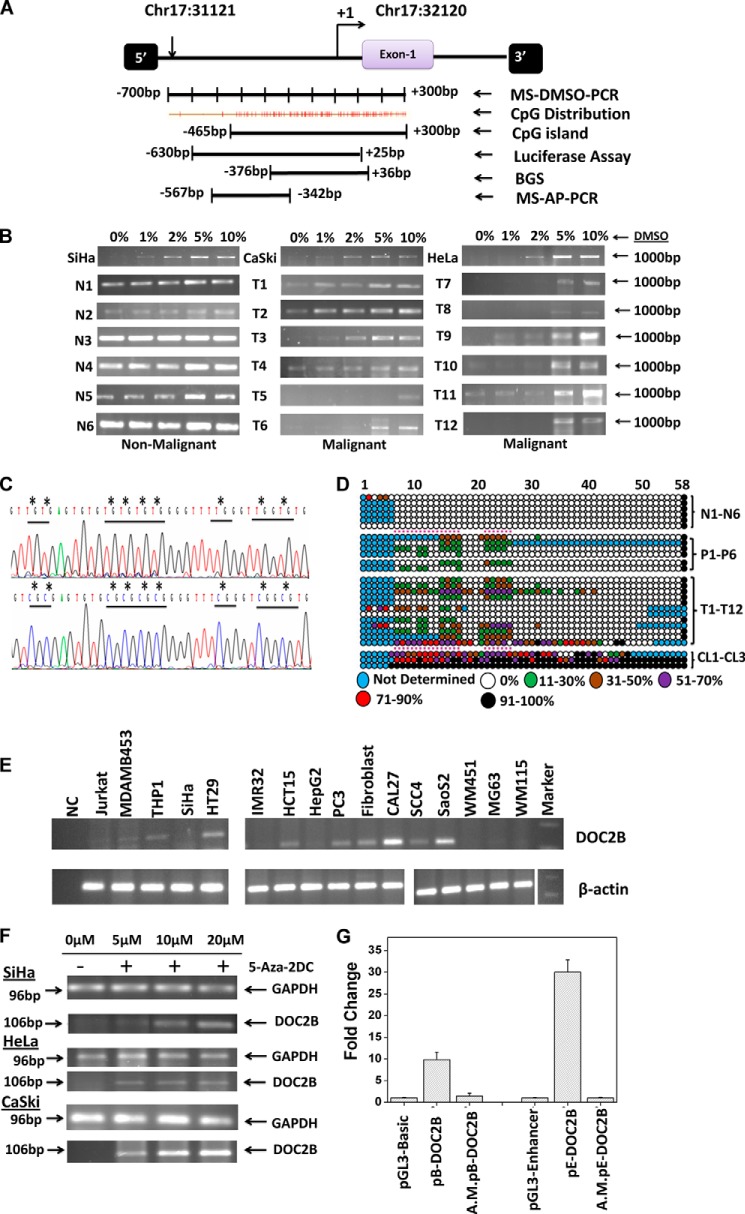
MS-AP-PCR was performed to identify differentially methylated regions in cervical cancer and non-malignant tissue samples (12, 13). We have isolated, cloned, and sequenced 15 hypermethylated fragments. A total of 10 fragments showed the characteristics of CpG islands and 6 fragments were found within the specific genes; whereas, the remaining fragments were found outside the gene coordinates. Nine hypermethylated fragments were associated with copy number variations regions (Table 2). The fragment 13 (Frg-13) was found to lie within the CpG island of the DOC2B gene promoter (−57 6bp to −342bp) and this was further analyzed for DNA methylation studies.
TABLE 2.
Summary of hypermethylated sequences identified by MS-AP-PCR
The presence of CpG Island was predicted using the following criteria: CG content greater than 50%, observed CpG/expected CpG greater than 0.60 and length greater than 200 bp. The copy number variation (CNV) analysis was performed by searching in genomic variant database.
| Clone | Fragment size | Gene | GC | Observed CpG/expected CpG | Chromosome location | CpG island | CNV |
|---|---|---|---|---|---|---|---|
| % | |||||||
| Frg1 | 238 bp | Myomesin2 | 55.4 | 8p23.3 | No | Yes | |
| Frg2 | 251 bp | hypothetical protein LOC441390 | 52.5 | 9p21.2 | No | No | |
| Frg3 | 312 bp | KLRG2 | 52.4 | 0.878 | 7q34 | Yes | No |
| Frg4 | 657 bp | IKBKG | 62.7 | 0.718 | Xq28 | Yes | Yes |
| Frg5 | 359 bp | ZBED4 | 55.6 | 0.752 | 22q13.33 | Yes | Yes |
| Frg6 | 406 bp | 187,243 bp at 5′ end: hypothetical protein | 55 | 0.6 | 17q24.2 | Yes | Yes |
| Frg7 | 455 bp | 440 bp at 5′ end: hypothetical protein | 57.5 | 0.607 | 15q26.3 | Yes | No |
| Frg8 | 362 bp | 838,601 bp at 5′ end: deleted in bladder cancer 1 | 50 | 1.083 | 9q33.2 | Yes | No |
| Frg9 | 399 bp | NXN | 51.6 | 0.603 | 17p13.3 | Yes | Yes |
| Frg10 | 613 bp | 23,224 bp at 5′ end: MMP16 | 51.1 | 0.657 | 8q21.3 | Yes | Yes |
| Frg11 | 501 bp | 2,332 bp at 3′ end: hypothetical protein LOC84262 | 53.4 | 7p22.3 | No | ||
| Frg12 | 287 | 3,315 bp at 3′ end: PBX/knotted 1 homeobox 2 | 51.5 | 0.601 | 11q24.2 | Yes | Yes |
| Frg13 | 225 bp | DOC2B promoter | 58.5 | 0.617 | 17p13.3 | Yes | Yes |
| Frg14 | 415 bp | 2,482 bp at 3′ end: proteasome assembling chaperone 3 | 61.8 | 0.64 | 7p22.3 | Yes | Yes |
| Frg15 | 689 bp | 95,927 bp at 3′ end: titin immunoglobulin domain protein | 37.5 | 5q31 | No | No |
Mapping of Hypermethylated CpG Loci
To examine the methylation status of fragment Frg-13 and determine its frequency of methylation in cervical cancer, MS-DMSO-PCR and BGS were performed. A total of 15 non-malignant, 30 tumor samples, and 3 cervical cancer cell lines were analyzed by MS-DMSO-PCR, which showed that DNA methylation indeed changed the sensitivity of amplification to the DMSO concentration in the PCR mixture (Fig. 1B). For each sample, four different DMSO concentrations were used. i.e. 1, 2, 5, and 10% and the reaction without DMSO acting as control. In the case of the hypermethylated samples, amplification was detected only at a DMSO concentration of 2% or higher, whereas unmethylated samples showed amplification in the absence of DMSO indicating that tumor samples were hypermethylated when compared with the non-malignant samples (Fig. 1B).
FIGURE 1.
Methylation profiling of the Frag-13 fragment in cervical samples. A, schematic representation of the region selected for validation of Frag-13 fragment by MS-AP-PCR, MS-DMSO-PCR, BGS, and characterization of the promoter region of the DOC2B gene by a luciferase assay. B, MS-DMSO-PCR showed that DNA methylation changed the sensitivity of amplification to the DMSO concentration in the PCR mixture. In the case of methylated samples, amplification was detected at a DMSO concentration of 2% or more, whereas unmethylated or less methylated samples showed amplification even in the absence of DMSO. N and T represent non-malignant and tumor samples, respectively. C, representative electropherogram of a portion of the 412-bp region of non-malignant and tumor samples showing methylated and unmethylated CpG sites. The differentially methylated CpG sites were highlighted by asterisks (*). D, methylation map of the DOC2B promoter fragment in non-malignant, pre-malignant, malignant, and cervical cancer cell lines. The open and filled circles represent the unmethylated and methylated CpG sites, respectively. The partially methylated CpG sites were filled with different colors depending on the extent of methylation. Each horizontal line represents single samples, and circles representing single CpG sites. N1-N6, P1-P6, T1-T12, and CL1-CL3 represent 6 non-malignant, 6 pre-malignant, 12 malignant cervical samples, and 3 cervical cancer cell lines namely SiHa, CaSki, and HeLa, respectively. E, expression analysis of DOC2B in various cell lines by semi-quantitative RT-PCR with β-actin as control. F, demethylation by 5-aza-2DC and subsequent reactivation of mRNA of the DOC2B gene. Cervical cancer cell lines SiHa, CaSki, and HeLa were used for demethylation and reactivation experiments. In all three cell lines, reactivation was observed at 5 μm or above of 5-aza-2-DC treatment. GAPDH was used as an internal control for the integrity of the cDNA. RT-PCR analysis shows the loss of DOC2B expression and treatment with the demethylating agent 5-aza-2-DC restores its expression. G, characterization of promoter activities of the DOC2B gene by transient transfection experiments using DOC2B promoter constructs in pGL3-Basic and pGL3-Enhancer vectors. Histograms represent mean ± S.D. for at least two independent experiments. pB-DOC2B and pE-DOC2B represent the DOC2B promoter constructs cloned in pGL3-Basic and pGL3-Enhancer vectors, whereas A.M.pE-DOC2B represents corresponding artificially methylated constructs as discussed under ”Experimental Procedures.“
To map the hypermethylated CpGs dinucleotides, BGS was performed for the (412 bp) region −376 to +36 bp of the DOC2B gene containing 58 CpG sites in 6 normal, 6 pre-malignant (4 low grade squamous intraepithelial lesion (LSIL), 2 high grade squamous intra epithelial lesion (HSIL)), 12 tumor samples, and 3 cervical cancer cell lines (SiHa, CaSki, and HeLa), respectively (Fig. 1, C and D). The CpG sites were unmethylated in normal samples, whereas hypermethylation was observed in 6 (of 6; 100%), 10 (of 12; 83.33%) and 3 (of 3; 100%) of pre-malignant, malignant, and cervical cancer cell lines, respectively (Table 3). Dense promoter methylation was observed in all the three cell lines tested, whereas partial to complete methylation of selected CpG sites were observed in LSIL, HSIL, and tumor samples (Fig. 1D). The promoter hypermethylation was found to be statistically significant between normal and tumor samples as well as between normal and LSIL/HSIL; and normal and malignant samples by one-way analysis of variance and Kruskal-Wallis test (p < 0.05), respectively. A significant association was found between HPV infection and DOC2B methylation (p < 0.005), however, more samples need to be screened before drawing any conclusion (Table 3). An independent association was observed between age and DOC2B promoter hypermethylation.
TABLE 3.
Mean percentage methylation of DOC2B fragment in normal, tumor and cervical cancer cell lines
The following abbreviations are used: N1–N6, normal; T1–T18, tumor; N, negative; P, positive; LSIL, low grade squamous intra epithelial lesions; HSIL, high grade squamous intra epithelial lesions; PD-SCC, poorly differentiated squamous cell carcinoma; UD-SCC, undifferentiated squamous cell carcinoma; MD-SCC, moderately differentiated squamous cell carcinoma; LC-NK-SCC, non-keratinizing squamous cell carcinoma.
| Samples | Tissue | Age | DOC2B methylation | HPV status |
|---|---|---|---|---|
| % | ||||
| N1 | Normal | 45 | 1.9 | N |
| N2 | Normal | 50 | 7 | N |
| N3 | Normal | 48 | 1.9 | N |
| N4 | Normal | 42 | 1.9 | N |
| N5 | Normal | 48 | 1.9 | N |
| N6 | Normal | 55 | 1.7 | N |
| Mean ± S.E. | 2.72 ± 0.86a | |||
| P1 | LSIL | 40 | 25 | N |
| P2 | LSIL | 46 | 42.85 | HPV16 |
| P3 | LSIL | 46 | 26.92 | HPV16 |
| P4 | LSIL | 54 | 13.46 | HPV18 |
| P5 | HSIL | 53 | 19.23 | HPV16 |
| P6 | HSIL | 74 | 11.53 | HPV11 |
| Mean ± S.E. | 23.17 ± 4.66a | |||
| T1 | PD-SCC (III-b) | 68 | 19.23 | N |
| T2 | UD-SCC (II-b) | 71 | 30.76 | HPV18 |
| T3 | LC-K-SCC (III-B) | 55 | 61.53 | HPV16 |
| T4 | MD-SCC (II-b) | 52 | 19.23 | HPV16 |
| T5 | LC-NK-SCC (I-b) | 40 | 1.9 | HPV16 |
| T6 | LC-NK-SCC (I-a | 57 | 24.48 | HPV16 |
| T7 | LC-NK-SCC (I-a) | 48 | 0 | N |
| T8 | UD-SCC (II-b) | 55 | 17.3 | N |
| T9 | PD-SCC (II-a) | 50 | 31.91 | HPV16 |
| T10 | MD-SCC (III-b) | 40 | 30.76 | HPV18 |
| T11 | MD-SCC (III-b) | 60 | 25 | HPV16 |
| T12 | LC-NK-SCC (II-b) | 49 | 86.66 | HPV16 |
| Mean ± S.E. | 29.06 ± 6.93a | |||
| SiHa | 91.1 | HPV16 | ||
| CaSki | 100 | HPV16 | ||
| HeLa | 100 | HPV18 |
a According to one-way analysis of variance, the percentage of methylation was found to be statistically significant between normal and malignant samples. It was also found to statistically significant between normal and pre malignant and normal and malignant when Kruskal-Wallis test was performed. p value less than p < 0.05 was considered as statistically significant.
Aberrant Methylation and Expression of DOC2B Gene in Cell Lines
We examined DOC2B expression status in a series of cancer cell lines and normal fibroblast using semi-quantitative RT-PCR and results are summarized in Fig. 1E. Significant reduction of DOC2B expression was observed in cancer cell lines of various origins but not in normal diploid fibroblast cells. The BGS analysis revealed that methylation is not confined to any specific region but it is spread uniformly throughout the DOC2B promoter in SiHa, CaSki, and HeLa cell lines (Fig. 1D and Table 3) leading to complete loss of DOC2B gene expression and was restored after treatment with 5-aza-2DC (5 μm and above) for 3 days in all three cervical cancer cell lines suggesting its transcriptional regulation by promoter-specific DNA methylation (Fig. 1F). Taken together, our results show that DOC2B expression is inhibited or reduced in different cancer cell lines, indicating its possible role as a negative regulator in multiple cancers.
Characterization of DOC2B Promoter by Transient Transfection Assay
The DOC2B promoter construct showed nearly 9-fold higher promoter activity when compared with vector alone (Fig. 1G). The heterologous enhancer-driven (SV40) DOC2B promoter construct was 30-fold more active than the respective control (p < 0.05). Upon artificial methylation the DOC2B promoter and enhancer-driven constructs showed diminished expression as opposed to mock methylated constructs in SiHa cells (Fig. 1G). These results confirmed that the DOC2B promoter was indeed under tight regulation by DNA methylation.
DOC2B and Tumor Suppressive Function
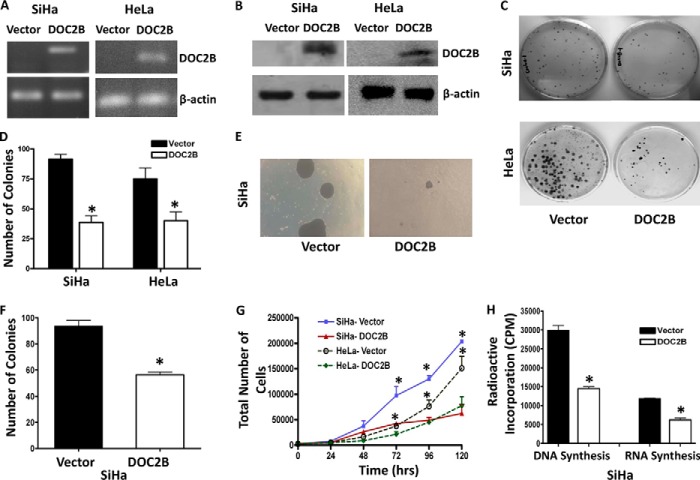
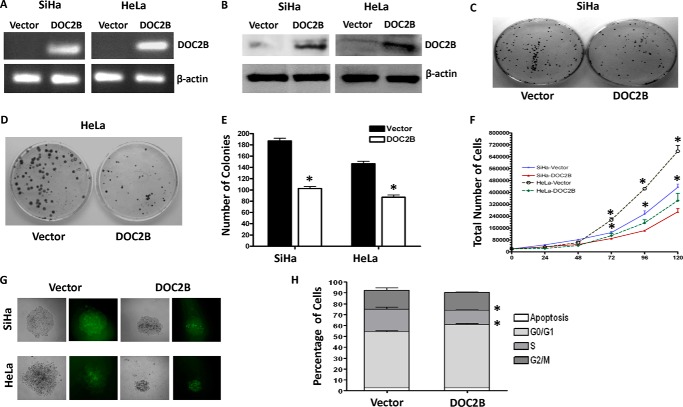
Frequent silencing of DOC2B in cervical cancer cell lines suggests it is likely to be associated with cell proliferation. To explore this, the DOC2B gene was overexpressed in SiHa cells, which do not inherently express DOC2B. After confirmations of ectopic DOC2B expression by RT-PCR (Fig. 2A) and Western blot analysis (Fig. 2B), SiHa and HeLa cells were subjected to colony formation, cell doubling, and growth curve assays. The colonies formed by DOC2B expressing cells were significantly less and smaller in size when compared with empty vector (p <0.005) transfected stable clones (Fig. 2, C and D). Moreover, DOC2B expression suppressed the colony size and decreased the number of colonies to 40% in anchorage-independent experiments (Fig. 2, E and F). Ectopic expression of DOC2B also inhibited cell proliferation and increased doubling time in both SiHa and HeLa, respectively (Fig. 2G). Overexpression of DOC2B showed a substantial increase in doubling time from 37.49 to 48.52 h in SiHa cells, whereas 22.79 to 27.88 h in HeLa cells. There was also a significant reduction in the rates of DNA and RNA synthesis (Fig. 2H) in DOC2B expressing cells as opposed to control cells (p < 0.05). Therefore, DOC2B expression may provide growth disadvantage function to cervical cancers. We have validated some of these findings by overexpressing DOC2B using pMX-IRES-GFP retroviral vector and confirmed by using RT-PCR and Western blot (Fig. 3, A and B) that DOC2B expression inhibited cell growth (Fig. 3C-3E) and proliferation (Fig. 3F) in SiHa and HeLa cells, respectively. The sizes of the colonies were smaller in DOC2B expressing cells when compared with control cells (Fig. 3G). Similar to single cell clones, polyclonal cells expressing retrovirally transduced DOC2B also inhibited cell cycle progression at the G0/G1 and S phases of cell cycle when compared with DOC2B-deficient control cells (Fig. 3H).
FIGURE 2.
Effect of ectopic expression of DOC2B on cell growth and proliferation. A and B, representative figures showing expression of the DOC2B gene upon transfection by RT-PCR and Western blot, respectively. DOC2B was detected by anti-DDK tag antibody. β-Actin was used as an internal control. C, DOC2B inhibits tumor growth in vitro in SiHa and HeLa cells, respectively. D, quantitative analysis of colony forming assay represented as mean ± S.D., *, p < 0.05, shows a significant decrease in colony number after ectopic expression of DOC2B. E, representative image of soft agar colony forming assay. F, quantitative analysis of colony number represented as mean ± S.D.; *, p < 0.05. G, represents the cell proliferation rate in DOC2B expressing stable clones in comparison with vector control. Ectopic expression of DOC2B significantly inhibited cell proliferation resulting in delayed cell doubling time. The cell doubling analysis was performed using the cell doubling time calculator. H, cell proliferation rate was significantly inhibited at both DNA and RNA levels in DOC2B expressing cells when compared with control cells by [3H]thymidine and [3H]uridine incorporation assays, respectively (mean ± S.D. from 3 independent experiments in duplicates). *, represents p < 0.05.
FIGURE 3.
Ectopic expression of DOC2B suppresses growth and proliferation in SiHa and HeLa cells. A and B, the DOC2B ORF was cloned into retroviral vector pMX-IRES-GFP to generate pMX-DOC2B-IRES-GFP and used to transduce SiHa and HeLa cells. The expression of DOC2B upon ectopic expression was confirmed by both RT-PCR (A) and Western blot by anti-DOC2B antibody (B). β-Actin was used as internal control. C and D, representative image of the colony formation assay in SiHa and HeLa, respectively. E, representive quantitative analysis of the colony numbers in SiHa and HeLa cells, respectively. The ectopic expression of DOC2B significantly reduced the colony numbers in both SiHa and HeLa cells, respectively. F, ectopic expression of DOC2B inhibits SiHa and HeLa cell proliferation, respectively. G, representative colonies from SiHa and HeLa, respectively. Both colony number and size decreased upon DOC2B expression. H, DOC2B expression induced arrest at the G0/G1 and S phases of the cell cycle. *, p < 0.05 by independent Student's t test was considered as statistically significant. The experiments were repeated 3 times in duplicate.
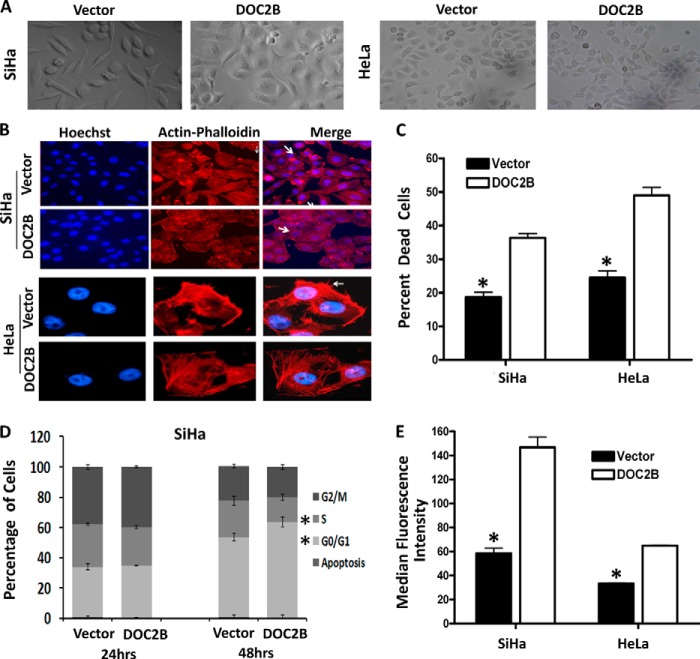
Ectopic Expression of DOC2B Changes Cell Morphology
DOC2B expressing cells showed distinct morphological changes compared with control cells such as increased cell-cell adhesion and decreased cell scattering (Fig. 4). Increased cell-cell adhesion and decreased cell scattering may have been a contributing factor toward the decreased ability of DOC2B expressing cells to migrate. To investigate the role of DOC2B in regulating the properties of tumor cell invasion and metastasis, actin-phalloidin staining was performed (Fig. 4B). Ectopic expression of DOC2B resulted in actin rearrangement and decreased lamellipodia formation when compared with control cells, indicating that DOC2B could induce cytoskeleton remodeling leading to inhibition of cervical cancer cell invasion (Fig. 4B).
FIGURE 4.
DOC2B expression changes cell morphology, induces remodeling of cytoskeletons, inhibits cell division, and increases intracellular calcium flux. A, representative cell morphology of DOC2B expressing and control cells in SiHa and HeLa cells, respectively (×40 magnification). B, representative images of actin staining showing rearrangement of actin fibers leading to increased cell to cell adhesion and decreased lamellipodia (indicated by arrows) in DOC2B expressing cells when compared with control cells (×400 magnifications). C, representative anoikis analysis for empty vector- and DOC2B-transfected SiHa and HeLa cells. Cells were cultured on poly-HEMA-coated tissue culture plates and cell death was analyzed by measuring the sub-G1 population of cells by propidium iodide staining and FACS analysis. Ectopic expression of DOC2B showed a significantly higher sub-G1 population of 18.66 versus 36.33% and 24.47 versus 48.96% between DOC2B and empty vector-transfected cells in both SiHa and HeLa cells, respectively. D, quantification of DNA content and cell cycle phase distribution in empty vector- and DOC2B-transfected SiHa cells at different time points as analyzed by a BrdU pulse-chase experiment. Ectopic expression of the DOC2B gene resulted in a significant cell cycle arrest at G0/G1 and S phases of the cell cycle. E, representative figures showing an increase in intracellular Ca2+ flux when compared with control cells. There was a significant increase in intracellular Ca2+ upon ectopic expression of DOC2B. Values were the median fluorescence intensities ± S.D. of at least three independent experiments performed in duplicates. The bar graph represents mean ± S.D. of triplicate experiments performed in duplicates. *, p < 0.05 by independent Student's t test. p value <0.05 was considered statistically significant.
DOC2B Expression Induces Anoikis in SiHa Cells
To determine the mechanism of DOC2B induced inhibition of colony formation, delay in cell doubling, and proliferation, we analyzed the effect on apoptosis and cell cycle progression. Normal cells may undergo cell death due to anoikis when detached from the extracellular matrix; whereas tumor cells escape anoikis due to oncogenetic transformation. Therefore we investigated apoptosis under anoikis conditions. Cells were cultured on poly-HEMA-coated tissue culture plates and cell death was analyzed by measuring the sub-G1 population of cells by propidium iodide staining and FACS analysis. Ectopic expression of DOC2B showed a significantly higher sub-G1 population 18.66 versus 36.33% and 24.47 versus 48.96% between DOC2B and empty vector-transfected cells in both SiHa and HeLa cells, respectively (Fig. 4C). These data suggest that DOC2B induces anoikis in SiHa and HeLa cells.
DOC2B Expression Regulates G0/G1-S Phase Transition and Increases Ca2+ Flux
DOC2B expression resulted in an increase in percent of G0/G1 cells and decrease in S phases cells (p < 0.05), respectively (Fig. 4E). Our result indicates that expression of DOC2B inhibits tumor cell growth by suppressing cell proliferation and influences delay in cell cycle progression and induces anoikis. Concurrently, expression of DOC2B showed an increase in intracellular Ca2+ flux (median fluorescence intensity; 153.2 versus 56.5, p < 0.05) when compared with control cells (Fig. 4F) as measured using Fluo-3-AM probe.
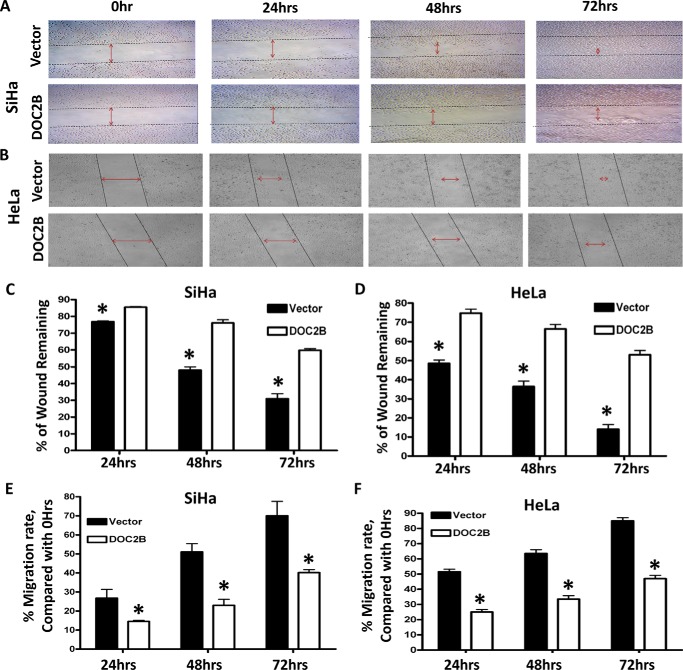
DOC2B Inhibits Migration and Invasion in Cervical Cancer Cells
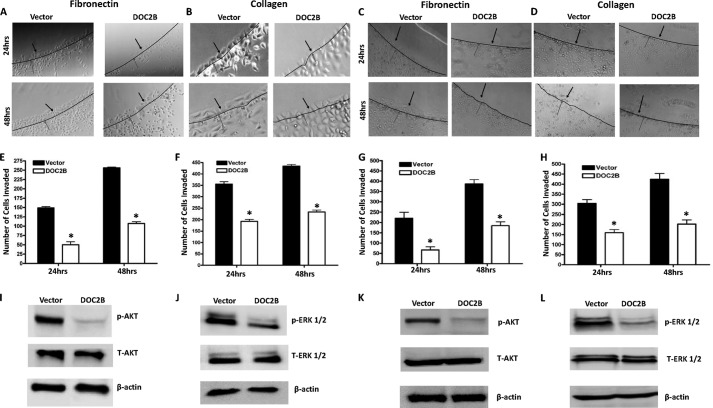
Expression of the DOC2B gene reduced cell migration (Fig. 5), which was evident from analysis of the migration index (p < 0.05) (Fig. 5, E and F). Quantitative analysis at 24, 48, and 72 h showed a progressive decrease in wound closure rate (14.5 versus 29.4%, 25.4 versus 53.9%, 40.1 versus 75.9%, and 25.26 versus 51.47%, 33.5 versus 63.6% and 47 versus 85.7%) between DOC2B and empty vector-transfected cells in both SiHa and HeLa, respectively (Fig. 5, C–F). DOC2B overexpression using pMX-IRES-GFP retroviral vector reduced cervical cancer cell migration in both SiHa and HeLa cells, respectively (Fig. 6). Moreover, at the end of 96 h, the wound was completely closed in vector-transfected cells, whereas it remained incomplete in DOC2B expressing cells. Regardless of DOC2B expressing clonal isolates or retroviral transduced cells, their invasion toward fibronectin and type I collagen-coated plates were significantly inhibited (p < 0.05) (Fig. 7, A–H). These results suggest that DOC2B may play a role as an inhibitor of invasion of cervical tumor cells in our experimental systems. Because DOC2B regulates cell survival, proliferation, growth, and cell cycle progression in SiHa cells, we examined the phosphorylation status of AKT1 and ERK1/2 proteins. DOC2B expressing cells inhibited AKT1 (Ser473) and ERK1/2 (Thr202/Tyr204) phosphorylation without showing significant changes in their total protein level (Fig. 7, I–L).
FIGURE 5.
DOC2B expression significantly suppresses the motility of cervical cancer cells. A and B, scratch assay was performed to assess the migration rate of SiHa and HeLa cells transfected with either vector or pCMV6-Entry-DOC2B. Photomicrographs were taken at the indicated time points under ×100 magnification. C and D, the percentage of wound closure in relationship to the time 0 h separation in SiHa and HeLa, respectively. E and F, the graph showing the rate of cell migration (migration index) in relationship to time 0 h separation in SiHa and HeLa, respectively. The migration index was calculated and quantified by measuring the area of the injured region in relationship to 0 h. The bar graph represents mean ± S.D. of triplicates experiments performed in duplicates. *, p < 0.05 by independent Student's t test. p value <0.05 was considered as statistically significant.
FIGURE 6.
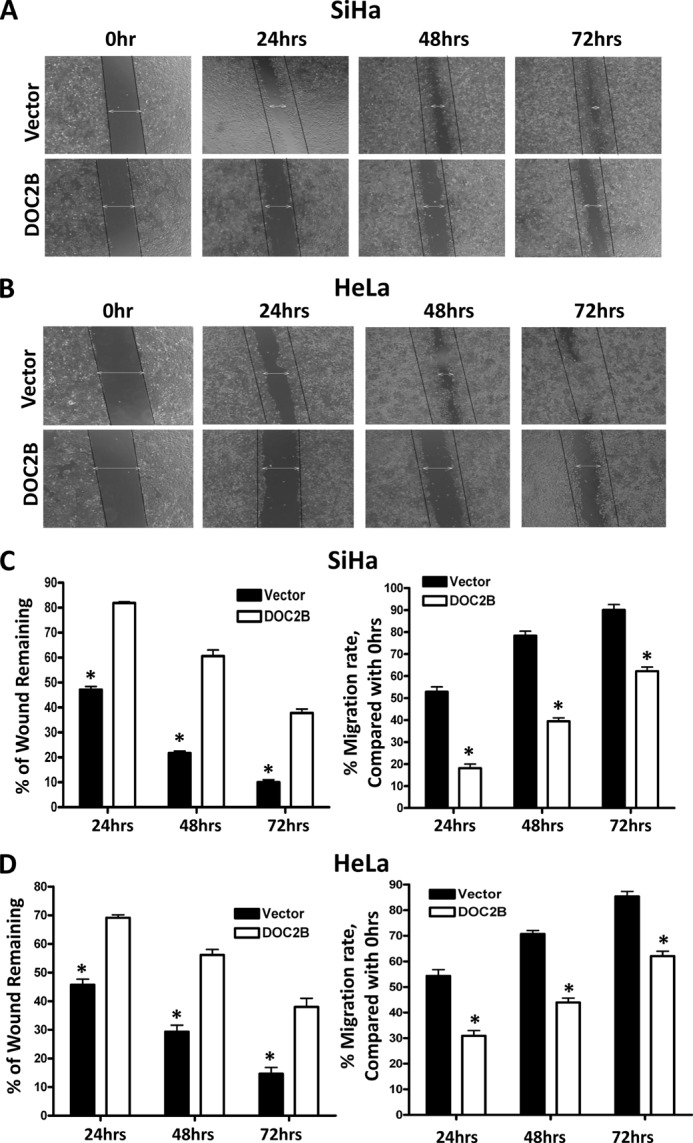
DOC2B expression inhibits migration of SiHa and HeLa cells in vitro. Wound healing assay was performed to identify the effect of DOC2B on cell migration. In brief cells were grown to confluence in 6-well plates, serum starved for 24 h, and a scratch was made using P-200 tip and would closure was monitored 72 h. A and B, representative image of wound healing assay. Our results show that the wound is completely closed in control cells by 72 h both in SiHa (left) and HeLa (right) cells, respectively. However, in DOC2B expressing SiHa and HeLa cells the wound was still intact even at the end of 72 h. C and D, representative graph showing the quantitative analysis of migration. Our experiments show that DOC2B expression inhibits migration of SiHa and HeLa cells. *, p < 0.05 by independent Student's t test was considered as statistically significant. The experiments were repeated 3 times in duplicates.
FIGURE 7.
DOC2B inhibits invasion and phosphorylation of AKT-1 and ERK1/2. A–D, representative images showing the inhibition of invasion onto the agarose spot containing fibronectin and type I collagen upon ectopic expression of DOC2B in a single cell clone (A and B) and retrovirally transduced polyclonal cells (C and D). E–H, quantitative analysis of a number of tumor cells invading fibronectin and type 1 collagen in a single cell clone (E and F) and retrovirally transduced polyclonal cells (G and H), respectively. The bar graph represents mean ± S.D. of triplicate experiments performed in duplicates. *, p < 0.05 by independent Student's t test was considered as statistically significant. I–L, ectopic expression of DOC2B regulated both AKT and ERK signaling. A Western blot was performed using antibodies against total AKT, phospho-AKT, total ERK1/2, and phospho-ERK1/2. β-Actin was used as internal control. DOC2B expression inhibited AKT phosphorylation, whereas there was a decrease in ERK1/2 phosphorylation when compared with vector-transfected control SiHa cells in both single cell clone (I and J) and polyclonal cells (K and L), respectively. Relative phosphorylation levels of AKT and ERK1/2 were determined by normalization with total AKT and total ERK1/2, respectively.
DOC2B Inhibits Tumor Growth in Vivo
Animal studies were conducted to evaluate the role of DOC2B in influencing tumor growth in athymic nude mice. The transfected cells were injected into the mice and progressive tumor growth was analyzed. The results demonstrated that cells in the control group formed a progressively growing tumor. In contrast, animals injected with DOC2B expressing cells produced a significantly smaller tumor (p < 0.05) (Fig. 8, A and B). Thus, these results indicate that, ectopic expression of DOC2B results in in vivo inhibition of tumor growth. The H&E staining of DOC2B expressing cells showed a decreased nucleus to cytoplasmic ratio, less abnormal nucleus and decreased tumor cell density, less pleomorphic (less atypical) and densely aggregated cells, and reduced the number of abnormal mitosis in contrast to DOC2B negative cells (Fig. 8C). Expression of the DOC2B gene may inhibit collagen degradation and/or synthesis, which is evident by Masson's trichrome staining of tumor xenografts grown in nude mice (Fig. 8D).
FIGURE 8.
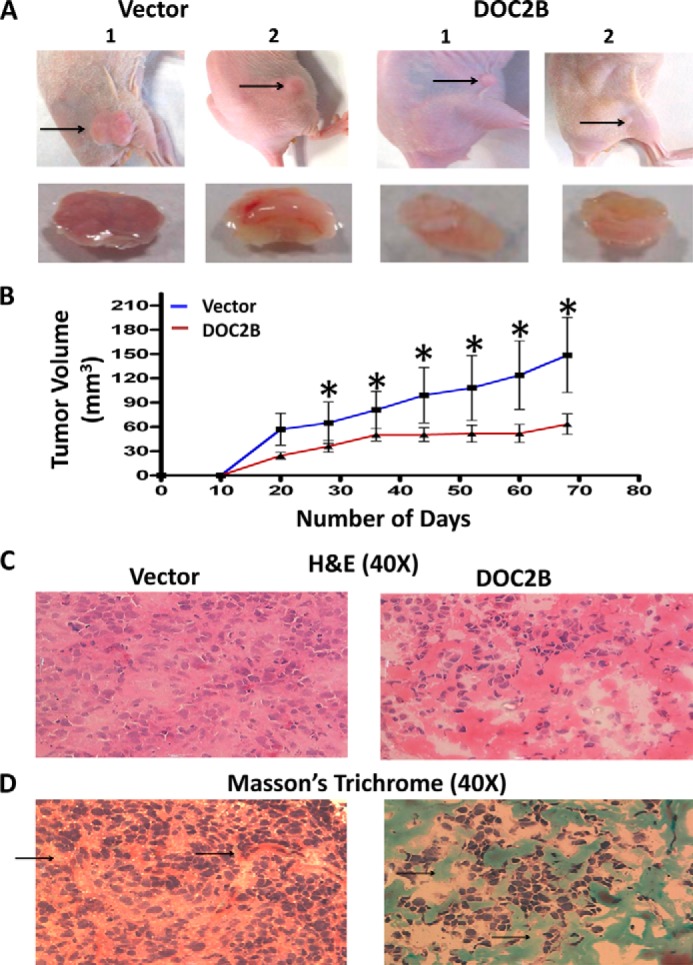
DOC2B inhibits cervical cancer growth in vivo in nude mice. SiHa cells (1 × 108 cells) transfected with pCMV-6-Entry vector or pCMV-6-DOC2B were injected subcutaneously into the flanks of female athymic nude (3 to 4 weeks old) mice (n = 6 each group). A, representative figure of tumor growth in nude mice inoculated subcutaneously with empty vector or DOC2B-transfected cells. B, tumor growth curves of DOC2B-expressing cells in nude mice were compared with control cells by a tumor xenograft experiment. The asterisk indicates statistical significance (*, p < 0.05). C, representative image of H&E staining of tumor xenografts with and without DOC2B expression. The H&E staining of DOC2B expressing cells showed a decreased nucleus to cytoplasmic ratio, less abnormal nucleus, and decreased tumor cell density. Furthermore, the cells were densely aggregated less in pleomorphic (atypical) cells and reduced in the number of abnormal mitosis. Stroma was also visible. In contrast to this, DOC2B negative cells were predominantly/increasingly pleomorphic, spindle morphology cells showing prominent nuclei with altered nucleus to cytoplasmic ratio and abnormal mitosis. D, representative image of Masson's trichrome staining of tumor xenografts with and without DOC2B expression. The Masson's trichrome staining showed significant degradation of collagen in the absence of DOC2B expression.
DISCUSSION
To investigate the role of differential methylation in cervical cancer and identify novel regions silenced in these tumors, we performed MS-AP-PCR that has previously been shown to frequently select methylated CpG islands (12, 13, 25). Although some of the CpG islands identified were near or within the genes such as IKBKG, KLRG2, Myomesin-2, and NXN, we chose DOC2B due to its proximity to functional promoter (Table 2). In silico analysis of −700 to +300 bp of the DOC2B by CpG Island searcher, TESS and AliBaba2.1 predicted a CpG island, E Box (CACGTG, −393/−387), and identification of binding sites for several methylation-sensitive transcription factor such as Sp-1, Ap-2, USF, and E2F; suggesting that transcription of DOC2B could be regulated by promoter methylation. Our experiment shows that methylation of the CpG island proximal to the transcription start site of the DOC2B gene leads to transcriptional repression in SiHa cells. Inactivation of the DOC2B gene by other mechanisms such as deletions, copy number variation, and mutations will also need to be evaluated to understand the role of the DOC2B gene in the pathogenesis of cancer in general and cervical cancer in particular.
DNA methylation is an epigenetic process that regulates expression of genes during a variety of physiological conditions (26, 27). Transcriptional repression of the tumor suppressor gene and activation of oncogenes via aberrant methylation is common in cancers. Interestingly, specific drugs have been shown to reactivate the down-regulated genes in cancers via inhibition of epigenetic machineries. We showed evidence for the first time that the DOC2B gene is frequently hypermethylated in cervical cancer leading to loss of its expression, which can be restored upon treatment with 5-aza-2DC, suggesting that promoter methylation directly contributes to DOC2B gene silencing. Thus we suggest that binding of chromatin remodeling complexes and associated proteins to the DOC2B promoter might lead to transcriptional repression by inhibiting the interaction with transcription factors (26, 27).
Our study shows that DOC2B may act as a pro-apoptotic tumor suppressor gene in the absence of adhesion to the matrix via inhibiting AKT/ERK signaling by inducing actin cytoskeleton remodeling and increase in intracellular Ca2+. Reduced cell-cell adhesion, inhibition of cell growth, proliferation, invasion, and migration, which all may be regulated by decreased phosphorylation of AKT/ERK proteins by interfering with their signaling pathways. Our study is the first to demonstrate the causes and consequences of DOC2B silencing in cervical cancer and supports the notion that DOC2B might be a putative tumor suppressor gene in multiple cancers.
In the present study, we showed that DOC2B inhibits growth, proliferation, migration, and invasion of cervical cancer cells in vitro. However, to begin to understand the molecular mechanism of the anti-tumor effect exerted by DOC2B, we focused on its effect on AKT1 and ERK1/2 signaling as it has been previously reported that the inhibition of both AKT and ERK activation by phosphorylation results in inhibition of cell growth, proliferation, migration, and invasion (28–31). In our study, phosphorylated ERK1/2 and AKT1 decreased upon DOC2B expression suggesting the role of ERK/MAPK and AKT signaling on its effects. It has been shown that the regulation of rearrangement of the actin cytoskeleton occurs via phosphorylating proteins such as EPLIN, MLCK, FAK vinexin by ERK1/2, and phosphorylation of girdin, fascin, and l-plastin by AKT1 leading to altered cell migration properties (28–35). Therefore, several actin-binding proteins may have been down-regulated due to reduced phosphorylation of ERK1/2 and AKT1 by DOC2B and may act in concert in regulation of actin dynamics and tumor cell motility. The anti-proliferative effect of DOC2B may be due to cell cycle arrest and anoikis-mediated cell death by inhibition of these critical signaling events. However, further detailed studies are required to identify the mechanism of the DOC2B-mediated effects.
HPV infection is one of the major etiological factors for cervical cancer and is shown to target the epigenetic machinery leading to transcriptional deregulation of genes (36, 37). Studies in keratinocytes transfected with HPV have shown a gradual increase of promoter hypermethylation of hTERT, TP73, ESR1, RARβ, and DAPK1 genes during cellular transformation. This could be attributed to oncogenic proteins encoded by HPV especially E6 and E7, which are the key deregulators of normal tissue homeostasis. A significant association was observed between HPV infection and DOC2B promoter DNA methylation (p < 0.005), which could be due to overactivation of epigenetic machinery by the oncogenic proteins of HPV. Our study shows that the DOC2B gene might be one target of HPV and more extensive work needs to be carried out to study the association between HPV and DOC2B. An independent association was observed between age and promoter methylation indicating that hypermethylation of the DOC2B promoter might be tumor specific in nature and may not be due to the methylation modifying factor(s) such as progressive age.
From clinical perspectives, the exfoliated cytology for screening of cervical cancer has reduced its incidence worldwide but new cases of cervical cancer continue to occur. Therefore, new screening modalities with high sensitivity and specificity need to be evaluated and pursued. Toward this, DNA methylation markers can be highly useful in screening early stages of cervical cancer (2, 38, 39). Hypermethylation of the DOC2B promoter and its low expression during the pre-malignant condition shows its potential as a marker for early screening of cervical cancer. In this direction, the role of DOC2B in early detection along with an additional panel of markers needs to be investigated. Restoration of DOC2B gene expression been has shown to inhibit tumor growth, proliferation, invasion, and metastasis both in vitro and in vivo, indicating its potential in management of cervical cancer. Although, the results of our study demonstrate the tumor growth inhibitory function of the DOC2B gene through several mechanisms, caveats may still exist as the in vivo experiments were performed only using cells derived from single cell clones. However, the in vitro experiments performed shows that DOC2B may act as an inhibitor of cancer cell growth, proliferation, invasion, and migration. Although, ectopic expression of DOC2B showed growth inhibitory properties in cervical cancer cell lines such as SiHa and HeLa, it may not be specific to cervical cancer as DOC2B showed varying levels of expression in several cancer cell lines tested. DOC2B expression increased intracellular calcium levels and inhibited ERK1/2 and AKT1 phosphorylation, which are key molecules of cell proliferation and survival pathways. Thus, additional studies are required to show whether the tumor suppressive function of DOC2B is specific to cervical cancer.
In conclusion, our study demonstrates for the first time that (i) promoter hypermethylation is a regulator of DOC2B gene transcription and is down-regulated in cervical cancer; (ii) DOC2B expression leads to an increase in intracellular Ca2+, remodeling of cytoskeleton, and inhibition of AKT and ERK phosphorylation to interfere with their downstream signaling indicating the involvement of multiple pathways; and (iii) expression of DOC2B may result in inhibition of key biological characteristics of tumor growth, migration, and invasion in cervical cancer cells. Therefore these characteristics and features of DOC2B can be exploited for therapeutic application in cervical cancer.
This work was supported by grants from TIFAC-CORE, Government of India, the Indian Council of Medical Research, Government of India, and the Manipal University.
- DOC2
- double C2-like domain
- MS-AP-PCR
- methylation-sensitive arbitrarily primed PCR
- BGS
- bisulfite genomic sequencing
- LSIL
- low grade squamous intraepithelial lesion
- HSIL
- high grade squamous intra epithelial lesion
- DMSO
- dimethyl sulfoxide
- HEMA
- 2-hydroxyethyl methacrylate
- HPV
- human papilloma virus.
REFERENCES
- 1. Shen H., Laird P. W. (2013) Interplay between the cancer genome and epigenome. Cell 153, 38–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dueñas-González A., Lizano M., Candelaria M., Cetina L., Arce C., Cervera E. (2005) Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol. Cancer 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Esteller M. (2008) Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 [DOI] [PubMed] [Google Scholar]
- 4. Duncan R. R., Shipston M. J., Chow R. H. (2000) Double C2 protein. A review. Biochimie 82, 421–426 [DOI] [PubMed] [Google Scholar]
- 5. Orita S., Sasaki T., Naito A., Komuro R., Ohtsuka T., Maeda M., Suzuki H., Igarashi H., Takai Y. (1995) Doc2: a novel brain protein having two repeated C2-like domains. Biochem. Biophys. Res. Commun. 206, 439–448 [DOI] [PubMed] [Google Scholar]
- 6. Sakaguchi G., Orita S., Maeda M., Igarashi H., Takai Y. (1995) Molecular cloning of an isoform of Doc2 having two C2-like domains. Biochem. Biophys. Res. Commun. 217, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 7. Ke B., Oh E., Thurmond D. C. (2007) Doc2β is a novel Munc18c-interacting partner and positive effector of syntaxin 4-mediated exocytosis. J. Biol. Chem. 282, 21786–21797 [DOI] [PubMed] [Google Scholar]
- 8. Malkinson G., Spira M. E. (2006) Calcium concentration threshold and translocation kinetics of EGFP-DOC2B expressed in cultured Aplysia neurons. Cell Calcium 39, 85–93 [DOI] [PubMed] [Google Scholar]
- 9. Fukuda N., Emoto M., Nakamori Y., Taguchi A., Miyamoto S., Uraki S., Oka Y., Tanizawa Y. (2009) DOC2B: a novel syntaxin-4 binding protein mediating insulin-regulated GLUT4 vesicle fusion in adipocytes. Diabetes 58, 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Groffen A. J., Martens S., Díez Arazola R., Cornelisse L. N., Lozovaya N., de Jong A. P., Goriounova N. A., Habets R. L., Takai Y., Borst J. G., Brose N., McMahon H. T., Verhage M. (2010) Doc2b is a high-affinity Ca2+ sensor for spontaneous neurotransmitter release. Science 327, 1614–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McMahon H. T., Kozlov M. M., Martens S. (2010) Membrane curvature in synaptic vesicle fusion and beyond. Cell 140, 601–605 [DOI] [PubMed] [Google Scholar]
- 12. Estécio M. R., Youssef E. M., Rahal P., Fukuyama E. E., Góis-Filho J. F., Maniglia J. V., Goloni-Bertollo E. M., Issa J. P., Tajara E. H. (2006) LHX6 is a sensitive methylation marker in head and neck carcinomas. Oncogene 25, 5018–5026 [DOI] [PubMed] [Google Scholar]
- 13. Gonzalgo M. L., Liang G., Spruck C. H., 3rd, Zingg J. M., Rideout W. M., 3rd, Jones P. A. (1997) Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res. 57, 594–599 [PubMed] [Google Scholar]
- 14. Kholod N., Boniver J., Delvenne P. (2007) A new dimethyl sulfoxide-based method for gene promoter methylation detection. J. Mol. Diagn. 9, 574–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balanathan P., Ball E. M., Wang H., Harris S. E., Shelling A. N., Risbridger G. P. (2004) Epigenetic regulation of inhibin alpha-subunit gene in prostate cancer cell lines. J. Mol. Endocrinol. 32, 55–67 [DOI] [PubMed] [Google Scholar]
- 16. Evans M. F., Adamson C. S., Simmons-Arnold L., Cooper K. (2005) Touchdown general primer (GP5+/GP6+) PCR and optimized sample DNA concentration support the sensitive detection of human papillomavirus. BMC Clin. Pathol. 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gravitt P. E., Peyton C. L., Alessi T. Q., Wheeler C. M., Coutlée F., Hildesheim A., Schiffman M. H., Scott D. R., Apple R. J. (2000) Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38, 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitamura T., Onishi M., Kinoshita S., Shibuya A., Miyajima A., Nolan G. P. (1995) Efficient screening of retroviral cDNA expression libraries. Proc. Natl. Acad. Sci. U.S.A. 92, 9146–9150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haraguchi M., Okubo T., Miyashita Y., Miyamoto Y., Hayashi M., Crotti T. N., McHugh K. P., Ozawa M. (2008) Snail regulates cell-matrix adhesion by regulation of the expression of integrins and basement membrane proteins. J. Biol. Chem. 283, 23514–23523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu X., Sui X., Li L., Huang X., Rong R., Su X., Shi Q., Mo L., Shu X., Kuang Y., Tao Q., He C. (2013) Protocadherin 17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J. Pathol. 229, 62–73 [DOI] [PubMed] [Google Scholar]
- 21. Kaneda A., Wakazono K., Tsukamoto T., Watanabe N., Yagi Y., Tatematsu M., Kaminishi M., Sugimura T., Ushijima T. (2004) Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 64, 6410–6415 [DOI] [PubMed] [Google Scholar]
- 22. Xu N., Zhang L., Meisgen F., Harada M., Heilborn J., Homey B., Grandér D., Ståhle M., Sonkoly E., Pivarcsi A. (2012) MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 287, 29899–29908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. June C. H., Moore J. S. (2004) Measurement of intracellular ions by flow cytometry. Curr. Protoc. Immunol. Chapter 5, Unit 5 5 [DOI] [PubMed] [Google Scholar]
- 24. Wiggins H., Rappoport J. (2010) An agarose spot assay for chemotactic invasion. BioTechniques 48, 121–124 [DOI] [PubMed] [Google Scholar]
- 25. Yu J., Ma X., Cheung K. F., Li X., Tian L., Wang S., Wu C. W., Wu W. K., He M., Wang M., Ng S. S., Sung J. J. (2010) Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene 29, 6464–6474 [DOI] [PubMed] [Google Scholar]
- 26. Egger G., Liang G., Aparicio A., Jones P. A. (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 [DOI] [PubMed] [Google Scholar]
- 27. Yoo C. B., Jones P. A. (2006) Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 5, 37–50 [DOI] [PubMed] [Google Scholar]
- 28. Enomoto A., Murakami H., Asai N., Morone N., Watanabe T., Kawai K., Murakumo Y., Usukura J., Kaibuchi K., Takahashi M. (2005) Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev. Cell 9, 389–402 [DOI] [PubMed] [Google Scholar]
- 29. Han M. Y., Kosako H., Watanabe T., Hattori S. (2007) Extracellular signal-regulated kinase/mitogen-activated protein kinase regulates actin organization and cell motility by phosphorylating the actin cross-linking protein EPLIN. Mol. Cell Biol. 27, 8190–8204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klemke R. L., Cai S., Giannini A. L., Gallagher P. J., de Lanerolle P., Cheresh D. A. (1997) Regulation of cell motility by mitogen-activated protein kinase. J. Cell Biol. 137, 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Viala E., Pouysségur J. (2004) Regulation of tumor cell motility by ERK mitogen-activated protein kinases. Ann. N.Y. Acad. Sci. 1030, 208–218 [DOI] [PubMed] [Google Scholar]
- 32. Cohan C. S., Welnhofer E. A., Zhao L., Matsumura F., Yamashiro S. (2001) Role of the actin bundling protein fascin in growth cone morphogenesis: localization in filopodia and lamellipodia. Cell Motil. Cytoskeleton 48, 109–120 [DOI] [PubMed] [Google Scholar]
- 33. Janji B., Giganti A., De Corte V., Catillon M., Bruyneel E., Lentz D., Plastino J., Gettemans J., Friederich E. (2006) Phosphorylation on Ser5 increases the F-actin-binding activity of L-plastin and promotes its targeting to sites of actin assembly in cells. J. Cell Sci. 119, 1947–1960 [DOI] [PubMed] [Google Scholar]
- 34. Yamakita Y., Ono S., Matsumura F., Yamashiro S. (1996) Phosphorylation of human fascin inhibits its actin binding and bundling activities. J. Biol. Chem. 271, 12632–12638 [DOI] [PubMed] [Google Scholar]
- 35. Zanet J., Jayo A., Plaza S., Millard T., Parsons M., Stramer B. (2012) Fascin promotes filopodia formation independent of its role in actin bundling. J. Cell Biol. 197, 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flanagan J. M. (2007) Host epigenetic modifications by oncogenic viruses. Br. J. Cancer 96, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uozaki H., Fukayama M. (2008) Epstein-Barr virus and gastric carcinoma: viral carcinogenesis through epigenetic mechanisms. Int. J. Clin. Exp. Pathol. 1, 198–216 [PMC free article] [PubMed] [Google Scholar]
- 38. Guenin S., Mouallif M., Deplus R., Lampe X., Krusy N., Calonne E., Delbecque K., Kridelka F., Fuks F., Ennaji M. M., Delvenne P. (2012) Aberrant promoter methylation and expression of UTF1 during cervical carcinogenesis. PLoS One 7, e42704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lai H. C., Lin Y. W., Huang T. H., Yan P., Huang R. L., Wang H. C., Liu J., Chan M. W., Chu T. Y., Sun C. A., Chang C. C., Yu M. H. (2008) Identification of novel DNA methylation markers in cervical cancer. Int. J. Cancer 123, 161–167 [DOI] [PubMed] [Google Scholar]