Background: The host substrate for Certhrax toxin is unknown.
Results: Certhrax toxin ADP-ribosylates vinculin at Arg-433, which disrupts focal adhesion complexes and host cell adhesion.
Conclusion: Certhrax toxin is the first bacterial toxin to add a post-translational modification to vinculin to disrupt the actin cytoskeleton.
Significance: This explains how Certhrax toxin can contribute to evasion of the host innate immune system.
Keywords: Actin, ADP-ribosylation, Bacillus, Bacterial Toxins, Microscopic Imaging
Abstract
Bacillus cereus is often associated with mild to moderate gastroenteritis; however, some recent isolates cause inhalational anthrax-like diseases and death. These potential emerging human pathogens express multiple virulence factors. B. cereus strain G9241 expresses anthrax toxin, several polysaccharide capsules, and the novel ADP-ribosyltransferase, Certhrax. In this study, we show that Certhrax ADP-ribosylates Arg-433 of vinculin, a protein that coordinates actin cytoskeleton and extracellular matrix interactions. ADP-ribosylation of vinculin disrupted focal adhesion complexes and redistributed vinculin to the cytoplasm. Exogenous vinculin rescued these phenotypes. This provides a mechanism for strain G9241 to breach host barrier defenses and promote bacterial growth and spread. Certhrax is the first bacterial toxin to add a post-translational modification to vinculin to disrupt the actin cytoskeleton.
Introduction
Bacillus cereus is typically considered an opportunistic pathogen associated with “food poisoning” or infections in immune compromised individuals; however, several B. cereus strains have recently been reported to cause severe or fatal “anthrax-like” disease in humans and non-human primates (1–4). These virulent strains often harbor virulence plasmids similar to Bacillus anthracis, whereas retaining B. cereus diagnostic phenotypes (5, 6). The severity of clinical disease suggests that these virulent B. cereus strains may be emerging human pathogens.
The B. cereus group consists of several Gram-positive, spore-forming Bacillus species including B. anthracis, Bacillus thuringiensis, and B. cereus. Notably, group members share high genomic similarity (>99%) but elicit different disease phenotypes, ranging from mild gastroenteritis to death. B. anthracis, the etiologic agent of anthrax, is the most pathogenic member of the Bacillus group to humans, with virulence modulated by two megaplasmids, pXO1 and pXO2. pXO1 encodes the genes for anthrax toxin, an AB toxin comprised of edema factor (EF),2 lethal factor, and protective antigen (PA) (7, 8). EF is an adenylate cyclase, lethal factor is a potent zinc metalloprotease targeting mitogen-activated protein kinase kinases, and PA binds to the host cell receptors, ANTXR-1 and ANTXR-2. After PA hepta- or octamerization, 3 or 4 molecules of EF/lethal factor bind PA and are delivered into the host cytosol (9–11). pXO2 encodes enzymes that produce the anti-phagocytic poly-d-glutamic acid capsule (12).
B. cereus strain G9241 was isolated from a patient who was admitted to the hospital with an anthrax-like fulminant pneumonia (3, 4). G9241 contains two megaplasmids, pBCXO1 and pBC210, which are analogous to the toxin and capsule virulence plasmids pXO1 and pXO2 present in B. anthracis (13). pBCXO1 encodes the three anthrax toxin components, as well as an operon responsible for producing a hyaluronic acid capsule (14, 15). pBC210 encodes a polysaccharide capsule operon, a PA homolog (60% identity), and a bacterial ADP-ribosyltransferase, Certhrax (13–15).
As found in B. anthracis, G9241 requires both pBCXO1 and pBC210 for maximum virulence (15). However, despite the fact that B. anthracis and G9241 both produce encapsulated bacilli and anthrax toxin, studies in mice and rabbits demonstrate that G9241 exhibits virulence similar to the attenuated, un-encapsulated B. anthracis Sterne (pXO1+/pXO2−) strain. Additionally, G9241 genetic knockouts of the two PA homologs demonstrated that pBCXO1-encoded PA is required for virulence in mice (16). This is consistent with the pathogenesis of G9241 being toxin-mediated, whereas the absence of the poly-d-glutamic acid capsule may account for the virulence attenuation when compared with the fully virulent B. anthracis Ames strain (15). Interestingly, in vitro studies indicate that Certhrax possesses an 80-fold lower LD50 than B. anthracis lethal factor toward macrophages (17). Although toxin production is necessary for G9241 pathogenesis and Certhrax possesses a remarkable potency, the role of Certhrax during infection remains to be determined.
Certhrax was previously identified as a bacterial ADP-ribosyltransferase with homology to the actin-modifying binary ADP-ribosyltransferase toxins (18–20). Recently, Certhrax was shown to ADP-ribosylate a single 120-kDa protein in HeLa cell lysates, and intracellular expression of Certhrax resulted in cell death and rearrangement of the actin cytoskeleton (18). In this study, we show that Certhrax ADP-ribosylates vinculin, a cytoskeleton adaptor protein responsible for linking the actin cytoskeleton and extracellular matrix (ECM) at focal adhesion sites (21–23). Vinculin is a multidomain matrix protein that links the actin cytoskeleton to the ECM through direct interactions with focal adhesion proteins, like talin, and actin stress fibers (24). Vinculin exists in two conformations, a cytoplasmic inactive conformation and a membrane-associated active confirmation. Vinculin activation results in release of an autoinhibitory “tail” domain from a protein binding “head” domain, allowing vinculin to interact with actin or other focal adhesion proteins (25). Activity is regulated through phosphorylation and binding to several other focal adhesion proteins, primarily talin (25–27). The ADP-ribosylation of vinculin by Certhrax is a novel mechanism by a bacterial toxin to modulate actin stress fibers and extends our appreciation of how bacterial pathogens deregulate the cytoskeleton to make the host more prone to bacterial invasion and colonization.
MATERIALS AND METHODS
Plasmid Vectors and Mutagenesis
DNA encoding the ADP-ribosyltransferase domain of Certhrax (residues 226–476, CerADPr) or CerADPr(E431D) was subcloned into pET15b (Novagen) or pEGFP-C3 (Clontech) as previously described (18). DNA encoding human vinculin (residues 1–1066, ATCC-MGC number 21734) was subcloned with the hemagglutinin epitope (HA) fused to the 3′ terminus, using primers 5′-GATCGATCGCTAGCATGCATCATCATCATCATCACAGCAGCGGCCCAGTGTTTCATACGCGCACG and 3′-GAAAGACTCCCTGGTACCAGACCCATACGACGTACCAGACTACGCAGAAGCAACGTATCCGTATGATGTACCGGATTATGCATAGGTCGACGATCGATC into pET-28a or pEGFP-N1. DNA encoding human paxillin (residues 1–591,Thermo Scientific, MHS6278-213245146) was subcloned into pET-28a or pEGFP-N1. Plasmids were sequenced to confirm the appropriate DNA and then transformed into Escherichia coli (TG1).
Expression and Purification of Recombinant Proteins
pET15b-CerADPr and pET-15b-CerADPr(E431D) were purified as previously described (17). pET-28a-Vinculin and pET28a-Paxillin were transformed into E. coli BL21(DE3). Cells were plated overnight at 37 °C with 250 μg/ml of ampicillin. Cells were added to 400 ml of LB broth (Miller salts, BD Biosciences) with ampicillin and cultured for 2 h at 30 °C when 1 mm isopropyl 1-thio-β-d-galactopyranoside was added and the cells were cultured overnight at 16 °C. Cells were harvested, suspended in binding buffer (20 mm Tris-HCl (pH 7.9), 500 mm NaCl, and 5 mm imidazole) with bacterial protease inhibitor mixture (Sigma), RNase (20 μg/ml), and DNase (20 μg/ml) and broken by French Press. The lysate was centrifuged (30,000 × g for 20 min) and the soluble fraction passed through a 0.45-μm filter. The filtrate was subjected to Ni2+ affinity resin chromatography (Qiagen). His6 fusion proteins were eluted in binding buffer with 250 mm imidazole. The eluate was subjected to size-exclusion fractionation by Sephacryl S200 (Sigma) in 10 mm Tris-HCl (pH 7.6) and 20 mm NaCl. Peak fractions were dialyzed into 10 mm Tris-HCl (pH 7.6), 20 mm NaCl, and 40% glycerol, and stored at −80 °C. A typical purification yielded ∼4 mg of CerADPr/liter of culture.
HeLa Cell Growth and Lysate Harvesting
HeLa cells (ATCC, CCL-2) were cultured in MEM (Invitrogen) supplemented with 10% fetal calf serum, non-essential amino acids, sodium pyruvate, sodium bicarbonate, and penicillin/streptomycin and maintained humidified at 37 °C in 5% CO2. Confluent plates (150 mm) of HeLa cells were washed (PBS) and scraped into 1 ml of HB buffer (200 mm sucrose, 3 mm imidazole, mammalian protease inhibitor mixture (Sigma). Cells were broken by 25–30 passes through a 26-gauge needle, and then centrifuged at 1000 × g to pellet unbroken cells and nuclei. Protein concentration was determined by BCA assay (Pierce).
In Vitro ADP-ribosylation Assay
Reactions contained: 10 mm Tris-HCl (pH 7.6), 20 mm NaCl, 50 μg of HeLa lysate or 2 μg of recombinant vinculin or paxillin protein, 4 μm 6-biotin-17-NAD (Trevigen), and the indicated amount of Certhrax. Reactions were incubated at room temperature (RT) for the indicated time and stopped with Laemmli buffer and boiling. Reactions were subjected to SDS-PAGE and transferred to polyvinylidene fluoride membranes (PVDF, Millipore). Membranes were subjected to the following analyses.
Detection of Biotin-ADP-ribose
Membranes were blocked for 30 min in PBS + 3% BSA and incubated with streptavidin-HRP (SA-HRP) conjugate (1:80,000 final dilution; Pierce) for 1 h at RT. Membranes were then incubated with Pico Super Signal (Pierce) and luminescence was measured.
Detection of Proteins in HeLa Cell Lysate
Membranes were blocked for 30 min in TBST + 3% milk and then incubated with α-GFP IgG (mouse, 1:2,000; Covance), α-tubulin IgG (rabbit, 1:5,000; Abcam), α-vinculin IgG (rabbit, 1:5,000; Abcam), or α-HA IgG (rat, 1:2,000; Sigma). Primary antibodies were detected with goat α-mouse-HRP IgG (1:30,000; Pierce), goat α-rat-HRP IgG (1:35,000; Pierce), or goat α-rabbit-HRP IgG (1:35,000; Pierce). Membranes were probed with Pico Super Signal (Pierce) and luminescence was measured with a CCD camera and AlphaView SA software (Cell Biosciences Inc.). Images were cropped in Photoshop (Adobe). Quantification of luminescence was performed with AlphaView software from a minimum of three independent experiments. An unpaired t test was performed with GraphPad Prism 5 software (GraphPad Software Inc.) (* = p < 0.05; ** = p < 0.01).
Mass Spectroscopy
A band comprising the 120-kDa protein ADP-ribosylated by CerADPr was excised and subjected to in-gel trypsinization, followed by LC-MS/MS on a linear ion trap-Orbitrap hybrid mass spectrometer (University of Massachusetts-Worcester Proteomic Center). Scaffold 3 (Proteome Software) was used to assign peptides. Analysis of common proteins from two independent biological replicates showed human vinculin (accession number P18206) as the most prevalent protein (Table 1).
TABLE 1.
Mass spectroscopic analysis of the 120-kDa protein ADP ribosylated by CerADPr
Biotin-ADP-ribosylated 120-kDa protein band was excised and subjected to LC-MS/MS analysis (University of Massachusetts-Worcester Proteomic Center). The 8 most prevalent protein scores from two separate experiments are shown, ordered by total number of unique spectra. Scaffold Viewer was used to visualize results.
| Identified proteins Homo sapiens | Accession number (human) | Molecular mass | Score (unique spectra) |
|---|---|---|---|
| kDa | |||
| Analysis 1 | |||
| Vinculin GN = VCL PE = 1 SV = 4 | VINC | 124 | 253 |
| ATP-citrate synthase GN = ACLY PE = 1 SV = 3 | ACLY | 121 | 236 |
| Eukaryotic translation initiation factor 3 subunit B GN = EIF3B PE = 1 SV = 3 | EIF3B | 92 | 84 |
| Kinesin-1 heavy chain GN = KIF5B PE = 1 SV = 1 | KINH | 110 | 73 |
| Eukaryotic translation initiation factor 3 subunit A GN = EIF3A PE = 1 SV = 1 | EIF3A | 167 | 70 |
| Integrin β-1 GN = ITGB1 PE = 1 SV = 2 | ITB1 | 88 | 62 |
| α-Actinin-4 GN = ACTN4 PE = 1 SV = 2 | ACTN4 | 105 | 51 |
| Integrin α-V GN = ITGAV PE = 1 SV = 2 | ITAV | 116 | 46 |
| Analysis 2 | |||
| Vinculin GN = VCL PE = 1 SV = 4 | VINC | 124 | 107 |
| Myosin-9 GN = MYH9 PE = 1 SV = 4 | MYH9 | 227 | 88 |
| Desmoplakin GN = DSP PE = 1 SV = 3 | DESP | 332 | 82 |
| Plectin GN = PLEC PE = 1 SV = 3 | PLEC | 532 | 73 |
| Filamin-A GN = FLNA PE = 1 SV = 4 | FLNA | 281 | 49 |
| Eukaryotic translation initiation factor 3 subunit A GN = EIF3A PE = 1 SV = 1 | EIF3A | 167 | 35 |
| Myosin-Ib GN = MYO1B PE = 2 SV = 3 | MYO1B | 132 | 34 |
| Ankycorbin GN = RAI14 PE = 1 SV = 2 | RAI14 | 110 | 33 |
Streptavidin-Ultralink Precipitation
HeLa cell lysates were incubated with CerADPr in the presence (+biotin) or absence (−biotin) of 6-biotin-17-NAD (Trevigen) for 1 h when Streptavidin-Ultralink beads (Pierce) were added for 2 h at RT, with rotation, to bind biotin-ADP-ribosylated proteins. The beads were washed 4 times with an equal volume of TBS and bound proteins were eluted with a mixture of 40 μm biotin and 1% SDS and boiling. Load, flow-through, and elution fractions were resolved by SDS-PAGE and transferred to PVDF membrane. Biotin-ADP-ribose and vinculin were detected by immunoblot as described above.
Cell Culture and Transfection
HeLa cells were cultured to ∼70% confluence in 6-well plates and mock transfected (Mock) or co-transfected with 0.5 μg of pEGFP (EGFP), pEGFP-vinculin (VCL), or pEGFP-paxillin (PXN) and 0.5 μg of pEGFP-CerADPr (Cer) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. At the indicated times, cells were washed twice with PBS and processed for microscopy analyses as described below.
HeLa cell lysates were cultured to ∼70% confluence in 6-well plates and transfected with 0.5 μg of the indicated plasmid in Lipofectamine 2000 for 20 h, scraped into 1 ml of HB, and lysed by 25–30 passages through a 26-gauge needle. Cell lysates were incubated with 0.1 μg of CerADPr and 4 μm 6-biotin-17-NAD for 1 h at RT to assay for ADP-ribosylation, or the lysates were immediately resolved by SDS-PAGE and transferred to PVDF membranes for cellular protein immunoblot analysis.
Cytotoxicity and Adherence of HeLa Cells
Cytotoxic potential of CerADPr was measured in HeLa cells using a trypan blue exclusion assay (28). Cells were mock transfected (Mock) or co-transfected with either 0.5 μg of pEGFP (EGFP) or pEGFP-vinculin (VCL) and 0.5 μg of pEGFP-CerADPr (Cer) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. At 5 or 20 h post-transfection (HPT), media was removed, and cells were washed and stained for 5 min with 0.4% trypan blue (Invitrogen). Unbound trypan blue was removed, cells were washed in Opti-MEM low serum media (Invitrogen), and color images were obtained using a ×10 objective and Retiga-2000R CCD camera (QImaging) with NIS Elements software (Nikon). Percent cytotoxicity was determined by counting the number of trypan blue-positive HeLa cells/total number of HeLa cells per field. Adherence was determined by counting the total number of cells remaining on the plate after removal of growth media. Five random fields were assayed for each transfection condition. A minimum of 10 fields of view from at least two independent biological replicates were quantified. Statistical significance was determined by one-way ANOVA analysis, performed in GraphPad Prism 5 software (GraphPad Software Inc.) (** = p < 0.01).
Immunofluorescence Microscopy
HeLa cells were washed in PBS (2 times) and fixed with 4% paraformaldehyde in PBS (15 min at RT). Next, cells were permeabilized in PBS + 0.1% Triton X-100 and 4% formaldehyde (15 min) and then incubated in 150 mm glycine (15 min). Wells were blocked with PBS + 2.5% BSA, 10% FBS, 0.05% Tween 20, and 0.1% Triton X-100 (1 h at RT) and incubated with primary antibody (1 h at RT). Alexa Fluor® 647-phalloidin (Invitrogen; 1:1000) labeled the actin cytoskeleton, and mouse α-vinculin IgG (Sigma; 1:1000) followed by goat α-mouse IgG Alexa Fluor® 568 (Invitrogen; 1:500) labeled vinculin. Antibodies were diluted in PBS + 1% BSA, 5% FBS, 0.05% Tween 20, and 0.1% Triton X-100. After secondary antibody incubation, cells were washed with PBS (3 times) and mounted with ProLong Gold antifade reagent (Invitrogen). Cells were viewed with a ×60 oil immersion objective. Images were taken with a CoolSnap HQ CCD camera (Photometrics) and NIS-Elements AR software (Nikon), which were processed with ImageJ software (NIH).
Measurement of Vinculin Subcellular Fractionation and Focal Adhesion Quantification
Quantification of focal adhesion complexes were visualized by immunofluorescence with mouse α-vinculin IgG vinculin (Sigma; 1:1000) and rabbit α-talin IgG (Sigma; 1:2000) antibodies. Images were taken of EGFP-CerADPr or EGFP-CerADPr(E431D) positive cells and scored for the presence or absence of a minimum of 10 focal adhesions double positive for both talin and vinculin. At least 50 cells were scored from biological replicates. Statistical significance was determined by one-way ANOVA analysis, performed in GraphPad Prism 5 software (GraphPad Software Inc.) (** = p < 0.01; *** p < 0.001).
RESULTS
CerADPr ADP-ribosylates Human Vinculin
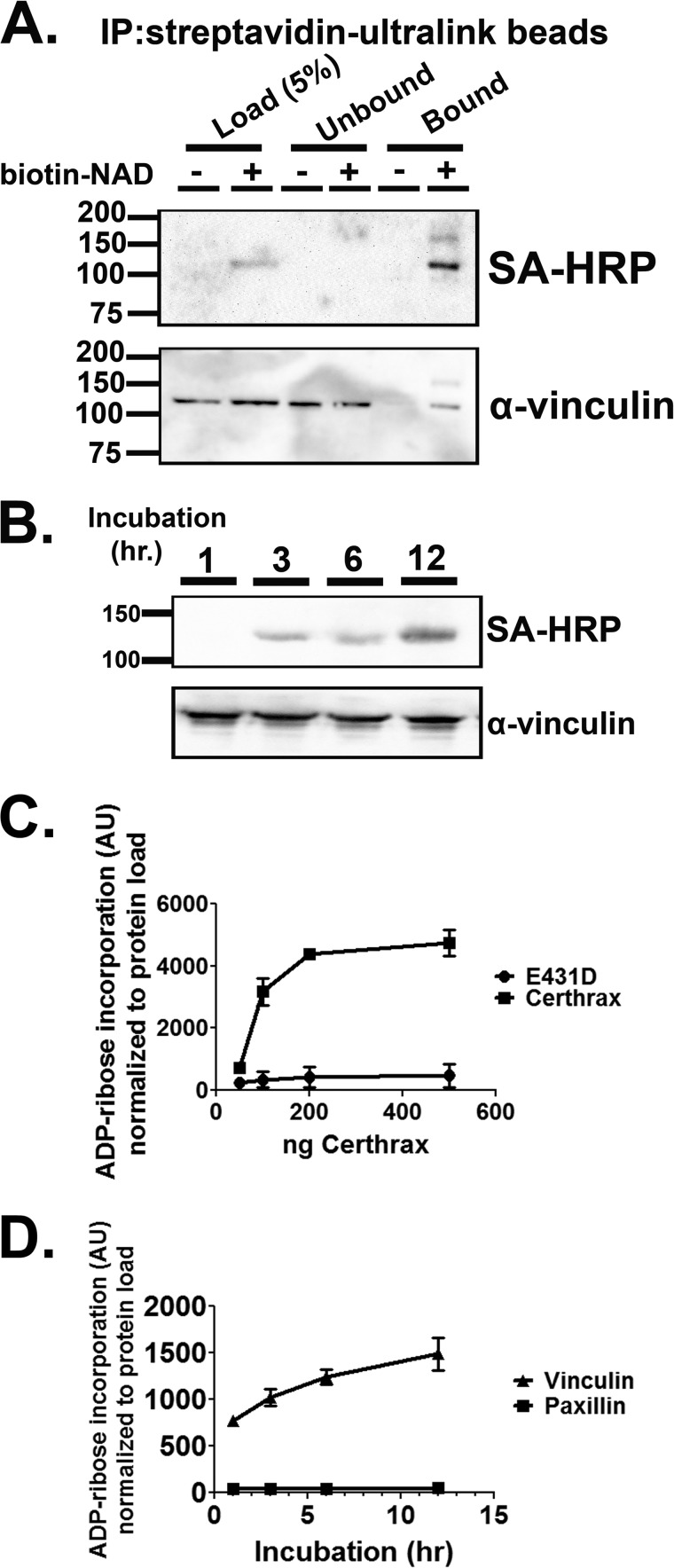
Earlier studies showed that recombinant CerADPr (rCerADPr) ADP-ribosylated an ∼120 kDa protein in HeLa cell lysates (18). Two independent mass spectroscopic analyses identified the 120-kDa protein as human vinculin protein (VCL), with an average of 55% peptide coverage (Table 1). To test this identification, rCerADPr + biotin-NAD-treated HeLa cell lysate was incubated with Streptavidin-Ultralink beads to precipitate biotin-ADP-ribose-linked protein. The precipitate, resolved by SDS-PAGE, contained a 120-kDa band that reacted with both streptavidin and α-vinculin antibody (Fig. 1A). Immunoprecipitation of lysate in the absence of biotin supported the specificity of the streptavidin and α-vinculin antibody reactivity band, as no vinculin nonspecifically bound the beads in the absence of biotin-NAD.
FIGURE 1.
Precipitation of biotin-NAD-labeled substrate with immobilized streptavidin pulls down vinculin protein. A, HeLa lysate was incubated with Certhrax and biotin-NAD for 1 h. Biotinylated protein (ADP-ribosylated) was then immunoprecipitated with Streptavidin-Ultralink beads. The beads were washed with TBS and bound material was eluted with free biotin and SDS. The reactions were resolved by SDS-PAGE and transferred to PVDF membrane. Immunoblot with α-vinculin or streptavidin-HRP (SA-HRP) conjugate detected vinculin and biotin-ADP-ribose, respectively. B, recombinant vinculin purified from E. coli was incubated with CerADPr and biotin-NAD for the indicated times. The reaction was stopped by addition of Laemmli buffer and boiling. The reaction was resolved by SDS-PAGE and transferred to PVDF. Biotin-ADP-ribose was detected with streptavidin-HRP conjugate. Total vinculin was detected by α-vinculin immunoblot. C, recombinant vinculin was incubated with increasing doses of CerADPr or catalytically inactive CerADPr(E431D). After 3 h, the reaction was stopped by addition of Laemmli buffer and boiling. The reaction was resolved by SDS-PAGE and transferred to PVDF. Biotin-ADP-ribose was detected with streptavidin-HRP conjugate and total protein was detected with α-vinculin antibody. Biotin signal was normalized to total vinculin (VCL) signal and expressed in arbitrary units (AU). D, recombinant vinculin or recombinant paxillin were incubated with CerADPr for the indicated times. At each time point, an aliquot was removed and the reaction was stopped by addition of Laemmli buffer and boiling. The reaction was resolved by SDS-PAGE and transferred to PVDF. Biotin-ADP-ribose was detected with streptavidin-HRP conjugate and total protein was detected with α-vinculin antibody. Biotin signal was normalized to total protein load and expressed in arbitrary units.
CerADPr ADP-ribosylates Recombinant Human Vinculin
To further characterize the CerADPr-substrate interaction, recombinant human vinculin was ADP-ribosylated by rCerADPr in a time- (Fig. 1B) and dose-dependent (Fig. 1C) manner. Kinetic assessment showed that CerADPr ADP-ribosylated recombinant human vinculin ∼20-fold slower than vinculin present in the HeLa lysate (not shown). The rate of ADP-ribosylation was salt sensitive, indicating that the CerADPr-vinculin interaction is likely mediated by electrostatic interactions. Catalytically inactive rCerADPr(E431D) showed no appreciable ADP-ribosylation activity toward recombinant vinculin, indicating a specific ADP-ribosyltransferase reaction (Fig. 1C). Finally, CerADPr did not ADP-ribosylate a control focal adhesion substrate, paxillin, over a 24-h time course, demonstrating that CerADPr has specific affinity for vinculin (Fig. 1D).
Recombinant Vinculin Is a Preferred Substrate for CerADPr in HeLa Cells
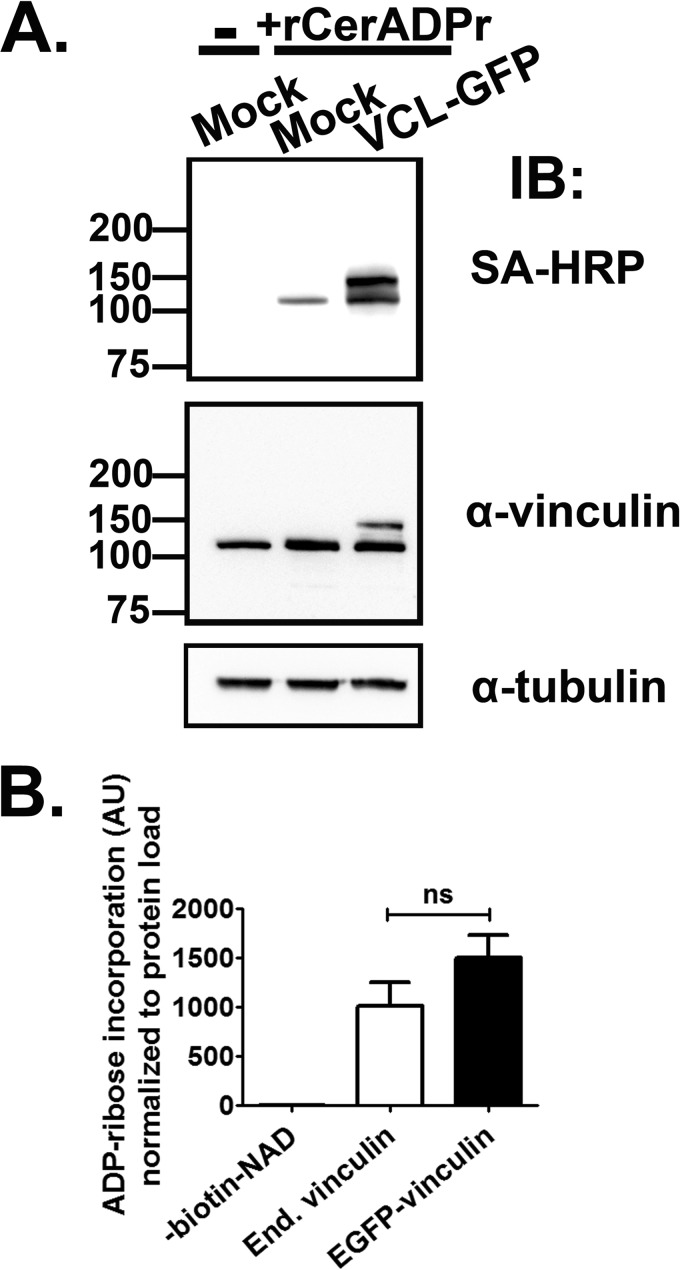
Because recombinant vinculin produced in E. coli was ADP-ribosylated at a slower rate than native vinculin in HeLa lysate, we hypothesized that CerADPr preferred vinculin when either bound to another host protein or when post-translationally modified, such as through a phosphorylation event required for signaling. It is also possible that CerADPr forms a stable complex with the E. coli-produced recombinant vinculin, which could account for the decreased rate of ADP-ribosylation. However, immunoprecipitation of recombinant vinculin protein does not co-immunoprecipitate CerADPr, which does not favor the formation of a stable complex (not shown). Additionally, whereas a eukaryotic cofactor could be necessary for full activation, addition of limiting amounts of HeLa lysate to an in vitro ADP-ribosylation assay did not result in a synergistic increase in ADP-ribosylation activity toward recombinant vinculin. To test the possibility that CerADPr preferentially ADP-riboyslates eukaryotic produced vinculin, human vinculin was expressed as an EGFP or HA fusion protein in HeLa cells. Fluorescence microscopic analysis localized the vinculin fusion proteins within focal adhesion complexes, co-localized with talin and near actin stress fibers, indicative of active and functional vinculin (not shown). To control for off-target effects of overexpressing focal adhesion proteins, expression of a second focal adhesion scaffold and signaling protein, paxillin, was used as a control. HeLa lysates from cells mock transfected or transiently transfected with pEGFP-vinculin were incubated with recombinant CerADPr and biotin-NAD (Fig. 2). rCerADPr ADP-ribosylated both endogenous vinculin (lower band), and transiently expressed EGFP-vinculin, which migrates at a higher molecular weight (upper band) than endogenous vinculin. Immunoblotting with α-vinculin antibody confirmed that the ADP-ribosylation signal was ADP-ribosylated vinculin, both the endogenous and transiently expressed proteins. Importantly, no other ADP-ribosylated proteins were detected, showing that CerADPr has a single eukaryotic target, vinculin. The rate of ADP-ribosylation of EGFP-vinculin was slightly faster than that of endogenous vinculin, although not a statistically significant increase (Fig. 2B). The increased rate of ADP-ribosylation of eukaryotic produced vinculin supports the hypothesis that a discrete form of vinculin, either post-translationally modified or associated with other host proteins, is the preferred substrate of CerADPr.
FIGURE 2.
Certhrax ADP-ribosylates mammalian-expressed vinculin. A, HeLa cells were transfected with empty vector (Mock) or pEGFP-vinculin (VCL-GFP). Cells were incubated for 20 h, lysed, and incubated with (+rCerthrax) or without (−) recombinant CerADPr and biotin-NAD for 1 h. The reaction was stopped with boiling and resolved by SDS-PAGE and transferred to PVDF membrane. ADP-ribosylation was detected by immunoblot with streptavidin-HRP and vinculin was detected with α-vinculin antibody. α-Tubulin immunoblot was used as a load control. B, ADP-ribose incorporation into endogenous (End.) or EGFP-vinculin was measured by densitometry. The amount of biotin-ADP-ribose signal from each protein was normalized to the vinculin signal from each protein and displayed as arbitrary units (AU). No significant (ns) difference was detected between endogenous and recombinant EGFP-vinculin by one-way ANOVA analysis (p value: 0.19).
Overexpression of Vinculin Rescues Cells from the Cytotoxic Effects of CerADPr
As previously shown, Certhrax elicits a cytotoxic effect in both cultured epithelial cells (18) and macrophages (17). HeLa cells mock transfected or transfected with pEGFP-vinculin-HA, alone or with pEGFP-CerADPr, were scored at 5 and 20 HPT for trypan blue exclusion. At 5 HPT, the transfected cells did not show appreciable levels of cytotoxicity. By 20 HPT, cells mock transfected showed few (∼10%) trypan blue-positive cells, whereas co-expression with CerADPr resulted in a significant increase in cell death, ∼45% (not shown). However, co-expression of vinculin and CerADPr resulted in only ∼30% of adherent cells showing trypan blue uptake, indicating a protective effect of vinculin overexpression.
Overexpression of Vinculin Reverses the Cell Detachment Elicited by CerADPr
Earlier studies showed that CerADPr stimulated actin depolymerization and detachment of HeLa cells from the growth surface (18). Assessment of cell adherence showed that at 5 HPT, CerADPr did not have an effect on cell adhesion, with an average of 180–200 cells/field observed (not shown). By 20 HPT, cell detachment was observed in cells transfected with CerADPr, with ∼80–90 cells per field observed (Fig. 3). Co-expression of vinculin and CerADPr significantly reduced the number of detached cells relative to CerADPr-transfected cells (Fig. 3, compare Cer/VCL versus Cer). Cell detachment was not influenced by expression of EGFP or vinculin alone. Importantly, overexpression of paxillin did not rescue CerADPr-induced cell detachment, indicating that rescue due to vinculin overexpression is a specific effect, not a global increase in focal adhesion signaling.
FIGURE 3.
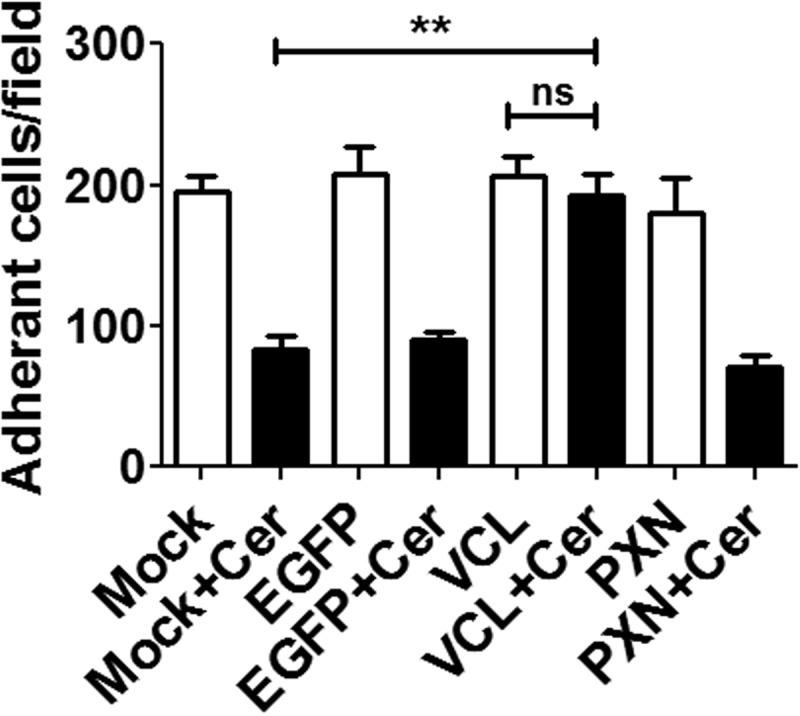
Transient expression of vinculin partially rescues Certhrax-induced cell detachment. HeLa cells were transfected with pEGFP, pEGFP-vinculin, or pEGFP-paxillin ± pCerADPr. 20 h HPT, cells were washed 4 times in Opti-MEM and color micrographs taken. Integrity of cell adherence was measured by counting the cells remaining attached to the growth surface after 4 washes. Adherent cells were scored from 10 random fields for each transfection. A one-way ANOVA was performed on all columns. **, p < 0.01.
ADP-ribosylation of Vinculin Stimulates Disruption of Focal Adhesions
To determine whether CerADPr also disrupted focal adhesion complexes, HeLa cells were either mock transfected or transfected with plasmids encoding vinculin-HA and either CerADPr or catalytically inactive CerADPr(E431D). At 20 HPT, cells were fixed and stained for the actin cytoskeleton (Fig. 4, phalloidin, blue) and vinculin (α-vinculin, red). Emission at 488 nm visualized EGFP-CerADPr or EGFP-CerADPr(E431D) expressing cells (green). Cells expressing CerADPr(E431D) showed normal cytoskeleton architecture, with prominent actin stress fibers and visible focal adhesions (Fig. 4A, top row). In contrast, cells transfected with the catalytically active CerADPr showed loss of stress fibers and aberrant actin staining, cell rounding, and did not have apparent focal adhesions, with vinculin redistributed to the cytoplasm (Fig. 4A, second row; compare EGFP-CerADPr+ cell to neighbors). Cells co-expressing CerADPr and vinculin displayed a more typical morphology, with visible stress fibers and focal adhesion complexes (Fig. 4A, bottom row). Overexpression of an EGFP-paxillin construct alone did not effect cell morphology, with EGFP-paxillin localized to focal adhesion complexes and cells having flattened cell morphology (Fig. 4B, left panel). However, cells co-transfection of paxillin and CerADPr displayed rounded morphology and did not have visible focal adhesion complexes (Fig. 4B, right panel). This indicates that overexpression of focal adhesion signaling proteins alone is not sufficient to rescue CerADPr-induced effects. Quantification of the number of cells displaying >10 focal adhesion complexes, as indicated by vinculin and talin immunofluorescence co-staining, showed that whereas <25% of CerADPr expressing cells showed >10 focal adhesions, co-expression of vinculin-HA and CerADPr resulted in more than 75% of cells with >10 focal adhesions (Fig. 4C). These data indicate that CerADPr expression results in loss of focal adhesion complexes, likely due to re-localization of vinculin to the cytoplasm, and that overexpression of vinculin inhibited this morphological phenotype. Extending the incubation showed that the protective effects of overexpression of vinculin on CerADPr disruption of focal adhesions was transient, consistent with the observed effect being due to increased vinculin expression, and not an off-target effect of vinculin overexpression (not shown).
FIGURE 4.
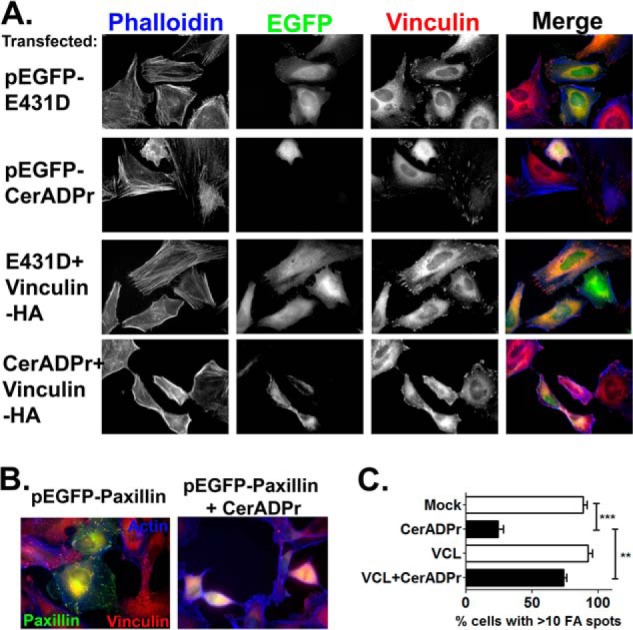
Overexpression of vinculin rescues Certhrax-induced cytoskeleton disruption. A, HeLa cells were transfected with pEGFP-Certhrax or pEGFP-Certhrax (E431D) in the presence or absence of pCMV-vinculin-HA. Cells were fixed and stained with phalloidin-647 (actin) and α-vinculin antibody. EGFP-Certhrax expression was visualized in the EGFP channel. Expression of CerADPr alone results in cell rounding, loss of actin stress fibers, and relocalization of vinculin from focal adhesions to the cytosol. Co-expression of CerADPr and vinculin results in changes to cell morphology, but no cell rounding. Vinculin is still present in focal adhesions. Expression of the E431D mutant shows no phenotype. B, HeLa cells were transfected with pEGFP-paxillin ± pCerADPr. Cells were fixed and stained with phalloidin-647 (actin) and α-vinculin antibody. EGFP-paxillin expression does not rescue CerADPr-induced morphological changes. C, quantification of the number of cells for each transfection condition displaying >10 focal adhesions, as measured by vinculin and talin staining. At least 50 cells were counted for each condition from biological replicates and displayed graphically as a percentage of total transfected cells counted. A one-way ANOVA was performed on all columns. **, p < 0.01; ***, p < 0.001.
CerADPr ADP-ribosylates Vinculin at Arg-433
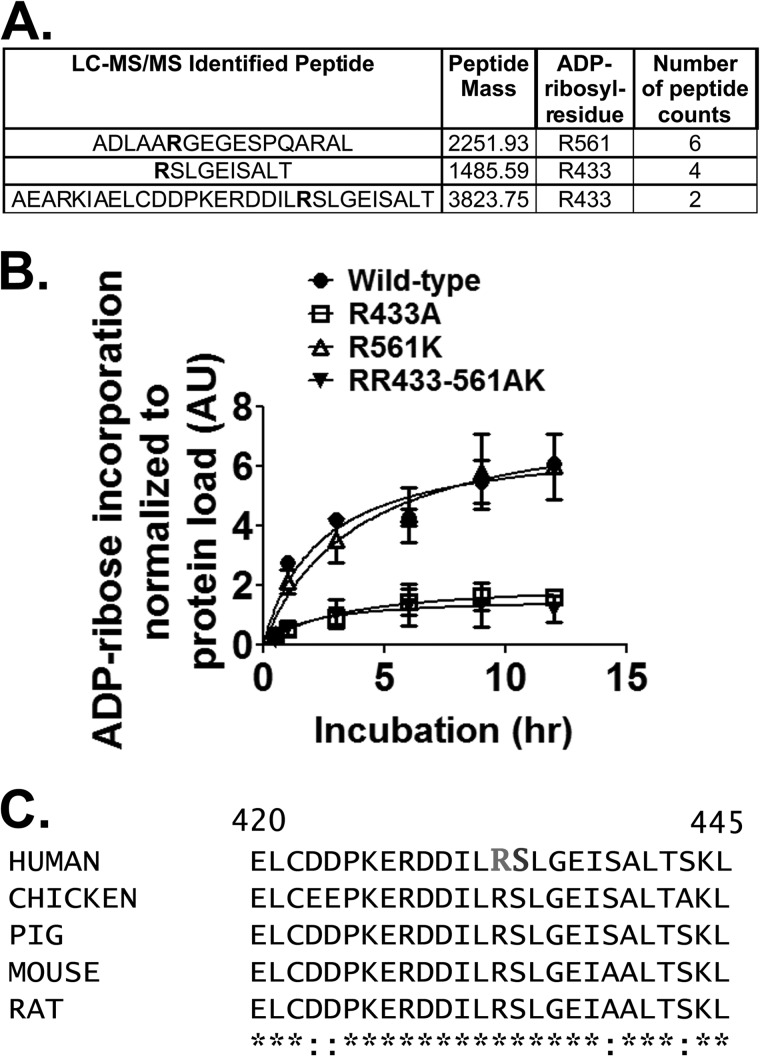
Next the site of ADP-ribosylation on vinculin was determined. Proteolysis experiments had previously narrowed the ADP-ribose to a region of the vinculin head domain, which is responsible for interactions with talin and other focal adhesion signaling molecules (not shown). However, mass spectrometry experiments using the biotin-ADP-ribose adduct were unsuccessful in identification of the modified residue. Instead, recombinant vinculin was ADP-ribosylated to saturation with unlabeled NAD+, subjected to partial trypsinization, and resolved by SDS-PAGE. ADP-ribosylated vinculin was in-gel proteolyzed and LC-MS/MS analyzed, which identified three candidate arginine residues in both protease-treated samples analyzed (Fig. 5A). Vinculin was subjected to site-directed mutagenesis at Arg-433 and Arg-561, the two residues with the highest spectral counts. In vitro analysis showed that CerADPr ADP-ribosylated vinculin(R433A) less efficiently than wild-type vinculin (Fig. 5B), whereas Cer ADPr ADP-ribosylated vinculin(R561K) at a similar rate similar to wild-type vinculin. CerADPr ADP-ribosylated vinculin(R433A/R561K) at the same rate similar to vinculin(R433A). This showed that CerADPr ADP-ribosylates vinculin at Arg-433.
FIGURE 5.
CerADPr ADP-ribosylates vinculin at Arg-433. A, recombinant vinculin was ADP-ribosylated to saturation and subjected to mass spectrometry peptide sequencing. Three candidate arginine residues that had mass increases consistent with addition of ADP-ribose are shown in bold in the peptide sequences. B, site-directed mutagenesis identified Arg-433 as the site of ADP-ribosylation on vinculin. Wild-type vinculin (circles), vinculin(R433A) (squares), vinculin(R561K) (open triangles), and vinculin(R433A-R561K) (closed triangles) were ADP-ribosylated by CerADPr over the indicated time course as described under “Materials and Methods.” Plotted is the amount of ADP-ribosylated vinculin per total vinculin in arbitrary units (AU). C, amino acid sequence of selected vinculin isoforms is conserved. The primary amino acid sequences of several mammalian- and chicken-vinculin (accession numbers: human, AAA61283; chicken, NP_990772; pig, NP_999099; mouse, NP_033528.3; and rat, NP_001100718.1) surrounding Arg-433 and Ser-434 (residues 420–425) are shown. Human vinculin Arg-433 and Ser-434 are in bold.
DISCUSSION
As emerging human pathogens are identified through bioinformatics or human infection, characterization of their virulence factors and mechanisms of activity becomes important from both preventative and therapeutic perspectives. In this work, vinculin was identified as the substrate for the recently identified bacterial ADP-ribosyltransferase, Certhrax, from B. cereus G9241. Identification of the molecular action of Certhrax will help define how Certhrax contributes to the anthrax-like disease caused by G9241. The current study shows that ADP-ribosylated vinculin is disassociated from focal adhesion complexes, leading to cytotoxicity and cell detachment. This is a new target of bacterial ADP-ribosyltransferase activity and a novel mechanism of bacterial pathogenesis.
Bacterial ADP-ribosyltransferases that target the actin cytoskeleton are typically grouped into three major categories defined by the targeted substrate: the Rho family GTPases (C3-like toxins), heterotrimeric G-proteins (cholera-like toxins), and actin (C2-like toxins). Based on initial sequence analysis, Certhrax was predicted to be a C2-like transferase, with highest similarity to Iota toxin of Clostridium perfringens (29). However, Certhrax did not ADP-ribosylate actin or the Rho proteins (17, 18). Additionally, although both actin and vinculin are highly α-helical proteins with partial structural alignment in the head domain, tryptic fragments of vinculin are not substrates for Certhrax.3 This suggests that Certhrax requires multiple structural elements of vinculin for proper substrate recognition and ADP-ribosylation.
Loss of vinculin is embryonic lethal in mice, whereas a targeted cellular knock-out of vinculin results in cell rounding, increased cytoskeleton remodeling, and loss of adhesion to ECM proteins (30, 31). These phenotypes can be linked to the prominent role vinculin plays in organizing focal adhesion complexes, linking the ECM to the actin cytoskeleton. This connection allows non-motile epithelial cells to remain adherent to the ECM and form a coherent physical barrier (32–34) and also allows motile cells, like macrophages, to form protrusions allowing for directed movement (35–37). Vinculin signals through direct interactions with the actin cytoskeleton and focal adhesion proteins like talin, as well as through recruitment of other proteins responsible for maintaining cytoskeletal cohesion. Critically, expression of paxillin, an additional scaffold and signaling molecule recruited to focal adhesions, does not abrogate the CerADPr-induced phenotypes, demonstrating a vinculin-specific effect.
Targeting the cytoskeleton is not unique for bacterial effectors, as many bacterial toxins target cytoskeletal proteins to promote bacterial invasion or cell death (38–40). These targeted proteins include the RhoGTPases or actin, which are upstream of vinculin in the cytoskeletal signal transduction cascade and exert pathogenic effects through blockade of actin polymerization. Disruption of focal adhesion proteins or complexes has been demonstrated for other pathogenic organisms (41–44) as a mechanism to dampen immune cell responses (45) and impair bacterial phagocytosis (46, 47). Vinculin is a target of other bacterial pathogens, with Helicobacter pylori CagA and Shigella flexneri IpaA proteins interfering with vinculin function through dephosphorylation (CagA), resulting in loss of focal adhesions (48), or robust activation of vinculin signaling (IpaA), which promotes bacterial invasion (49). However, Certhrax appears to be the first example of a bacterial toxin adding a post-translational modification to disrupt vinculin function.
The finding that CerADPr had ∼20-fold higher affinity for vinculin produced in mammalian cells compared with vinculin produced in E. coli suggests that a post-translational modification or protein-protein interaction is required for optimal vinculin ADP-ribosylation. Although CerADPr may require a eukaryotic cofactor to attain full catalytic activity, as has been shown for other bacterial ADPr (50–52), these enzymes also require the cofactor for NAD-glycohydrolase activity, which is not the case for CerADPr (17, 18). Thus, Certhrax may prefer vinculin already incorporated into focal adhesions, targeting an active conformation to produce the most robust toxic effect. Supporting this model are cellular studies in which disruption of vinculin signaling results in loss of cell adherence and promotion of cell motility and invasion (31, 53). Vinculin directly interacts with actin cytoskeleton and the focal adhesion protein talin, which in turn binds the β3-integrin tail to promote cell attachment to the ECM and stabilization of cellular architecture (54). Loss of integrin signaling through focal adhesions results in initiation of cell death pathways in many adherent cells (55, 56). Gram-positive and Gram-negative bacterial pathogens use secreted virulence factors to disrupt focal adhesions or the epithelial barrier to promote pathogen entry and survival (42, 44, 57–60). G9241 may utilize Certhrax for a similar function.
A proposed mechanism of Certhrax-mediated cytotoxicity and cytoskeletal rearrangement is detailed (Fig. 6). Talin recruits vinculin to focal adhesion complexes (A) (22, 24, 27). Vinculin association with talin or actin leads to the activation of vinculin, typically through phosphorylation (61–63); otherwise, vinculin is released back into the cytosol in an inactive conformation (B). Vinculin binding allows talin to stabilize an active conformation of integrins at focal adhesions, promoting cell adherence and polymerization of actin stress fibers (C). Upon G9241 infection, Certhrax enters through a protective antigen-mediated entry mechanism (17) and traffics to the plasma membrane by an unidentified pathway to ADP-ribosylate vinculin at Arg-433 in focal adhesions (D). ADP-ribosylated vinculin loses affinity for actin or other focal adhesion proteins and released into the cytoplasm (D). The inability of ADP-ribosylated vinculin to signal collapses actin stress fibers, which leads to loss of cell adherence and integrin signaling, culminating in cell detachment and death. Vinculin is conserved among mammals and chicken and Arg-433 is present in chicken and mammalian vinculin. Arg-433 is adjacent to Ser-434, a bioinformatics identified phosphorylation site. Thus, ADP-ribosylated Arg-433 might block the phosphorylation of Ser-434 through steric hindrance or charge repulsion. This is under investigation.
FIGURE 6.
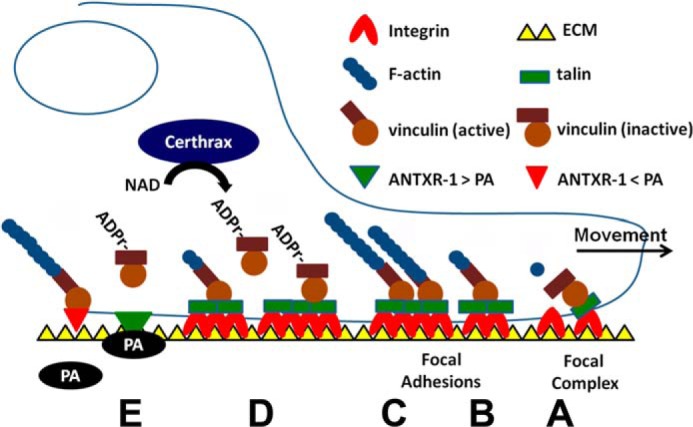
How Certhrax may stimulate actin depolymerization, cell detachment, and potential synergy with anthrax toxin. A, in a normal cell, talin recruits inactive vinculin to focal complexes. B, vinculin associates with PIP2, actin, or other binding partners, which leads to complete activation of vinculin; otherwise, vinculin is released back into the cytosol. C, vinculin binding to talin stabilizes an active conformation of integrins in focal adhesions, which allow vinculin to form a coherent contact between the ECM and promote polymerization of actin stress fibers. D, upon Certhrax intoxication, vinculin is ADP-ribosylated, and ADP-r-vinculin is lost from the focal adhesion site, which stimulates focal adhesion complex disruption. E, actin depolymerization may enhance PA affinity for the anthrax toxin receptor, increasing G9241 virulence. (Modified with permission from Humphries et al. (24).) Vinculin controls focal adhesion formation by direct interactions with talin and actin.
During the course of G9241 infection, Certhrax disruption of vinculin signaling resulting in loss of cell adherence could lead to disruption of the epithelial barrier lining the lung alveoli. Alternatively, Certhrax could play a general anti-phagocytic role, as seen for other pathogens, which disrupt focal adhesions. Certhrax may act in concert with G9241 lethal factor, which in B. anthracis pathogenesis is thought to primarily target macrophages (64), to promote efficient G9241 colonization of the lung through dual mechanisms of physical barrier disruption and disruption of the innate immune system. However, because Certhrax also promotes toxicity in macrophages (17), Certhrax may also have a general cytopathogenic role in G9241 pathogenesis. Decreased association of actin with ANTXR-1 increases the affinity of PA for ANTXR-1 (65, 66). Thus, Certhrax may enhance the potency of G9241 anthrax toxin through disruption of the actin cytoskeleton (E). Further investigations into potential toxin synergy may open a new frontier in bacterial pathogenesis and lead to improvements in preventing emerging pathogens from spreading into the human populace.
Acknowledgment
We acknowledge the assistance of Amanda Przedpelski for protein production.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI031062 and U54 AI057153.
N. C. Simon and J. T. Barbieri, unpublished data.
- EF
- edema factor
- PA
- protective antigen
- ECM
- extracellular matrix
- HPT
- hours post-transfection
- VCL
- human vinculin protein
- ANOVA
- analysis of variance.
REFERENCES
- 1. Miller J. M., Hair J. G., Hebert M., Hebert L., Roberts F. J., Jr., Weyant R. S. (1997) Fulminating bacteremia and pneumonia due to Bacillus cereus. J. Clin. Microbiol. 35, 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klee S. R., Ozel M., Appel B., Boesch C., Ellerbrok H., Jacob D., Holland G., Leendertz F. H., Pauli G., Grunow R., Nattermann H. (2006) Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Côte d'Ivoire and Cameroon. J. Bacteriol. 188, 5333–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoffmaster A. R., Hill K. K., Gee J. E., Marston C. K., De B. K., Popovic T., Sue D., Wilkins P. P., Avashia S. B., Drumgoole R., Helma C. H., Ticknor L. O., Okinaka R. T., Jackson P. J. (2006) Characterization of Bacillus cereus isolates associated with fatal pneumonias: strains are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J. Clin. Microbiol. 44, 3352–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avashia S. B., Riggins W. S., Lindley C., Hoffmaster A., Drumgoole R., Nekomoto T., Jackson P. J., Hill K. K., Williams K., Lehman L., Libal M. C., Wilkins P. P., Alexander J., Tvaryanas A., Betz T. (2007) Fatal pneumonia among metalworkers due to inhalation exposure to Bacillus cereus containing Bacillus anthracis toxin genes. Clin. Infect. Dis. 44, 414–416 [DOI] [PubMed] [Google Scholar]
- 5. Bottone E. J. (2010) Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23, 382–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turnbull P. C., Sirianni N. M., LeBron C. I., Samaan M. N., Sutton F. N., Reyes A. E., Peruski L. F., Jr. (2004) MICs of selected antibiotics for Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, and Bacillus mycoides from a range of clinical and environmental sources as determined by the Etest. J. Clin. Microbiol. 42, 3626–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okinaka R., Cloud K., Hampton O., Hoffmaster A., Hill K., Keim P., Koehler T., Lamke G., Kumano S., Manter D., Martinez Y., Ricke D., Svensson R., Jackson P. (1999) Sequence, assembly and analysis of pX01 and pX02. J. Appl. Microbiol. 87, 261–262 [DOI] [PubMed] [Google Scholar]
- 8. Okinaka R. T., Cloud K., Hampton O., Hoffmaster A. R., Hill K. K., Keim P., Koehler T. M., Lamke G., Kumano S., Mahillon J., Manter D., Martinez Y., Ricke D., Svensson R., Jackson P. J. (1999) Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181, 6509–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Milne J. C., Furlong D., Hanna P. C., Wall J. S., Collier R. J. (1994) Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J. Biol. Chem. 269, 20607–20612 [PubMed] [Google Scholar]
- 10. Mogridge J., Cunningham K., Collier R. J. (2002) Stoichiometry of anthrax toxin complexes. Biochemistry 41, 1079–1082 [DOI] [PubMed] [Google Scholar]
- 11. Kintzer A. F., Thoren K. L., Sterling H. J., Dong K. C., Feld G. K., Tang I. I., Zhang T. T., Williams E. R., Berger J. M., Krantz B. A. (2009) The protective antigen component of anthrax toxin forms functional octameric complexes. J. Mol. Biol. 392, 614–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Green B. D., Battisti L., Koehler T. M., Thorne C. B., Ivins B. E. (1985) Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49, 291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmaster A. R., Ravel J., Rasko D. A., Chapman G. D., Chute M. D., Marston C. K., De B. K., Sacchi C. T., Fitzgerald C., Mayer L. W., Maiden M. C., Priest F. G., Barker M., Jiang L., Cer R. Z., Rilstone J., Peterson S. N., Weyant R. S., Galloway D. R., Read T. D., Popovic T., Fraser C. M. (2004) Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. U.S.A. 101, 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oh S.-Y., Budzik J. M., Garufi G., Schneewind O. (2011) Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol. Microbiol. 80, 455–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson M. K., Vergis J. M., Alem F., Palmer J. R., Keane-Myers A. M., Brahmbhatt T. N., Ventura C. L., O'Brien A. D. (2011) Bacillus cereus G9241 makes anthrax toxin and capsule like highly virulent B. anthracis Ames but behaves like attenuated toxigenic nonencapsulated B. anthracis Sterne in rabbits and mice. Infect. Immun. 79, 3012–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oh S.-Y., Maier H., Schroeder J., Richter G. S., Elli D., Musser J. M., Quenee L. E., Missiakas D. M., Schneewind O. (2013) Vaccine protection against Bacillus cereus-mediated respiratory anthrax-like disease in mice. Infect. Immun. 81, 1008–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Visschedyk D., Rochon A., Tempel W., Dimov S., Park H.-W., Merrill A. R. (2012) Certhrax toxin, an anthrax-related ADP-ribosyltransferase from Bacillus cereus. J. Biol. Chem. 287, 41089–41102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simon N. C., Vergis J. M., Ebrahimi A. V., Ventura C. L., O'Brien A. D., Barbieri J. T. (2013) Host cell cytotoxicity and cytoskeleton disruption by CerADPr, an ADP-ribosyltransferase of Bacillus cereus G9241. Biochemistry 52, 2309–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aktories K., Braun U., Rösener S., Just I., Hall A. (1989) The rho gene product expressed in E. coli is a substrate of botulinum ADP-ribosyltransferase C3. Biochem. Biophys. Res. Commun. 158, 209–213 [DOI] [PubMed] [Google Scholar]
- 20. Han S., Arvai A. S., Clancy S. B., Tainer J. A. (2001) Crystal structure and novel recognition motif of Rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. J. Mol. Biol. 305, 95–107 [DOI] [PubMed] [Google Scholar]
- 21. Carisey A., Ballestrem C. (2011) Vinculin, an adapter protein in control of cell adhesion signalling. Eur. J. Cell Biol. 90, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng X., Nelson E. S., Maiers J. L., DeMali K. A. (2011) in International Review of Cell and Molecular Biology (Kwang W. J., ed) pp. 191–231, Academic Press, New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geiger B. (1979) A 130K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells. Cell 18, 193–205 [DOI] [PubMed] [Google Scholar]
- 24. Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J., Ballestrem C. (2007) Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., Liddington R. C. (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586 [DOI] [PubMed] [Google Scholar]
- 26. Bois P. R., O'Hara B. P., Nietlispach D., Kirkpatrick J., Izard T. (2006) The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J. Biol. Chem. 281, 7228–7236 [DOI] [PubMed] [Google Scholar]
- 27. Ziegler W. H., Liddington R. C., Critchley D. R. (2006) The structure and regulation of vinculin. Trends Cell Biol. 16, 453–460 [DOI] [PubMed] [Google Scholar]
- 28. Pederson K. J., Barbieri J. T. (1998) Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol. Microbiol. 30, 751–759 [DOI] [PubMed] [Google Scholar]
- 29. Fieldhouse R. J., Turgeon Z., White D., Merrill A. R. (2010) Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLoS Comput. Biol. 6, e1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodríguez Fernández J. L., Geiger B., Salomon D., Ben-Ze'ev A. (1993) Suppression of vinculin expression by antisense transfection confers changes in cell morphology, motility, and anchorage-dependent growth of 3T3 cells. J. Cell Biol. 122, 1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu W., Baribault H., Adamson E. D. (1998) Vinculin knockout results in heart and brain defects during embryonic development. Development 125, 327–337 [DOI] [PubMed] [Google Scholar]
- 32. Maddugoda M. P., Crampton M. S., Shewan A. M., Yap A. S. (2007) Myosin VI and vinculin cooperate during the morphogenesis of cadherin cell–cell contacts in mammalian epithelial cells. J. Cell Biol. 178, 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davies J. A., Garrod D. R. (1997) Molecular aspects of the epithelial phenotype. BioEssays 19, 699–704 [DOI] [PubMed] [Google Scholar]
- 34. Wehrle-Haller B. (2012) Assembly and disassembly of cell matrix adhesions. Curr. Opin. Cell Biol. 24, 569–581 [DOI] [PubMed] [Google Scholar]
- 35. Owen K. A., Pixley F. J., Thomas K. S., Vicente-Manzanares M., Ray B. J., Horwitz A. F., Parsons J. T., Beggs H. E., Stanley E. R., Bouton A. H. (2007) Regulation of lamellipodial persistence, adhesion turnover, and motility in macrophages by focal adhesion kinase. J. Cell Biol. 179, 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gardel M. L., Schneider I. C., Aratyn-Schaus Y., Waterman C. M. (2010) Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26, 315–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu K., Ji L., Applegate K. T., Danuser G., Waterman-Storer C. M. (2007) Differential transmission of actin motion within focal adhesions. Science 315, 111–115 [DOI] [PubMed] [Google Scholar]
- 38. Aktories K., Lang A. E., Schwan C., Mannherz H. G. (2011) Actin as target for modification by bacterial protein toxins. FEBS J. 278, 4526–4543 [DOI] [PubMed] [Google Scholar]
- 39. Haglund C. M., Welch M. D. (2011) Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 195, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schiavo G., van der Goot F. G. (2001) The bacterial toxin tool kit. Nat. Rev. Mol. Cell Biol. 2, 530–537 [DOI] [PubMed] [Google Scholar]
- 41. Cappello R. E., Estrada-Gutierrez G., Irles C., Giono-Cerezo S., Bloch R. J., Nataro J. P. (2011) Effects of the plasmid-encoded toxin of enteroaggregative Escherichia coli on focal adhesion complexes. FEMS Immunol. Med. Microbiol. 61, 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Black D. S., Bliska J. B. (1997) Identification of p130Cas as a substrate of Yersinia YopH (Yop51), a bacterial protein-tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. EMBO J. 16, 2730–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kogan T. V., Jadoun J., Mittelman L., Hirschberg K., Osherov N. (2004) Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J. Infect. Dis. 189, 1965–1973 [DOI] [PubMed] [Google Scholar]
- 44. Wilke G. A., Bubeck Wardenburg J. (2010) Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. U.S.A. 107, 13473–13478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sauvonnet N., Lambermont I., van der Bruggen P., Cornelis G. R. (2002) YopH prevents monocyte chemoattractant protein 1 expression in macrophages and T-cell proliferation through inactivation of the phosphatidylinositol 3-kinase pathway. Mol. Microbiol. 45, 805–815 [DOI] [PubMed] [Google Scholar]
- 46. Alrutz M. A., Isberg R. R. (1998) Involvement of focal adhesion kinase in invasin-mediated uptake. Proc. Natl. Acad. Sci. U.S.A. 95, 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andersson K., Carballeira N., Magnusson K.-E., Persson C., Stendahl O., Wolf-Watz H., Fällman M. (1996) YopH of Yersinia pseudotuberculosis interrupts early phosphotyrosine signalling associated with phagocytosis. Mol. Microbiol. 20, 1057–1069 [DOI] [PubMed] [Google Scholar]
- 48. Moese S., Selbach M., Brinkmann V., Karlas A., Haimovich B., Backert S., Meyer T. F. (2007) The Helicobacter pylori CagA protein disrupts matrix adhesion of gastric epithelial cells by dephosphorylation of vinculin. Cell. Microbiol. 9, 1148–1161 [DOI] [PubMed] [Google Scholar]
- 49. Tran Van Nhieu G., Ben-Ze'ev A., Sansonetti P. J. (1997) Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 16, 2717–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Coburn J., Kane A. V., Feig L., Gill D. M. (1991) Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J. Biol. Chem. 266, 6438–6446 [PubMed] [Google Scholar]
- 51. Liu S., Yahr T. L., Frank D. W., Barbieri J. T. (1997) Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J. Bacteriol. 179, 1609–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bobak D. A., Bliziotes M. M., Noda M., Tsai S. C., Adamik R., Moss J. (1990) Mechanism of activation of cholera toxin by ADP-ribosylation factor (ARF): both low- and high-affinity interactions of ARF with guanine nucleotides promote toxin activation. Biochemistry 29, 855–861 [DOI] [PubMed] [Google Scholar]
- 53. Mierke C. T., Kollmannsberger P., Zitterbart D. P., Diez G., Koch T. M., Marg S., Ziegler W. H., Goldmann W. H., Fabry B. (2010) Vinculin facilitates cell invasion into three-dimensional collagen matrices. J. Biol. Chem. 285, 13121–13130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., Campbell I. D. (2007) Structural basis of integrin activation by talin. Cell 128, 171–182 [DOI] [PubMed] [Google Scholar]
- 55. Chen C. S., Mrksich M., Huang S., Whitesides G. M., Ingber D. E. (1997) Geometric control of cell life and death. Science 276, 1425–1428 [DOI] [PubMed] [Google Scholar]
- 56. Ruoslahti E. (1997) Stretching is good for a cell. Science 276, 1345–1346 [DOI] [PubMed] [Google Scholar]
- 57. Kim M., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Sasakawa C. (2010) Bacterial interactions with the host epithelium. Cell Host Microbe 8, 20–35 [DOI] [PubMed] [Google Scholar]
- 58. Eisele N. A., Anderson D. M. (2011) Host defense and the airway epithelium: frontline responses that protect against bacterial invasion and pneumonia. J. Pathog. 2011, 249802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lehmann M., Noack D., Wood M., Perego M., Knaus U. G. (2009) Lung epithelial injury by B. anthracis lethal toxin is caused by MKK-dependent loss of cytoskeletal integrity. PLoS One 4, e4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Langer M., Duggan E. S., Booth J. L., Patel V. I., Zander R. A., Silasi-Mansat R., Ramani V., Veres T. Z., Prenzler F., Sewald K., Williams D. M., Coggeshall K. M., Awasthi S., Lupu F., Burian D., Ballard J. D., Braun A., Metcalf J. P. (2012) Bacillus anthracis lethal toxin reduces human alveolar epithelial barrier function. Infect. Immun. 80, 4374–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Golji J., Lam J., Mofrad M. R. (2011) Vinculin activation is necessary for complete talin binding. Biophys. J. 100, 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Küpper K., Lang N., Möhl C., Kirchgessner N., Born S., Goldmann W. H., Merkel R., Hoffmann B. (2010) Tyrosine phosphorylation of vinculin at position 1065 modifies focal adhesion dynamics and cell tractions. Biochem. Biophys. Res. Commun. 399, 560–564 [DOI] [PubMed] [Google Scholar]
- 63. Golji J., Wendorff T., Mofrad M. R. (2012) Phosphorylation primes vinculin for activation. Biophys. J. 102, 2022–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hanna P. C., Acosta D., Collier R. J. (1993) On the role of macrophages in anthrax. Proc. Natl. Acad. Sci. U.S.A. 90, 10198–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Garlick K. M., Batty S., Mogridge J. (2012) Binding of filamentous actin to anthrax toxin receptor 1 decreases its association with protective antigen. Biochemistry 51, 1249–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Go M. Y., Chow E. M., Mogridge J. (2009) The cytoplasmic domain of anthrax toxin receptor 1 affects binding of the protective antigen. Infect. Immun. 77, 52–59 [DOI] [PMC free article] [PubMed] [Google Scholar]