Background: Symbiotic protists of the termite gut contribute to lignocellulosic biomass degradation.
Results: A novel protistan β-mannanase efficiently degrades glucomannan and displays glucose/mannose binding properties when complexed with gluco-manno-oligosaccharide.
Conclusion: Specific recognition and accommodation of glucose at the distal - subsites provides the structural basis for activity against glucomannan.
Significance: The mechanism underlying heteropolysaccharide recognition by mannanase has been clarified.
Keywords: Crystal Structure, Glycoside Hydrolases, Plant Cell Wall, Polysaccharide, Symbiosis, Mannanase, Symbiotic Protist, Termite
Abstract
Termites and their symbiotic protists have established a prominent dual lignocellulolytic system, which can be applied to the biorefinery process. One of the major components of lignocellulose from conifers is glucomannan, which comprises a heterogeneous combination of β-1,4-linked mannose and glucose. Mannanases are known to hydrolyze the internal linkage of the glucomannan backbone, but the specific mechanism by which they recognize and accommodate heteropolysaccharides is currently unclear. Here, we report biochemical and structural analyses of glycoside hydrolase family 26 mannanase C (RsMan26C) from a symbiotic protist of the termite Reticulitermes speratus. RsMan26C was characterized based on its catalytic efficiency toward glucomannan, compared with pure mannan. The crystal structure of RsMan26C complexed with gluco-manno-oligosaccharide(s) explained its specificities for glucose and mannose at subsites −5 and −2, respectively, in addition to accommodation of both glucose and mannose at subsites −3 and −4. RsMan26C has a long open cleft with a hydrophobic platform of Trp94 at subsite −5, facilitating enzyme binding to polysaccharides. Notably, a unique oxidized Met85 specifically interacts with the equatorial O-2 of glucose at subsite −3. Our results collectively indicate that specific recognition and accommodation of glucose at the distal negative subsites confers efficient degradation of the heteropolysaccharide by mannanase.
Introduction
Termites are destructive insect pests that cause serious economic damage. The termite genus Reticulitermes includes the significant pest species of North America (Reticulitermes flavipes complex), Europe (Reticulitermes lucifugus complex), and Asia, including Japan, (Reticulitermes speratus) (1). Traditional Japanese houses are predominantly built of softwood in which the major component of hemicellulose is (galacto)glucomannan (10–30% of total cell wall weight (2)), indicating that glucomannan-degrading enzymes play an important role in the termite digestive system as well as the cellulolytic system. Termites thrive on dead plant biomass with the aid of microbial symbionts (3) and possess two cellulolytic systems: endogenous cellulases (4, 5) and prokaryotes and flagellated protists (single cell eukaryotes) in the termite hindgut (6). The dual cellulolytic system that is well established in lower termites enables almost complete decomposition of the cellulose (74–99%) and hemicellulose (65–87%) components of ingested plant biomass (3). Because the plant cell wall constitutes a complex network of polysaccharides, this efficient lignocellulolytic system of termites and their symbionts may be applicable to the industrial processes of biorefinery (7). Gut protists take up the partially digested wood particles into food vacuoles via phagocytosis (8) and degrade them with cellulases and hemicellulases to produce acetate, which is absorbed by termites as their energy and carbon source (9). Therefore, the symbiotic protists provide a novel enzymatic resource for efficient degradation of plant biomass. Indeed, protists possess both cellulases and hemicellulases but termites harbor only endo-glucanases and β-glucosidases (10). To determine the lignocellulolytic mechanism of symbiotic protists of termites, we previously constructed a cDNA library involved in lignocellulose digestion from the symbiotic protist community in the hind gut of the lower termite, R. speratus (11) and identified numerous genes encoding putative cellulases and hemicellulases, including endo-1,4-β-mannanases.
Mannan is one of the main components of the plant cell wall, composed of a β-1,4-linked backbone containing mannose (Man)2 or a combination of glucose (Glc) and Man moieties, which can be substituted with α-1,6-galactosyl side chains (12) and acetylated at the O-2 and/or O-3 positions (13, 14). Endo-1,4-β-mannanases (EC 3.2.1.78) hydrolyze the internal β-1,4-linkage of the mannan backbone by a retaining double-displacement mechanism, and are classified into glycoside hydrolase families 5, 26, and 113 in the CAZy (carbohydrate-active enzyme database) (15). All mannanases have a canonical (β/α)8-barrel fold in clan GH-A with two catalytic glutamic acids located at the C terminus of β-strands 4 (acid/base) and 7 (nucleophile) (16, 17). Some mannanases hydrolyze heteropolymers of β-1,4-linked Glc and Man more efficiently than homopolymers of Man (18). These enzymes possibly use a specific mechanism for recognition or accommodation of Glc and Man. Recent studies have shown that whereas GH5 mannanases generally show relaxed specificity for Glc or Man at the −2 subsite, GH26 mannanases display specificity for Man at both subsites −2 and −1 (19). The Man at subsite −1 adopts the B2,5 transition conformation with the key specificity determinants being the pseudoequatorial O-2 and pseudoaxial O-3 (20, 21). However, the structural mechanism underlying the recognition and accommodation of heteropolysaccharides is currently unclear.
Here, we have performed biochemical and structural analyses of a GH26 mannanase, Man26C, from a symbiotic protist of the lower termite, R. speratus. Biochemical characterization experiments revealed that RsMan26C displays the highest activity toward glucomannan among the structurally different mannans. To elucidate how RsMan26C recognizes or accommodates heteropolysaccharides comprising both Glc and Man, the crystal structure of RsMan26C complexed with a gluco-manno-oligosaccharide was solved. The topology of the open catalytic cleft revealed Glc-specific recognition and accommodation at the distal negative subsites in the RsMan26C structure.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A cDNA fragment encoding the putative mature region of RsMan26C (DDBJ accession no. AB684778) was inserted between the EcoRI and NotI sites of the expression vector pPICZα-A (Invitrogen), and N-terminal c-myc epitope tag with polyhistidine tag, cleaved from the Pichia pastoris expression vector pBGP2 (described elsewhere (22)) between the XhoI and EcoRI sites, was also inserted into the same plasmid. The constructed plasmid was named pPICZα-mhRsMan26C. P. pastoris KM71H strain was transformed according to the instruction manual of the EasySelectTM Pichia expression kit (Invitrogen). The recombinant RsMan26C was produced using a mini jar-fermentor (TSC-M5L; Takasugi Seisakusho, Tokyo, Japan) equipped with a DO controller (DJ-1033; ABLE Corp., Tokyo, Japan) according to the Pichia fermentation process guidelines (Invitrogen). After 3 days in methanol-fed batch culture, the medium was collected by centrifugation (4 °C, 30 min, 8,000 × g). The culture supernatant was ultrafiltrated with a Kvick Lab Cassette 100 kDa (GE Healthcare, Buckinghamshire, England) and concentrated with a Kvick Lab Cassette 5 kDa (GE Healthcare).
The enzyme solution was purified on a HiTrap Phenyl FF column (5 ml; GE Healthcare) by linear reverse gradient of 30–0% ammonium sulfate in 50 mm Tris-HCl (pH 7.5). The sample was then fractionated on a HiTrap DEAE FF column (5 ml; GE Healthcare) by linear gradient of 0–1.0 m NaCl in 50 mm Tris-HCl (pH 7.5). The protein was then treated with endoglycosidase H (New England Biolabs) according to the manufacturer's instructions. After the deglycosylation, the solution was applied on a HiLoad 16/60 Superdex 75 prep grade column (120 ml; GE Healthcare) and eluted with 20 mm Tris-HCl (pH 7.5) containing 150 mm NaCl. The purified protein was finally dialyzed with 5 mm Tris-HCl (pH 8.0). Protein concentration was determined using Bio-Rad protein assay (Bio-Rad) using bovine serum albumin as the standard.
Enzyme Assays
Polysaccharides and oligosaccharides used in the enzyme assays are as follows: manno-oligosaccharides, konjac glucomannan, β-mannan (prepared from carob galactomannan with removal of all α-galactosyl residues by α-galactosidase), and ivory nut mannan (Megazyme Intl.); locust bean gum, Avicel, carboxymethylcellulose, and birchwood xylan (Sigma-Aldrich); guar gum and chitin (Wako Pure Chemical Industries, Osaka, Japan); and cello-oligosaccharides (Seikagaku Biobusiness, Tokyo, Japan).
β-Mannanase assay was conducted at 30 °C by adding 5 μl of enzyme solution to 100 μl of 0.5–5% (w/v) substrate in 50 mm sodium acetate (pH 5.5). After incubation for 15 min, the reaction was stopped by boiling for 5 min. The reducing sugars produced were measured with tetrazolium blue reagent (23) by the method described previously (24). A standard curve was drawn using the solution containing various concentrations of Man. The reaction products released from manno-oligosaccharides were separated on a TLC Silica gel 60 plate (Merck KGaA) with a solvent system containing n-propanol-ethanol-water (7:1:2) and visualized by staining with 2.5% (v/v) anisaldehyde, 3.4% (v/v) sulfinic acid, and 1.0% (v/v) acetic acid in ethanol and baking at 100 °C for 5 min. The spots on the TLC plate were quantified by Multi Gauge (version 3.1, Fujifilm, Tokyo, Japan). The soluble products released from 1,4-β-d-mannan (Megazyme) were analyzed on a HPLC system equipped with a CoronaTM Charged Aerosol DetectorTM (ESA Biosciences, Inc., Chelmsford, MA). The supernatant was separated on a Shodex Asahipak NH2P-50 4E column (Showa Denko K.K., Tokyo, Japan) equipped with a guard column NH2P-50G 4A (Showa Denko) using the following elution conditions: 0–10 min, a linear gradient of acetonitrile/H2O (60/40 to 50/50, v/v); 10–15 min, acetonitrile/H2O (50/50, v/v); and 15–20 min acetonitrile/H2O (60/40, v/v).
Crystallography
Crystals of RsMan26C were grown at 20 °C for 1 week, using a sitting-drop vapor diffusion method. Crystals were obtained by mixing 1 μl of protein solution comprised of 10 mg/ml protein, 5 mm mannopentaose, and 5 mm Tris-HCl (pH 8.0) and 1 μl of reservoir solution comprised of 25% (w/v) PEG 3350, 0.2 m MgCl2, and 0.1 m Bis-Tris (pH 5.5). X-ray diffraction data for the RsMan26C native form and ligand-bound form were collected at the BL-17A station (λ = 1.0 Å) (Photon Factory, High Energy Accelerator Research Organization, Tsukuba, Japan). Complex crystals were prepared by soaking in the ligand solution as follows: 5 ml of 0.5% (w/v) glucomannan in 5 mm Bis-Tris (pH 5.5) was incubated for 1 h with 50 μl of appropriately diluted RsMan26C. The reaction products were freeze-dried for 24 h and then dissolved in 100 μl of water, which was used for soaking crystals. Crystals were cryo-protected in the reservoir solution supplemented with 20% glycerol and were flash-cooled at 95 K in a stream of nitrogen gas. Data were processed using HKL2000 (25). Molecular replacement was performed with Balbes (26). Manual model building and refinement were performed using Refmac5 (27) and Coot (28). Data collection and refinement statistics are provided in Table 1.
TABLE 1.
Data collection and refinement statistics
| Native | Gluco-manno-oligosaccharide | |
|---|---|---|
| Data collection | ||
| PDB code | 3WDQ | 3WDR |
| Beamline | BL17A | BL17A |
| Space group | P21 | P21 |
| Unit cell | a = 45.5, b = 61.4, and c = 57.8 Å; β = 92.7° | a = 45.5, b = 61.6, and c = 58.1 Å; β = 92.3° |
| Resolution (Å) | 50–1.3 (1.32–1.30) | 50–1.4 (1.42–1.40) |
| Total reflections | 570,178 | 235,046 |
| Unique reflections | 77,766 (3846) | 62,697 (3085) |
| Completeness (%) | 99.9 (99.7) | 99.2 (98.5) |
| Rsym (%) | 6.8 (48.0) | 6.9 (30.7) |
| I/σ(I) | 34.6 (2.3) | 20.2 (4.3) |
| Redundancy | 7.3 (6.8) | 3.8 (3.7) |
| Refinement | ||
| Resolution (Å) | 28.87–1.30 | 36.51–1.40 |
| R/Rfree (%) | 14.4/16.4 | 13.7/16.0 |
| No. of reflections | 73,833 | 59,428 |
| Number of atoms | 3,135 | 3,192 |
| r.m.s.d. from ideal valuesa | ||
| Bond length (Å) | 0.011 | 0.011 |
| Bond angles | 1.477° | 1.537° |
a r.m.s.d., root mean square deviation.
RESULTS
Biochemical Properties of RsMan26C
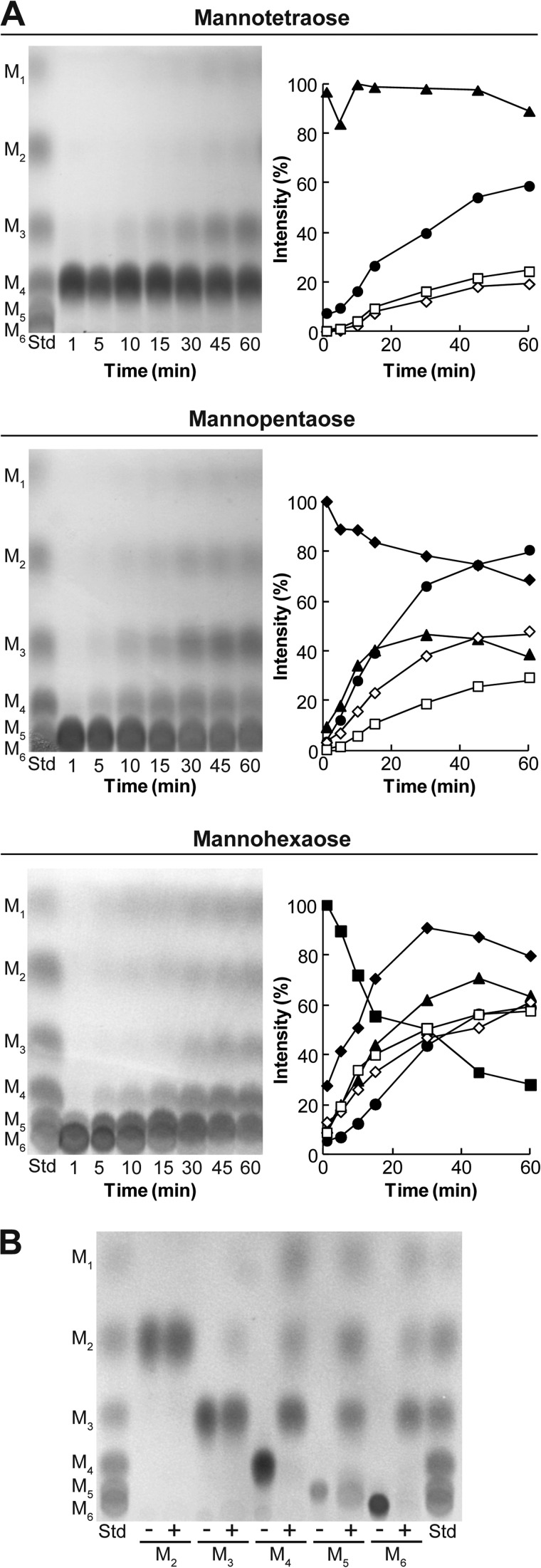
To elucidate the biochemical properties of RsMan26C, we analyzed the hydrolysis products of manno-oligosaccharides. Recombinant RsMan26C randomly hydrolyzed the β-1,4-linkage of manno-oligosaccharides and generated reaction products of various sizes (Fig. 1A), a typical feature of endo-acting enzymes. Specifically, RsMan26C generated mannose, mannobiose, and mannotriose as final reaction products from mannotetraose, mannopentaose, and mannohexaose (Fig. 1B). In addition, the enzyme displayed partial activity against mannotriose, but failed to cleave mannobiose after incubation for 24 h. No transglycosylation activity was observed. Therefore, RsMan26C is a typical endo-mode mannanase that cleaves the internal β-1,4-linkage of manno-oligosaccharides with a degree of polymerization higher than three.
FIGURE 1.
Hydrolysis of manno-oligosaccharides. A, the reaction products of mannotetraose, mannopentaose, and mannohexaose catalyzed by RsMan26C were sequentially monitored using TLC (left) and subjected to densitometric quantification (right). B, final reaction products of manno-oligosaccharides incubated with RsMan26C for 24 h. M1, mannose (open squares); M2, mannobiose (open diamonds); M3, mannotriose (solid circles); M4, mannotetraose (solid triangles); M5, mannopentaose (diamonds); M6, mannohexaose (solid squares); Std, manno-oligosaccharide standard.
The substrate specificity of RsMan26C was analyzed to elucidate the hydrolysis profiles of polysaccharides (Table 2). RsMan26C displayed the highest activity against glucomannan (kcat/Km = 9.1 × 103 ml mg−1 min−1), which was ∼1.1-fold and 6.1-fold higher than those against galactomannan from locust bean gum (kcat/Km = 8.5 × 103 ml mg−1 min−1) and β-mannan (kcat/Km = 1.5 × 103 ml mg−1 min−1), respectively. RsMan26C additionally degraded and released reducing sugars from crystalline ivory nut mannan and galactomannan from guar gum (Fig. 2), but activities against these substrates were too weak to measure the kinetic parameters. No activity against β-1,4-xylan, carboxymethylcellulose, Avicel, chitin, and cello-oligosaccharides was observed.
TABLE 2.
Biochemical properties of RsMan26C
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| mg/ml | min−1 | ml min−1 mg−1 | |
| Glucomannana | 5.1 ± 1.3×10−4 | 4.7×104 ± 1.0×10−3 | 9.1×104 ± 1.0×10−3 |
| β-Mannana | 10.7 ± 1.0×10−3 | 1.6×104 ± 1.4×10−2 | 1.5×104 ± 1.4×10−2 |
| Locust bean guma | 2.5 ± 1.3×10−4 | 2.1×104 ± 2.2×10−3 | 8.5×104 ± 2.2×10−3 |
| Guar guma | NDb | NDb | NDb |
| Ivory nut mannana | NDb | NDb | NDb |
a The polysaccharide substrates were prepared as follows: 0.5% glucomannan (konjac), β-mannan (prepared from carob galactomannan with removal of all α-galactosyl residues by α-galactosidase), galactomannan (locust bean gum), 2% galactomannan (guar gum), or 5% ivory nut mannan were dissolved in 50 mm sodium acetate buffer (pH 5.5).
b ND indicates not detected.
FIGURE 2.
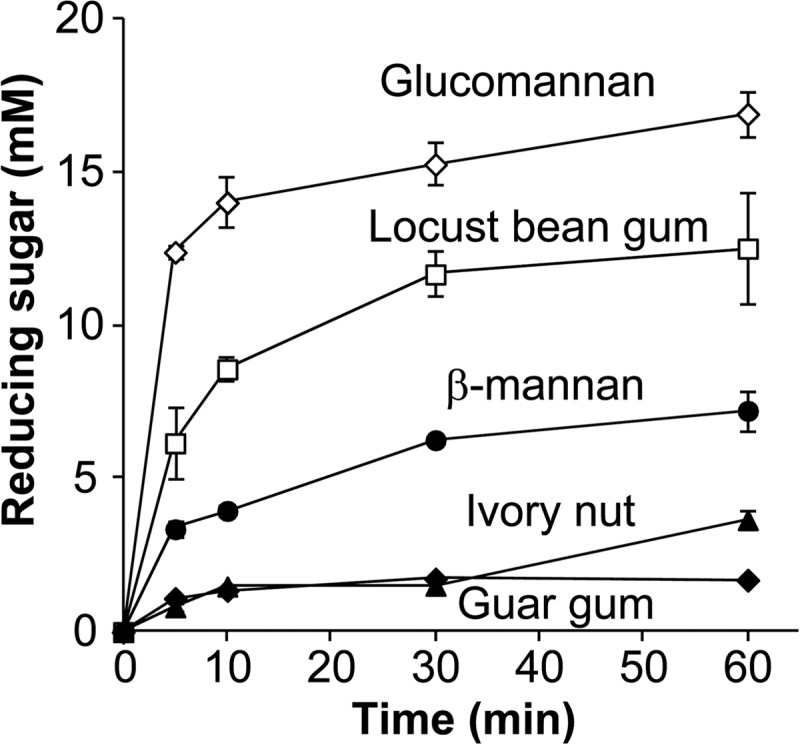
Substrate specificities of RsMan26C. Reducing sugar released from various substrates by RsMan26C was sequentially determined. 0.5% glucomannan (open diamonds), 0.5% locust bean gum (open squares), 0.5% β-mannan (solid circles), 5% ivory nut (solid triangles), and 2% guar gum (solid diamonds) are dissolved in 50 mm sodium acetate buffer (pH 5.5).
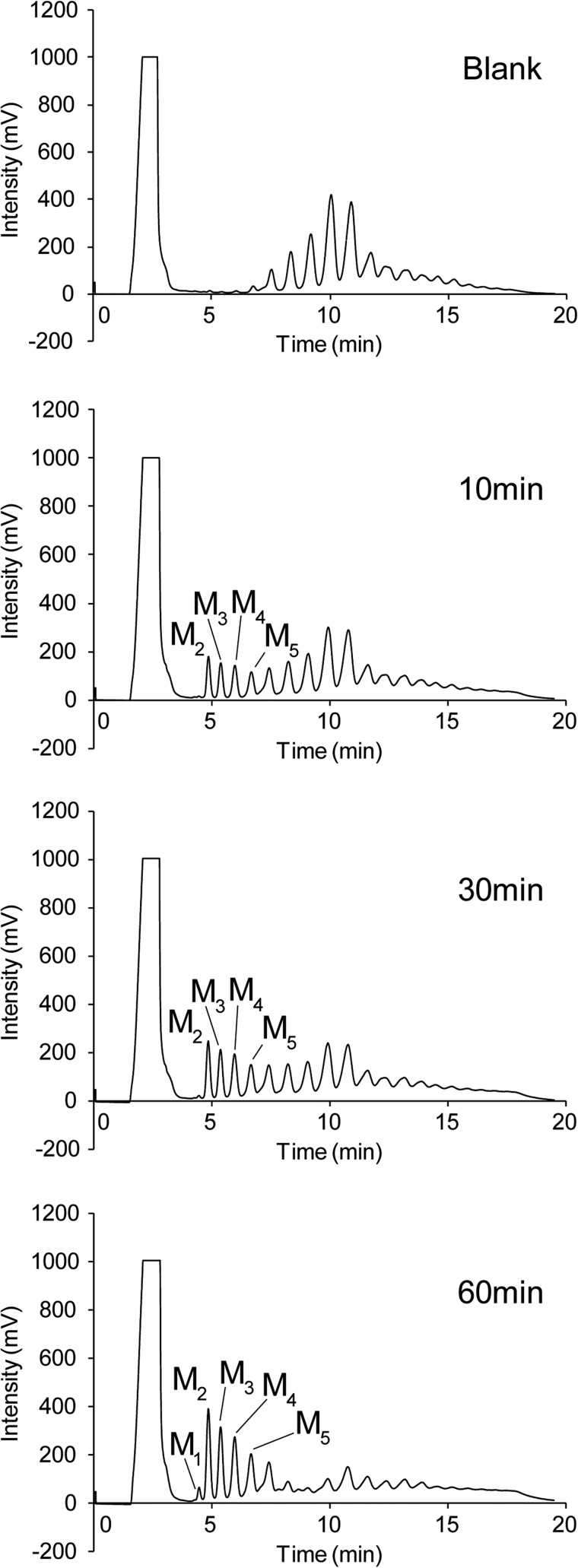
Sequential detection of the reaction products of RsMan26C with β-mannan as substrate using HPLC revealed dramatic breakdown of substrate. Long chain manno-oligosaccharides of various sizes were detected before initiation of the reaction (Fig. 3, Blank). Following addition of RsMan26C, the degree of substrate polymerization was decreased and mannose, mannobiose, mannotriose, mannotetraose, and mannopentaose were generated after 60 min of reaction (Fig. 3, 10–60 min). This result clearly signifies an endo mode of action of RsMan26C, consistent with the results of manno-oligosaccharide hydrolysis (Fig. 1).
FIGURE 3.
Hydrolysis of β-mannan by RsMan26C. Hydrolysis products of β-mannan incubated without (blank) or with RsMan26C for 10 to 60 min were determined using HPLC. M1, mannose; M2, mannobiose; M3, mannotriose; M4, mannotetraose; M5, mannopentaose.
Three-dimensional Structure of RsMan26C
The crystal structure of RsMan26C was determined using x-ray crystallography at 1.3 Å resolution (Table 1). The initial phases were determined by molecular replacement using Bacillus subtilis β-mannanase BsMan26A (PDB code 3CBW (19)) as a search model. The structure of native RsMan26C was determined at 1.30 Å resolution and refined to R/Rfree = 14.4/16.4%, with one molecule in the asymmetric unit. The resolved structure contained residues from Gln15 to Trp344 of full-length RsMan26C. RsMan26C displays a canonical (β/α)8-barrel topology with the catalytic nucleophile, Glu288, and the acid/base, Glu191, located at the end of β-strands 4 and 7, respectively, typical of the GH26 family (Fig. 4A). Despite the presence of mannopentaose in crystal growth conditions, no mannopentaose or hydrolyzed manno-oligosaccharides were detected in the enzyme structure.
FIGURE 4.
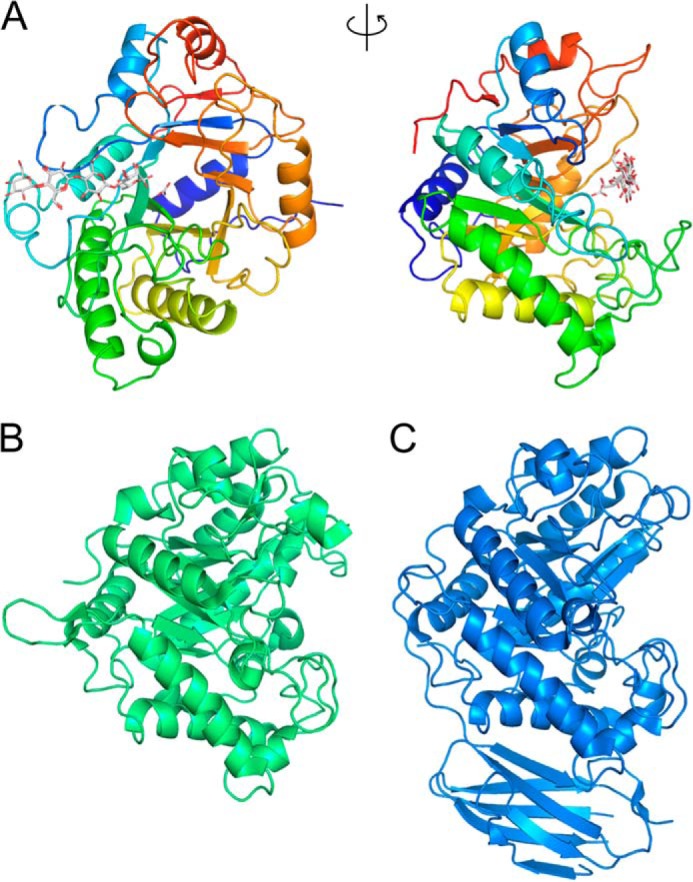
Three-dimensional structures of RsMan26C and GH26 family enzymes. A, overall structure of the RsMan26C ligand complex: front view (left) and side view (right). B and C, overall structures of CjMan26C (PDB code 2VX6, green (21)) (B) and CfMan26A (PDB code 2BVT, blue (16)) (C).
Complex Structure of RsMan26C with a Gluco-manno-oligosaccharide
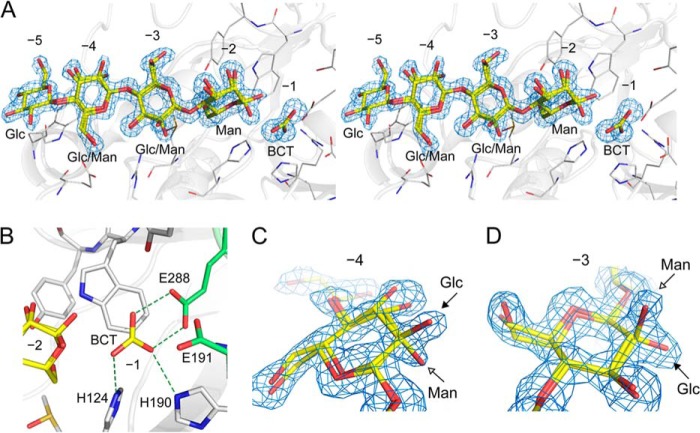
As shown from biochemical analysis, RsMan26C exhibited highest specific activity against glucomannan among all the Man-containing substrates (Table 2). To determine the mechanism by which RsMan26C recognizes and degrades this heteropolysaccharide, a complex crystal was prepared by soaking in the limit digest of glucomannan with the degree of polymerization ranging approximately from 1 to 6 (described in detail under “Experimental Procedures”). The complex structure was successfully determined via x-ray crystallography at 1.4 Å resolution. The ligand was a tetrasaccharide comprising a combination of Glc and Man (gluco-manno-oligosaccharide), and bound at subsites −5 to −2. The electron density map of the sugars at subsites −4 and −3 displayed both equatorial O-2 of Glc and axial O-2 of Man, forming double conformers of Glc/Man (Figs. 5, A–D). Because B-factor/temperature factor and occupancy are highly correlated in protein x-ray crystallography, it is difficult to precisely estimate the occupancy of atoms under ordinary resolution. The double conformers of Glc/Man at subsites −4 and −3 were refined on the presumption that occupancy of each epimer is 0.5. The temperature factor of O-2 of Glc/Man was estimated as follows: Glc/Man = 17.62/19.34 Å2 at subsite −4 and Glc/Man = 11.88/14.80 Å2 at subsite −3. This result indicates that comparable amounts of Glc and Man are bound to both subsites. Furthermore, Glc at subsite −5 and Man at subsite −2 were observed in the electron density map (Fig. 5A). The Man at subsite −2 was a mixture of α-Man and β-Man and refined with 0.5:0.5 occupancies.
FIGURE 5.
Structure of RsMan26C complexed with its ligand. A, stereo view of the 2|Fo| − |Fc| map (1.0σ) of the ligand at the active site. The RsMan26C structure is complexed with gluco-manno-oligosaccharide at subsites −5 to −2 and a bicarbonate ion (BCT) at subsite −1. B, interactions with a bicarbonate ion at subsite −1. A bicarbonate ion trapped at subsite −1 forms three hydrogen bonds with His124, His190, and the catalytic nucleophile, Glu288. C and D, enlarged view of the 2|Fo| − |Fc| map (1.0σ) of the ligand showing mixture of Glc and Man at subsites −4 (C) and −3 (D). Equatorial O-2 of Glc and axial O-2 of Man are indicated by arrows.
At subsite −1, a bicarbonate ion was trapped in the gluco-manno-oligosaccharide-bound form (Fig. 5A), whereas no ligand was observed in the native form. This ligand forms three hydrogen bonds with His124, His190, and the catalytic nucleophile, Glu288 (Fig. 5B), which suggests that the central carbon atom of the ligand should be surrounded by three oxygen atoms but not methyl groups. Because activity was not inhibited by adding bicarbonate ion into the reaction solution (data not shown), we assume that the bicarbonate ion is not an inhibitor of the enzyme, but accidentally trapped in the complex crystal structure. Detail of the interactions between the ligands and RsMan26C protein is summarized in Table 3.
TABLE 3.
Interactions between the ligands and RsMan26C protein
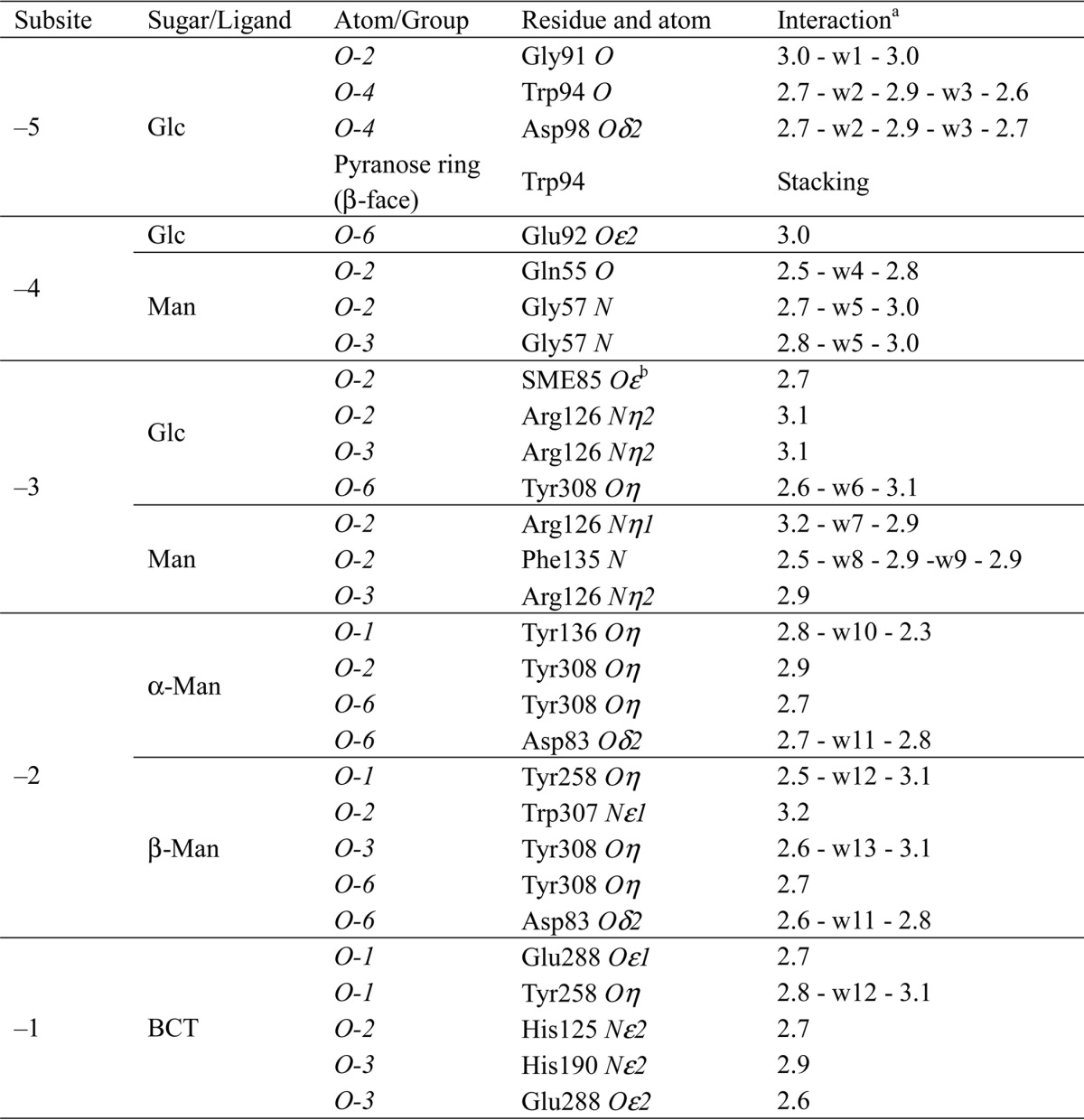
a Numbers indicate hydrogen bond distances from the ligand atom to the protein atom in Å. Mediating water molecules are designated by “w” with numbers.
b Oxidized methionine side chains are indicated.
Structural Comparison of RsMan26C with GH26 Family Enzymes
The structure of RsMan26C was compared with those of other GH26 enzymes using Dali server search. RsMan26C showed the highest similarity to Podospora anserina mannanase PaMan26A (29), followed by B. subtilis mannanase BsMan26A (Table 4) (19). Among the ligand-bound structures of GH26, RsMan26C was most similar to Cellvibrio japonicus mannanase CjMan26C (21) (Z-score, 32.6; root mean square deviation, 2.3 Å), followed by Cellulomonas fimi mannanase CfMan26A (16) (Z-score, 32.0; root mean square deviation, 2.4 Å). The overall structures of RsMan26C, CjMan26C, and CfMan26A were compared (Fig. 4, B and C). Most α-helices and β-strands could be superimposed, whereas the loop regions had some structural deviations. The topology of the active center cleft of RsMan26C (Fig. 6A) was compared with those of CjMan26C and CfMan26A (Fig. 6, B and C). The active cleft of RsMan26C displays open ends, which confer endo activity and allow binding to the internal chain of hemicellulose. An extended loop (Leu82–Lys103) forms the distal negative subsites and contains an aromatic residue (Trp94) at subsite −5 (described in detail below). The equivalent loop (Gly76–Thr86) of CfMan26A extends outside the active cleft and forms long negative subsites (Fig. 6C). In contrast, the active cleft of CjMan26C is restricted at one end by an extended loop, which renders unique exomannobiohydrolase activity (Fig. 6B). The equivalent loop is not conserved in the RsMan26C and CfMan26A structures.
TABLE 4.
Structural comparisons with GH26 β-mannanases by Dali server
r.m.s.d., root mean square deviation.
| Enzyme | Ligand | Z score | r.m.s.d. (Å) (no. of residues) | PDB code | Ref. |
|---|---|---|---|---|---|
| P. anserina PaMan26A | 41.6 | 1.6 (300) | 3ZM8 | 29 | |
| B. subtilis BsMan26A | 34.2 | 2.3 (292) | 3CBW (2WHK) | 19 | |
| B. subtilis BCman | 34.0 | 2.3 (291) | 2QHA | 38 | |
| C. japonicus CjMan26C | Gal1Man4 | 32.7 | 2.2 (291) | 2VX6 | 21 |
| C. fimi CfMan26A | Mannotriose | 32.2 | 2.4 (296) | 2BVT | 16 |
FIGURE 6.
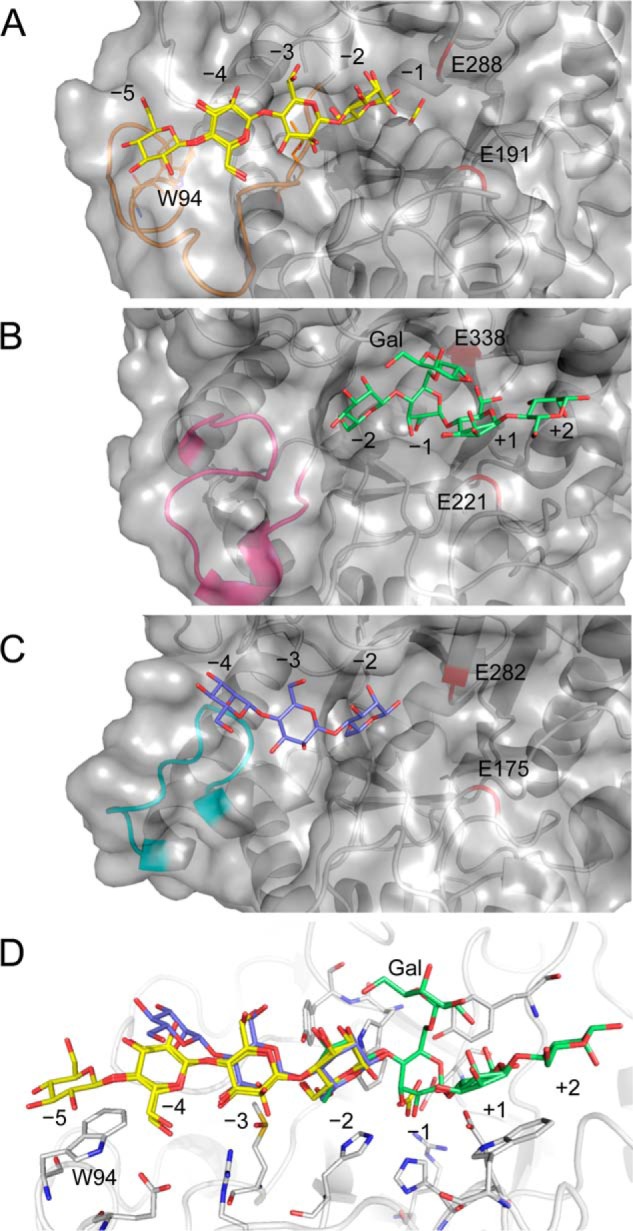
Surface view of the active center cleft of GH26 β-mannanases. A, active cleft of RsMan26C complexed with gluco-manno-oligosaccharide (yellow). An extended loop (Leu82–Lys103) forming the distal negative subsite is colored orange. B, active cleft of CjMan26C (PDB code 2VX6 (21)) complexed with Gal1Man4 (green). An extended loop (Leu118–Asp133) surrounding subsite −2 is colored magenta. C, active cleft of CfMan26A (PDB code 2BVT (16)) complexed with mannotriose (blue). An extended loop (Gly76–Thr86) forming the distal negative subsite is colored cyan. Catalytic acid/base (Glu191, Glu221, and Glu175 in RsMan26C, CjMan26C, and CfMan26A, respectively) and nucleophile (Glu288, Glu338, and Glu282 in RsMan26C, CjMan26C, and CfMan26A, respectively) are shown in red. D, active center of RsMan26C complexed with gluco-manno-oligosaccharide (yellow) is superimposed with Gal1Man4 (green) from the CjMan26C structure and mannotriose (blue) from the CfMan26A structure.
Substrate Recognition in the Active Site of RsMan26C
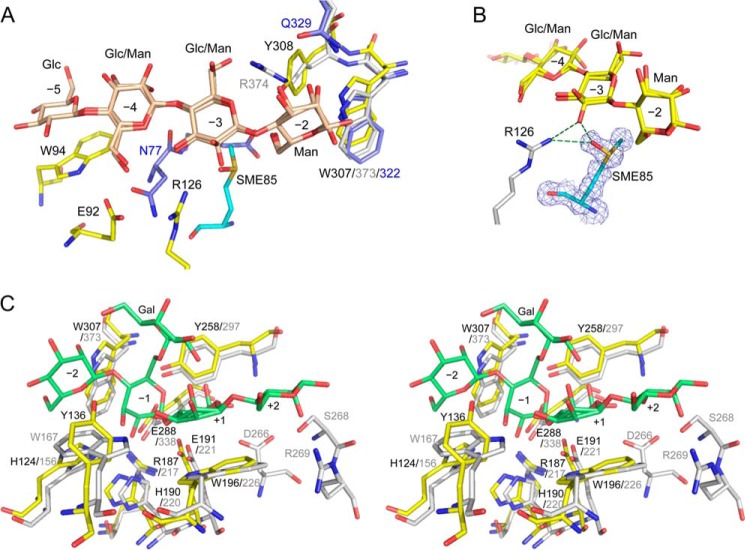
In the RsMan26C complex structure, the Glc moiety binds to subsite −5. Trp94 generates a hydrophobic platform by stacking against the sugar ring. This tryptophan residue is specific for RsMan26C, but not conserved among other GH26 mannanases, e.g. CjMan26C (21) and CfMan26A (16). In the case of PaMan26A (29), the corresponding Trp is also absent at subsite −5, whereas two aromatic residues, Trp244 and Trp245, exist at subsite −4, proposed to stabilize the mannopyranose ring in this region. Notably, subsite −5 of RsMan26C only interacts with Glc and not Man, in contrast to subsites −4 and −3, which are bound to both Man and Glc (see below). The equatorial O-2 of Glc forms a water-mediated hydrogen bond in RsMan26C. Owing to the absence of steric hindrance or interactions with the axial O-2 of Man around subsite −5, this region may accommodate both Man and Glc.
As shown in the electron density map, a mixture of Glc and Man was clearly observed at subsite −4 (Fig. 5C). There are no polar interactions with the equatorial O-2 of Glc but the axial O-2 of Man forms two water-mediated hydrogen bonds in RsMan26C. In CfMan26A, Asn77 forms a hydrogen bond with O-2 of Man (Fig. 7A). This finding suggests that subsite −4 of RsMan26C can accommodate both Glc and Man. In addition, Glu92 forms a hydrogen bond with O-6 of the sugar. The residues corresponding to Trp244 and Trp245 at subsite −4 in PaMan26A are absent in RsMan26C.
FIGURE 7.
Active site structures of RsMan26C. A, the active center structure of RsMan26C (yellow) complexed with gluco-manno-oligosaccharide (beige) at subsites −5 to −2 superimposed with those of CjMan26C (PDB code 2VX6, gray (21)) and CfMan26A (PDB code 2BVT, blue (16)). SME85 is colored cyan. B, the 2|Fo| − |Fc| map (1.0σ) of SME85 (green) at subsite −3. C, the active site structure of RsMan26C (yellow) is superimposed with that of CjMan26C (gray) complexed with Gal1Man4 (green) at subsites −2 to +2 in the stereo view.
In addition to subsite −4, we observed specific recognition of Glc at subsite −3 to which a mixture of Glc and Man was bound (Fig. 5D). The electron density map revealed that the sulfur atom of Met85 is oxidized, forming a methionine sulfoxide (SME) in both the native and complex structures (Fig. 7, A and B). This oxidized Met85 (termed “SME85”) specifically interacts with the equatorial O-2 of Glc. The distance between the oxygen atom of the sulfoxide group of SME85 and equatorial O-2 is 2.7 Å. To our knowledge, this is the first case among the protein structures determined so far demonstrating that an oxidized methionine side chain interacts specifically with the substrate. No other residues, including methionine, were oxidized, as evident from the electron density map. Another possibility is that Met85 displays an alternative side chain conformation. However, the distance between the side chain of Met85 (here, we presumed methionine sulfoxide) and amino group of Arg126 is 2.9 Å, indicating that these two molecules interact via a hydrogen bond. This structural feature strongly suggests that the amino group of Arg126 should connect to the oxygen of SME85 but not the methyl side chain of the Met85 alternative conformation. The equatorial O-2 of Glc also displays a polar interaction with Arg126 of RsMan26C. This arginine residue additionally interacts with O-3 of the sugar. The structural equivalents of SME85 and Arg126 in RsMan26C are present in PaMan26A as non-oxidized Met195 and Arg201, but are not widely conserved in other GH26 enzymes. In RsMan26C, two water-mediated hydrogen bonds were observed with the axial O-2 of Man, suggesting that Man can be also accommodated at subsite −3.
Man at subsite −2 of CjMan26C was shown to be tightly recognized by Arg374 and Gln385, equivalent to Gln329 in CfMan26A (Fig. 7A). The structural equivalent in RsMan26C is Tyr308, which makes direct hydrogen bonds with the axial O-2 of Man. Electron density of Glc was not observed at subsite −2 of RsMan26C, suggesting specificity for the Man unit. Man at subsite −2 also displays several interactions with the enzyme. Nϵ1 of Trp307 forms a hydrogen bond with the axial O-2 of Man. Trp307 is conserved in CjMan26C (Trp373), CfMan26A (Trp322), and other GH26 structures. In addition, Tyr308 forms a direct hydrogen bond with O-6 of Man and occupy the extra space for α-1,6-linked galactosyl decoration.
No sugar binding was observed at subsite −1 and the positive subsites of RsMan26C. To determine substrate recognition at these subsites, the RsMan26C structure was superimposed with CjMan26C complexed with 6163-Gal-Man4 at subsites −2 to +2 (Fig. 7C). Superimposition of the −1 subsite showed that almost all residues surrounding Man, which forms the B2,5 transition state described as typical for the GH26 family, are highly conserved. In RsMan26C, the catalytic acid/base, Glu191, and catalytic nucleophile, Glu288, are located at the C termini of β-strands 4 and 7, respectively, in common with clan GH-A enzymes, e.g. CjMan26C (Glu221 and Glu338). Other interactions between Man and the enzyme are also invariant in RsMan26C, specifically, His124 interacting with O-3, His190 interacting with O-2, and Trp307 making a hydrophobic contact with the pyranose ring. The equivalents in CjMan26C are His156, His220, and Trp373, respectively. In addition, Arg187 and Tyr258 stabilize the position of the catalytic nucleophile, Glu288, whereas Trp196 stabilizes the catalytic acid/base, Glu191, equivalent to Arg217, Tyr297, and Trp226 in CjMan26C, respectively.
Superimposition of the positive subsites of RsMan26C with those of CjMan26C revealed a functional subsite +1 and non-functional subsite +2. Trp196 forms a stacking interaction with the sugar ring at subsite +1 in RsMan26C equivalent to Trp226 in CjMan26C. In contrast to subsite +1, RsMan26C is likely to make poor contact with the substrate at subsite +2. However, CjMan26C displays several interactions with the substrate. Specifically, Ser268 interacts with O-2 and Arg269 with endocyclic oxygen and O-2, which are not conserved in RsMan26C.
DISCUSSION
This study focuses on how mannanase from a symbiotic protist of termite recognizes and accommodates heteromannan composed of Glc and Man. The GH26 mannanases characterized to date generally display tight Man specificity at both subsites −2 and −1, whereas GH5 mannanases show relaxed specificity for Glc or Man at subsite −2 (19). Considering their catalytic specificity for Man, it is proposed that GH26 mannanases prefer attacking homopolymers of Man. Indeed, the RsMan26C complex structure with gluco-manno-oligosaccharide revealed that the axial O-2 of Man is specifically recognized by Tyr308 at subsite −2. Man-specific recognition at subsite −1 may also be conserved in RsMan26C in addition to other GH26 mannanases because the catalytic apparatus at this site is highly conserved. In contrast, the gluco-manno-oligosaccharide complex structure reveals unique Glc-specific recognition at subsite −3 and accommodation at subsites −5 and −4. The oxidized Met85 (described below in detail) and Arg126 at subsite −3 make direct hydrogen bonds with equatorial O-2 of Glc, whereas water-mediated hydrogen bonds with the axial O-2 of Man are observed.
Binding properties of RsMan26C were further clarified with an electron density map of gluco-manno-oligosaccharide in the active cleft. The almost equal occupancies of Glc and Man at both subsites −4 and −3 reflect the ratio of Glc and Man (∼40:60) in the glucomannan substrate (from Konjac). Interestingly, subsite −5 binds to Glc, but not Man. The sequence of the bound oligosaccharide could also reflect the sequence frequency of Glc and Man in konjac glucomannan in addition to subsite specificity of the enzyme. In summary, our structural analysis suggests subsite specificity and accommodation of Glc/Man in the active site of RsMan26C as follows: (Glc(/Man))-(Glc/Man)-(Glc/Man)-Man-Man from subsites −5 to −1, respectively.
Biochemical data on RsMan26C demonstrate that this mannanase efficiently attacks heteropolysaccharides, especially glucomannan. Recently, some GH26 mannanases as well as RsMan26C were reported to display higher activity on glucomannan than pure mannan, although GH26 mannanases are known to specifically select Man at both subsites −2 and −1. The P. anserina mannanase, PaMan26A, exhibited the highest specificity toward konjac glucomannan among the structurally different mannan polysaccharides, including galactomannan, glucomannan, and β-1,4-mannan (30). Kulcinskaja and co-workers (31) reported that the presence of backbone Glc in konjac glucomannan possibly does not restrict the activity of Bifidobacterium adolescentis mannanase, BaMan26A. These features were consistent for CjMan26A from C. japonicus (21), ManB from Bacillus licheniformis (32), and BsMan26A from B. subtilis (19) in terms of catalytic efficiency, whereas CjMan26C from C. japonicus (21) was less active on glucomannan than β-mannan, indicating significant variations in substrate specificity within members of the GH26 family. Together with the substrate binding property of RsMan26C, we propose that the distal negative subsites (−5 to −3) play a key role in recognition and accommodation of the backbone Glc and potentiate catalytic efficiency toward glucomannan.
The crystal structure of RsMan26C revealed that the unique oxidized methionine at position 85 contributes to specific recognition of the substrate. Oxidative modification of methionine has been observed in several protein structures solved to date, e.g. structural analysis of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 suggests that oxidation of the methionine adjacent to the catalytic serine alters the substrate specificities of the enzyme (33). However, to our knowledge, direct substrate recognition by methionine sulfoxide has not been described previously. In the RsMan26C structure, the sulfoxide oxygen of SME85 besides Arg126 displays a specific polar interaction with O-2 of Glc at subsite −3. This oxidation of methionine may have occurred during heterologous production of the recombinant protein in P. pastoris, which is fermented in highly aerated culture conditions. Intriguingly, however, no residues are oxidized other than SME85. It is unclear whether this methionine is oxidized in vivo. RsMan26C is a symbiotic protist living in the anaerobic environment of the termite gut, although the presence of steep gradients of oxygen in the termite gut has been also reported (34). Notably, equivalent residues of Met85 are highly conserved in six other GH26 mannanase sequences identified together with RsMan26C in the cDNA library of the symbiotic protist community of R. speratus (11). Therefore, the issue of whether Met85 oxidation is an experimental artifact or a biologically functional modification contributing to specific substrate recognition remains controversial.
The dual digestion system of termites and their symbiotic protists is essential for the efficient degradation of lignocellulose in lower termites, such as R. speratus. Symbiotic protists in the termite gut take up wood particles or lignocellulosic fragments partially digested by endogenous termite cellulases into food vacuoles (8) in which cellulolytic enzymes of the protists are secreted (35). Because glucomannan is the major hemicellulose component of the coniferous cell wall, RsMan26C is likely to target glucomannan and not mannan as its natural substrate. This theory is supported by the topological features of the substrate-binding cleft of mannanase. RsMan26C displays a long open groove harboring a unique hydrophobic platform (Trp94) at subsite −5, which may facilitate enzyme binding to polysaccharides, rather than oligosaccharides. Furthermore, specific recognition and accommodation of Glc by RsMan26C is observed in the three of the distal negative subsites (−5 to −3), which may be optimized for utilizing glucomannan as the preferred substrate. Notably, no cellulases isolated from protists possess a carbohydrate-binding module, regardless of glycoside hydrolase classification, including RsMan26C (36). One of the general roles of the carbohydrate-binding module is to enhance the concentration of enzyme molecules on the substrate surface (37). However, RsMan26C would be secreted at high concentrations around the substrate within food vacuoles as well as other cellulolytic enzymes of protists (36), which compensates for the lack of carbohydrate-binding module and facilitates adsorption of the enzymes on the surface of polysaccharides. Our results clearly imply that RsMan26C preferentially targets heteropolysaccharides, particularly glucomannan, in wood components.
In conclusion, this study provides valuable insights into how mannanase from a symbiotic protist of the termite recognizes and binds to glucomannan heteropolysaccharides consisting of Glc and Man. Although Man specificity is suggested at both subsites −2 and −1 in RsMan26C, specific recognition and accommodation of Glc at the distal negative subsites provides high substrate specificity for glucomannan. These findings indicate that both subsites proximal to the catalytic center and distal binding sites are important to define the substrate specificity of mannanase and may be effectively applied to construct an efficient degradation and utilization system of lignocellulose, which constitutes a highly complex network of diverse polysaccharides.
Acknowledgments
We thank the staff of the Photon Factory for the x-ray data collection.
This work was supported in part by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN) (to M. A.) and by a Grant-in-Aid for Innovative Areas 24114001 and 24114008 (to K. I.) from the Japanese Ministry of Education, Culture, Sports, and Technology (MEXT).
The atomic coordinates and structure factors (codes 3WDQ and 3WDR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Man
- mannose
- Glc
- glucose
- RsMan26C
- GH26 β-mannanase C from a symbiotic protist of Reticulitermes speratus
- PDB
- Protein Data Bank
- SME
- methionine sulfoxide.
REFERENCES
- 1. Cameron S. L., Whiting M. F. (2007) Mitochondrial genomic comparisons of the subterranean termites from the genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae). Genome 50, 188–202 [DOI] [PubMed] [Google Scholar]
- 2. Scheller H. V., Ulvskov P. (2010) Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289 [DOI] [PubMed] [Google Scholar]
- 3. Ohkuma M. (2003) Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl. Microbiol. Biotechnol. 61, 1–9 [DOI] [PubMed] [Google Scholar]
- 4. Watanabe H., Tokuda G. (2001) Animal cellulases. Cell Mol. Life Sci. 58, 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Watanabe H., Tokuda G. (2010) Cellulolytic systems in insects. Annu. Rev. Entomol. 55, 609–632 [DOI] [PubMed] [Google Scholar]
- 6. Hungate R. E. (1938) Studies on the nutrition of zootermopsis II: The relative Importance of the termite and the protozoa in wood digestion. Ecology 19, 1–25 [Google Scholar]
- 7. Sims R. E., Mabee W., Saddler J. N., Taylor M. (2010) An overview of second generation biofuel technologies. Bioresour. Technol. 101, 1570–1580 [DOI] [PubMed] [Google Scholar]
- 8. Yamaoka I., Nagatani Y. (1977) Cellulose digestion system in the termite, Reticulitermes speratus (Kolbe). II. Ultra-structural changes related to the ingestion and digestion of cellulose by the flagellate, Trichonympha agilis. Zool. Mag. 86, 34–42 [Google Scholar]
- 9. Breznak J. A. (1994) Acetogenesis from carbon dioxide in termite guts in Acetogenesis, pp. 551–565, Springer US, New York [Google Scholar]
- 10. Hongoh Y. (2011) Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol. Life Sci. 68, 1311–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Todaka N., Moriya S., Saita K., Hondo T., Kiuchi I., Takasu H., Ohkuma M., Piero C., Hayashizaki Y., Kudo T. (2007) Environmental cDNA analysis of the genes involved in lignocellulose digestion in the symbiotic protist community of Reticulitermes speratus. FEMS Microbiol. Ecol. 59, 592–599 [DOI] [PubMed] [Google Scholar]
- 12. Moreira L. R., Filho E. X. (2008) An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79, 165–178 [DOI] [PubMed] [Google Scholar]
- 13. Pawar P. M., Koutaniemi S., Tenkanen M., Mellerowicz E. J. (2013) Acetylation of woody lignocellulose: significance and regulation. Front. Plant Sci. 4, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teleman A., Nordström M., Tenkanen M., Jacobs A., Dahlman O. (2003) Isolation and characterization of O-acetylated glucomannans from aspen and birch wood. Carbohydr. Res. 338, 525–534 [DOI] [PubMed] [Google Scholar]
- 15. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le Nours J., Anderson L., Stoll D., Stålbrand H., Lo Leggio L. (2005) The structure and characterization of a modular endo-β-1,4-mannanase from Cellulomonas fimi. Biochemistry 44, 12700–12708 [DOI] [PubMed] [Google Scholar]
- 17. Hogg D., Woo E. J., Bolam D. N., McKie V. A., Gilbert H. J., Pickersgill R. W. (2001) Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding. J. Biol. Chem. 276, 31186–31192 [DOI] [PubMed] [Google Scholar]
- 18. Han Y., Dodd D., Hespen C. W., Ohene-Adjei S., Schroeder C. M., Mackie R. I., Cann I. K. (2010) Comparative analyses of two thermophilic enzymes exhibiting both β-1,4 mannosidic and β-1,4 glucosidic cleavage activities from Caldanaerobius polysaccharolyticus. J. Bacteriol. 192, 4111–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tailford L. E., Ducros V. M., Flint J. E., Roberts S. M., Morland C., Zechel D. L., Smith N., Bjørnvad M. E., Borchert T. V., Wilson K. S., Davies G. J., Gilbert H. J. (2009) Understanding how diverse β-mannanases recognize heterogeneous substrates. Biochemistry 48, 7009–7018 [DOI] [PubMed] [Google Scholar]
- 20. Ducros V. M., Zechel D. L., Murshudov G. N., Gilbert H. J., Szabó L., Stoll D., Withers S. G., Davies G. J. (2002) Substrate distortion by a β-mannanase: snapshots of the Michaelis and covalent-intermediate complexes suggest a B(2,5) conformation for the transition state. Angew. Chem. Int. Ed Engl. 41, 2824–2827 [DOI] [PubMed] [Google Scholar]
- 21. Cartmell A., Topakas E., Ducros V. M., Suits M. D., Davies G. J., Gilbert H. J. (2008) The Cellvibrio japonicus mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J. Biol. Chem. 283, 34403–34413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uchima C. A., Arioka M. (2012) Expression and one-step purification of recombinant proteins using an alternative episomal vector for the expression of N-tagged heterologous proteins in Pichia pastoris. Biosci. Biotechnol. Biochem. 76, 368–371 [DOI] [PubMed] [Google Scholar]
- 23. Jue C. K., Lipke P. N. (1985) Determination of reducing sugars in the nanomole range with tetrazolium blue. J. Biochem. Biophys. Methods 11, 109–115 [DOI] [PubMed] [Google Scholar]
- 24. Inoue T., Murashima K., Azuma J., Sugimoto A., Slaytor M. (1997) Cellulose and xylan utilisation in the lower termite Reticulitermes speratus. J. Insect Physiol. 43, 235–242 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Couturier M., Roussel A., Rosengren A., Leone P., Stålbrand H., Berrin J. G. (2013) Structural and biochemical analyses of glycoside hydrolase families 5 and 26 β-(1,4)-mannanases from Podospora anserina reveal differences upon manno-oligosaccharide catalysis. J. Biol. Chem. 288, 14624–14635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Couturier M., Haon M., Coutinho P. M., Henrissat B., Lesage-Meessen L., Berrin J. G. (2011) Podospora anserina hemicellulases potentiate the Trichoderma reesei secretome for saccharification of lignocellulosic biomass. Appl. Environ. Microbiol. 77, 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kulcinskaja E., Rosengren A., Ibrahim R., Kolenová K., Stålbrand H. (2013) Expression and characterization of a bifidobacterium adolescentis β-mannanase carrying mannan-binding and cell association motifs. Appl. Environ. Microbiol. 79, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Songsiriritthigul C., Buranabanyat B., Haltrich D., Yamabhai M. (2010) Efficient recombinant expression and secretion of a thermostable GH26 mannan endo-1,4-β-mannosidase from Bacillus licheniformis in Escherichia coli. Microb. Cell Fact. 9, 20–2859- 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nonaka T., Fujihashi M., Kita A., Saeki K., Ito S., Horikoshi K., Miki K. (2004) The crystal structure of an oxidatively stable subtilisin-like alkaline serine protease, KP-43, with a C-terminal β-barrel domain. J. Biol. Chem. 279, 47344–47351 [DOI] [PubMed] [Google Scholar]
- 34. Brune A., Emerson D., Breznak J. A. (1995) The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61, 2681–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamaoka I., Nagatani Y. (1975) Cellulose digestion system in the termite, Reticulitermes speratus (Kolbe). I. Producing sites and physiological significance of two kinds of cellulase in the worker. Zool. Mag. 84, 23–29 [Google Scholar]
- 36. Inoue T., Moriya S., Ohkuma M., Kudo T. (2005) Molecular cloning and characterization of a cellulase gene from a symbiotic protist of the lower termite, Coptotermes formosanus. Gene 349, 67–75 [DOI] [PubMed] [Google Scholar]
- 37. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan X. X., An X. M., Gui L. L., Liang D. C. (2008) From structure to function: insights into the catalytic substrate specificity and thermostability displayed by Bacillus subtilis mannanase BCman. J. Mol. Biol. 379, 535–544 [DOI] [PubMed] [Google Scholar]