Abstract
Mitochondrial transcription factor A (TFAM) is one of the key regulators of the transcription of mtDNA. In diabetes, despite increase in gene transcripts of TFAM, its protein levels in the mitochondria are decreased and mitochondria copy numbers become subnormal. The aim of this study is to investigate the mechanism(s) responsible for decreased mitochondrial TFAM in diabetes. Using retinal endothelial cells, we have investigated the effect of overexpression of cytosolic chaperone, Hsp70, and TFAM on glucose-induced decrease in mitochondrial TFAM levels, and the transcription of mtDNA-encoded genes, NADH dehydrogenase subunit 6 (ND6) and cytochrome b (Cytb). To investigate the role of posttranslational modifications in subnormal mitochondrial TFAM, ubiquitination of TFAM was accessed, and the results were confirmed in the retina from streptozotocin-induced diabetic rats. While overexpression of Hsp70 failed to prevent glucose-induced decrease in mitochondrial TFAM and transcripts of ND6 and Cytb, overexpression of TFAM ameliorated decrease in its mitochondrial protein levels and transcriptional activity. TFAM was ubiquitinated by high glucose, and PYR-41, an inhibitor of ubiquitination, prevented TFAM ubiquitination and restored the transcriptional activity. Similarly, TFAM was ubiquitinated in the retina from diabetic rats, and it continued to be modified after reinstitution of normal glycemia. Our results clearly imply that the ubiquitination of TFAM impedes its transport to the mitochondria resulting in subnormal mtDNA transcription and mitochondria dysfunction, and inhibition of ubiquitination restores mitochondrial homeostasis. Reversal of hyperglycemia does not provide any benefit to TFAM ubiquitination. Thus, strategies targeting posttranslational modification could provide an avenue to preserve mitochondrial homeostasis, and inhibit the development/progression of diabetic retinopathy.
Keywords: diabetic retinopathy, mitochondria, mitochondrial transcription factor A, mtDNA, posttranslational modifications, ubiquitination
1. Introduction
In diabetes, retinal mitochondria become dysfunctional, their biogenesis is compromised, DNA (mtDNA) is damaged, and the proteins encoded by mtDNA are decreased (Madsen-Bouterse, Mohammad et al. 2010; Santos and Kowluru 2011; Santos, Mohammad et al. 2011; Santos, Tewari et al. 2011). Since mtDNA encodes proteins which are important in the electron transport chain, this transport system becomes dysfunctional resulting in increased superoxide levels, and activation of the apoptosis machinery (Madsen-Bouterse, Zhong et al. 2010; Madsen-Bouterse, Mohammad et al. 2010; Santos, Tewari et al. 2011). Apoptosis of retinal cells, including capillary cells, is accelerated, and capillaries begin to degenerate resulting in the histopathology which is the hallmark of diabetic retinopathy (Kern, Tang et al. 2000; Frank 2004).
Mitochondria are equipped with a small circular DNA which transcribes only 13 proteins (Scarpulla 2006; Scarpulla 2008), and the majority of proteins required for mitochondria biogenesis and ATP generation are encoded by nuclear DNA. Nuclear encoded proteins required for mitochondria homeostasis are synthesized in the cytosol and transported to the mitochondria via chaperons, such as heat shock protein 70 (Hsp70), and into the mitochondria via mitochondrial membrane transporters (Scarpulla 2006; Scarpulla 2008; Schmidt, Pfanner et al. 2010). In diabetes, the association of cytosolic Hsp70 with mitochondrial transcription factor A (TFAM) is decreased, and that with matrix metalloproteinase-9 (MMP-9), a protein associated with damage of mitochondria and capillary cell apoptosis, is increased in the retina (Scarpulla 2008; Kowluru, Mohammad et al. 2011; Santos and Kowluru 2011; Santos, Tewari et al. 2011).
TFAM is one of the key factors with many important roles; it binds to the regulatory area of the mtDNA, the displacement loop (D-loop), to initiate the transcription of the proteins encoded by mtDNA (Scarpulla 2006; Scarpulla 2008). In diabetes, although the gene expression of TFAM is increased in the retina, its total protein levels remain unchanged, but mitochondrial protein levels are decreased (Santos and Kowluru 2011; Santos, Tewari et al. 2011). In addition, diabetes also compromises mitochondria membrane transport systems in the retina (Zhong and Kowluru 2011). Diabetic environment favors posttranslational modifications, and these modifications are considered to play important role in its complications (Goldberg, Whiteside et al. 2006; Reddy and Natarajan 2011; Zhong and Kowluru 2011; Song, Peng et al. 2013; Zhong and Kowluru 2013). Posttranslational modifications, in addition to increasing the functional diversity of the proteome and facilitating proteolytic cleavage of the entire proteins, can also alter the localization and interaction of proteins with other cellular molecules (Piantadosi and Suliman 2006; Bergink and Jentsch 2009; Madsen-Bouterse, Mohammad et al. 2010). Ubiquitination of mitochondrial proteins is shown to be vital for the maintenance of mitochondrial homeostasis by removing the dysfunctional mitochondria (Neutzner, Benard et al. 2008). The role of posttranslational modification in the impaired translocation of TFAM into the mitochondria in diabetes remains to be investigated.
Epidemiologic and experimental studies have demonstrated that prior exposure to hyperglycemia has long-lasting consequences, and diabetic retinopathy continues to progress even after the hyperglycemic insult is terminated, suggesting a ‘metabolic memory’ phenomenon (Engerman and Kern 1987; Diabetes Control and Complications Trial Research Group 1993; Diabetes Control and Complications Trial Research Group 1998; Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group 2000; Kowluru 2003). Mitochondria remain dysfunctional, and mtDNA continues to be damaged with impaired biogenesis and subnormal protein levels of TFAM in the mitochondria even after re-institution of normal glycemic control in diabetic rats (Madsen-Bouterse, Mohammad et al. 2010; Santos and Kowluru 2011). Furthermore, we have shown that the retinal proteins continue to be posttranslationally modified (Kanwar and Kowluru 2009; Madsen-Bouterse, Mohammad et al. 2010). The role of posttranslational modification of TFAM in the metabolic memory phenomenon is not understood.
The aim of this study is to investigate the mechanism(s) responsible for decreased TFAM protein levels in the retinal mitochondria in diabetes. Using retinal endothelial cells, the cells associated with histopathology of diabetic retinopathy, we have investigated the effect of posttranslational modifications of TFAM on mtDNA transcription, and confirmed the in vitro results in the retina from rodent model of diabetic retinopathy. The role of posttranslational modification of TFAM in the metabolic memory phenomenon was also investigated, both in vivo and in vitro models of diabetic retinopathy.
2. Methods
2.1 Retinal endothelial cells
Retinal endothelial cells isolated from bovine eyes (BRECs) were cultured on polystyrene culture plates coated with 0.1% gelatin in a humidified incubator at 37°C in an atmosphere of 5% CO2 and 95% air, as routinely performed in our laboratory (Kowluru and Abbas 2003; Madsen-Bouterse, Mohammad et al. 2010; Santos and Kowluru 2011; Tewari, Zhong et al. 2012). The cells from 4th-6th passage were incubated in Dulbecco's modified Eagle medium (DMEM) containing 2% heat-inactivated fetal bovine serum, 10% Nu serum, 50μg/ml heparin, 1μg/ml endothelial growth factor, and antibiotic/antimycotic, supplemented with 5 or 20mM glucose for 4 days. To evaluate the effect of inhibition of ubiquitination on mitochondrial transcription, the cells were pre-incubated with 5μM PYR-41 (Sigma Aldrich, St. Louis, MO) for 4 hours (Guan and Ricciardi 2012) before incubating in 5mM glucose or 20mM glucose for 4 days. Each experiment included an osmotic control in which the cells were incubated with 20mM mannitol instead of 20mM glucose.
To investigate the effect of overexpression of cytosolic Hsp70, and that of increasing overall TFAM, on glucose-induced decrease in mtDNA transcription, these proteins were overexpressed using 2μg/ml HSPA1A (gene that encodes the cytosolic HSP70) or GFP-tagged TFAM plasmid with TurboFectin 8.0 from OriGene Technologies (Rockville, MD). After transfection, cells were rinsed with DMEM, and incubated in 5mM or 20mM glucose media for 4 days. In parallel, incubation with only the transfection reagent was carried out (Mock). The transfection efficiency was verified by microscopy by assessing the red fluorescence with mouse monoclonal antibody Hsp70 (Santa Cruz Biotechnology, Santa Cruz, CA) followed by Image J software quantification, or by quantifying green fluorescence of GFP-tagged TFAM. Cells were washed with PBS mounted with Vecta Shield containing DAPI (Vector Laboratories Burlingame, CA) and examined under a Zeiss ApoTome using 40X magnification (Carl Zeiss Inc.) (Tewari, Santos et al. 2012; Santos and Kowluru 2013).
To examine the effect of reversal of high glucose insult on posttranslational modifications of TFAM, the cells from 4th-6th passage were incubated in 20mM glucose for 4 days followed by 5mM glucose for 4 additional days (20-5). Parallel controls included cells incubated in continuous 5mM glucose or in 20mM glucose for the entire duration of the experiment. The cells received fresh media every 48 hours. In the 20-5 group, at the end of the initial 4 days of 20mM glucose, the cells were rinsed with DMEM before changing to 5mM glucose medium (Zhong and Kowluru 2011; Zhong and Kowluru 2013).
2.2 Rats
Wistar rats (male, body weight 200g) were randomly assigned into two groups: normal or streptozotocin-induced diabetic (55 mg/kg body weight). Diabetic rats were either allowed to remain in poor glycemic control for 8 months (PC); in PC for 4 months, followed by good glycemic control for 4 additional months (Rev) or in good glycemic control for 8 months (GC).The rats in poor glycemic control received 1-2 IU insulin 4-5 times a week to prevent ketosis and weight loss and the rats in which good glycemic control received insulin twice daily (5-7 IU/day) to maintain a steady gain in body weight and blood glucose values below 150 mg/dl. These procedures are routinely performed in our laboratory (Kowluru 2003; Santos and Kowluru 2011; Zhong and Kowluru 2011; Santos and Kowluru 2013). Each group had 10 or more rats, and at the end of the desired experimental duration, the animals were euthanized by CO2 inhalation, and the retina were immediately isolated. Treatment of animals conformed to the Association for Research in Vision and Ophthalmology's Resolution on Treatment of Animals in Research (National Institutes of Health) and the Institutional guidelines.
2.3 Sample preparation
Mitochondria and cytosol were isolated from retina using mitochondria isolation kit from Invitrogen (Carlsbad, CA, USA), according to the vendor's protocols. As reported previously, mitochondria prepared by this method are largely devoid of contaminations (Madsen-Bouterse, Mohammad et al. 2010; Santos, Tewari et al. 2011; Tewari, Santos et al. 2012). To isolate the cytosol fraction, the homogenate was centrifuged at 105,000×g for 90 minutes. Protein was estimated by the bicinchoninic acid protein assay (Sigma-Aldrich, St Louis, MO).
2.4 Protein expression
Protein (20-60 g cytosol, mitochondrial) was separated on an 8–19% SDS–PAGE, transferred to a nitrocellulose membrane, and blocked with 5% non-fat milk for 1 hour. The membranes were incubated with antibodies against TFAM (Santa Cruz Biotechnology, Santa Cruz-CA) or Hsp70 (Abcam, Cambridge, MA) overnight at 4°C. β-actin (Sigma-Aldrich) was used as a loading control for cytosolic fraction, and Cox IV (Santa Cruz Biotechnology) for the mitochondria fraction.
2.5 Gene expression
Total RNA was extracted from BRECs with Trizol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). mRNA levels of Hsp70 and mtDNA encoded protein NADH dehydrogenase 1 and 6 (ND1 and ND6) were quantified by real-time quantitative RT-PCR (qPCR) using the SYBR green assay using the conditions routinely employed in our laboratory (Santos and Kowluru 2011; Santos, Tewari et al. 2011; Santos, Tewari et al. 2013). Due to some concerns about proper amplification, gene transcripts of Cytochrome oxidase b (Cytb) were quantified by conventional qPCR using the conditions previously reported by us (Madsen-Bouterse, Mohammad et al. 2010). Primer list is presented in Table I. Relative amplification was quantified by normalizing the gene-specific amplification to that of β-actin in each sample. Changes in mRNA abundance were calculated using the ΔΔCt, and for Cytb the products were analyzed on an agarose gel and the ratio Cytb and β-actin was calculated as reported by us previously (Madsen-Bouterse, Mohammad et al. 2010).
Table I.
Primer for the target genes
| BRECs | Product size | ||
|---|---|---|---|
| Hsp70 | forward | 5’-TGGACAGGACTCCCCCACTGGA-3′ | 227bp |
| reverse | 5’-TGGCGCGGGCATTTCCTCTC-3’ | ||
| ND1 | forward | 5’-AGGACCATTTGCCCTCTTCT-3’ | 297bp |
| reverse | 5’-GGTGGGATGCCTGATGTAAG-3’ | ||
| ND6 | forward | 5’-CGTGATAGGTTTTGTGGGGT-3’ | 250bp |
| reverse | 5’-GCCAGTAACAAATGCCCCTA-3’ | ||
| Cytb | forward | 5’-CGATACATACACGCAAACGG-3’ | 298bp |
| reverse | 5’-AGAATCGGGTAAGGGTTGCT-3’ | ||
| β-actin | forward | 5’-CCTCTATGCCAACACAGTGC-3’ | 205bp |
| reverse | 5’-CATCGTACTCCTGCTTGCTG-3’ | ||
| Rat | |||
| Hsp70 | forward | 5’-TCCTCGTGGACGGTCGTCCG-3’ | 101bp |
| Reverse | 5’-CTGCACACGGAACCCGACGG-3’ | ||
| β-actin | forward | 5’-CCTCTATGCCAACACAGTGC-3’ | 220bp |
| Reverse | 5’-CATCGTACTCCTGCTTGCTG-3’ |
2.6 Ubiquitination of TFAM
Ubiquitination of TFAM was performed by 2-3 independent techniques. For immunoprecipitation, ~150μg protein (BRECs or retina) was incubated overnight at 4°C with 1μg of ubiquitin or TFAM antibody, followed by 1 hour with 20μl Protein A and G Plus agarose immunoprecipitation beads reagent (pre-washed and suspended in the lysis buffer). The beads were washed 4 times with lysis buffer, and the proteins were separated by SDS-PAGE. The membrane was immunoblotted with anti-TFAM or ubiquitin respectively (Santa Cruz Biotechnology) and developed by chemiluminescence (Kowluru, Mohammad et al. 2011; Santos and Kowluru 2011).
Ubiquitination of TFAM was also assessed by immunofluorescence microscopy. The cells grown on 12-mm-diameter coverslips coated with 0.1% gelatin were incubated with 5 or 20mM glucose for 4 days. At the end of the treatment, coverslips were rinsed with PBS and the cells were fixed with cold methanol (-20oC) for 15 minutes. They were blocked in 5% BSA for 1 hour and incubated with rabbit anti-TFAM antibody and mouse anti-ubiquitin antibody (Santa Cruz Biotechnology) overnight. After washing the cells with PBS, they were incubated with anti-mouse (Texas Red conjugated; Molecular Probes) and anti-rabbit (FITC green conjugate; Sigma) secondary antibodies for 1 hour in the dark. Cells were washed with PBS mounted with Vecta Shield mounting solution (Vector Laboratories) and examined under a Zeiss ApoTome using 40X magnification (Madsen-Bouterse, Zhong et al. 2010; Tewari, Santos et al. 2012; Tewari, Zhong et al. 2012; Santos and Kowluru 2013).
2.7 Statistics analysis
All of the data presented here was statistically analyzed using Sigma Stat software. The Shapiro-Wilk test was performed to test for normal distribution. For results with normal distribution, T-test was performed for analysis of two groups, and ANOVA followed by Bonferroni test for variables with more than two groups. Data that did not present normal distribution was analyzed by Mann-Whitney U for variable containing two groups and Kruskal-Wallis test followed by Dunn's for more than two groups. Data are expressed as means ± standard deviation, and p value <0.05 was considered statistically significant.
3. Results
3.1 Retinal endothelial cells
3.1.1 Role of Hsp70 in decreased mitochondrial protein levels of TFAM, and in mtDNA transcription
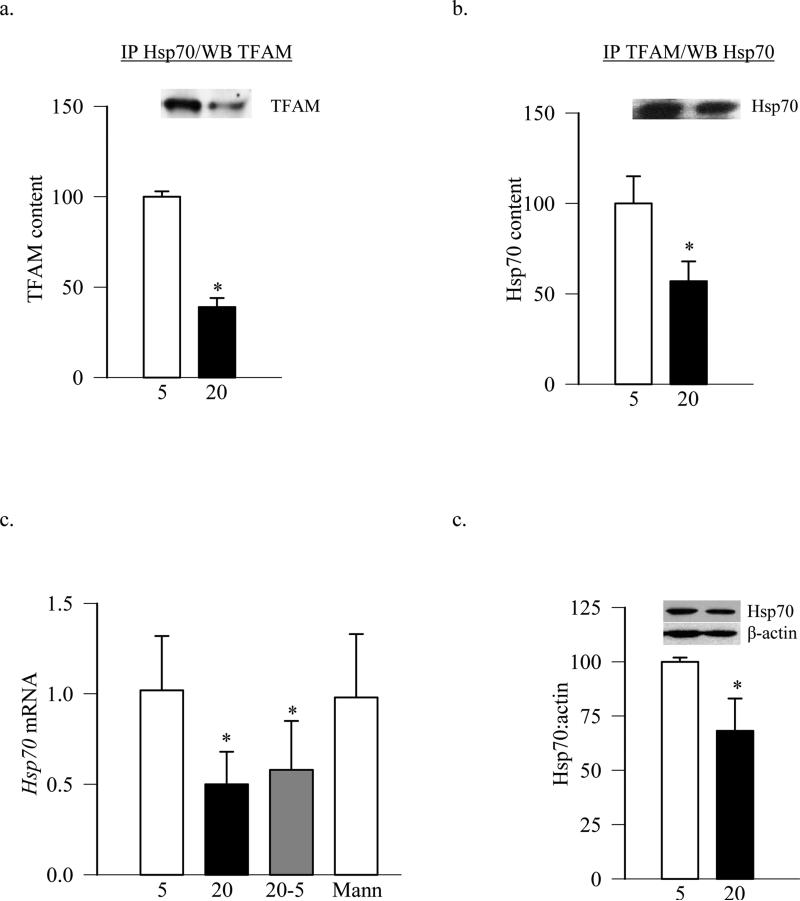
Our previous work has shown that despite increased gene expression of TFAM and no change in its total protein levels in hyperglycemic milieu, TFAM levels in the mitochondria are decreased (Santos and Kowluru 2011; Santos, Tewari et al. 2011). Cytosolic Hsp70 is a chaperone which carries mitochondrial proteins from the cytosol to the mitochondria surface. To investigate its role in transporting TFAM from cytosol to the mitochondria, association of Hsp70 with TFAM was determined by co-immunoprecipitating either Hsp70 or TFAM followed by western blotting for TFAM or Hsp70 respectively. Figures 1a and 1b show that the amount of TFAM associated with Hsp70 was less in the cells exposed to high glucose compared to the cells exposed to normal glucose. Furthermore, while high glucose significantly decreased Hsp70 mRNA levels, 20mM mannitol had no effect (Figure 1c). Consistent with the decrease in Hsp70 mRNA levels, its cytosolic protein levels were also decreased by ~35% in the cells exposed to high glucose (Figure 1d).
Figure 1.
Exposure of retinal endothelial cells to high glucose decreases the association of TFAM with Hsp70. Association of Hsp70 with TFAM was quantified by immunoprecipitation of (a) Hsp70 or (b) TFAM in the cells (150μg protein) incubated in 5mM or 20mM glucose, followed by western blot analysis for TFAM or Hsp70 respectively. (c) Hsp70 mRNA level was measured by real time q-PCR using β-actin as internal control. (d) Cytosolic expression of Hsp70 was quantified by western blot using the expression of β-actin as a loading control. The values are presented as means ± SD of 3-4 experiments, each done in duplicate. 5 = 5mM of glucose; 20 = 20mM glucose; 20-5= 4 days in 20mM glucose followed by 4 days in 5mM glucose; Mann= 20mM of Mannitol. *p<0.05 compared to 5mM glucose.
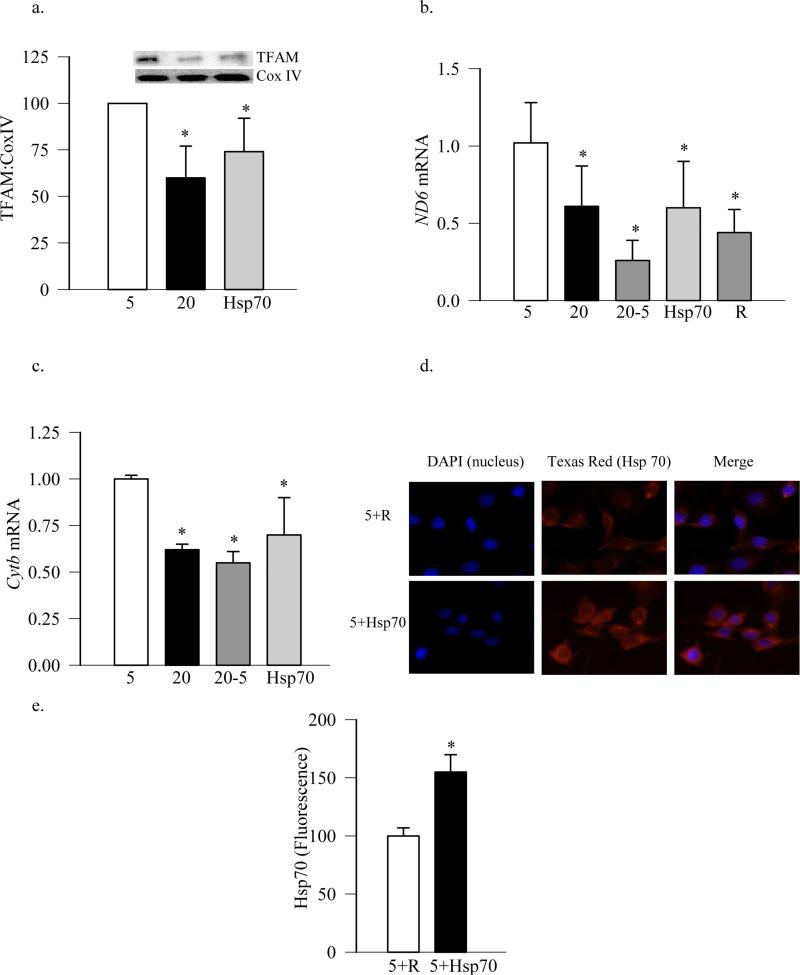
To further evaluate the role of cytosolic Hsp70 in the translocation of TFAM, cells overexpressing Hsp70 were used. Overexpression of Hsp70 did not ameliorate glucose-induced decrease in mitochondrial TFAM levels, the values obtained from Hsp70 transfected and untransfected cells exposed to high glucose were similar (Figure 2a). Since the transcription of mtDNA is dependent on TFAM protein levels in the mitochondria, and the gene expression of mtDNA-encoded proteins, e.g., Cytb, ND6, represents an indirect marker of the mitochondrial accumulation of TFAM, the effect of Hsp70 overexpression on mtDNA transcription was evaluated. As shown in figure (2b & c) overexpression of Hsp70 had no beneficial effect on glucose-induced decrease in mtDNA-encoded ND6 and Cytb; their mRNA levels remained 30-50% lower compared to the cells incubated in normal glucose. Consistent with this, the transfection reagent alone did not increase high glucose-induced decrease in ND6 mRNA levels (Figure 2b). Figures 2d&e are included to show the transfection efficiency of Hsp70. Quantification of the Hsp70 fluorescence using Image J software shows ~50% more Hsp70 in the cells transfected with Hsp70 plasmids compared to the cells incubated with the transfection regent alone.
Figure 2.
Overexpression of Hsp70 in endothelial cells does not affect TFAM levels in the mitochondria. (a) TFAM levels in the mitochondria were quantified by western blot technique using Cox IV as internal control. Gene transcripts of (b) ND6 and (c) Cytb were assessed by qPCR using β-actin as a housekeeping gene. (d) Transfection efficiency of Hsp70 was determined by microscopy using Texas Red conjugated secondary antibodies for Hsp70, and the cells were examined under a Zeiss ApoTome at 40X magnification. (e) The intensity of fluorescence was quantified in the cells transfected with Hsp70 or just with the reagent, and values obtained from the cells with the reagent were considered as 100%. Each measurement was made in duplicate using three different cell preparations. The values are presented as means ± SD. 5= 5mM glucose; 20=20mM glucose; Hsp70= cells overexpressing Hsp70 and incubated in 20mM glucose; 20-5= cells incubated in 20mM glucose for 4 days followed by 5mM glucose for 4 additional days; R=cells transfected with reagent alone and incubated in 20mM glucose respectively; 5+R and 5+Hsp70=cells incubated with regent alone or transfected with Hsp70 respectively, followed by incubation in 5mM glucose. *p<0.05 compared to 5mM glucose or R
3.1.2 Effect of overexpression of TFAM on mtDNA biogenesis
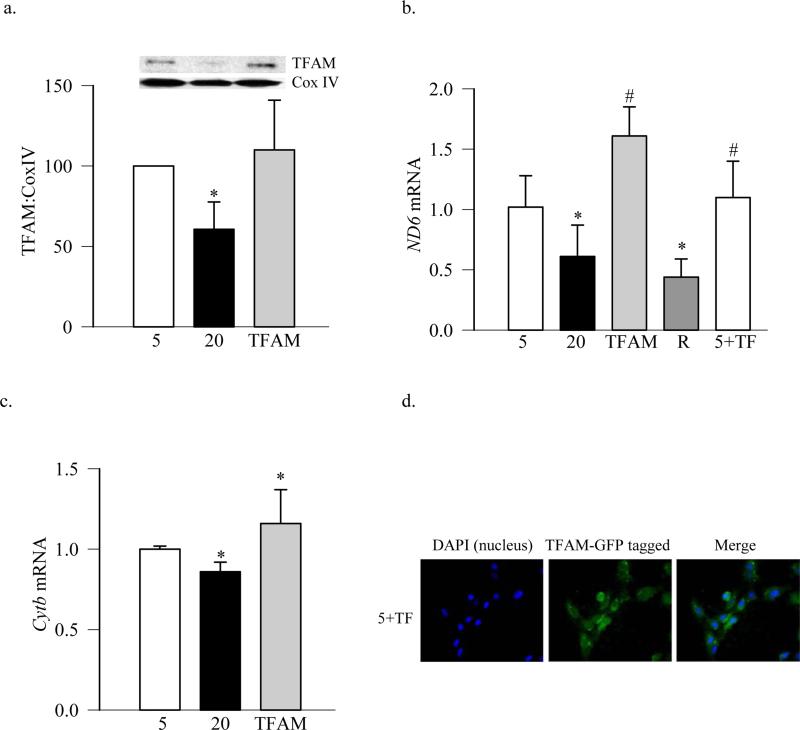
TFAM plays an important role in mitochondria biogenesis, and its levels are decreased in the retinal mitochondria in diabetes (Scarpulla 2008; Santos and Kowluru 2011; Santos, Tewari et al. 2011). To investigate the effect of TFAM overexpression on the mitochondria biogenesis, the cells were transfected with the GFP-tagged TFAM plasmids. Overexpression of TFAM prevented glucose-induced decrease in its levels in the mitochondria (Figure 3a), and also restored mtDNA transcription, as shown by increased expression of mtDNA-encoded ND6 and Cytb compared to untransfected cells exposed to high glucose (Figures 3b & c). In contrast, when the cells were incubated with the transfection reagent alone, without any plasmids, the expression of ND6 remained subnormal. Cells overexpressing TFAM, incubated in normal or high glucose had similar mtDNA transcription, as evidenced by similar mRNA levels of ND6 (Figure 3b). Figure 3d is included to show that the cells transfected with GFP-tagged TFAM plasmids had high levels of TFAM-GFP.
Figure 3.
Overexpression of TFAM ameliorates glucose-induced decrease in its mitochondrial protein levels. BRECs transfected with GFP-tagged TFAM plasmids were incubated in 5mM or 20mM glucose for 4 days. (a) Levels of TFAM in the mitochondria were quantified by western blot technique using Cox IV as internal control. Gene transcripts of (b) ND6 and (c) Cytb were quantified by real time PCR using β-actin as a housekeeping gene. (d) Transfection efficiency of TFAM was determined by microscopy using GFP-tagged on TFAM plasmid, and the cells were examined under a Zeiss ApoTome at 40X magnification. Each measurement was made in duplicate using three different cell preparations. The values are presented as means ± SD. 5= 5mM glucose; 20=20mM glucose; TFAM and 5+TF = TFAM transfected cells incubated in 20mM and 5mM glucose respectively; R= cells transfected with reagent alone and incubated in 20mM glucose. *p<0.05 compared to 5mM glucose, and #p<0.05 to 20mM glucose.
3.1.3 Ubiquitination of TFAM
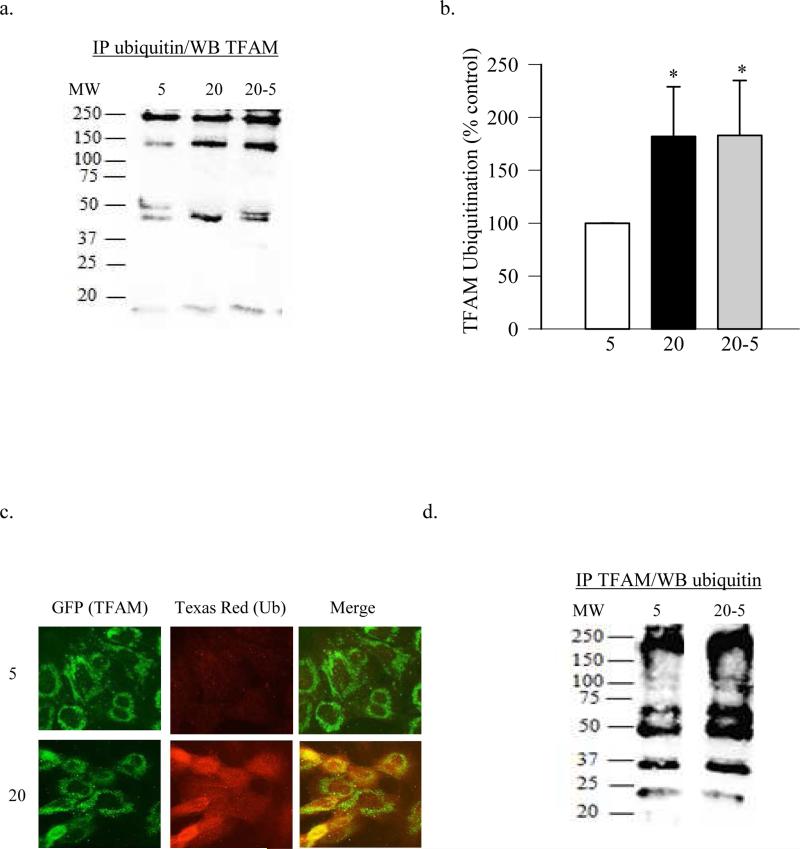
Ubiquitination, a post translational modification which is initiated by ubiquitin-activating enzyme UBA1 (E1), covalently attaches ubiquitin to the proteins labeling them for destruction (Rosenbaum, Fredrickson et al. 2011). To investigate if increase in ubiquitination has any effect on the decreased TFAM protein levels in the mitochondria, ubiquitination of TFAM was quantified by immunoprecipitation, and confirmed by fluorescence microscopy. The expression of TFAM in the ubiquitin immunoprecipitate was increased in the cells exposed to high glucose compared to normal glucose (Figures 4a). Ubiquitination is a complex process in which proteins can be mono, multi-mono or poly ubiquitinated, and the polyubiquitination could be homotypic or heterotypic (Komander 2009). The presence of a high molecular weight band, instead of ~25KD band, suggests that high glucose polyubiquinated TFAM. In support others have shown tetra-ubiquitination of TFAM in sperm extracts during porcine gametogenesis (Antelman, Manandhar et al. 2008). Figure 4b presents the quantification of the ubiquitinated TFAM. TFAM ubiquitination was confirmed by fluorescence microscopy; figure 4c shows that the cells incubated in high glucose had significantly high ubiquitin levels compared to the cells incubated in normal glucose as evidenced by increased red staining. When TFAM (green) was co-localized with ubiquitin (red), increased yellowish-orange staining was observed in high glucose conditions compared to normal glucose.
Figure 4.
High glucose ubiquitinates proteins in retinal endothelial cells. (a) Ubiquitination of TFAM was performed by immunoprecipitating ubiquitin and western blotting for TFAM, and (b) represents quantification of TFAM ubiquitination. (c) Ubiquitination was also confirmed by fluorescence microscopy using GFP-tagged TFAM and Texas Red-tagged ubiquitin antibodies. The cover slips were mounted with Vecta Shield and examined under a Zeiss ApoTome at 40X magnification. (d) Ubiquitination was quantified by immunoprecipitating TFAM, followed by western blotting for ubiquitin. MW=molecular weight marker; 5= 5mM glucose; 20=20mM glucose; 20-5= 4 days in 20mM glucose followed by 4 days in 5mM glucose.
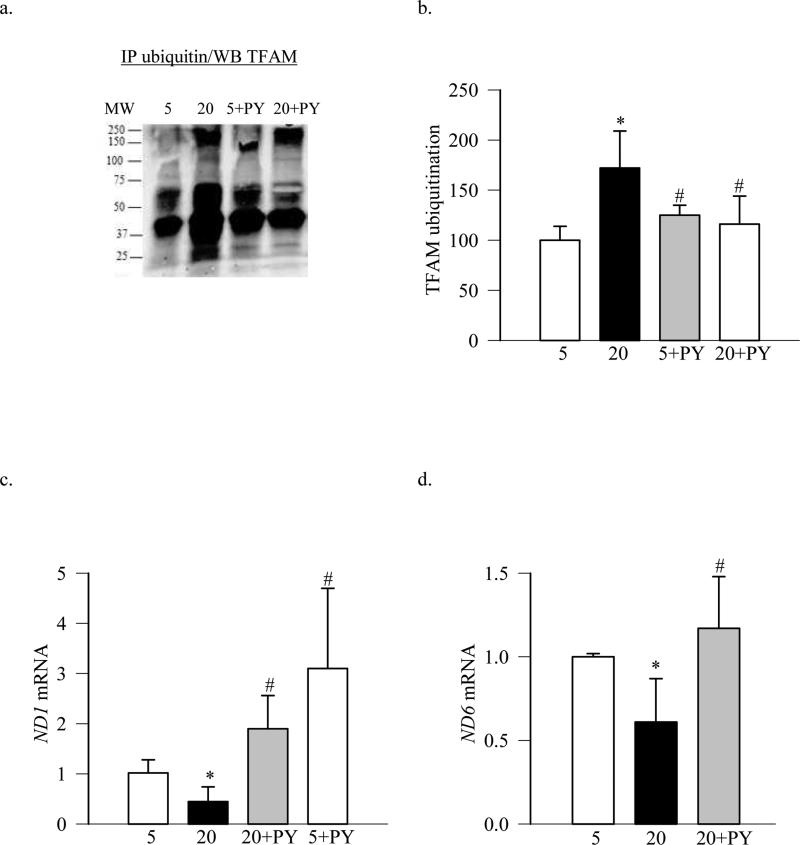
To confirm the role of ubiquitination of TFAM in its impaired transport into the mitochondria, the effect of an irreversible inhibitor of E1, PYR-41 (Guan and Ricciardi 2012), on mitochondria transcription was evaluated. As shown in Figure 5a, PYR-41 inhibited glucose-induced ubiquitination of TFAM and the values obtained from the PYR-41 treated cells incubated in 5mM or 20mM glucose were not significantly different from each other (Figure 5b). Furthermore, PYR-41 also prevented glucose-induced decrease in the transcription of mtDNA- encoded proteins ND1 and ND6 (Figure 5c & d), suggesting amelioration of glucose-induced increase in mitochondrial ROS levels.
Figure 5.
Inhibition of ubiquitination attenuates high glucose-induced TFAM ubiquitination and impairment in mtDNA transcription. BRECs were pre-incubated with 5μM PYR-41 for 4 hours before incubating in 5mM and 20mM glucose media. (a) Ubiquitination of TFAM was performed by immunoprecipitating ubiquitin and western blotting for TFAM; (b) shows the quantification of ubiquitination. Gene transcripts of (c) ND1 and (d) ND6 were quantified by real time RT-PCR using β-actin as a housekeeping gene. 5= 5mM glucose; 20=20mM glucose; 20+PY and 5+PY= cells pre-incubated with PYR-41 for 4 hours followed by in 20mM and 50mM glucose media, respectively. *p<0.05 compared to 5mM glucose, and #p<0.05 to 20mM glucose.
3.2 Rat Retina
3.2.1 Effect of diabetes on ubiquitination of TFAM
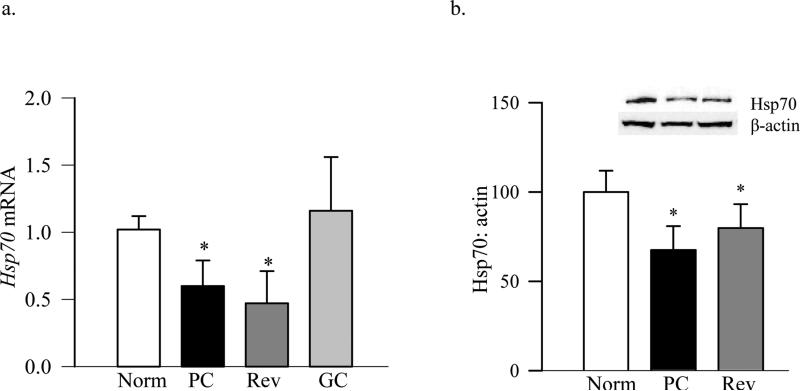
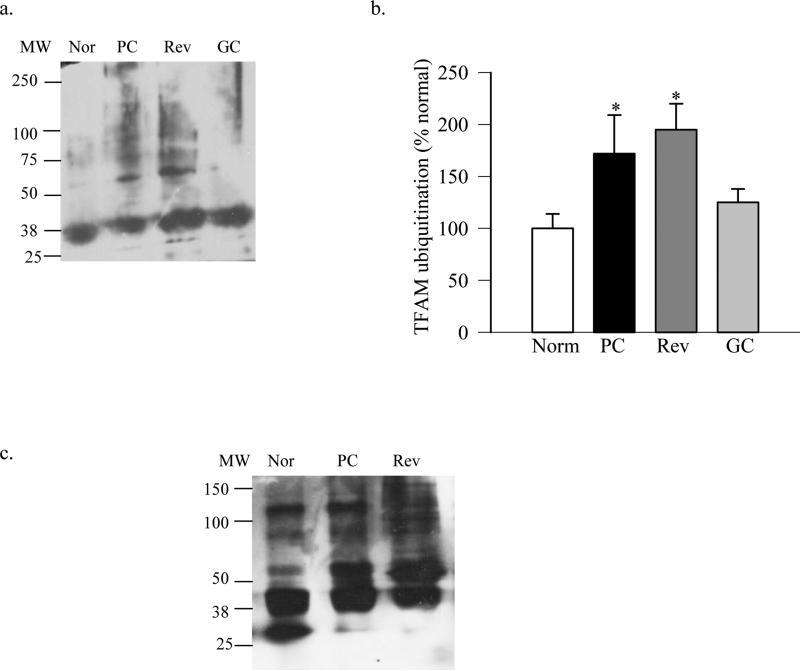
Consistent with the results from endothelial cells, diabetes decreased the expression of Hsp70 in the retina; its mRNA levels were decreased by ~50% and protein expression by 30% in diabetic rats compared to normal rats (Figure 6a & b). In the same retina, ubiquitination of TFAM, as estimated by immunoprecipitating either ubiquitin, or TFAM, followed by western blotting for TFAM or ubiquitin respectively (Figures 7a-c), was increased by over 70% in the retina from diabetic rats compared to the values from normal rat retina.
Figure 6.
Diabetes decreases Hsp70 expression in the retina. Hsp70 (a) gene expression was quantified in rat retina by qPCR and (b) protein expression by western blot technique using β-actin as a loading control. Each experiment was performed in duplicate in 5-6 rats in each group. Nor= Normal rats, PC= rats in 8 months of continuous poor glycemic control, Rev=rats in 4 months of poor glycemic control followed by 4 months of good glycemic control, GC= rats with 8 months of continuous good glycemic control. *p<0.05 vs normal
Figure 7.
Ubiquitination of retinal proteins is increased in diabetes. (a)TFAM ubiquitination was performed by immunoprecipitating ubiquitin, followed by western blot analysis for TFAM. The blots are representative from 4-5 assays, and (b) the expression of ubiquitinated TFAM was quantified. (c) Ubiquitination of TFAM was performed by immunoprecipitating TFAM, followed by western blot analysis for ubiquitin. MW=molecular weight marker, Nor= Normal rats, PC= rats in 8 months of continuous poor glycemic control, Rev=rats in 4 months of poor glycemic control followed by 4 months of good glycemic control, GC= rats with 8 months of continuous good glycemic control.
3.2.2 Reversal of hyperglycemia and ubiquitination
Reinstitution of good control in diabetic rats, which has followed poor control, fails to reverse mtDNA damage in the retina and mtDNA-encoded proteins remain subnormal with decreased protein levels of TFAM in the mitochondria (Madsen-Bouterse, Mohammad et al. 2010; Santos and Kowluru 2011). To investigate the mechanism responsible for decreased TFAM in the mitochondria, the effect of reversal of hyperglycemia on the posttranslational modification of TFAM was investigated. In addition to providing no beneficial effect of re-institution of good glycemic control on the Hsp70 expression (Figure 6), retinal proteins continued to be ubiquitinated. As shown in figure 7, TFAM remained polyubiquitinated, and the values of TFAM ubiquitination in the retina from rats in PC and Rev groups were significantly higher compared to those from normal group. However, in contrast, when good glycemic control was initiated soon after induction of diabetes, expression of Hsp70 and ubiquitination of retinal TFAM was similar to that observed in the age-matched normal rat retina (Figures 6&7).
The role of the posttranslational modification of TFAM in the metabolic memory was further confirmed in retinal endothelial cells. Gene expression of Hsp70 remained subnormal even after 4 days of normal glucose that followed 4 days of high glucose (Figure 1c). Consistent with our previous reports (Madsen-Bouterse, Zhong et al. 2010), the gene transcripts of mtDNA-encoded ND6 and Cytb continued to be subnormal (Figure 2b&c). In the same cell preparations, reversal of high glucose insult had no beneficial effect on the ubiquitination of TFAM, as confirmed by both immunoprecipitating ubiquitin or TFAM, followed by western blotting for TFAM or ubiquitin respectively (Figures 4a,b&d).
4. Discussion
Apoptosis of capillary cells precedes the development of histopathology that is considered the hallmark of diabetic retinopathy, and mitochondria dysfunction plays a pivotal role in the accelerated apoptosis of retinal capillary cells (Kern, Tang et al. 2000; Kowluru and Abbas 2003; Madsen-Bouterse, Zhong et al. 2010; Madsen-Bouterse, Mohammad et al. 2010; Santos, Tewari et al. 2011; Tewari, Santos et al. 2012; Santos and Kowluru 2013). Mitochondrial ROS are increased in the retina in diabetes impairing mtDNA biogenesis, and mitochondria copy numbers are decreased (Kanwar, Chan et al. 2007; Madsen-Bouterse, Mohammad et al. 2010; Santos, Tewari et al. 2011). TFAM is an essential transcriptional factor in the biogenesis of mitochondria, and in diabetes its mRNA levels are increased in the retina, but its mitochondrial levels are decreased (Scarpulla 2008; Santos and Kowluru 2011; Santos, Tewari et al. 2011). Here, we show that TFAM is posttranslationally modified, and the chaperone which carries TFAM to the mitochondria surface, Hsp70, appears to play less significant role, as its overexpression does not alleviate hyperglycemia-induced decrease in mitochondrial protein levels of TFAM. However, pharmacological inhibition of ubiquitination of TFAM helps restore normal mtDNA transcription and mitochondrial homeostasis. Furthermore, reversal of hyperglycemia by normal glycemia does not attenuate TFAM ubiquitination, and it continues to be polyubiquitinated suggesting the role of TFAM ubiquitination in the metabolic memory phenomenon associated with the continued progression of diabetic retinopathy.
TFAM binds to the mtDNA to regulate transcription and mtDNA, but to reach to the mitochondria, it requires a complex mechanism (Ohgaki, Kanki et al. 2007; Malarkey, Bestwick et al. 2012). Cytosolic Hsp70 is one of the key chaperones which transports most of the proteins to the mitochondria membrane (Atalay, Oksala et al. 2009). We have shown that the amount of Hsp70 associated with MMP-9 is significantly increased in the retina, while that with TFAM is decreased (Kowluru, Mohammad et al. 2011; Santos and Kowluru 2011; Santos, Tewari et al. 2011). In addition to the decreased mitochondrial Hsp70 (Kowluru, Mohammad et al. 2011), here our data show that the levels of cytosolic Hsp70 are also decreased, suggesting subnormal chaperon activity. However, when cytosolic Hsp70 is overexpressed, it fails to ameliorate glucose-induced impaired mtDNA biogenesis, clearly implying that cytosolic Hsp70 might not be the major factor responsible for chaperoning TFAM to the mitochondria.
Using multiple independent methods, including copy number and transcription, mtDNA density and mtDNA replication, we have shown that retinal mitochondria biogenesis is decreased in diabetes. However, despite decrease in biogenesis, the gene expression of transcription factors and proteins important in mtDNA biogenesis, e.g., peroxisome proliferator-activated receptor γcoactivator-1α, nuclear regulatory factor and TFAM are increased (Santos and Kowluru 2011; Santos, Tewari et al. 2011; Santos, Tewari et al. 2012). Diabetes also downregulates mitochondrial transport system, and the association of TFAM with Tom70 and Tim44 of the mitochondria transport system is decreased (Santos and Kowluru 2013). Here we show that the overexpression of TFAM protects the cells from high glucose-induced decrease in TFAM protein levels in the mitochondria and revives its transcriptional capacity; these results suggests that posttranslational modification of TFAM could be contributing to in its decreased transport to the mitochondria. In support, diabetic environment facilitates posttranslational modifications of proteins and histones (Goldberg, Whiteside et al. 2006; Suarez, Hu et al. 2008; Kanwar and Kowluru 2009; Reddy and Natarajan 2011; Zhong and Kowluru 2011; Harcourt, Penfold et al. 2013; Song, Peng et al. 2013; Zhong and Kowluru 2013). Furthermore, in support of the role of posttranslational modification in decreased protein levels of TFAM in the mitochondria, our previous study has shown that the posttranslational modifications of glyceraldehyde dehydrogenase favor its translocation into the nucleus (Kanwar and Kowluru 2009; Madsen-Bouterse, Mohammad et al. 2010) .
Ubiquitination, an enzymatic posttranslational modification, targets proteins for destruction, and regulates protein stability, subcellular distribution, DNA-binding affinity and transcriptional activity (Neutzner, Benard et al. 2008; Bergink and Jentsch 2009). Ubiquitination of mitochondrial proteins is considered vital for the maintenance of mitochondrial homeostasis, and in removing the dysfunctional mitochondria (Neutzner, Benard et al. 2008). This proteasomepathway helps eliminate misfolded proteins that are generated under stress conditions, and is initially activated by E1 enzyme which activates ubiquitin. In diabetes, ubiquitin-proteasome pathway is increased in the kidney affecting its insulin activity (Gao, Chen et al. 2013), and ubiquitination of sarcoplasmic reticulum calcium ATPase 2 is implicated in diabetes-induced diastolic dysfunction (Takada, Miki et al. 2012). Here, our results suggest that polyubiquitination of TFAM could be one of the mechanisms responsible for its subnormal levels in the mitochondria. Consistent with our results, others have shown that polyubiquitination of TFAM in sperm extracts during porcine gametogenesis induces mitochondria degradation (Antelman, Manandhar et al. 2008). To further support the role of ubiquitination, inhibition of E1 by PYR-41 alleviates posttranslational modification of TFAM and restores mitochondria transcription machinery. Since impaired mitochondria homeostasis is important in diabetic retinopathy (Kowluru and Abbas 2003; Madsen-Bouterse, Zhong et al. 2010; Madsen-Bouterse, Mohammad et al. 2010; Santos and Kowluru 2011; Santos, Tewari et al. 2011; Tewari, Santos et al. 2012; Tewari, Zhong et al. 2012; Santos and Kowluru 2013), our data presented here clearly implicate that the inhibition of ubiquitination has potential to prevent its development by maintaining mitochondria homeostasis and TFAM's transcriptional activity. The focus of our study was to understand the role of posttranslational modifications in impaired mtDNA biogenesis, and we cannot rule out the role of Ubiquitin-26S proteasome system in decreased levels of TFAM in the mitochondria. However, since diabetes does not decrease the total protein expression of TFAM suggesting that the degradation of TFAM in the cytosol might not be responsible for its decreased levels in the mitochondria. Furthermore, phosphorylation of TFAM is also shown to decrease its protein levels in mitochondria and reduce its subnormal transcriptional activity (Lu, Lee et al. 2013). Although the present study investigated ubiquitination of TFAM, the role of other posttranslational modifications, including phosphorylation, nitration and glycation, in its impaired transport to the mitochondria in diabetes, however, cannot be ruled out.
It is well established that the progression of diabetic retinopathy does not halt once the glycemic insult is terminated, and the previous severity and the duration of glycemic control dictates the outcome of the glycemic control that follows suggesting a metabolic memory or the legacy effect (Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group 2000; Kowluru 2003). Retinal mitochondria remain dysfunctional, and their DNA continues to be damaged and transcription impaired with subnormal levels of TFAM in the mitochondria (Santos and Kowluru 2011; Tewari, Zhong et al. 2012). The results presented here show that the TFAM remains polyubiquitinated, possibly contributing to its continued decreased mitochondrial protein levels and the transcription of mtDNA. Consistent with this, posttranslational modifications are postulated to play a critical role in the metabolic memory associated with diabetic complications, including retinopathy (Kowluru 2003; Kanwar and Kowluru 2009; Reddy and Natarajan 2011; Zhong and Kowluru 2011; Zhong and Kowluru 2013). However, if good glycemic control is initiated soon after induction of diabetes, TFAM escapes polyubiquitination, suggesting that once TFAM is posttranslationally modified, de-ubiquitination of TFAM becomes difficult even if the glycemic insult is reversed.
5. Conclusion
In conclusion, we have provided novel data suggesting that due to ubiquitination of TFAM in diabetes, its transport to the mitochondria (the site of its action) is impaired, resulting in subnormal mtDNA biogenesis. Furthermore, reversal of hyperglycemic insult by normal glycemia does not provide any benefit and TFAM continues to be posttranslationally modified, and retinopathy continues to progress. Thus, modulation of ubiquitination by pharmacological means could have potential to maintain mitochondrial homeostasis, and inhibit/retard the development of diabetic retinopathy.
Highlights.
TFAM is one of the key regulators of mtDNA transcription.
TFAM ubiquitination in diabetes impedes its transport to the retinal mitochondria.
Decreased TFAM levels in the mitochondria attenuate retinal mtDNA biogenesis.
Preserving TFAM will protect retinal mitochondria and inhibit diabetic retinopathy.
Acknowledgement
Authors thank Doug Putt and Mangayarkarasi Thandampallayam for their technical assistance. This study was supported in parts by grants to RAK from the National Institutes of Health (EY014370, EY017313 and EY022230), Juvenile Diabetes Research Foundation (5-2012-313) and the Thomas Foundation, and unrestricted funds to the Department of Ophthalmology from Research to Prevent Blindness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Author Disclosure and contribution
Authors declare that they do not have any competing financial interests. JMS researched data and wrote the manuscript, MM researched data, and RAK wrote and edited the manuscript. RAK is the guarantor of this work and, as such, has full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- Antelman J, Manandhar G, et al. Expression of mitochondrial transcription factor A (TFAM) during porcine gametogenesis and preimplantation embryo development. J Cell Physiol. 2008;217:529–543. doi: 10.1002/jcp.21528. [DOI] [PubMed] [Google Scholar]
- Atalay M, Oksala N, et al. Heat shock proteins in diabetes and wound healing. Curr Protein Pept Sci. 2009;10:85–95. doi: 10.2174/138920309787315202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458(7237):461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Diabetes Control and Complications Trial Research Group The effect of intensive treatment of diabetes on the development of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- Diabetes Control and Complications Trial Research Group Early worsening of diabetic retinopathy in the diabetes control and complication trial. Arch. Ophthalm. 1998;116:874–886. doi: 10.1001/archopht.116.7.874. [DOI] [PubMed] [Google Scholar]
- Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. N Engl J Med. 2000;342:381–389. doi: 10.1056/NEJM200002103420603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engerman RL, Kern TS. Progression of incipient diabetic retinopathy during good glycemic control. Diabetes. 1987;36:808–812. doi: 10.2337/diab.36.7.808. [DOI] [PubMed] [Google Scholar]
- Frank RN. Diabetic Retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- Gao C, Chen G, et al. Impact of high glucose and proteasome inhibitor MG132 on histone H2A and H2B ubiquitination in rat glomerular mesangial cells. J Diabetes Res. 2013;2013:589474. doi: 10.1155/2013/589474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg HJ, Whiteside CI, et al. Posttranslational, reversible O-glycosylation is stimulated by high glucose and mediates plasminogen activator inhibitor-1 gene expression and Sp1 transcriptional activity in glomerular mesangial cells. Endocrinology. 2006;147:222–231. doi: 10.1210/en.2005-0523. [DOI] [PubMed] [Google Scholar]
- Guan H, Ricciardi RP. Transformation by E1A oncoprotein involves ubiquitin-mediated proteolysis of the neuronal and tumor repressor REST in the nucleus. J Virol. 2012;86:5594–5602. doi: 10.1128/JVI.06811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt BE, Penfold SA, et al. Coming full circle in diabetes mellitus: from complications to initiation. Nat Rev Endocrinol. 2013;9(2):113–123. doi: 10.1038/nrendo.2012.236. [DOI] [PubMed] [Google Scholar]
- Kanwar M, Chan PS, et al. Oxidative damage in the retinal mitochondria of diabetic mice: possible protection by superoxide dismutase. Invest Ophthalmol Vis Sci. 2007;48:3805–3811. doi: 10.1167/iovs.06-1280. [DOI] [PubMed] [Google Scholar]
- Kanwar M, Kowluru R. Role of glyceraldehyde 3-phosphate dehydrogenase in the development and progression of diabetic retinopathy. Diabetes. 2009;58:227–234. doi: 10.2337/db08-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern TS, Tang J, et al. Response of capillary cell death to aminoguanidine predicts the development of retinopathy: Comparison of diabetes and galactosemia. Invest Ophthalmol Vis Sci. 2000;41:3972–3978. [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- Kowluru RA. Effect of re-institution of good glycemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes. 2003;52:818–823. doi: 10.2337/diabetes.52.3.818. [DOI] [PubMed] [Google Scholar]
- Kowluru RA, Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Inves Ophthalmol Vis Sci. 2003;44:5327–5334. doi: 10.1167/iovs.03-0353. [DOI] [PubMed] [Google Scholar]
- Kowluru RA, Mohammad G, et al. Abrogation of MMP9 gene protects against the development of retinopathy in diabetic mice by preventing mitochondrial damage. Diabetes. 2011;60:3023–3033. doi: 10.2337/db11-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Lee J, et al. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell. 2013;49:121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen-Bouterse S, Mohammad G, et al. Glyceraldehyde 3 phosphate dehydrogenase in retinal microvasculature: Implications for the development and progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2010;51:1765–1772. doi: 10.1167/iovs.09-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen-Bouterse S, Zhong Q, et al. Oxidative damage of mitochondrial DNA in diabetes, and its protection by manganese superoxide dismutase. Free Rad Research. 2010;44:313–321. doi: 10.3109/10715760903494168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen-Bouterse SA, Mohammad G, et al. Role of mitochondrial DNA damage in the development of diabetic retinopathy, and the metabolic memory phenomenon associated with its progression. Antioxid Redox Signal. 2010;13:797–805. doi: 10.1089/ars.2009.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkey CS, Bestwick M, et al. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A, Benard G, et al. Role of the ubiquitin conjugation system in the maintenance of mitochondrial homeostasis. Ann N Y Acad Sci. 2008;1147:242–253. doi: 10.1196/annals.1427.012. [DOI] [PubMed] [Google Scholar]
- Ohgaki K, Kanki T, et al. The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J Biochem. 2007;141:201–211. doi: 10.1093/jb/mvm020. [DOI] [PubMed] [Google Scholar]
- Piantadosi CA, Suliman HB. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J Biol Chem. 2006;281:324–233. doi: 10.1074/jbc.M508805200. [DOI] [PubMed] [Google Scholar]
- Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res. 2011;90:421–429. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JC, Fredrickson EK, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Kowluru RA. Role of mitochondria biogenesis in the metabolic memory associated with the continued progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2011;52:8791–8798. doi: 10.1167/iovs.11-8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Kowluru RA. Impaired transport of mitochondrial transcription factor A (TFAM) and the metabolic memory phenomenon associated with the progression of diabetic retinopathy. Diabetes Metab Res Rev. 2013;29:204–213. doi: 10.1002/dmrr.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Mohammad G, et al. Diabetic retinopathy, superoxide damage and antioxidant. Curr Pharm Biotechnol. 2011;12:352–361. doi: 10.2174/138920111794480507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Tewari S, et al. Mitochondria biogenesis and the development of diabetic retinopathy. Free Rad Biol Med. 2011;51:1849–1860. doi: 10.1016/j.freeradbiomed.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Tewari S, et al. A compensatory mechanism protects retinal mitochondria from initial insult in diabetic retinopathy. Free Rad Biol Med. 2012;53:1729–1737. doi: 10.1016/j.freeradbiomed.2012.08.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Tewari S, et al. Interrelationship between activation of matrix metalloproteinases and mitochondrial dysfunction in the development of diabetic retinopathy. Biochem Biophys Res Commun. 2013;438:760–764. doi: 10.1016/j.bbrc.2013.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, et al. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Song R, Peng W, et al. Central role of E3 ubiquitin ligase MG53 in insulin resistance and metabolic disorders. Nature. 2013;494:375–379. doi: 10.1038/nature11834. [DOI] [PubMed] [Google Scholar]
- Suarez J, Hu Y, et al. Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am J Physiol Cell Physiol. 2008;296:C1561–C1568. doi: 10.1152/ajpcell.00076.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A, Miki T, et al. Role of ER stress in ventricular contractile dysfunction in type 2 diabetes. PLoS One. 2012;7:e39893. doi: 10.1371/journal.pone.0039893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari S, Santos JM, et al. Damaged mitochondrial DNA replication system and the development of diabetic retinopathy. Antioxid Redox Signal. 2012;17:492–504. doi: 10.1089/ars.2011.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari S, Zhong Q, et al. Mitochondria DNA replication and DNA methylation in the metabolic memory associated with continued progression of diabetic retinopathy. Inves Ophthalmol Vis Sci. 2012;53:4881–4888. doi: 10.1167/iovs.12-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Kowluru RA. Diabetic retinopathy and damage to mitochondrial structure and transport machinery. Inves Ophthalmol Vis Sci. 2011;52:8739–8746. doi: 10.1167/iovs.11-8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Kowluru RA. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. 2011;60:1304–1313. doi: 10.2337/db10-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Kowluru RA. Epigenetic modification of Sod2 in the development of diabetic retinopathy and in the metabolic memory: Role of histone methylation. Invest Ophthalmol Vis Sci. 2013;54:244–250. doi: 10.1167/iovs.12-10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Kowluru RA. Regulation of matrix metalloproteinase-9 by epigenetic modifications and the development of diabetic retinopathy. Diabetes. 2013;62:2559–2568. doi: 10.2337/db12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]