Background: Large numbers of peptides encoded in human short open reading frames have been recently identified but not yet functionally characterized.
Results: A peptide interacts with the Ku heterodimer and stimulates nonhomologous end-joining DNA repair.
Conclusion: Newly discovered cellular peptides can be functionally characterized by identifying their interaction partners.
Significance: Short ORF-encoded polypeptides participate in essential cellular processes.
Keywords: DNA Repair, Peptide Interactions, Peptides, Protein-Protein Interactions, Proteomics, Short ORF
Abstract
The recent discovery of numerous human short open reading frame (sORF)-encoded polypeptides (SEPs) has raised important questions about the functional roles of these molecules in cells. Here, we show that a 69-amino acid SEP, MRI-2, physically interacts with the Ku heterodimer to stimulate DNA double-strand break ligation via nonhomologous end joining. The characterization of MRI-2 suggests that this SEP may participate in DNA repair and underscores the potential of SEPs to serve important biological functions in mammalian cells.
Introduction
Interest in the complete coding potential of the human genome has recently exploded due to technical advances permitting detection of translation of nonannotated sequences. In particular, widespread translation of short open reading frames (1) (sORFs)3 producing peptides <150 amino acids has been reported in mammalian cells by ribosome profiling (2, 3), proteomics (4), and peptidomics (5–8). The apparent prevalence of sORF translation raises the question of whether sORF-encoded polypeptides (SEPs) are functional biomolecules.
Although sORF translation is prevalent, few mammalian SEPs have been demonstrated to have biological functions. SEPs regulating limb morphogenesis (9, 10) and cardiac function (11) have been characterized in insects, and a few bioactive yeast (12) and plant (13–15) SEPs have been reported. In humans, the only well characterized SEP is the peptide humanin, which inhibits amyloid β-induced neuronal cell death (16, 17). The biological relevance of SEP translation can only be addressed if general strategies for their functional annotation can be developed. Here, we report a functional proteomics strategy to identify the biological functions of SEPs, and use it to identify the SEP MRI-2 as a novel Ku interaction partner.
EXPERIMENTAL PROCEDURES
Cloning and Gene Synthesis
MRI-2, Ku70, and Ku80 coding sequences were obtained from Open Biosystems. The MRI-2 coding sequence was subcloned with a C-terminal FLAG epitope tag into pcDNA3, and with a C-terminal His6 tag into pET21a, for mammalian and bacterial expression, respectively. The MRI-1 coding sequence with a C-terminal FLAG tag was synthesized by GenScript and subcloned into pcDNA3. The MRI-3-FLAG coding sequence was subcloned from this construct into pcDNA3. The Ku80 coding sequence was subcloned into pcDNA3 with a C-terminal HA tag. The Ku70 coding sequence was subcloned into pcDNA3 with a C-terminal c-Myc tag.
Co-immunoprecipitation and Proteomics
FLAG-tagged MRI-1–3 in pcDNA3 (or empty pcDNA3) was transfected into a 10-cm dish of HEK 293T cells using Lipofectamine 2000 and Opti-MEM (Life Technologies). 24 h after transfection, cells were harvested and lysed using Tris-buffered saline with 1% Triton X-100 (TBS-T) and Roche Applied Science Complete protease inhibitor cocktail tablets. Cells were lysed on ice for 20 min followed by centrifugation at 14,000 rpm, 4 °C, 15 min. Anti-FLAG agarose (clone M2, Sigma) was washed with TBS-T, collected by centrifugation, and then suspended in the cell lysate. Bead suspensions were rotated at 4 °C for 1 h and then washed three times with TBS-T. Bound protein was eluted with 3× FLAG peptide (Sigma) at 4 °C for 1 h. For proteomics, samples were analyzed by SDS-PAGE with Coomassie Brilliant Blue stain, and bands elevated in the IP samples were identified and excised. For quantitation via spectral counting, the gel was cut straight across with a clean razor to excise the same molecular weight band in both the sample and the negative control.
Protein-containing gel slices were digested with trypsin overnight. The resulting peptide mixtures were extracted from the gel and run on an Orbitrap Velos (Thermo Fisher Scientific) with 90-min liquid chromatography and tandem mass spectrometry (LC-MS/MS) using a standard TOP20 method as described previously (7).
Mass spectra were analyzed using our in-house Proteome Browser System against the UniProt_human database. Carbamidomethylated cysteines were set as a fixed modification, and methionine oxidation and N-terminal acetylation were set as variable modifications. A mass error of 20 ppm was set for precursor ions, and 0.6 Da was set for MS/MS peaks. Two missed cleavages were allowed. Maximum False Discovery Rates were set to 0.01 on both peptide and protein levels. Minimum required peptide length was five amino acids. Interaction candidates were identified by spectral counting as proteins elevated >1.7-fold in the IP relative to the negative control, with an abundance cutoff of 20 spectral counts in the immunoprecipitate.
Western Blotting
SDS-PAGE gels and transfers were performed as described previously (7). Immunoblots were blocked with Rockland Immunochemicals fluorescent blocking buffer and then probed with the following primary antibodies at a 1:1000 dilution in the same buffer: mouse anti-FLAG (clone M2, Sigma), mouse anti-XRCC6 (Ku70, clone 4C2-1A6, Sigma), mouse anti-XRCC5 (Ku80, clone 3D8, Sigma). Secondary antibodies, goat anti-mouse IR dye 800 and goat anti-rabbit IR dye 680 (LI-COR), were applied at a dilution of 1:4000 in Rockland Immunochemicals fluorescent blocking buffer. Infrared imaging was performed on a LI-COR Odyssey.
Immunofluorescence and Confocal Imaging
Cells were grown, transfected, fixed, and permeabilized as described previously (7). Etoposide (Sigma) was applied to cells at 50 μm for 4 h. Cells were stained with primary antibodies: mouse anti-FLAG (clone M2, Sigma); rabbit anti-HA (Rockland Immunochemicals); and/or chicken anti-c-Myc (Aves Labs). Secondary antibodies (Life Technologies) were goat anti-mouse Alexa Fluor 568, goat anti-rabbit Alexa Fluor 488, and goat anti-chicken Alexa Fluor 647. All antibodies were applied at a 1:1000 dilution in fluorescence blocking buffer (Rockland Immunochemicals) for at least 1 h at 4 °C followed by three phosphate-buffered saline (PBS) washes after fixation with formalin and nuclear counterstain with Hoechst 33342.
Coverslips were inverted and imaged in PBS in MatTek imaging dishes. Confocal imaging was performed on a Zeiss AxioObserver inverted confocal microscope with 40× oil immersion objective, Yokogawa spinning disk confocal head, quad-band notch dichroic mirror (405/488/568/647), and 405 (diode), 491 (diode pumped solid-state laser), 561 (diode pumped solid-state laser), and 640 nm (diode) lasers (all 50 milliwatt). Hoechst 33342 was imaged with 405 nm laser excitation, 445/40 nm emission; Alexa Fluor 488 with 491 nm laser excitation, 528/38 nm emission; Alexa Fluor 568 with 561 nm laser excitation, 617/73 nm emission; and Alexa Fluor 647 with 640 nm laser excitation, 700/75 nm emission. Differential interference contrast (DIC) images were collected. All image collection and analysis was performed with SlideBook software (Intelligent Imaging Innovations). Fluorophore channels in each experiment were normalized to the same intensity ranges for presentation.
To determine the nuclear fraction of MRI-2, regions of interest were manually drawn on whole cells, as defined by MRI-2 staining and the DIC image, and on the nucleus, as defined by Hoechst 33342 staining. Background was defined for whole cell and nucleus using an untransfected cell. For each cell, the ratio of the background-corrected nuclear sum intensity to the background-corrected whole cell sum intensity was used to calculate the nuclear fraction. Single cell data were plotted as histograms, and population averages were calculated. Statistical analysis of significance was by two-tailed Student's t test, and error bars represent S.E.
MRI-2 Purification
His6-tagged MRI-2 in pET21a was transformed into BL21 Star DE3 cells. A saturated 10-ml overnight culture in Luria Bertani broth (LB) with 100 μg/ml ampicillin was diluted into a 2-liter LB-ampicillin culture and grown at 37 °C with shaking to optical density 0.6. Protein expression was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside, and cells were grown at 30 °C with shaking for 4 h. Cells were harvested by centrifugation at 8000 rpm for 10 min at 4 °C and stored at −80 °C until use. Protein was purified using a protocol previously reported for Ku (18). Pure MRI-2 (assayed by SDS-PAGE) was dialyzed into 50 mm Tris-HCl, pH 8.0, 20% glycerol, 50 mm NaCl, and 1 mm DTT, aliquoted, and flash-frozen. Protein concentration was determined with Bradford assay (Pierce) with reference to BSA.
Nonhomologous End Joining Assay
Ramos whole cell extract was prepared as described previously (19). In vitro nonhomologous end joining (NHEJ) assays were carried out essentially as described previously (19–22). Briefly, reactions (10 μl) were carried out in 50 mm HEPES, pH 8.0, 100 mm KOAc, 0.5 mm Mg(OAc)2, 1 mm ATP, 1 mm DTT, 0.1 mg/ml bovine serum albumin (BSA), and HindIII-linearized 5′-32P-labeled pBluescript DNA (10 ng) with 20 μg of whole cell extract. Wortmannin (Sigma) was added at 3 μm, and anti-XRCC4 antibody (Abcam) was used at 1:50 final dilution. Four replicate experiments were performed; statistical analysis of significance was by two-tailed Student's t test, and error bars represent S.E.
RESULTS
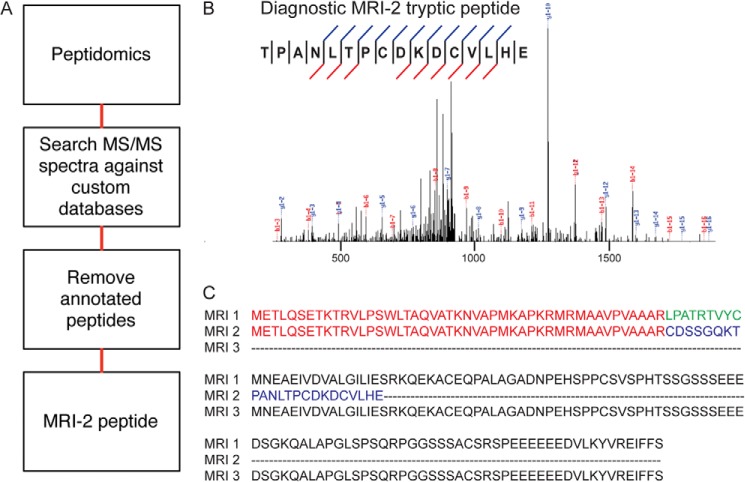
We focused our study on a 69-amino acid SEP from the C7orf49 gene identified in our previous peptidomics profiling of K562 cells (Fig. 1A) (6, 7). Because this SEP was annotated as a predicted protein in the NCBI nonredundant protein database, we did not include it in the list of reported SEPs (7); however, our evidence is the first experimental detection of this polypeptide in cells. Identifying a protein or polypeptide from a single MS/MS spectrum requires the highest spectral quality, and this spectrum passes our criteria (7), with a continuous y ion series of 11 fragments and an Sf score (23) of 0.88 (Fig. 1B). This provides the first evidence for endogenous expression of this SEP in a human cell line.
FIGURE 1.
Detection of C7ORF49 isoform 2 (MRI-2) in K562 cells. A, peptidomics workflow to identify nonannotated short ORFs. The K562 cellular peptidome was isolated, fractionated, and subjected to liquid chromatography-mass spectrometry. Nonannotated peptides were identified by SEQUEST search against a K562 RNA deep sequencing library and subsequent removal of annotated sequences. B, MS/MS spectrum of the unique C7ORF49/MRI-2 C-terminal tryptic peptide, with detected fragment ions marked in blue (b ions) and red (y ions). C, multiple sequence alignment (ClustalW2) of the three MRI isoforms reveals that isoforms 1 and 2 have identical N-terminal sequences of 46 amino acids (red) before a frameshift generates a unique C-terminal sequence for isoform 2 (blue for isoform 2, green for isoform 1). Isoform 3 is identical to isoform 1, but lacks the N-terminal 46 amino acids.
This SEP represented a good candidate for functional characterization for two reasons. First, C7orf49 is conserved in mammals (24), suggesting that this gene has undergone functional selection. Second, the alternatively spliced C7orf49 gene is predicted to produce three polypeptide isoforms with distinct sequences, one of which has a known function (Fig. 1C). Isoform 1, a 157-amino acid polypeptide, was identified as a modulator of retrovirus infection in a phenotypic screen (24), although its molecular mechanism remains unclear. C7orf49 isoform 1 was therefore named modulator of retrovirus infection homolog 1 (MRI-1). The SEP we detect corresponds to MRI isoform 2 (MRI-2), and the third isoform is referred to as MRI-3, neither of which has been previously characterized. Taken together, we characterized the MRI-2 SEP because of the conservation and known cellular function of the MRI/C7orf49 gene.
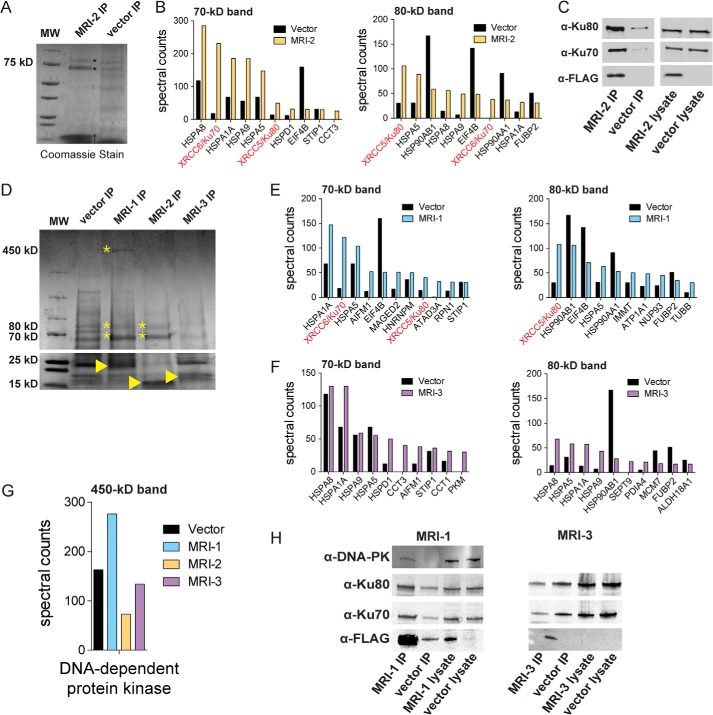
SEPs are small and, therefore, most likely to function via interactions with other biomolecules than to have catalytic activity. Identifying SEP-protein interactions using co-immunoprecipitation (co-IP) proteomics represents a general strategy for SEP functional characterization. Co-IP of MRI-2 from HEK293T cells enriched two bands at 70 and 80 kDa (Fig. 2A). Semiquantitative proteomics via spectral counting revealed these proteins to be Ku70 and Ku80, the two subunits of the heterodimeric DNA end-binding protein Ku (Fig. 2B). We validated these interactions by Western blotting using antibodies against Ku70 and Ku80 (Fig. 2C). We note that enrichment of heat shock protein 70 family members (e.g. HSPA8, HSPA1A, HSPA9), which function as chaperones, is not likely to be functionally relevant as these proteins are the most commonly detected nonspecific interaction partners detected in co-IP experiments (25).
FIGURE 2.
MRI-2 interacts with Ku. A, MRI-FLAG was co-immunoprecipitated (IP) from HEK 293T cells and analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining. The co-IP of MRI-2 (†) enriched two bands (*), which were identified as Ku70 (XRCC6) and Ku80 (XRCC5) by mass spectrometry (data not shown). MW, molecular weight markers. B, semiquantitative proteomics by spectral counting was used to confirm MRI-2 interaction partners. Bands of interest were excised from experimental and control samples by cutting straight across the gel and subjected to in-gel digest and proteomics. The top 10 most abundant proteins in each experimental sample were plotted in terms of spectral counts and compared with the negative control. Ku70 and Ku80 were enriched 3–10-fold. C, confirmation of MRI-2 interaction with Ku70 and Ku80 by Western blotting. FLAG-tagged MRI-2 or empty vector was immunoprecipitated from HEK 293T cells and then analyzed by Western blotting with antibodies against Ku70 and Ku80. Lysates were simultaneously probed to confirm equal loading. Enrichment of Ku70 and Ku80 by the MRI-2 co-IP as compared with vector background was observed. D, co-immunoprecipitation of MRI-1, MRI-2, and MRI-3 (indicated by arrowhead on gel) from HEK293T, with SDS-PAGE analysis and Coomassie Brilliant Blue staining. The four molecular mass regions analyzed were 70, 80, and 450 kDa (bands of interest indicated by *). E and F, semiquantitative proteomics by spectral counting demonstrates enrichment of Ku70 and Ku80 by MRI-1 (E) but not MRI-3 (F). G, spectral counting for the 450-kDa band for all three proteins shows enrichment of DNA-PK by MRI-1. H, co-IP and Western blotting confirm interactions of MRI-1 with Ku and DNA-PK and the absence of interactions with MRI-3. Lysates were analyzed to demonstrate equal loading.
MRI-1 and -3 may serve related functions, so we repeated the co-IP for the additional isoforms. Co-IP of MRI-1 and -3, followed by quantitative proteomics and Western blotting, revealed that MRI-1 also binds to Ku, as well as DNA-dependent protein kinase catalytic subunit (DNA-PKcs), whereas MRI-3 does not enrich any proteins (Fig. 2, D–H). MRI-1 and -2 likely interact with Ku through their shared N-terminal 46 amino acids, which are missing in MRI-3 (Fig. 1C).
Ku and DNA-PKcs are key participants in the NHEJ pathway of DNA double-strand break (DSB) repair. DSBs, caused by ionizing radiation, DNA-damaging drugs, and free radicals (26), are so toxic that a single unrepaired lesion can lead to cell death (27); alternatively, unrepaired DSBs can lead to chromosomal instability and/or translocations (27). NHEJ is the predominant DSB repair pathway in mammalian cells (28, 29). The first protein to bind the DSB is the Ku heterodimer, a ring-shaped DNA end-binding protein (30) that acts as the DNA-binding subunit of the DNA-dependent protein kinase (DNA-PK) (31). Binding of Ku and subsequent assembly of DNA-PK are followed by recruitment of additional repair factors that resect and ligate the DSB. The interaction of MRI-2 with Ku suggests that MRI-2 may participate in NHEJ.
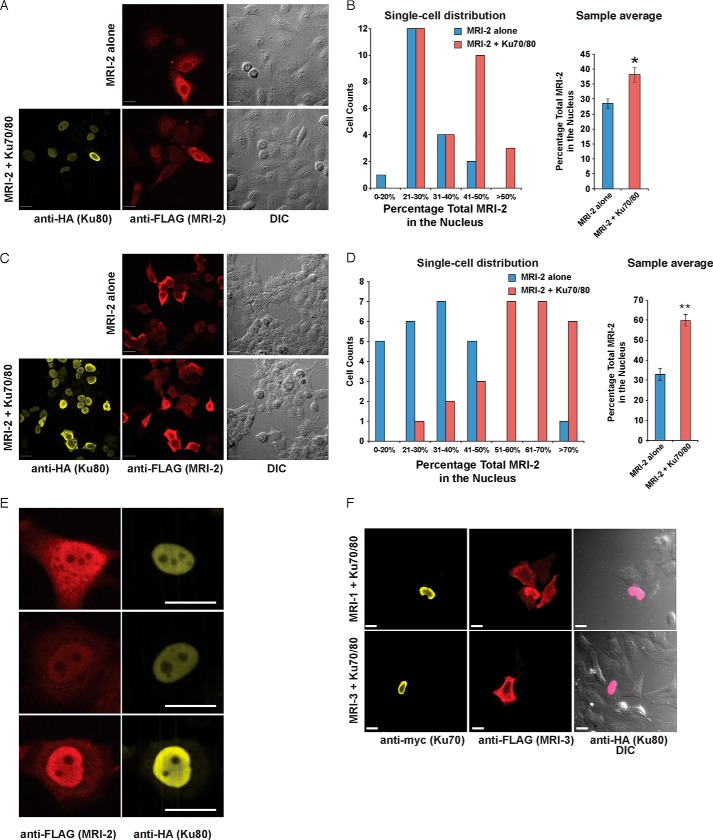
MRI-2 expressed in HeLa and HEK293T cells was observed in both nuclear and cytoplasmic compartments (Fig. 3, A and C, top). The nuclear fraction of MRI-2 in HeLa ranged from 19 to 45% of the total protein, with a population average of 29 ± 7% (S.E.) (Fig. 3A, top, and 3C). Similarly, in HEK293T cells, nuclear MRI-2 ranged from 14 to 80%, with a population average of 33 ± 3% (Fig. 3B, top, and 3D).
FIGURE 3.
MRI-1–3 nuclear localization requires Ku. A, the subcellular localization of MRI-2 was assessed by transfection into HeLa cells and detection with anti-FLAG immunofluorescence (red) and confocal microscopy. DIC images were acquired to analyze cell morphology. When transfected alone (top), MRI-2 partitions between the nucleus and cytoplasm; when co-transfected with Ku70 and Ku80 (bottom, Ku80-HA, yellow), the nuclear fraction of MRI-2 increases. B, single cell plots and population averages of the percentage of total MRI-2 immunofluorescence in the nucleus reveal a 38% increase in the presence of Ku (*, p < 0.05). Each data set represents >21 cells from four fields of view. An identical analysis in HEK293T cells (C and D) reveals a stronger enhancement of MRI-2 nuclear localization of 82% in the presence of Ku in this cell line (**, p < 0.005). Each data set represents >24 cells from two fields of view. E, the subnuclear distribution of MRI-2 (anti-FLAG, red) is identical to that of co-transfected Ku80 (anti-HA, yellow). F, top, MRI-1 (anti-FLAG, red) is also recruited to the nucleus by Ku co-expression (Ku70-Myc, yellow, and Ku80-HA, pink), but MRI-3 is not (bottom). Therefore, nuclear localization of MRI-1 and -2 may be mediated by their interaction with Ku. All scale bars, 20 μm. Statistical analysis was by two-tailed Student's t test, and error bars represent S.E.
Although a significant fraction of MRI-2 is nuclear, MRI-2 does not contain an obvious nuclear localization sequence. This SEP is small enough to passively diffuse through the nuclear pore, which is permeable to proteins <20–40 kDa (32). We hypothesize that the mechanism of MRI-2 nuclear localization depends upon its passive diffusion into nuclei followed by association with Ku, which is almost exclusively nuclear (33). For factors that depend upon protein-protein interactions for localization, overexpression can saturate interaction partners, leading to aberrant cellular distribution (34–36). This suggests that the cell-to-cell variation in MRI-2 nuclear localization might be caused by different levels of MRI-2 expression.
We therefore examined whether increased Ku expression, through co-expression of Ku70 and Ku80, increased nuclear localization of MRI-2. Co-expression of Ku resulted in enhanced nuclear localization of MRI-2, which ranged from 23 to 85% (average 38 ± 2%) in HeLa and from 27 to 85% (average 60 ± 3%) in HEK293T (Fig. 3, A and B, bottom, and C and D). This represents a Ku-mediated increase in MRI-2 nuclear localization of 33% in HeLa (p = 0.005) and 82% in HEK23T (p = 8 × 10−8). Inspection of nuclei of co-transfected cells revealed co-localization of MRI-2 and Ku80 (Fig. 3E). We then tested the effect of Ku co-expression on the subcellular distribution of MRI-1 and -3. Nuclear localization of MRI-1 was enriched by Ku co-expression, whereas MRI-3 was unaffected (Fig. 3F). These data are consistent with our hypothesis that nuclear enrichment of MRI-1 and -2 is dependent upon direct interaction with Ku.
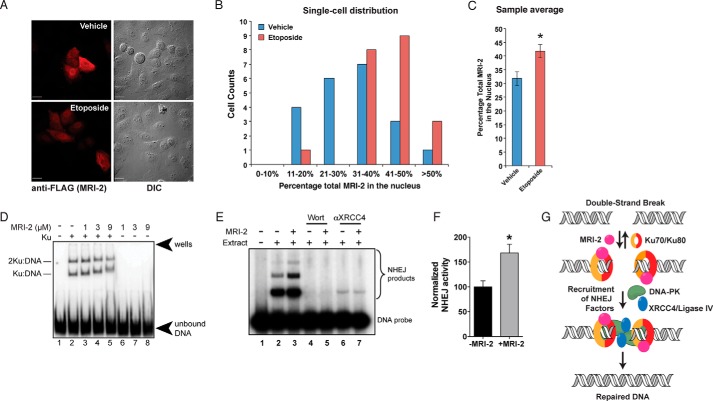
We then examined the nuclear localization of MRI-2 after induction of DSBs using the topoisomerase inhibitor etoposide, which creates DSBs and induces NHEJ. Etoposide treatment increased the nuclear localization of MRI-2 relative to vehicle-treated cells (Fig. 4A), evident in both single cell histograms of the percentage of MRI-2 in the nucleus (Fig. 4B) and the population average of MRI-2 nuclear localization, which increases by ∼31% (p = 0.007, Fig. 4C). Although the magnitude of this effect is small, nuclear recruitment of MRI-2 by etoposide treatment is consistent with a role for this protein in NHEJ in cells.
FIGURE 4.
MRI-2 nuclear localization during NHEJ, binding to Ku-DNA complexes, and stimulation of NHEJ in vitro support its functional role in DSB repair. A, representative images of HeLa cells expressing MRI-2 treated either with vehicle (top) or with 50 μm etoposide (bottom) for 4 h. This drug treatment elicits DSBs and activates NHEJ. B and C, single cell plots (B) and population averages (C) of the percentage of total MRI-2 in the nucleus demonstrate an etoposide-mediated increase of 31% (*, p < 0.05). Statistical analysis was by two-tailed Student's t test, and error bars represent S.E. Each data set represents 21 cells from five fields of view. D, MRI-2 interaction with a Ku-DNA complex was assayed by electrophoretic mobility shift. Purified Ku70 and Ku80 were preincubated with a radiolabeled DNA duplex, and increasing amounts of purified MRI-2 were added. Complexes of one and two Ku heterodimers bound to DNA are resolved by gel electrophoresis, and both exhibit progressive, dose-dependent supershift due to binding of multiple copies of MRI-2. E, NHEJ activity was assayed by adding radiolabeled double-stranded DNA probe to NHEJ-competent cell lysates and assaying formation of ligation products (dimers and trimers, bracket) by separation on a gel (top) in the absence or presence of 5 μm purified MRI-2. Wort, Wortmannin. F, quantitation of end-joining activity in four replicate experiments shows an increase in NHEJ efficiency (percentage of DNA ends joined) of 68% as compared with the basal reaction (p = 0.02, Student's t test). G, model of the potential interaction(s) between MRI-2 and the NHEJ machinery. The Ku heterodimer binds to DSBs and recruits other NHEJ factors. MRI-2 binds to Ku-DNA to stimulate this process.
Using an electrophoretic mobility shift assay, we confirmed that MRI-2 interacts with Ku bound to DNA in vitro. Radiolabeled double-stranded DNA is incubated with purified Ku70 and Ku80, which results in formation of Ku-DNA or Ku2-DNA complexes that can be resolved by gel electrophoresis. The addition of purified MRI-2 results in the decreased mobility of the complex, confirming tripartite complex formation (Fig. 4D). Although we do not have independent confirmation that the lower mobility species contain MRI-2, this conclusion is consistent with the co-IP data. Furthermore, the progressive mobility shift suggests that multiple molecules of MRI-2 bind to each Ku-DNA complex.
Finally, we measured the effect of purified MRI-2 on NHEJ efficiency in vitro using a double-stranded DNA ligation assay in cell extracts (18, 22, 37). Purified recombinant MRI-2 was added to NHEJ-competent cell extracts, and the conversion of a radiolabeled DNA duplex to higher molecular weight oligomeric ligation products was monitored. Comparison of the basal NHEJ reaction to a reaction containing added MRI-2 (5 μm) demonstrated an increase in product formation of 68% (p = 0.02) (Fig. 4E). Controls employing a DNA-PK inhibitor, wortmannin, as well as an antibody against the required NHEJ protein, XRCC4, eliminate end joining in these extracts, confirming that the ligation activity observed is due to bona fide NHEJ. This demonstrates that MRI-2 stimulates DSB ligation via NHEJ.
DISCUSSION
In this study, we have identified MRI-2 as an interaction partner of Ku. MRI-2 is recruited to the nucleus by Ku overexpression and by induction of DSBs, and enhances the rate of NHEJ in vitro. MRI-2 is therefore a novel NHEJ factor (Fig. 4F), which may stimulate NHEJ in a number of ways, such as enhancing DSB binding by Ku, promoting assembly of the repair complex, and/or releasing Ku from DNA to promote multiple turnovers. Future mechanistic studies will reveal the molecular role of this SEP in DSB repair.
The MRI gene produces three polypeptides, MRI-1–3, and although we focused on characterization of the interaction partners and functions of MRI-2, we also investigated the interactions of MRI-1 and -3. MRI-1 interacts with Ku as well as DNA-PK, suggesting that MRI-1 and -2 may both function in NHEJ. MRI-3 does not bind to Ku, suggesting that this isoform may be inactive. These varying interaction specificities identify a Ku binding sequence in MRI-1 and -2, and also suggest that alternative splicing of MRI transcripts could differentially affect NHEJ efficiency inside cells.
MRI-1 was originally identified as a factor that enabled retroviral infection of mammalian cells (24), based on genetic complementation of retroviral infection-resistant cells by overexpression of an MRI-1 cDNA clone. This study also reported that RNAi-mediated silencing of MRI decreases retroviral infection sensitivity. Because treatment of the cells with a proteasome inhibitor phenocopied MRI-1 overexpression, the authors concluded that MRI-1 acts on the proteasome. Our work, however, suggests a different mechanism for the involvement of the MRI gene in retroviral infection. It is well established that NHEJ in the host cell is utilized during retroviral infection, promoting stable viral genome integration by resolving linear DNA fragments of this process (38–41). Therefore, MRI-1 may enhance retroviral infection via its interaction with Ku, and MRI-2 may also be involved in this process. Future work to determine the role of NHEJ during the rescue of retroviral expression by MRI-1 and -2 will test this hypothesis.
In conclusion, hundreds of newly discovered SEPs have now been reported in human cells (1, 6–8), but few have been functionally characterized. The identification of the MRI-2-Ku interaction suggests that functional proteomics is a general method to identify the cellular and biochemical roles of newly discovered SEPs. The involvement of MRI-2 in NHEJ, a central process required for cell survival, suggests that many more SEPs may have important cellular functions and demonstrates the continued need to discover and functionally characterize all SEPs produced from the human genome.
Acknowledgment
We thank Alice Ting for the use of a confocal microscope.
This work was supported by a National Research Service Award postdoctoral fellowship (1F32GM099408) (to S. A. S.) and National Institutes of Health Grant (R01GM102491) through the NIGMS (to A. S.).
- sORF
- short open reading frame
- SEP
- sORF-encoded polypeptide
- MRI
- modulator of retroviral infection
- NHEJ
- nonhomologous end joining
- DIC
- differential interference contrast
- DSB
- double-strand break
- DNA-PK
- DNA-dependent protein kinase
- co-IP
- co-immunoprecipitation
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1. Andrews S. J., Rothnagel J. A. (2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15, 193–204 [DOI] [PubMed] [Google Scholar]
- 2. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ingolia N. T., Lareau L. F., Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappé J., Gevaert K., Van Damme P. (2013) Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12, 1780–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vanderperre B., Lucier J. F., Bissonnette C., Motard J., Tremblay G., Vanderperre S., Wisztorski M., Salzet M., Boisvert F. M., Roucou X. (2013) Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One 8, e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma J., Ward C. C., Jungreis I., Slavoff S. A., Schwaid A. G., Neveu J., Budnik B. A., Kellis M., Saghatelian A. (2014) Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J. Proteome Res. 13, 1757–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slavoff S. A., Mitchell A. J., Schwaid A. G., Cabili M. N., Ma J., Levin J. Z., Karger A. D., Budnik B. A., Rinn J. L., Saghatelian A. (2013) Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwaid A. G., Shannon D. A., Ma J., Slavoff S. A., Levin J. Z., Weerapana E., Saghatelian A. (2013) Chemoproteomic discovery of cysteine-containing human short open reading frames. J. Am. Chem. Soc. 135, 16750–16753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galindo M. I., Pueyo J. I., Fouix S., Bishop S. A., Couso J. P. (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., Kageyama Y. (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 9, 660–665 [DOI] [PubMed] [Google Scholar]
- 11. Magny E. G., Pueyo J. I., Pearl F. M., Cespedes M. A., Niven J. E., Bishop S. A., Couso J. P. (2013) Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341, 1116–1120 [DOI] [PubMed] [Google Scholar]
- 12. Kastenmayer J. P., Ni L., Chu A., Kitchen L. E., Au W. C., Yang H., Carter C. D., Wheeler D., Davis R. W., Boeke J. D., Snyder M. A., Basrai M. A. (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 16, 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casson S. A., Chilley P. M., Topping J. F., Evans I. M., Souter M. A., Lindsey K. (2002) The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14, 1705–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rohrig H., Schmidt J., Miklashevichs E., Schell J., John M. (2002) Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. U.S.A. 99, 1915–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Coninck B., Carron D., Tavormina P., Willem L., Craik D. J., Vos C., Thevissen K., Mathys J., Cammue B. P. (2013) Mining the genome of Arabidopsis thaliana as a basis for the identification of novel bioactive peptides involved in oxidative stress tolerance. J. Exp. Bot. 64, 5297–5307 [DOI] [PubMed] [Google Scholar]
- 16. Hashimoto Y., Ito Y., Niikura T., Shao Z., Hata M., Oyama F., Nishimoto I. (2001) Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein. Biochem. Biophys. Res. Commun. 283, 460–468 [DOI] [PubMed] [Google Scholar]
- 17. Hashimoto Y., Niikura T., Tajima H., Yasukawa T., Sudo H., Ito Y., Kita Y., Kawasumi M., Kouyama K., Doyu M., Sobue G., Koide T., Tsuji S., Lang J., Kurokawa K., Nishimoto I. (2001) A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Aβ. Proc. Natl. Acad. Sci. U.S.A. 98, 6336–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanakahi L. A. (2007) 2-Step purification of the Ku DNA repair protein expressed in Escherichia coli. Protein Expr. Purif. 52, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baumann P., West S. C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. U.S.A. 95, 14066–14070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanakahi L. A., Bartlet-Jones M., Chappell C., Pappin D., West S. C. (2000) Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell 102, 721–729 [DOI] [PubMed] [Google Scholar]
- 21. Cheung J. C., Salerno B., Hanakahi L. A. (2008) Evidence for an inositol hexakisphosphate-dependent role for Ku in mammalian nonhomologous end joining that is independent of its role in the DNA-dependent protein kinase. Nucleic Acids Res. 36, 5713–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smeaton M. B., Miller P. S., Ketner G., Hanakahi L. A. (2007) Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 35, e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Christofk H. R., Vander Heiden M. G., Wu N., Asara J. M., Cantley L. C. (2008) Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 [DOI] [PubMed] [Google Scholar]
- 24. Agarwal S., Harada J., Schreifels J., Lech P., Nikolai B., Yamaguchi T., Chanda S. K., Somia N. V. (2006) Isolation, characterization, and genetic complementation of a cellular mutant resistant to retroviral infection. Proc. Natl. Acad. Sci. U.S.A. 103, 15933–15938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jackson S. P. (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23, 687–696 [DOI] [PubMed] [Google Scholar]
- 28. Chapman J. R., Taylor M. R., Boulton S. J. (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 [DOI] [PubMed] [Google Scholar]
- 29. Lieber M. R., Ma Y., Pannicke U., Schwarz K. (2003) Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4, 712–720 [DOI] [PubMed] [Google Scholar]
- 30. Walker J. R., Corpina R. A., Goldberg J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 31. Smith G. C., Jackson S. P. (1999) The DNA-dependent protein kinase. Genes Dev. 13, 916–934 [DOI] [PubMed] [Google Scholar]
- 32. Fried H., Kutay U. (2003) Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60, 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bertinato J., Schild-Poulter C., Haché R. J. (2001) Nuclear localization of Ku antigen is promoted independently by basic motifs in the Ku70 and Ku80 subunits. J. Cell Sci. 114, 89–99 [DOI] [PubMed] [Google Scholar]
- 34. Gross G. G., Junge J. A., Mora R. J., Kwon H. B., Olson C. A., Takahashi T. T., Liman E. R., Ellis-Davies G. C., McGee A. W., Sabatini B. L., Roberts R. W., Arnold D. B. (2013) Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gibson T. J., Seiler M., Veitia R. A. (2013) The transience of transient overexpression. Nat. Methods 10, 715–721 [DOI] [PubMed] [Google Scholar]
- 36. Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M. R. (2001) Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanakahi L. (2011) Effect of the inositol polyphosphate InsP6 on DNA-PK-dependent phosphorylation. Mol. Cancer Res. 9, 1366–1376 [DOI] [PubMed] [Google Scholar]
- 38. Daniel R., Greger J. G., Katz R. A., Taganov K. D., Wu X., Kappes J. C., Skalka A. M. (2004) Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J. Virol. 78, 8573–8581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee S. H., Oshige M., Durant S. T., Rasila K. K., Williamson E. A., Ramsey H., Kwan L., Nickoloff J. A., Hromas R. (2005) The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc. Natl. Acad. Sci. U.S.A. 102, 18075–18080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li L., Olvera J. M., Yoder K. E., Mitchell R. S., Butler S. L., Lieber M., Martin S. L., Bushman F. D. (2001) Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20, 3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jeanson L., Subra F., Vaganay S., Hervy M., Marangoni E., Bourhis J., Mouscadet J. F. (2002) Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 300, 100–108 [DOI] [PubMed] [Google Scholar]