Background: ELMOD proteins are atypical GAPs for the ARF family with links to deafness.
Results: ELMOD proteins have distinct specific activities for ARF proteins and bind the non-opioid sigma-1 receptor.
Conclusion: Different specific activities of ELMOD1–3 are predicted to underlie different cellular functions.
Significance: Specificities and regulation of ELMODs contribute to understanding their roles in cell signaling and the inner ear.
Keywords: ARF, Enzyme Kinetics, GTPase, Sigma Receptor, Signal Transduction, ARL2, ELMOD, GTPase-activating Protein (GAP), HEK Cell Expression, Sigma-1 Receptor (S1R)
Abstract
The ARF family of regulatory GTPases, within the RAS superfamily, is composed of ∼30 members in mammals, including up to six ARF and at least 18 ARF-like (ARL) proteins. They exhibit significant structural and biochemical conservation and regulate a variety of essential cellular processes, including membrane traffic, cell division, and energy metabolism; each with links to human diseases. We previously identified members of the ELMOD family as GTPase-activating proteins (GAPs) for ARL2 that displayed crossover activity for ARFs as well. To further characterize the GAP activities of the three human ELMODs as GAPs we developed new preparations of each after overexpression in human embryonic kidney (HEK293T) cells. This allowed much higher specific activities and enhanced stability and solubility of the purified proteins. The specificities of ELMOD1–3 as GAPs for six different members of the ARF family were determined and found to display wide variations, which we believe will reveal differences in cellular functions of family members. The non-opioid sigma-1 receptor (S1R) was identified as a novel effector of GAP activity of ELMOD1–3 proteins as its direct binding to either ELMOD1 or ELMOD2 resulted in loss of GAP activity. These findings are critical to understand the roles of ELMOD proteins in cell signaling in general and in the inner ear specifically, and open the door to exploration of the regulation of their GAP activities via agonists or antagonists of the S1R.
Introduction
GTPase-activating proteins (GAPs)2 bind to the activated (GTP-bound) conformations of regulatory GTPases within the RAS superfamily to speed the rate of GTP hydrolysis and consequently inactivate the signaling function. Regulators of G protein signaling proteins provide comparable functions for the trimeric G proteins. These regulators of GTPase biology were originally viewed as solely terminators of signal propagation but more recently have come to be appreciated as providing essential spatial and temporal regulation to the biology of each GTPase. In addition, many GAPs are themselves effectors and essential downstream components in the pathway controlled by the GTPase (1).
GAPs for different families of GTPase use a number of different mechanisms but one common one is the insertion of a “catalytic arginine,” provided by the GAP, into the nucleotide-binding site near the β-γ phosphoryl bond to help neutralize the negative charge in the transition state of the hydrolysis reaction (2, 3). GAPs are capable of speeding the rate of GTP hydrolysis by the GTPase as much as 5 orders of magnitude. Members of the ARF family lack any detectible intrinsic GTPase activity so are completely dependent upon GAPs to provide timing or termination to signaling (4). To date, all GAPs that work on ARF family members employ a catalytic arginine mechanism and mutation of that single residue, even to the conservative lysine, results in a substantial loss of GAP activity (5, 6).
GAPs can speed GTP hydrolysis by multiple GTPases, although to date only within one family of GTPases. But within that family one GAP may act on GTPases with distinct functions, thereby creating a potential functional linkage between signaling pathways. Thus, the identification of the GAP(s) acting on any GTPase and the specificities of GAPs for substrates are central questions to the generation of accurate models of cell signaling by regulatory GTPases.
Shortly after RAS and related GTPases were found to play central roles in oncogenesis and human disease, their GAPs were found to play similar roles, consistent with their roles as effectors and modulators of GTPase signaling. Thus, mutations in RAS GAPs (e.g. NF1 and RASAL2) (7–10) are oncogenic. Diseases have also been linked to the regulators of G protein signaling/GAP proteins acting on heterotrimeric G proteins (11). More recently, two of the three members of the ELMOD (cell engulfment and motility domain containing) family of ARL2 GAPs have been linked to deafness in mice and humans: ELMOD1 (12) and ELMOD3 (13), respectively.
ARF-like 2 (ARL2), within the ARF family, is notable for having established roles in the regulation of tubulin folding and microtubule dynamics, but is also found inside mitochondria and in the nucleus (14, 15). The diversity of functions adds to the complexity in deconvolving its signaling pathways but also supports the idea that ARL2 has the potential to serve as a mediator between these essential cell functions. In studies of ARL2 biology we purified and identified the ARL2 GAP, ELMOD2 (16). The closest paralogs, ELMOD1 and ELMOD3, share GAP activity and a conserved ELMO domain, a ∼160 residue domain also present in the three human ELMO proteins, ELMO1–3 (17, 18). The ELMODs are ancient and likely present in the last eukaryotic common ancestor, whereas the ELMOs arose later and are found only in metazoans and fungi (19). The ELMO proteins often contain additional domains (e.g. a single PH domain) and may be obligate heterodimers with members of the DOCK family of unconventional Rac1 guanine nucleotide exchange factors (20). In contrast, the ELMODs possess the single ELMO domain and no obligate or other partners have been identified. We earlier identified a highly conserved arginine, absent in the ELMO proteins, which is critical for GAP activity of ELMOD1 and ELMOD2 (19). Our earlier studies were hampered by the fact that human ELMODs expressed in bacteria were insoluble or partially soluble but very unstable. Thus, we often had to use lysates of HeLa cells overexpressing each ELMOD protein to perform basic characterizations of the protein. Such assays suffer from high backgrounds of non-GAP-mediated GTP hydrolysis and uncertain amounts of the GAP being assayed. Thus, the absence of a stable and active preparation of mammalian ELMODs has been a clear hindrance to further progress in characterizing their activities modeling their cellular roles. Here we describe the expression, purification, and characterization of the mammalian cell expressed recombinant human ELMOD1–3.
EXPERIMENTAL PROCEDURES
Plasmids
The human ELMOD1–3 complete open reading frames (ORFs) were amplified by PCR, using primers that introduced KpnI and SphI restriction sites. These were used to subclone into the pLEXm-GST vector (the generous gift of Dr. James Hurley, NIH) resulting in pLEXm-GST-ELMOD1–3. The ELMOD1 and ELMOD2 ORFs were inserted into the pET28 and pCold-TF bacterial expression vectors resulting in N-terminal maltose-binding protein (MBP) and trigger factor (TF) fusion proteins, respectively. DNA sequencing of each plasmid generated through the use of PCR confirmed the following sequences: ELMOD1 (NM_018712.3, encodes a protein of 334 residues), ELMOD2 (NM_153702, encodes a protein of 293 residues), and ELMOD3, isoform b (NM_001135021.1, encodes a protein of 381 residues). In addition, mouse ARL13B (NM_026577.3, encodes a protein of 427 residues) was also subcloned into the pLEMm-GST vector. The plasmids used for expression of MBP-ELMOD1 and ELMOD2 were described previously (16, 19). Vectors used for bacterial expression of mouse ELMO1 and ELMO2 were provided by Dr. Kodi Ravichandran (University of Virginia) and included the entire ORFs in the pGEX-4T-2 plasmid. A plasmid containing the rat non-opioid sigma-1 receptor (S1R) cDNA subcloned into pFLAG-CMV-5a was provided by Dr. Tsung Ping Su (21).
Bacterial Expression and Purification
The complete ORFs of the human ARF family members (ARL1, ARL2, ARL3, ARF1, and ARF6) were cloned into pET3C for bacterial expression and purified as described previously (22, 23). Briefly, expression of GTPases was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside in Luria Broth (LB) medium (24, 25) or through the use of autoinduction medium (26). Bacteria were lysed with a French press and homogenates were clarified by centrifugation at 100,000 × g for 1 h. Recombinant proteins were purified on sequential ion exchange and gel filtration columns, as previously described (23). Purity was typically ∼90%, as estimated by visual inspection of Coomassie Blue-stained SDS gels. The MBP fusion proteins were expressed and purified as described by Bowzard et al. (16). TEV was expressed and purified as described previously (27, 28).
TF fusion proteins were expressed in bacteria grown at 37 °C to mid-log (A600 = 0.5) in LB medium supplemented with 100 μg/ml of ampicillin. The culture was then chilled to 4 °C for 20 min without shaking, and protein expression was induced by addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Induction continued for 16 h at 15 °C with agitation at 250 rpm. Cells were collected, pelleted by centrifugation at 6000 rpm, and suspended in lysis buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, and 5 mm imidazole). The cells were lysed with a French press and homogenates were clarified by centrifugation at 100,000 × g. His6 tags in the TF fusion were used for purification using a 5-ml HiTrap Ni-NTA column (GE Healthcare). The column was washed with 50 ml of wash buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 50 mm imidazole), and eluted with 15 ml of elution buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 250 mm imidazole). The proteins were concentrated by ultrafiltration using a unit with a 30-kDa cutoff (Millipore), and analyzed for purity in SDS gels.
Mammalian Cell Expression and Purification
Human embryonic kidney 293T (HEK) cells were grown in 10-cm plates in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen), supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), at 37 °C in a humidified environment gassed with 5% CO2. The medium was switched to DMEM with 2% FBS and cells were transfected with 1 μg of DNA/ml of medium at a cell density of ∼90% using 3 μg of polyethyleneimine (PEI Max; Polysciences, catalog number 24765-2)/ml of medium. Time course testing suggested that 2 days was optimal for protein expression. Thus, cells were harvested 48 h after transfection, pelleted by centrifugation at 3,000 × g, frozen in liquid nitrogen, and stored at −80 °C until used for protein purification.
Cells were thawed on ice and lysed in 5 volumes of lysis buffer (50 mm HEPES, pH 7.4, 100 mm NaCl, 1% CHAPS, with protease inhibitor mixture (Sigma S8830), and 10 μg/ml of deoxyribonuclease I (DNase, Sigma D4263)). The cells were maintained on ice for 30 min before cell debris was removed by centrifugation at 14,000 × g for 10 min at 4 °C. The proteins were purified with GST- or FLAG-affinity chromatography. Specifically, 500 μl of glutathione-Sepharose 4B (GE Healthcare, catalog number 17-0756-01) beads or 100 μl of anti-FLAG M2 affinity gel (Sigma F2426) was added to 120 mg of protein lysate and incubated at 4 °C for 2 h. The beads were then washed three times with 5 column volumes of lysis buffer. The proteins were eluted from the beads with 3 column volumes of elution buffer (25 mm HEPES, pH 7.4 100 mm NaCl, 10 mm glutathione) or 50 ng/μl of FLAG peptide (Sigma F4799), respectively.
GST Cleavage
On-column cleavage of the GST-ELMOD2 was accomplished by addition of up to 30% (mg/mg of protein) recombinant TEV protease to the beads bound with the GST-ELMOD2 and samples were incubated overnight at 4 °C with agitation. Proteins were washed from the beads with the wash buffer (25 mm HEPES, pH 7.4, 100 mm NaCl).
Nucleotide Binding Assay
This assay was used to quantify nucleotide binding to GTPases. Each GTPase (1 μm) was assayed in a buffer consisting of 20 mm HEPES, pH 7.4, 100 mm NaCl, 0.5–2.5 mm MgCl2 (varies with GTPases), 1 mm EDTA, and 10 μm nucleotide ([35S]GTPγS, [γ-32P]GTP, or [3H]GDP (PerkinElmer Life Sciences) at specific activities of 5000–8000 cpm/pmol. Detergent (0.1% Triton X-100) was included in some assays for those GTPases displaying a lipid/detergent dependence. Binding at 30 °C at time points up to 1 h was determined after stopping the reaction by addition of 2 ml of ice-cold TNMD (20 mm Tris, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol), and filtration through BA85 nitrocellulose filters (0.45 μm, 25 mm (Whatman)). Scintillation fluid (Fisher Scientific) was added to filters and binding was quantified using a liquid scintillation counter. Bound nucleotide was quantified by normalization to specific activity after subtraction of blanks, as previously described (16).
GAP Assays
GAP activities were determined using one of two different assays, depending upon the off-rate of guanine nucleotides from the GTPase used in the assay. The “charcoal assay” was used for ARL2 because dissociation of bound nucleotides is too rapid to allow quantification after filtration on nitrocellulose filters. For other GTPases the “filter-binding assay” was used. Each assay measures a single round of GTP hydrolysis as they each require pre-binding with radiolabeled nucleotide and the GAP incubation is performed in the presence of a vast excess of unlabeled nucleotide to prevent re-binding.
Both charcoal and filter-binding assays involved pre-loading the GTPase (1 μm) with [γ-32P]GTP at 30 °C for 30 min in a total volume of 100 μl of loading buffer (ARL2 = 25 mm HEPES, pH 7.4, 100 mm NaCl, 2.5 mm MgCl2, 1 mm EDTA; other GTPases = 25 mm HEPES, pH 7.4, 100 mm NaCl, 0.5 mm MgCl2, 1 mm EDTA, 0.1% Triton X-100). The GAP reaction was performed in a total volume of 50 μl. The reaction was initiated by the addition of 5 μl of pre-loaded GTPase, the assay buffer consisted of 25 mm HEPES, pH 7.4, 2.5 mm MgCl2, 1 mm dithiothreitol, 1.6 mm ATP, and 2.5 mm GTP. These non-radiolabeled nucleotides are present to decrease nonspecific and non-GAP-dependent hydrolysis, thereby lowering the overall background. Reactions were incubated for 4 min at 30 °C. For the charcoal assays, reactions were stopped by adding 750 μl of ice-cold charcoal suspension (5% in 50 mm NaH2PO4) to the tubes with mixing. Charcoal, with bound nucleotides, was pelleted at 4 °C by centrifugation at 3,000 × g for 10 min. Half (400 μl) of each sample was counted in the liquid scintillation counter to determine the amount of GAP-dependent, released 32Pi. For the filter-binding assays, reactions were stopped by adding 2 ml of TNMD buffer (20 mm Tris, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm DTT), and filtration through BA85 nitrocellulose filters (0.45 μm, 25 mm (Whatman)).
In parallel, the following control reactions were performed: GTPase with pre-bound [32P]GTP without GAP to monitor any intrinsic GTPase activity (mock), and GAP plus [32P]GTP but without GTPase to monitor any GTPase activity present in the GAP sample. Thus, only the GAP-dependent activities are described, and specific activities reflect conversion from counts per minute to picomole of GTP hydrolyzed, normalized to the amount of GAP protein and time of the assay (16).
Stable Isotope Labeling with Amino Acids in Cell Culture
HeLa cells infected with lentivirus encoding Tet-On inducible expression of human ELMOD1-HA, as previously described (19), were cultured for 11 days in DMEM supplemented with 200 mg/ml of proline and 10% FBS without (lite medium) or with 13C- and 15N-labeled arginine and lysine (heavy medium; Dundee Cell). On the 10th day, ELMOD1-HA expression was induced with 2 μg/ml of doxycycline for 12 h, where indicated. Cells were harvested and lysed in 25 mm HEPES, pH 7.4, 100 mm NaCl, 1% CHAPS, and protease inhibitor mixture (Sigma). Lysates were cleared by centrifugation at 14,000 × g for 5 min. A total of 1.5 mg of protein was incubated with 1 μl of HA antibody (Covance MMS-101P) for 1 h at 4 °C. Protein G-Sepharose 4 Fast Flow beads (GE Healthcare) were washed twice with lysis buffer and 30 μl of the slurry was incubated with each lysate for 1 h at 4 °C. Beads were collected at 14,000 × g for 15 s and washed twice with lysis buffer. The final pellets were resuspended in 40 μl of Laemmli SDS sample buffer. A sample from cells cultured in heavy medium and induced for ELMOD1-HA expression was combined with an equal volume from a sample from uninduced cells cultured in lite medium and vice versa. Samples were run 0.6 cm into an 11% polyacrylamide SDS gel and the gel was excised for tandem MS/MS analysis at the Emory Mass Spectrometry Core Facility. Mass spectrometry, bioinformatics, and quantitation of peptide fold-enrichment were performed as described previously (29, 30).
Co-immunoprecipitation
HEK293T cells were grown in 6-well plates to ∼90% confluence. Medium was changed to DMEM with 2% FBS and the cells were transfected with 6 μg of PEI using 1 μg each of the following plasmids: GST-ELMOD and FLAG-vector, GST-ELMOD and FLAG-S1R, FLAG-S1R and GST vector. After 48 h the cells were collected and lysed with 25 mm HEPES, pH 7.4, 100 mm NaCl, 1% CHAPS, supplemented with protease inhibitor mixture (Sigma S8830) and DNase I. Lysates were clarified at 13,000 × g for 30 min at 4 °C. The supernatants were collected and incubated with glutathione-Sepharose 4B beads or anti-FLAG M2 affinity gel (Sigma F2426) at 4 °C for 2 h. Beads were then washed 4 times with 5 column volumes of lysis buffer. The proteins were eluted in 2× SDS buffer, boiled for 3 min, and analyzed with SDS-PAGE gel electrophoresis and immunoblotting.
RESULTS
Previous work in our laboratory had documented that all three mammalian ELMOD proteins, ELMOD1–3, possess GAP activity against ARL2 and that ELMOD2 also displayed weaker GAP activity for ARFs (13, 16). However, different preparations used in earlier work yielded proteins that were unstable to even one cycle of freeze/thaw or maintenance at 4 °C. Most also displayed highly variable, unstable, and disparate specific activities that were for the most part well below that described for the ARL2 GAP activity purified from bovine testes (16) (also see Table 1). Thus, a soluble, stable and active preparation of the ELMOD proteins was sought, to allow detailed characterizations of the activities and specificities of this novel family of cell regulators with predicted effector functions.
TABLE 1.
Specific activities of bacterially or mammalian cell expressed ELMOD proteins as ARL2 GAPs
ARL2 was present at a concentration of 0.2 μm in the assay. The specific activity of each ELMOD preparation was obtained from titration curves that included at least eight concentrations between 0 and 4 μm GAP. The specific activity values were calculated from the average of at least three points, each performed in duplicate, within the linear region of the curve obtained in two experiments using different preparations of each GAP. Variations within duplicates and between experiments were less than 15%.
| Protein | SA |
|
|---|---|---|
| HEK | Escherichia coli | |
| nmol min−1 mg−1 | ||
| GST-ELMOD1 | 0.76 | NDa |
| GST-ELMOD2 | 30.0 | ND |
| GST-ELMOD3 | 0.03 | ND |
| TF-ELMOD1 | ND | 0.0003 |
| TF-ELMOD2 | ND | 0.007 |
| TF-ELMOD3 | ND | NAb |
| CL ELMOD2c | 22 | ND |
| BVN ELMOD2d | 23 | ND |
a ND, not determined.
b NA, not active, indicates no detectable difference between the ELMOD sample and buffer control in the GAP assay.
c CL, ELMOD2 that has had its GST tag cleaved with TEV protease.
d BVN ELMOD2, ELMOD2 purified from bovine testis.
Bacterial Expression
We reported earlier that bacterially expressed MBP-ELMOD2 was mostly insoluble but that the soluble fraction displayed ARL2 GAP activity that was ∼1,000-fold lower than the preparation containing ELMOD2 that was purified from bovine testes (16). This preparation was also very unstable, likely contributing to the low specific activity. A number of different bacterial expression systems were assessed. Attempts at refolding protein from the insoluble pellet or inducing protein expression in bacteria under a variety of conditions, which included lower temperatures and concentrations of inducer, were not successful at increasing the yield or activity, with one exception. Insertion of the open reading frames of human ELMODs into the pCOLD-TF vector allowed expression of N-terminal fusion proteins with trigger factor, a 48-kDa prokaryotic chaperone. Each of the three human ELMODs were expressed as trigger factor fusion proteins, TF-ELMOD1–3, that contained a His6 motif and thrombin cleavage site between the trigger factor and ELMOD sequences to allow rapid purification and cleavage of the tag.
TF-ELMOD1–3 were expressed to high levels in bacteria and purified using Ni-chelate chromatography, as described under “Experimental Procedures” (Fig. 1A). Yields were typically ∼80 mg/liter for TF-ELMOD1 or TF-ELMOD2, and ∼20 mg of protein/liter of bacterial culture for TF-ELMOD3. In each case the TF-ELMOD proteins were largely soluble, remaining in solution after lysis of bacteria using a French press and centrifugation at 100,000 × g. Solubility was retained after freeze/thaw cycles or storage at 4 °C.
FIGURE 1.
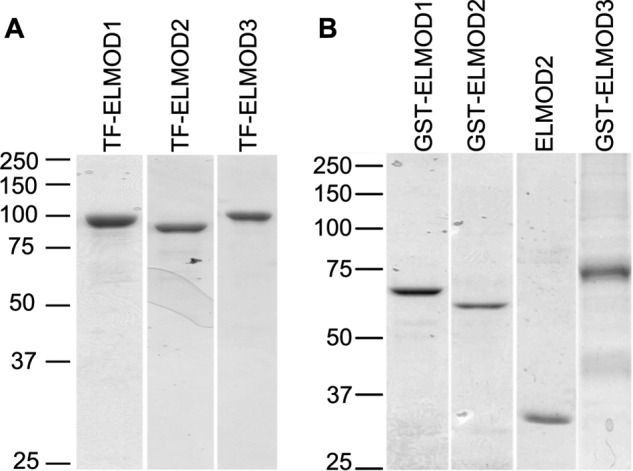
A, TF-ELMOD1–3 proteins purified from bacteria. Each of the human ELMOD proteins was expressed in bacteria as N-terminal TF fusion proteins and purified as described under “Experimental Procedures.” Proteins (1 μg each) were resolved in 11% acrylamide SDS gels and stained with Coomassie Blue. B, GST-ELMOD1–3 proteins and ELMOD2 after cleavage of the GST tag, purified from HEK293T cells. Proteins were expressed in HEK cells, purified, and one (ELMOD2) was cleaved with TEV, as described under “Experimental Procedures” prior to loading samples (1 μg each) onto SDS-PAGE gels and staining with Coomassie Blue.
Assessing Activity of Bacterially Expressed Proteins
Specific activities in all cases reported herein were determined by titration of the GAP at a fixed concentration of the substrate in the GAP assay. The rates of GTP hydrolysis as a function of GAP concentration were then plotted and saturation curves resulted. The linear portion of the curve, including a minimum of three points, each determined in duplicate, was used to determine the specific activities. Specific activities determined in this way were found to be quite consistent between preparations and with some variation in substrate concentrations.
GAP assays of the purified TF fusion proteins using ARL2·[32P-γ]GTP as substrate yielded specific activities of 0.0003 and 0.007 nmol of GTP hydrolyzed/min/mg of protein for TF-ELMOD1 and TF-ELMOD2, respectively (Table 1). No activity was detected with up to 10 μg (∼1 μm)/assay of TF-ELMOD3. We had earlier purified ELMOD2 from bovine testes and determined its specific activity in the same assay to be 23.0 nmol/min/mg (16). Thus, TF-ELMOD2 was found to be ∼3,000-fold less active than bovine ELMOD2 in the same assay. We considered the possibility that the TF fusion protein at the N terminus interfered with activity, despite the earlier finding that truncation of the N-terminal 80 residues did not alter GAP activity of recombinant ELMOD2 (16). To test this, thrombin was added to Ni-NTA-purified protein at final concentrations of up to 5 units/mg of TF-ELMOD2, and incubated for up to 24 h at 20 °C in efforts to cleave the tag. Cleavage was monitored with SDS-PAGE and found to be incomplete even at the highest concentrations of protease used and after prolonged incubations. In addition, the cleaved proteins were not resolved with either gel filtration or Ni-NTA chromatography, suggesting interactions between the proteins even after cleavage. Thus, despite the fact that the TF fusions of the human ELMODs express to high levels in bacteria, are readily purified, and remain in solution after purification, freeze/thaw cycles and storage at 4 or −80 °C, these preparations have very low specific activities and are not readily purified away from the large TF fusion protein. We have found these preparations useful for some purposes (e.g. as antigens in raising antibodies in mice and rabbits) but not in studies of their functional, enzymatic properties.
HEK Cell Expression of ELMODs
Adherent HEK cells were tested for use in the expression of recombinant human ELMOD proteins, using variations of the methods described in Ariescu et al. (31, 32). PEI was used as a lower cost alternative to lipid-based transfection reagents for large scale transfections. Trials were performed in which the amounts of PEI and DNA, and the ratio of the two were varied to determine the optimal conditions for each protein expressed. We also varied the time after transfection for optimal protein expression. We found that collecting cells 48 after transfection of ∼90% confluent HEK cells in a 10-cm plate (∼107 cells) with 30 μg of PEI and 10 μg of DNA was optimal for all three human ELMOD proteins in the pLEXm-GST vector.
After collection, the cells are lysed by resuspension into buffer containing 1% CHAPS (50 mm HEPES, pH 7.4, 100 mm NaCl, 1% CHAPS, DNase I, and protease inhibitor mixture) and proteins were affinity purified using glutathione-Sepharose beads, as described under “Experimental Procedures.” The yields of purified GST-ELMOD1–3 proteins from HEK cells prepared in this way were typically ∼1 mg/liter of medium for GST-ELMOD1 or GST-ELMOD2, and ∼0.35 mg/liter of medium for GST-ELMOD3. The purified proteins were stable to freezing and storage for at least a few months at −80 °C with no apparent loss of activity (see below). However, these preparations do lose activity when stored for days at 4 °C. We found that ELMOD2 lost about half of its GAP activity after 6 days of storage at 4 °C so are typically used within 24 h of thawing for all key experiments.
Preparations of GST-ELMOD proteins from HEK cells were typically at least 50% pure, as judged by Coomassie Blue staining of SDS gels, with the GST-ELMOD3 the lowest (Fig. 1B). The GST-ELMOD proteins were the predominant band but a major contaminating band was also observed, migrating at ∼24 kDa, by comparison to standards. This contaminant was found in similar amounts after purification of six different N-terminal GST-tagged proteins from HEK cells and is not immunoreactive with our GST antibodies in immunoblots. Thus, we do not believe it represents a breakdown product of our fusion proteins. Rather, we believe this protein to be an endogenous HEK cell glutathione-binding protein, as described earlier in insect cells by Bichet et al. (33). However, in contrast to that published contaminant we were not able to readily resolve the ELMODs and major contaminant protein by modifying the concentration of glutathione used for elution. The contaminant was effectively removed from our GST-ELMOD preparations using either gel filtration chromatography or a Spincon concentrator with 30,000 cutoff, with repeated cycles of dilution with buffer and concentration. Either of these methods is sufficient to increase the purity of the GST-ELMOD1 or -2 to at least an estimated 90%. GST-ELMOD3 is somewhat less pure, perhaps related to its lower level of expression in HEK cells.
HEK cell-expressed ELMOD1–3 were assayed in the ARL2 GAP assay and compared with our previous preparations. Titration curves were performed for each of the human ELMOD preparations (Fig. 2), from which specific activities were derived. As seen in Table 1, ELMOD2 purified from HEK cells as a GST fusion was comparable in specific activity to the preparation of ELMOD2 from bovine testes and roughly 3 orders of magnitude more active than those purified from bacteria. Thus, we believe that this is a faithful representative of the native protein. ELMOD1 and ELMOD3 proteins have never been purified from cells or tissues so comparison to a preparation from a mammalian tissue is not possible but we conclude by analogy and because of the higher specific activities from HEK cells versus bacteria that these too are appropriate for detailed biochemical analyses.
FIGURE 2.
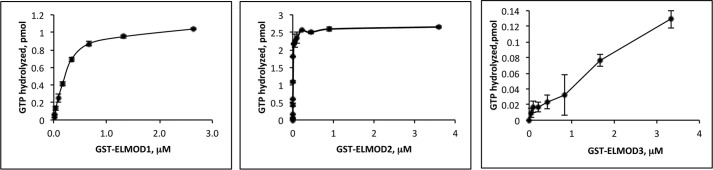
ELMOD1–3 GAP activities were determined using ARL2·[γ-32P]GTP as substrate with the charcoal GAP assay. A fixed amount of substrate was used in these assays and the amount of GAP was varied to generate the dose-response curves shown. Duplicate time points were taken and averaged, with error bars representing mean ± S.E. Note the differences in the y axes with GST-ELMOD1 (left), GST-ELMOD2 (center), and GST-ELMOD3 (right) due to the large differences in specific activities of the three GAPs for this one GTPase.
Despite GST-ELMOD2 having very similar specific activity to the bovine testes preparation, we were concerned that the presence of the N-terminal tag may interfere with activities. To test this, the GST tag was cut from ELMOD2 with TEV protease. After testing a series of TEV concentrations for cleavage efficiencies, either in solution or while bound to glutathione beads, we found cleavage on beads with 30% (mg/mg protein) purified recombinant TEV to yield the most complete cleavage. The uncleaved and cleaved ELMOD2 were then compared in ARL2 GAP assays and found to have specific activities of 30 and 22 nmol/min/mg, respectively (Table 1). We conclude from this that the presence of GST at the N terminus does not interfere with ELMOD2 activities as a GAP.
We consistently found large differences in specific activities of the three human ELMODs for ARL2, with GST-ELMOD2 > GST-ELMOD1 > GST-ELMOD3 (Fig. 2, Table 2). The specific activities for the three ELMODs using ARL2 as substrate (30, 0.76, and 0.03 nmol of GTP hydrolyzed/min/mg of protein) reveal that ELMOD2 is almost 40-fold more active than ELMOD1, which is 25-fold more active than ELMOD3. This reveals a ∼1,000-fold difference between GST-ELMOD2 and GST-ELMOD3 for ARL2·GTP.
TABLE 2.
Specific activities for GST-ELMOD proteins against six different ARF family GTPases
Specific activity values (nmol min−1 mg−1) for GST fusion proteins of the human ELMODs against each of the indicated ARF family members. Specific activities were determined as described under “Experimental Procedures.” GTPases (1 μm) were pre-loaded with [γ-32P]GTP as described under “Experimental Procedures” and diluted into the assay to begin the reaction. Specific activities were determined as described in the legend to Table I, using titration curves that included at least 8 concentrations of each ELMO domain protein between 0 and 4 μm. Variations within duplicates and between experiments were less than 15%.
| ARL2 | ARF1 | ARF6 | ARL1 | ARL3 | ARL13B | |
|---|---|---|---|---|---|---|
| ELMOD1 | 0.76 | 0.06 | 93 | 10 | 0.001 | NAa |
| ELMOD2 | 30.0 | 255 | 422 | 1107 | 67 | NA |
| ELMOD3 | 0.03 | 0.02 | NA | 0.07 | 0.02 | NA |
| ELMO1 | NA | NA | NA | NA | NA | NA |
a NA, not active, indicates no detectable difference between the ELMOD sample and buffer control in the GAP assay.
Specificities of ELMOD1–3 as GAPs for ARF Family Members
The remarkably large variation in specific activities between the ELMODs led us to speculate that three members of this family of ARF family GAPs have diverged over evolutionary time to use different members of the family. To compare the specificities of the different ELMOD proteins as GAPs we examined a broad spectrum of ARF family members as substrates. However, the different members of the ARF family have different optimal conditions for binding GTP and the purified, recombinant proteins bind guanine nucleotides to different stoichiometries. Thus, some differences in reaction conditions were necessary to accommodate differences in binding preferences. Differences in GTP binding stoichiometries led to the concern that product inhibition may contribute to differences in activities. That is, because we have shown previously that ARF1 co-purified from bacteria with stoichiometric amounts of GDP (4), it is possible that the ARF1 that failed to bind GTP in the loading step would be present in the GAP assay as ARF1·GDP, the product of the GAP reaction, and effectively compete for the GTPase binding site on the ELMOD. To test for this potential product inhibition we compared GAP activities of ARL2 in the presence of an increasing concentration of ARF1·GDP. We found no effect of up to 10 μm ARF1·GDP on ARL2 GAP activity, which included 0.1 μm ARL2. Thus, we conclude that product inhibition is not a concern, at least in the concentration ranges used in our studies.
Six members of the human ARF family were purified from bacterial (ARL1–3, ARF1, and ARF6) or HEK (GST-ARL13B) cells and tested as substrates in GAP assays with the three human ELMODs. GAP activities were determined using a range of ELMODs appropriate for each substrate, as shown in one example (Fig. 3) of GST-ELMOD2. Surprisingly, even though ELMOD2 was purified from bovine tissues as an ARL2 GAP, several other family members proved to be better substrates. ARL1 was the best, with a specific activity of 1107 nmol/min/mg, or 37-fold higher activity than for ARL2 (Table 2). Even the ARFs (ARF1 and ARF6) had higher specific activities in these assays than did ARL2. These results differ from our earlier finding that ARF1 and ARF6 had only 15 and 5%, respectively, of the ARL2 GAP activity of bacterially expressed MBP-ELMOD2 (16). Similarly, ARL3 was less active (5%) than ARL2 with MBP-ELMOD2 earlier but comparable with ARL2 with the GST-ELMOD2 preparation. These differences are not readily explained but given the overall low specific activities of the bacterially expressed MBP-ELMODs we suspect that those earlier data were compromised by overall low and likely variable activities, often pushing the lower limits of the assay, and the instability of the purified proteins.
FIGURE 3.
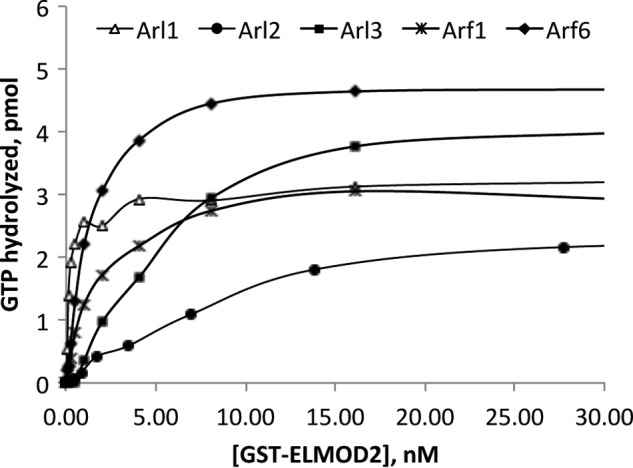
GAP activities of GST-ELMOD2 against different ARF family GTPases. GAP activities against each of five different GTPases within the ARF family were determined using fixed concentrations of substrates and varying the amount of GST-ELMOD2, as described under “Experimental Procedures” and in the legend to Fig. 2. Note that the specific activities were calculated from the lower concentrations that fell along the straight part of each curve.
We completed the comparison of substrate specificities for all three GST-ELMODs as GAPs for all six ARF family members (Table 2). None of the ELMODs displayed any GAP activity for GST-ARL13B, the most divergent member of the family tested. GST-ELMOD2 was consistently found to be the most active ELMOD against any of these five ARF family members that were substrates for at least one of the GAPs. In contrast, GST-ELMOD3 displayed only minimal specific activities against these GTPases, ranging from undetectable for ARF6 to 0.07 for ARL1 (Table 2). This very low activity explains our earlier conclusion that ELMOD3 lacks GAP activity for ARF family members (16). Thus, GST-ELMOD2 is 1,000- to >15,000-fold more active than is GST-ELMOD3 against different substrates. The data with GST-ELMOD1 is perhaps the most interesting as the great variation in activities, ranging from 0.01 (ARL3) to 93 (ARF6), yielding a 9,300-fold difference, is most consistent with different specificities among ARF family members.
We also performed preliminary testing for an accessory protein or lipid activator of the GAP activity of ELMOD3, due to its relatively low specific activity with each of the substrates tested. We assayed the ARL2 GAP activity of GST-ELMOD3 in the presence and absence of a range of single, di-, and tri-phosphorylated phosphatidylinositol phosphates (PIP) that were sonicated in the presence of 3 mm dimyristoyl phosphatidylcholine. Alternatively, we obtained a series of large unilamellar vesicles (kindly provided by Dr. Paul Randazzo, NCI, National Institutes of Health) containing the same assortment of PIPs and prepared as described earlier (34, 35). In neither case did we observe any effect upon the GAP activity. In contrast, preliminary testing of a range of bovine tissue homogenates suggests the possible existence of a factor capable of increasing the specific activity of GST-ELMOD3, although the nature of this factor is still unknown.
Homologous Arginine Residues Are Essential to GAP Activity in All Three ELMODs
Phylogenetic analyses and primary sequence alignments allowed us to identify a highly conserved arginine residue in the middle of the most conserved region of the ELMO domain that we predicted acts as the catalytic arginine in canonical GAP proteins (19, 36). This was tested in our earlier study (19) by comparing the ARL2 GAP activities of TF-ELMOD1 to TF-ELMOD1(R174K) and TF-ELMOD2 to TF-ELMOD2(R167K). Each of these conservative point mutations resulted in the near complete loss of GAP activity, although activities of even the wild type TF fusion proteins were very low. For that reason we earlier also expressed ELMOD1 and ELMOD2 as C-terminal myc/His6 fusion proteins in HeLa cells and assayed them and Arg to Lys point mutants in cell homogenates. In this system we also observed the loss of activity resulting from the arginine mutations, although since we were assaying cell lysates the backgrounds were quite high. Given the very low specific activities of the fusion proteins and high background seen in HeLa cell homogenates, we decided to re-examine this question. We generated the three homologous arginine to lysine mutants to generate purified human GST-ELMOD1(R174K), GST-ELMOD2(R167K), and GST-ELMOD3(R211K). These were assayed for ARL2 GAP activity in comparison to their wild type counterparts. We found essentially a complete loss of activity for the ELMOD1 and ELMOD2 mutants, with ∼80% loss of activity for ELMOD3(R211K) (Fig. 4). The activity seen for purified GST-ELMOD3(R211K) was near the lower limit of the assay and had large error bars as a result. These results are consistent with the previous evidence yet provide substantially stronger evidence that these homologous and highly conserved arginines are critical to ARL2 GAP activity.
FIGURE 4.
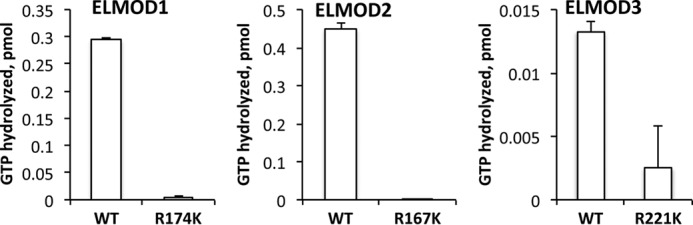
Mutation of the putative catalytic arginines reduces GAP activity of ELMOD1–3 against ARL2. ELMOD (1 μm) wild type and arginine to lysine point mutants were tested for activity as GAPs for ARL2 using the standard GAP assay in duplicate, as described under “Experimental Procedures.” This experiment was repeated three times, and the results averaged as the amount of GTP hydrolyzed. The error bars represent mean ± S.E. values.
FLAG-tagged Sigma-1 Receptor (S1R-FLAG) Binds ELMOD Proteins and Inhibits Their ARL2 GAP Activity
In a search for ELMOD1 binding partners, we performed co-IP experiments using C-terminal HA-tagged ELMOD1 (ELMOD1-HA), as described under “Experimental Procedures.” Stable isotope labeling with amino acids in cell culture was used to quantify fold-enrichment for potential binding partners. Lentivirus directing expression of doxycycline-inducible ELMOD1-HA was used to infect HeLa cells prior to use. Induced versus uninduced cell lysates were treated identically with HA antibodies in the parallel co-IPs, prior to pooling and analysis by LC-MS/MS. This one-step affinity enrichment resulted in a 44.9-fold enrichment of ELMOD1-HA when the induced culture was in heavy medium and 10.6-fold enrichment when grown in lite medium. The protein showing the next largest fold-enrichment was the S1R, at 24.9- and 7.1-fold enrichment, respectively. In addition, an unrelated C-terminal HA-tagged protein (ARL13B) was used for co-IP at the same time and no peptides from S1R were found. In a repeat experiment we found even larger fold-enrichments, with ELMOD1-HA increased 205- and 10-fold in heavy and lite medium, respectively, with S1R again the next most highly enriched, at 167- and 14-fold enrichment over controls. We identified four unique tryptic peptides derived from human S1R (1MQWAVGR7, 40EEIAQLAR47, 48QYAGLDHELAFSR60, and 212LELTTYLFGQDP223), which included a total of 40 residues from the total of 223 residues, or coverage of 18%. This represents the longest human splice variant in the NCBI database for the human message as both N- and C-terminal tryptic peptides were found in the LC-MS/MS dataset.
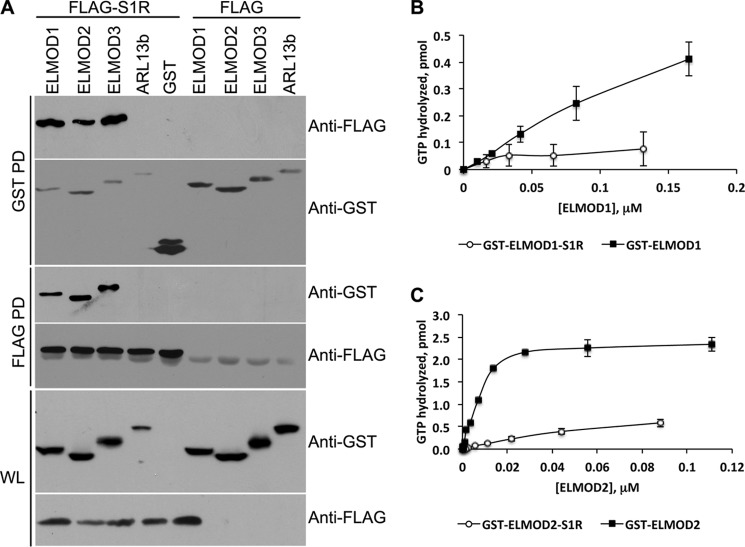
We believe that ELMOD1 and S1R are binding directly as the fold-enrichments are comparable and no other proteins were found to be enriched comparably. Nevertheless, we further tested this conclusion by co-expression in HEK cells of our GST-ELMOD1 construct with C-terminal FLAG-tagged full-length rat S1R (NM_030996; encoding all 223 residues; gift of Dr. Tsung Ping Su, National Institutes of Health) and performed reciprocal co-IPs (Fig. 5A). In addition, in these studies we also compared the three different ELMOD paralogs for the ability to bind S1R-FLAG. Controls in this study include comparison of S1R-FLAG to FLAG tag alone, GST-ELMODs to GST alone, and GST-ELMODs to GST-ARL13B, as an unrelated protein. Fig. 5A shows the results of the co-IPs, with the proteins co-expressed indicated at the top (S1R-FLAG or FLAG with each of the GST proteins underneath). The top four panels are immunoblots from pulldowns with either glutathione-Sepharose (GST PD) or anti-FLAG (FLAG PD), indicated on the left. The antibodies used to develop each immunoblot are indicated on the right. The bottom two panels in Fig. 5A show immunoblotting data from whole cell lysates using GST or FLAG antibodies. These confirm that each of the proteins was expressed where indicated.
FIGURE 5.
FLAG-S1R binds to GST-ELMODs and inhibits the ARL2 GAP activity of GST-ELMOD1 or GST-ELMOD2. A, reciprocal co-immunoprecipitations of GST-ELMOD and FLAG-S1R reveals specific binding of S1R to all three ELMODs. All proteins were tagged (FLAG-S1R or GST all others) and co-expressed in HEK293T cells (as indicated along the top), and used in reciprocal pulldown assays using glutathione-Sepharose or anti-FLAG antibodies (as indicated on the left side of each panel). Each panel shows an immunoblot, using the antibody indicated at the right. WL, whole cell lysate; PD, pulldown. The key panels are the top panel, which shows GST pulldowns from each ELMOD expressed also enriches for FLAG-S1R, and the third panel down, which shows FLAG pulldowns each of the GST-ELMODs. GST and GST-ARL13B are used as negative controls for the ELMOD proteins; note the lack of interactions with FLAG-S1R. The empty FLAG vector is indicated as FLAG at the top but because of the small size of the recombinant protein does not appear in any of the immunoblots. A nonspecific band is seen in all lanes using the FLAG antibody, running right below the FLAG-S1R. B, ARL2 GAP assays for GST-ELMOD1 or GST-ELMOD1-S1R are shown as a function of increasing GST-ELMOD1 concentrations. Equal amounts of GST-ELMOD1, alone or complexed with S1R, were assayed. C, the same as panel B, except GST-ELMOD2 is used instead of GST-ELMOD1.
The affinity matrices, glutathione-Sepharose or anti-FLAG beads, were each efficient at pulling down the appropriate tagged proteins from 1% CHAPS homogenates of HEK cells. Probing the GST-ELMOD1 pulldown from lysates of HEK cells expressing both proteins with FLAG antibody revealed that a subset, estimated at ∼5% of total, of the S1R-FLAG was brought down. In controls run in parallel there was no S1R-FLAG detected in the pulldown performed in the absence of GST-ELMOD1 expression or with GST only. We performed the reciprocal co-IP by pulling down with the FLAG antibody and probing immunoblots with the GST antibody and found specific co-IP of GST-ELMOD1. We obtained essentially the same results with co-expression of FLAG-S1R and any of the human GST-ELMOD proteins, but not with GST alone or with GST-ARL13B. Thus, we conclude that each of the ELMOD proteins is capable of specific binding to S1R-FLAG.
As a further and stringent test of binding and functional consequences, we asked whether the binding of S1R-FLAG to GST-ELMOD1 or GST-ELMOD2 had any effect upon their GAP activities. The relevant proteins were co-expressed in HEK cells and the complex was purified using FLAG antibodies, yielding a preparation highly enriched for GST-ELMOD1 or GST-ELMOD2 and with an excess of S1R-FLAG. The amount of GST-ELMOD1 or GST-ELMOD2 in the co-IP was determined by comparative immunoblotting, using a dilution series of the purified proteins as standards (data not shown). We then determined the ARL2 GAP activities of equal amounts of GST-ELMOD1 or GST-ELMOD2 alone or when complexed with S1R-FLAG. As seen in Fig. 5B, binding of S1R-FLAG resulted in a potent inhibition of the ARL2 GAP activity of GST-ELMOD1 or GST-ELMOD2.
DISCUSSION
Cell regulation by GTPases provides spatial and temporal resolution to signaling but also allows for cross-talk between pathways regulated by distinct GTPases; e.g. through the use of regulators (GEFs and GAPs) with shared specificities or the recruitment by one activated GTPase of an activator or inhibitor of another. Thus, definition of the specificities and binding partners of GEFs and GAPs is essential to the construction of integrated models of cell signaling. Efforts to define these characteristics for the new family of ARF family GAPs, the ELMODs, have been hampered by problems of stability and activity with the recombinant sources. One possible explanation for the poor solubility and stability of bacterially expressed proteins is the absence of an obligate binding partner. This possibility seemed likely for the ELMODs as their closest paralogs, the ELMOs, are found in cells bound to Dock180, an atypical Rac GEF. Here we describe the use of a mammalian cell expression system in HEK cells to generate preparations of all three human ELMODs that are fully functional, as defined by specific activities compared with the sole preparation purified from mammalian tissue, bovine ELMOD2. When tested against a collection of six ARF family members we found these three GAPs to have remarkably variable specific activities, with ELMOD2 the most active and ELMOD3 almost inactive. Also surprising was that the highest specific activity for ELMOD2 was against ARL1 and for ELMOD1 was against ARF6. Thus, as is not uncommon in this field, the original description of the ELMOD family as ARL2 GAPs may well prove to be misleading from their cellular roles. Finally, we identify the non-opioid sigma-1 receptor as a specific binding partner for all three ELMODs and show that binding leads to loss of ARL2 GAP activity of ELMOD1 and ELMOD2. The implications of these interactions are discussed below.
Purification of a biochemical activity from mammalian tissue remains perhaps the best way to identify novel regulators of cell signaling but also typically provides insufficient material for extensive functional or structural characterization. Protein preparations from tissues are also more likely to provide accurate information on the activities of the properly folded protein. Thus, we have been concerned that preparations of ELMOD2 proteins from bacterial sources are unusually unstable to storage at 4 °C or one cycle of freeze/thaw and have specific activities many fold lower than that purified from bovine testes. The use of HEK cells for expression and source of protein purification appears to have eliminated these concerns in allowing the generation of milligram amounts of GST-ELMOD2 and ELMOD2 (after cleavage) that is much more stable and displays specific activities as an ARL2 GAP that is indistinguishable from the bovine preparation (30 versus 23 nmol/min/mg). Clearly, solubility alone is not a sufficient criterion for proper folding and activity of the ELMODs as the TF fusions proteins were ∼1,000-fold lower in specific activity (Table 1). Because the GST fusions of ELMOD1 and ELMOD3 purified from HEK cells have higher specific activities than the bacterially expressed proteins, and have solubility and stability profiles similar to that of ELMOD2 we believe they offer the best preparation available and represent the native proteins.
The increased specific activity seen with the mammalian cell expression system may result from one or more post-translational modification(s) that is important to activity. We are currently using mass spectrometry to identify such modifications and will test for effects on activities. Similarly, it is possible that a small molecule, e.g. a lipid, may bind to one or more ELMODs to allosterically regulate activity. Based upon homology to the ARF and ARF GAP proteins we began testing for effects of PIPs, particularly on ELMOD3 as its activity is so low. However, to date we have found no effects of added PIPs on the GAP activity of ELMOD3. Preliminary data from testing of a broader screen of tissue homogenates suggests the possibility that a factor exists capable of increasing the specific activity of our purified ELMOD3 and we are currently pursuing this activity through purification. Thus, we believe that the HEK cell expression system offers a number of benefits over the bacterial system that we hope will allow us to identify the means of regulation of the GAP activities in mammalian cells.
The use of the HEK cell preparations has also allowed us to re-examine many of our initial findings of the GAP activities of the three human ELMODs. For example, we now provide much stronger experimental support for our conclusion that mutation of the conserved arginine to lysine in each of the ELMODs results in complete or nearly complete loss of GAP activity (Fig. 4). Future structural studies are still needed to confirm this as the catalytic arginine and mechanism of hydrolysis by this novel family of ARF family GAPs.
When a collection of ARF family members were tested as substrates for the three human GST-ELMODs we found a surprising amount of variation in specific activities (>55,000-fold between highest and lowest; see Table 2) both between the different ELMODs for one GTPase and for one ELMOD for different GTPases. In general, we found that ELMOD2 is more active as a GAP against every GTPase tested than is ELMOD1, than is ELMOD3. The finding that ELMOD2 has >35-fold higher specific activity against ARL1 than ARL2 suggests a possible role in membrane traffic at the Golgi; for which there is currently no evidence. Similarly, the finding that ELMOD1 is most active against ARF6, with a specific activity >100-fold higher than against ARL2, is suggestive of a role at the plasma membrane. Interestingly, ELMO1, which lacks GAP activity in our assays, has been proposed to act downstream of ARF6 and promote Rac activation through association with Dock 180 (37). Commercial and our own antibodies directed against the mammalian ELMODs have not proven to be as robust or specific as we would like, making localization of these proteins in cells difficult, with one exception. We have found that ELMOD2 is found inside mitochondria and in its knockdown phenocopies that of ARL2 in two of the three mitochondrial phenotypes described.3 Among the GTPases tested here only ARL2 is present inside mitochondria. Thus, it is very likely that specific localization of the GTPases and ELMODs in cells provides additional sources of specificity in signaling, as is generally true for GAPs. Nevertheless, we have begun to use the specific activity data, summarized in Table 2, to test for functions of specific ELMODs in different parts of the cell.
Although the high activities of ELMOD2 and ELMOD1 are suggestive of novel functions and sites of action, the very low activities found for ELMOD3 may lead one even to question whether it acts in cells as an ARF family GAP. Indeed, our initial testing of ARL2 GAP activity was negative, as we used the bacterially expressed proteins (16). One possibility that may explain these low specific activities for GST-ELMOD3 is that our assays are lacking a crucial cofactor or post-translational modification needed by ELMOD3 for maximal activity. The ARFs and ARF GAPs have a long history of strong functional links with specific lipids, most commonly phosphatidylinositols (38–45). Although initial testing of PIP stimulation has been negative we are continuing to vary conditions in the assay to examine this possibility.
One condition we found that clearly alters activity of at least ELMOD1 and ELMOD2 is binding to S1R. A direct comparison of GST-ELMOD1 or GST-ELMOD2 to the same amount of each bound to S1R showed a near complete loss of their ARL2 GAP activity (Fig. 5). The little residual activity may result from dissociation of some of the ELMOD from S1R so the magnitude of the inhibition will require more detailed studies. S1R is a very interesting but incompletely understood protein, implicated as a receptor for a number of clinically important drugs and a number of biological activities (46–51). S1R is a transmembrane ER protein that localizes predominantly to the interface between the ER and mitochondria, called the mitochondria-associated membrane, where it is proposed to regulate calcium signaling between the ER and mitochondria (21) and as part of cell survival pathways (52). The proposed mechanism of S1R in calcium regulation is as a molecular chaperone for inositol 1,4,5-trisphosphate receptors (IP3Rs) by directly binding the mature channel-forming IP3R tetramer to prevent its rapid ubiquitination and degradation upon activation with inositol 1,4,5-trisphosphate. S1R is also proposed to interact with inactive, monomeric IRE1 under conditions of ER stress to both prevent its degradation and regulate the temporal pattern of IRE1 activation/dimerization allowing for a longer lasting signal. Importantly, because S1R is a receptor for a number of drugs that are proposed to act as either agonists or antagonists of S1R its direct binding to ELMODs makes them potential effectors of one or more of these drugs. The finding that S1R binding to ELMOD1 or ELMOD2 decrease their GAP activities could be an indication that S1R is acting to prolong and thereby promote the actions of ELMODs as effectors. Alternatively, S1R may bind ELMODs to sterically occlude or allosterically decrease the binding of the substrate GTPase from the active site to decrease GTPase signaling. Thus, the inhibitory effects of S1R on the GAP activity of the ELMODs adds an intriguing layer of complexity to the regulation of these proteins and may be an indication of the cellular demand for tight regulation of the ELMODs and, by extension, their substrate GTPases.
Acknowledgments
We thank Drs. Kelley Moremen (University of Georgia) and James Hurley (NIH) and members of their laboratories for very helpful discussions regarding the use of HEK cells for protein expression, as well as the latter for the pLEXm-GST parent plasmid. We also appreciate and thank Drs. Paul Randazzo and Xiaoying Jian for the generous gift of a collection of large unilamellar vesicles containing different phosphatidylinositol phosphates.
This work was supported, in whole or in part, by National Institutes of Health Grants GM090158 and GM61268 (to A. A. I. and R. A. K.).
L. E. Newman, C. J. Zhou, and R. A. Kahn, unpublished data.
- GAP
- GTPase activating protein
- ARL
- ARF-like
- S1R
- non-opioid sigma-1 receptor
- ELMOD
- cell engulfment and motility domain-containing protein
- MBP
- maltose-binding protein
- TF
- trigger factor
- TEV
- tobacco etch virus
- Ni-NTA
- nickel-nitrilotriacetic acid
- PEI
- polyethyleneimine
- PIP
- phosphatidylinositol phosphate
- ER
- endoplasmic reticulum
- co-IP
- co-immunoprecipitation
- GEF
- guanine nucleotide exchange factor.
REFERENCES
- 1. East M. P., Kahn R. A. (2011) Models for the functions of Arf GAPs. Semin. Cell Dev. Biol. 22, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Du X., Sprang S. R. (2009) Transition state structures and the roles of catalytic residues in GAP-facilitated GTPase of Ras as elucidated by 18O kinetic isotope effects. Biochemistry 48, 4538–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 4. Kahn R. A., Gilman A. G. (1986) The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP-binding protein. J. Biol. Chem. 261, 7906–7911 [PubMed] [Google Scholar]
- 5. Mandiyan V., Andreev J., Schlessinger J., Hubbard S. R. (1999) Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein β. EMBO J. 18, 6890–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldberg J. (1999) Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell 96, 893–902 [DOI] [PubMed] [Google Scholar]
- 7. Cichowski K., Jacks T. (2001) NF1 tumor suppressor gene function: narrowing the GAP. Cell 104, 593–604 [DOI] [PubMed] [Google Scholar]
- 8. Polakis P., McCormick F. (1992) Interactions between p21ras proteins and their GTPase activating proteins. Cancer Surv. 12, 25–42 [PubMed] [Google Scholar]
- 9. Maheshwar M. M., Cheadle J. P., Jones A. C., Myring J., Fryer A. E., Harris P. C., Sampson J. R. (1997) The GAP-related domain of tuberin, the product of the TSC2 gene, is a target for missense mutations in tuberous sclerosis. Hum. Mol. Genet. 6, 1991–1996 [DOI] [PubMed] [Google Scholar]
- 10. McLaughlin S. K., Olsen S. N., Dake B., De Raedt T., Lim E., Bronson R. T., Beroukhim R., Polyak K., Brown M., Kuperwasser C., Cichowski K. (2013) The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell 24, 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hollinger S., Hepler J. R. (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54, 527–559 [DOI] [PubMed] [Google Scholar]
- 12. Johnson K. R., Longo-Guess C. M., Gagnon L. H. (2012) Mutations of the mouse ELMO domain containing 1 gene (Elmod1) link small GTPase signaling to actin cytoskeleton dynamics in hair cell stereocilia. PloS One 7, e36074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaworek T. J., Richard E. M., Ivanova A. A., Giese A. P., Choo D. I., Khan S. N., Riazuddin S., Kahn R. A., Riazuddin S. (2013) An alteration in ELMOD3, an Arl2 GTPase-activating protein, is associated with hearing impairment in humans. PLoS Genet. 9, e1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharer J. D., Shern J. F., Van Valkenburgh H., Wallace D. C., Kahn R. A. (2002) ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol. Biol. Cell 13, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shern J. F., Sharer J. D., Pallas D. C., Bartolini F., Cowan N. J., Reed M. S., Pohl J., Kahn R. A. (2003) Cytosolic Arl2 is complexed with cofactor D and protein phosphatase 2A. J. Biol. Chem. 278, 40829–40836 [DOI] [PubMed] [Google Scholar]
- 16. Bowzard J. B., Cheng D., Peng J., Kahn R. A. (2007) ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 282, 17568–17580 [DOI] [PubMed] [Google Scholar]
- 17. Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., Nishiwaki K., Walk S. F., Nemergut M. E., Macara I. G., Francis R., Schedl T., Qin Y., Van Aelst L., Hengartner M. O., Ravichandran K. S. (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 [DOI] [PubMed] [Google Scholar]
- 18. Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., Ravichandran K. S. (2002) Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4, 574–582 [DOI] [PubMed] [Google Scholar]
- 19. East M. P., Bowzard J. B., Dacks J. B., Kahn R. A. (2012) ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for Arf protein family GTPases. J. Biol. Chem. 287, 39538–39553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grimsley C. M., Kinchen J. M., Tosello-Trampont A. C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O., Ravichandran K. S. (2004) Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 279, 6087–6097 [DOI] [PubMed] [Google Scholar]
- 21. Hayashi T., Su T. P. (2007) Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131, 596–610 [DOI] [PubMed] [Google Scholar]
- 22. Van Valkenburgh H., Shern J. F., Sharer J. D., Zhu X., Kahn R. A. (2001) ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J. Biol. Chem. 276, 22826–22837 [DOI] [PubMed] [Google Scholar]
- 23. Randazzo P. A., Weiss O., Kahn R. A. (1995) Preparation of recombinant ADP-ribosylation factor. Methods Enzymol. 257, 128–135 [DOI] [PubMed] [Google Scholar]
- 24. Studier F. W., Moffatt B. A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 25. Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60–89 [DOI] [PubMed] [Google Scholar]
- 26. Studier F. W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 27. Tropea J. E., Cherry S., Waugh D. S. (2009) Expression and purification of soluble His6-tagged TEV protease. Methods Mol. Biol. 498, 297–307 [DOI] [PubMed] [Google Scholar]
- 28. Phan J., Zdanov A., Evdokimov A. G., Tropea J. E., Peters H. K., 3rd, Kapust R. B., Li M., Wlodawer A., Waugh D. S. (2002) Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277, 50564–50572 [DOI] [PubMed] [Google Scholar]
- 29. Dammer E. B., Fallini C., Gozal Y. M., Duong D. M., Rossoll W., Xu P., Lah J. J., Levey A. I., Peng J., Bassell G. J., Seyfried N. T. (2012) Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PloS One 7, e38658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seyfried N. T., Gozal Y. M., Donovan L. E., Herskowitz J. H., Dammer E. B., Xia Q., Ku L., Chang J., Duong D. M., Rees H. D., Cooper D. S., Glass J. D., Gearing M., Tansey M. G., Lah J. J., Feng Y., Levey A. I., Peng J. (2012) Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards. J. Proteome Res. 11, 2721–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aricescu A. R., Assenberg R., Bill R. M., Busso D., Chang V. T., Davis S. J., Dubrovsky A., Gustafsson L., Hedfalk K., Heinemann U., Jones I. M., Ksiazek D., Lang C., Maskos K., Messerschmidt A., Macieira S., Peleg Y., Perrakis A., Poterszman A., Schneider G., Sixma T. K., Sussman J. L., Sutton G., Tarboureich N., Zeev-Ben-Mordehai T., Jones E. Y. (2006) Eukaryotic expression: developments for structural proteomics. Acta Crystallogr. D Biol. Crystallogr. 62, 1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aricescu A. R., Lu W., Jones E. Y. (2006) A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 62, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 33. Bichet P., Mollat P., Capdevila C., Sarubbi E. (2000) Endogenous glutathione-binding proteins of insect cell lines: characterization and removal from glutathione S-transferase (GST) fusion proteins. Protein Expr. Purif. 19, 197–201 [DOI] [PubMed] [Google Scholar]
- 34. Jian X., Brown P., Schuck P., Gruschus J. M., Balbo A., Hinshaw J. E., Randazzo P. A. (2009) Autoinhibition of Arf GTPase-activating protein activity by the BAR domain in ASAP1. J. Biol. Chem. 284, 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nie Z., Hirsch D. S., Luo R., Jian X., Stauffer S., Cremesti A., Andrade J., Lebowitz J., Marino M., Ahvazi B., Hinshaw J. E., Randazzo P. A. (2006) A BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr. Biol. 16, 130–139 [DOI] [PubMed] [Google Scholar]
- 36. Scheffzek K., Ahmadian M. R., Wittinghofer A. (1998) GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23, 257–262 [DOI] [PubMed] [Google Scholar]
- 37. Santy L. C., Ravichandran K. S., Casanova J. E. (2005) The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 15, 1749–1754 [DOI] [PubMed] [Google Scholar]
- 38. Randazzo P. A., Kahn R. A. (1994) GTP hydrolysis by ADP-ribosylation factor is dependent on both an ADP-ribosylation factor GTPase-activating protein and acid phospholipids. J. Biol. Chem. 269, 10758–10763 [PubMed] [Google Scholar]
- 39. Terui T., Kahn R. A., Randazzo P. A. (1994) Effects of acid phospholipids on nucleotide exchange properties of ADP-ribosylation factor 1. Evidence for specific interaction with phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 269, 28130–28135 [PubMed] [Google Scholar]
- 40. Kahn R. A., Terui T., Randazzo P. A. (1996) Effects of acid phospholipids on ARF activities: potential roles in membrane traffic. J. Lipid Mediat. Cell Signal. 14, 209–214 [DOI] [PubMed] [Google Scholar]
- 41. Zheng Y., Glaven J. A., Wu W. J., Cerione R. A. (1996) Phosphatidylinositol 4,5-bisphosphate provides an alternative to guanine nucleotide exchange factors by stimulating the dissociation of GDP from Cdc42Hs. J. Biol. Chem. 271, 23815–23819 [DOI] [PubMed] [Google Scholar]
- 42. Brown M. T., Andrade J., Radhakrishna H., Donaldson J. G., Cooper J. A., Randazzo P. A. (1998) ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 18, 7038–7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kam J. L., Miura K., Jackson T. R., Gruschus J., Roller P., Stauffer S., Clark J., Aneja R., Randazzo P. A. (2000) Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1. Evidence for the pleckstrin homology domain functioning as an allosteric site. J. Biol. Chem. 275, 9653–9663 [DOI] [PubMed] [Google Scholar]
- 44. Nie Z., Stanley K. T., Stauffer S., Jacques K. M., Hirsch D. S., Takei J., Randazzo P. A. (2002) AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J. Biol. Chem. 277, 48965–48975 [DOI] [PubMed] [Google Scholar]
- 45. Che M. M., Boja E. S., Yoon H. Y., Gruschus J., Jaffe H., Stauffer S., Schuck P., Fales H. M., Randazzo P. A. (2005) Regulation of ASAP1 by phospholipids is dependent on the interface between the PH and Arf GAP domains. Cell. Signal. 17, 1276–1288 [DOI] [PubMed] [Google Scholar]
- 46. Cobos E. J., Entrena J. M., Nieto F. R., Cendán C. M., Del Pozo E. (2008) Pharmacology and therapeutic potential of sigma(1) receptor ligands. Curr. Neuropharmacol. 6, 344–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hashimoto K., Ishiwata K. (2006) Sigma receptor ligands: possible application as therapeutic drugs and as radiopharmaceuticals. Curr. Pharm. Des. 12, 3857–3876 [DOI] [PubMed] [Google Scholar]
- 48. Maurice T., Su T. P. (2009) The pharmacology of sigma-1 receptors. Pharmacol. Therap. 124, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Niitsu T., Iyo M., Hashimoto K. (2012) Sigma-1 receptor agonists as therapeutic drugs for cognitive impairment in neuropsychiatric diseases. Curr. Pharm. Des. 18, 875–883 [DOI] [PubMed] [Google Scholar]
- 50. Skuza G., Wedzony K. (2004) Behavioral pharmacology of σ-ligands. Pharmacopsychiatry 37, S183–188 [DOI] [PubMed] [Google Scholar]
- 51. Tsai S. Y., Hayashi T., Mori T., Su T. P. (2009) Sigma-1 receptor chaperones and diseases. Cent. Nerv. Syst. Agents Med. Chem. 9, 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mori T., Hayashi T., Hayashi E., Su T. P. (2013) Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PloS One 8, e76941. [DOI] [PMC free article] [PubMed] [Google Scholar]