Background: Gdown1 modulates pausing in early elongation but also blocks binding of the essential initiation factor TFIIF to pol II.
Results: TFIIF can compete successfully with Gdown1 to support preinitiation complex assembly.
Conclusion: Gdown1 cannot functionally interact with already formed preinitiation complexes.
Significance: Pathways exist allowing Gdown1 to enter the transcription complex early in elongation, thus allowing Gdown1 to affect pausing.
Keywords: General Transcription Factors, RNA Polymerase II, Transcription, Transcription Elongation Factors, Transcription Initiation Factors, GRINL1A, Gdown1, POLR2M, TFIIF, Preinitiation Complex
Abstract
Gdown1, the substoichiometric 13th subunit of RNA polymerase II (pol II), has an important role in pausing during the initial stage of transcript elongation. However, Gdown1 quantitatively displaces the essential initiation factor TFIIF from free pol II and elongating pol II. Thus, it is not clear how or even if pol II can initiate in the presence of Gdown1. Using an in vitro transcription system with purified factors and pol II lacking Gdown1, we found that although Gdown1 is strongly inhibitory to transcription when prebound to pol II, a fraction of complexes do remain active. Surprisingly, when Gdown1 is added to complete preinitiation complexes (PICs), it does not inhibit initiation or functionally associate with the PICs. Gdown1 does associate with pol II during the early stage of transcript elongation but this association is competitive with TFIIF. By phosphorylating TFIIF, PICs can be assembled that do not retain TFIIF. Gdown1 also fails to functionally associate with these TFIIF-less PICs, but once polymerase enters transcript elongation, complexes lacking TFIIF quantitatively bind Gdown1. Our results provide a partial resolution of the paradox of the competition between Gdown1 and TFIIF for association with pol II. Although Gdown1 completely displaces TFIIF from free pol II and elongation complexes, Gdown1 does not functionally associate with the PIC. Gdown1 can enter the transcription complex immediately after initiation. Modification of TFIIF provides one pathway through which efficient Gdown1 loading can occur early in elongation, allowing downstream pausing to be regulated.
Introduction
A significant fraction (30–50%) of mammalian RNA polymerase II purified from tissue sources bears a 13th subunit called Gdown1 (1). Originally annotated as part of the GRINL1A transcription unit (2), Gdown1 is now designated as POLR2M to indicate its status as a substoichiometric subunit of pol II.2 Gdown1 binds to pol II with very high affinity and resists removal by high salt (3–5). Pol II that contains Gdown1 (pol IIG) supports transcript elongation in promoter-independent assays (1). However, Gdown1 strongly facilitates pausing during the early stages of transcript elongation for promoter-dependent pol II complexes assembled with nuclear extracts (3). Gdown1 may therefore be an essential part of establishing the poised polymerases that are found at ∼50 bp downstream of most active mammalian promoters (6–8). Consistent with this possibility, ChIP studies demonstrate that Gdown1 co-localizes with poised pol II, although Gdown1 is also present upstream of transcription start and along with pol II within transcription units (3).
An additional property of Gdown1, which is particularly important in the context of gene regulation is the competitive interaction of Gdown1 and TFIIF with pol II. TFIIF is a general transcription factor (GTF) required for the assembly of the pol II PIC (9). TFIIF can bind directly to pol II and was originally isolated based on its affinity for polymerase (10). TFIIF also strongly stimulates transcript elongation in vitro (11–14). However, Gdown1 displaces TFIIF from both free pol II and transcriptionally engaged polymerase. Thus, Gdown1 completely blocks the ability of TFIIF to increase elongation rates (3, 5). This is consistent with the significant overlap of the surfaces on pol II that are involved in the interaction of free polymerase with Gdown1 or TFIIF (5). The ability of Gdown1 to displace TFIIF from free pol II suggests that Gdown1 should inhibit transcription at the initiation stage. Results consistent with this idea have been reported using an assay with supercoiled plasmid templates that do not require the full set of GTFs (4).
Recent findings from in vitro studies point to a pathway that could allow loading of Gdown1 into the pol II PIC before the start of transcription. When TFIIF is phosphorylated by casein kinase 2 (CK2), the phosphorylated factor (P-IIF) retains the ability to bind to pol II and support efficient PIC assembly (15). However, P-IIF is relatively unstable within the PIC, and it can be removed with a gentle wash step (16). PICs, which have lost P-IIF, are nevertheless fully competent for transcript initiation. Thus, although TFIIF is required for PIC assembly, it need not remain in the complex at the initiation step, although loss of TFIIF from the PIC may partially destabilize TFIIB (16).
These observations suggest that after the departure of TFIIF from the PIC, Gdown1 could join the transcription complex, making Gdown1 available to mediate pol II pausing further downstream. This assumes that inclusion of Gdown1 in the PIC does not affect initiation, a possibility that has not been addressed. The ability of Gdown1 to displace TFIIF effectively from free pol II (5) and from the transcript elongation complex (3, 4) raises the additional question of whether Gdown1 can displace TFIIF from a PIC containing nonmodified TFIIF. A comparison of the location of the Ssl2p subunit of TFIIH in the yeast PIC (17) with the interaction sites of Gdown1 with pol II (5) suggests that XBP (the mammalian analog of Ssl2p) might also interfere with the binding of Gdown1 to pol II (but see also Refs. 18 and 19). This further complicates the interpretation of the inhibitory effect of Gdown1 at the stage of PIC assembly.
In the present work, we show that Gdown1 cannot inhibit initiation by preformed PICs and cannot functionally associate with complete PICs, including PICs that have lost TFIIF. Gdown1 does bind to the elongation complex and blocks TFIIF stimulation of transcript elongation, but at least early in elongation, TFIIF can compete with Gdown1 for association with pol II. The high affinity of Gdown1 for pol II does not prevent transcript initiation.
EXPERIMENTAL PROCEDURES
Reagents
DNA primers and oligonucleotides were purchased from Integrated DNA Technologies, NTPs and CK2 were from New England Biolabs, CpA was from TriLink, streptavidin-coated M280 Dyna Beads were from Invitrogen, RNasin Plus was from Promega, and 800 Ci/mmol [α-32P]CTP was from PerkinElmer Life Science.
DNA Templates
Templates contained either the adenovirus major late promoter (Ad ML) or the cytomegalovirus promoter (CMV) with overall lengths of 1199 and 1258 bp, respectively. Each template had 56 bp upstream of transcription start. Templates were generated by PCR amplification with the upstream primer biotinylated to enable bead attachment. The −9/+3 bubble/premelted template, which contains the Ad ML promoter, was prepared by annealing oligonucleotides with the nontemplate strand containing a biotin moiety at the 5′ end as described (20).
Proteins and Factors
Recombinant TATA box binding protein (TBP), TFIIB, and TFIIE were prepared as described (20, 21). Recombinant human TFIIF subunits, including the truncated 1–227 large subunit, were expressed, purified, and assembled as described (15). TFIIF was phosphorylated by CK2 as described (15), followed by dialysis into BB100 (20 mm Tris-HCl (pH 7.9), 0.05% Triton X-100, 1.5 mm β-mercaptoethanol, 0.2 mm PMSF, 20% glycerol, 100 mm KCl) containing 10 mm NaF, 2 mm β-glycerophosphate, and 2 mm sodium pyrophosphate. Phosphorylation and protein integrity were verified as described (15). TFIIH was purified from HeLa nuclear extract by chromatography on P11 phosphocellulose, DE52, and Mono Q as described (15). RNA polymerase II was purified from the initial chromatin pellets obtained during preparation of HeLa nuclear extract by chromatography on DE52, heparin Sepharose and Mono Q using the method of Maldonado et al. (22) with modifications as described (21). Recombinant His-tagged Gdown1 was expressed in E. coli and purified on nickel-nitrilotriacetic acid resin before purification on a Mono Q column (3). Gdown1 was stored and diluted in HGKEDP (25 mm Hepes, pH 7.6, 20% glycerol, 300 mm KCl, 0.1 mm EDTA, 1 mm DTT, and 0.1% PMSF) with 10 μg/ml BSA.
Assembly of Preinitiation Complexes and Transcript Initiation
Preinitiation complexes were assembled for 20 min at 30 °C with the following per 10-μl reaction: 50 ng of bead-attached template, 0.4 μl of pol II (∼6 ng, ∼12 fmol), 95 fmol of TBP, 72 fmol of TFIIB, 72 fmol of TFIIF, 3.3 fmol of TFIIE, and 1 μl of TFIIH (∼75% saturation of activity). Transcription activity was completely dependent on each of the GTFs. Final buffer for PIC assembly was 20 mm Tris-HCl (pH 7.9), 60 mm KCl, 8 mm MgCl2, 2 mm DTT, and 0.12 mg/ml BSA. In some cases, PICs were gently rinsed with BC100 (20 mm Tris-HCl, pH 7.9, 0.2 mm EDTA, 20% glycerol, 100 mm KCl) containing 1 mm DTT and 10 μg/ml BSA followed by a rinse with buffer M5 (20 mm Tris-HCl, pH 7.9, 0.25 mm EDTA, 65 mm KCl, 10 mm β-glycerophosphate, and 10 mm MgCl2) containing 1 mm DTT and 10 μg/ml BSA. Treatment of pol II or PICs with Gdown1 or HGKEDP buffer was performed for 5 min at room temperature. The pol II-Gdown1 incubations were performed at a final KCl concentration of 313 mm and the PIC-Gdown1 incubations at a final KCl concentration of 76 mm. Unless otherwise specified, 240 fmol Gdown1 was added per reaction, which represents a 20-fold molar excess over the amount of pol II present. When noted, PIC supernatant was removed after Gdown1 treatment and replaced with buffer M5 containing 1 mm DTT and 10 μg/ml BSA before initiation.
Transcription was initiated to generate early elongation complexes (EECs) on the Ad ML promoter in a 10-μl reaction volume with 0.25 mm CpA (initiating position −1), 50 μm dATP, 0.5 mm UTP, 0.7 μm [α-32P]CTP and 0.5 units/μl RNasin Plus for 5 min at 30 °C followed by incubation with 0.1 mm CTP for 2 min at 30 °C. EECs were generated on the CMV promoter in a 10-μl reaction volume with 0.25 mm CpA, 50 μm GTP, 50 μm ATP, 1 μm UTP, 0.7 μm [α-32P]CTP and 0.5 units/μl RNasin Plus for 1 min at 30 °C. PICs were prepared on the −9/+3 Ad ML bubble template as described above but with only 10 ng of template per reaction. In some cases, TFIIE and TFIIH were omitted from these reactions, as indicated in Fig. 7. EECs on the bubble template were generated in a 10-μl reaction volume with 0.25 mm CpA, 0.5 mm UTP, 0.7 μm [α-32P]CTP and 0.5 units/μl RNasin Plus for 5 min at 30 °C followed by incubation at 0.1 mm CTP for 2 min at 30 °C. Transcripts were resolved on 18% denaturing acrylamide gels and imaged on a Typhoon Trio; quantitation was performed with ImageQuantTL software.
FIGURE 7.
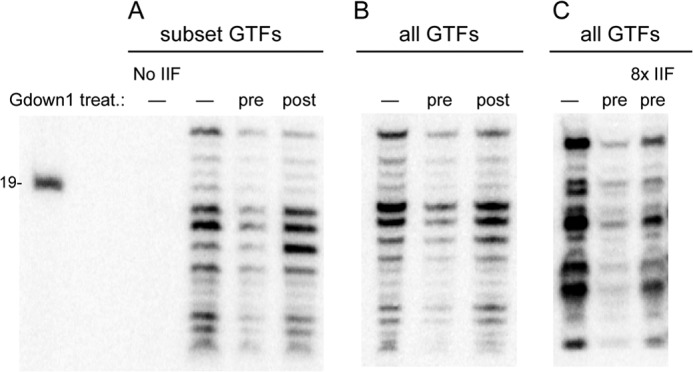
Pol IIG is inefficient in supporting transcription on a bubble template, but Gdown1 does not affect transcription when added to an existing bubble template PIC. A, PICs were formed on premelted templates using only a subset of GTFs (TBP, TFIIB, TFIIF) as described under “Experimental Procedures.” Gdown1 was either preincubated in 20-fold excess over pol II (pre) or added in 20-fold excess over pol II to the already assembled PIC (post), followed by pulse labeling as described under “Experimental Procedures”. The amount of Gdown1 (240 fmol) corresponds to 3.3-fold excess over TFIIF (72 fmol). Lane 1 contained all components for PIC formation except TFIIF to demonstrate TFIIF dependence for the initiation assay. The length of the size marker is shown on the left. B, as in A, except all GTFs were used. C, as described in B with Gdown1 pretreatment of 20-fold excess over pol II (pre) or with 0.62 pmol of IIF (8× standard amount of TFIIF used) incubated with Gdown1 and pol II during the pretreatment. treat., treatment.
Transcript Elongation
EECs were prepared as above and high salt-rinsed with a buffer containing 1.6 m KCl followed by a 40 mm KCl rinse to remove excess salt. When challenged with Gdown1, EECs were incubated with 240 fmol of Gdown1 per initial 10-μl reaction for 5 min at room temperature. Chases were performed in 12.5 μl of reaction volume in MEMDM buffer (20 mm Tris-HCl, pH 7.9, 8 mm MgCl2, 10 mm β-glycerophosphate, 0.5 mm EDTA, 10% glycerol, and 1 mm DTT) with a final KCl concentration of 52 mm for up to 1 min with 100 μm NTPs unless otherwise noted. When EECs were chased with TFIIF, 166 fmol of TFIIF or P-IIF were added with NTPs at the start of the chase per 10-μl initial reaction. Reactions were stopped with a buffer containing 10 mm Tris-HCl (pH 7.9), 100 mm NaCl, 10 mm EDTA, 1% sarkosyl, and 0.2 mg/ml tRNA. Transcripts were resolved on 12% denaturing acrylamide gels. For continuous transcription assays (see Fig. 5), PICs were prepared as above with full-length TFIIF or truncated 1–227 TFIIF without rinsing. (In 1–227 TFIIF, the largest subunit is truncated to only the 227 N-terminal amino acids.) After 60 s of pulse labeling, complexes were directly chased without rinsing by addition of 100 μm NTPs and, where noted, 166 fmol of TFIIF with final concentrations of 60 mm KCl and 6 mm MgCl2 in the chase buffer.
FIGURE 5.
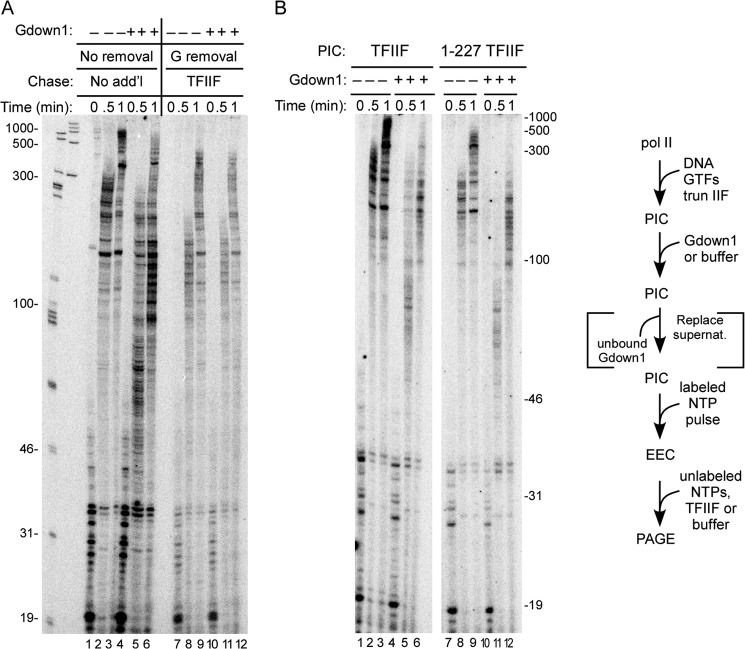
In a continuous transcription assay, Gdown1 does not inhibit stimulation of elongation by TFIIF until complexes are actively transcribing. PICs were prepared on the CMV template as described under “Experimental Procedures.” A, PICs were incubated with 240 fmol of Gdown1 (20-fold excess relative to pol II and 3.3-fold excess relative to TFIIF used to assemble PICs) or buffer. In lanes 7–12, the supernatant, including the unbound Gdown1, was removed before the pulse (inclusion of bracketed step in the schematic). PICs were then pulse-labeled and chased with no rinse step. In the lanes where the supernatant had been removed (lanes 7–12), additional (add'l) TFIIF (166 fmol) was added with the chase. The schematic of the assay is shown to the right, and the lengths of the size markers are shown to the left. B, as in the left of A, except in lanes 7–12, the large subunit of TFIIF was truncated to only the 227 N-terminal amino acids. Lengths of size markers are shown to the right.
Electrophoretic Mobility Shift Assay (EMSA)
High salt-rinsed complexes containing labeled 30-mer transcripts (EECs) were generated on the Ad ML promoter template pML20-40 31g (total template length ∼ 390 nt) as described (15). Briefly, PICs were assembled with the same conditions as described above for initiation assays and rinsed with BC100 and M5 buffer. Complexes were labeled in M5 buffer with 0.5 mm ATP, 50 μm UTP, and 2.5 μm [α-32P]CTP with 0.5 units/μl RNasin Plus for 2 min at 30 °C followed by a chase with unlabeled 50 μm CTP for 30 s at 30 °C. The resulting complexes, which contained predominantly 30-nt RNAs, were rinsed with a wash buffer containing 1.6 m KCl followed by a 40 mm KCl wash buffer to remove excess salt. Complexes were released from the beads by cleavage with Cac8I for 25 min at 30 °C in MEMDM buffer containing 40 mm KCl, followed by addition of 1.5 μg of salmon sperm DNA per μg template. The EECs were diluted 5-fold and treated with 173–230 fmol of TFIIF (25–33 fmol/μl) or 240 fmol of Gdown1 for 5 min at room temperature per initial 10-μl reaction followed in some cases by additional challenge by 231 fmol of TFIIF (33 fmol/μl) for 5 min at room temperature. Glycerol was added to a final volume of 10%, and samples were resolved on 4% Tris-glycine native acrylamide gels run at 7 watts for 4 h at 4 °C. Gels were dried and analyzed with a Typhoon Trio phosphorimager and ImageQuantTL software.
RESULTS
We investigated the effect of Gdown1 on pol II function using in vitro approaches. We employed immobilized DNA templates (double-stranded except for Fig. 7) containing the strong CMV or Ad ML TATA box promoters. Pol II PICs were assembled with recombinant (TBP, TFIIB, TFIIF, and TFIIE) and purified (TFIIH) human GTFs and purified human pol II. Western blots with Gdown1 antibody did not reveal any Gdown1 in our pol II preparation, when we would have detected 1% of the polymerases containing Gdown1 (data not shown). Transcription was initiated with a brief pulse of NTPs at limiting levels including 32P-labeled CTP, generating an EEC. As shown in Fig. 1, transcripts in the EECs were typically 20–30 nt in length with the CMV template (lane 1).
FIGURE 1.
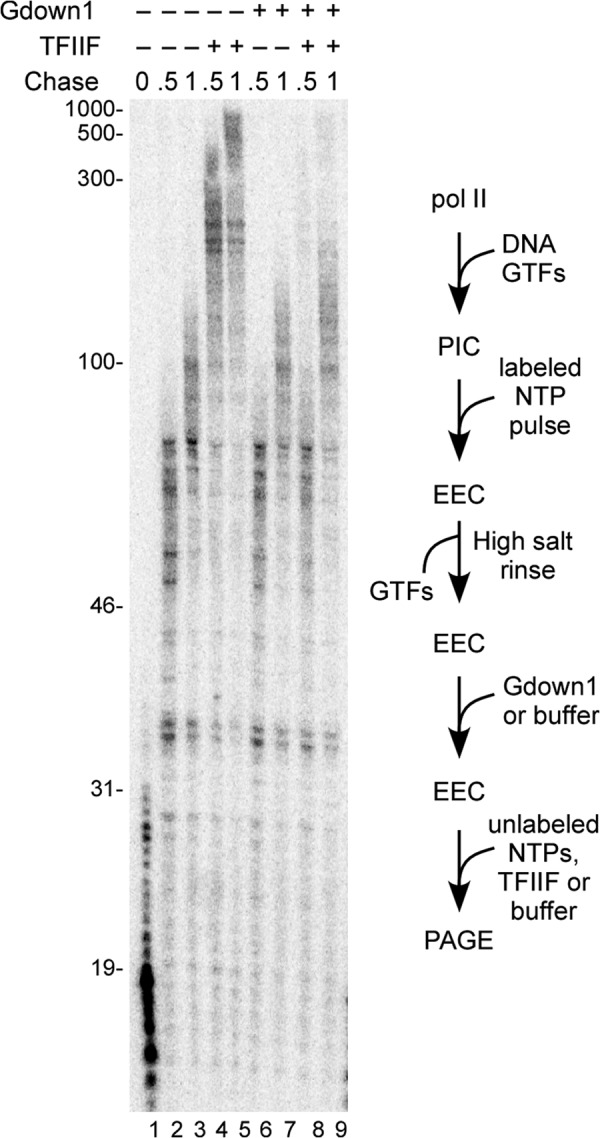
Addition of Gdown1 to early elongation complexes inhibits stimulation of elongation by TFIIF. EECs were generated on the CMV template, high salt-rinsed, and (except for lane 1) chased for 30 or 60 s in the presence or absence of 166 fmol of TFIIF as described under “Experimental Procedures.” In lanes 6–9, 240 fmol of Gdown1 were added to the EEC and incubated for 5 min before chase. (240 fmol is a 20-fold excess over pol II in the original PIC and 1.4-fold excess over TFIIF used during the chase.) Lengths of markers are shown on the left of the gel. A schematic of the assay is shown to the right of the gel.
To study the response of the EECs to elongation factors, we first rinsed them with 1.6 m KCl to remove any TFIIF. Addition of excess nonlabeled NTPs to these complexes allowed pol II to continue transcript elongation at ∼100 nt/min (Fig. 1, lanes 2 and 3), as expected from earlier work (15). Supplementing the elongation reactions with TFIIF stimulated the elongation rate by 3–4-fold (lanes 4 and 5), also as expected (15). We anticipated from earlier studies that Gdown1 would block the ability of TFIIF to stimulate elongation by pol II. In those experiments, the initial EECs were generated with nuclear extracts (3). To verify that EECs produced with GTFs respond similarly, we incubated recombinant human Gdown1 (in large excess over pol II) with high salt-rinsed EECs and then chased with nonlabeled NTPs, with or without TFIIF. As shown in Fig. 1, lanes 6–9, Gdown1 alone had no significant effect on elongation rate (as expected, see Ref. 3). Gdown1 completely blocked rate stimulation by TFIIF, indicating that Gdown1 fully associated with the high salt-rinsed EECs.
The ability of Gdown1 to displace TFIIF from free pol II (4, 5) strongly suggests that Gdown1 should prevent assembly of the pol II PIC, at least if assembly is attempted with pol II already bound with Gdown1 (pol IIG). Jishage et al. (4) demonstrated that Gdown1 can completely inhibit the ability of pol II to transcribe from a promoter on a supercoiled template, an assay that does not require TFIIE or TFIIH. We tested the effect of Gdown1 on functional PIC formation by pretreating pol II with a range of Gdown1 levels (from 5- to 50-fold excess of Gdown1 to pol II), followed by PIC assembly with the full complement of the GTFs on either the Ad ML (Fig. 2A) or CMV (Fig. 2B) templates. For these assays, transcription was limited to the initial pulse labeling step. Preincubation of pol II with excess Gdown1 did strongly inhibit transcription on both templates (Fig. 2, A and B). However, inhibition was never complete in these assays on either template. We repeated the 20-fold excess test seven times with the CMV promoter and found that 12% (±4%) of the control activity remained; at 50-fold excess on this promoter, 11% (±2%) remained (n = 5). Limited tests on the CMV promoter at a 100-fold excess of Gdown1 to pol II (data not shown) did not reveal any further significant reduction in RNA synthesis. For Ad ML, in four tests, we found 20% (±4%) of the control activity after preincubation of pol II with a 20-fold excess of Gdown1.
FIGURE 2.
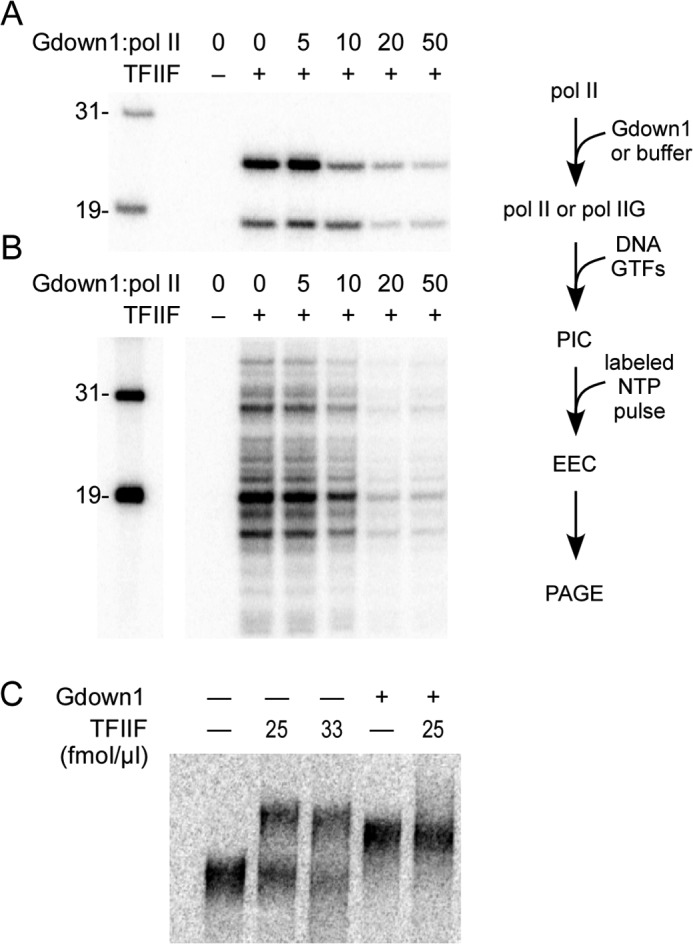
Pol IIG can support PIC assembly, but it is less effective than pol II alone. A, PICs were assembled on Ad ML templates using pol II preincubated with the indicated fold excesses of Gdown1 (20× contained 240 fmol of Gdown1), followed by assembly into PICs with GTFs, including 72 fmol of TFIIF per reaction and generation of EECs as described under “Experimental Procedures.” B, as described in A, except that the CMV template was used. The space between the markers and the rest of the gel separates non-adjacent lanes within the same gel. Lane 1 in both A and B contained all components for PIC formation except TFIIF to demonstrate TFIIF dependence for the initiation assay. Lengths are shown for the size markers in the left-most lane of A and B, and a schematic of the assay utilized for A and B is shown to the right. C, pol II elongation complexes containing labeled transcripts were resolved on a native gel after addition of Gdown1, TFIIF, or both factors as described under “Experimental Procedures.” No factors were added in the left-most lane.
The lack of complete inhibition by pol IIG suggests that TFIIF can at least partially compete with Gdown1 to function in PIC formation. However, one might alternatively suppose that a small fraction of our pol II preparation is unable to bind Gdown1. To test for this possibility, we took advantage of the shift in mobility caused by Gdown1 binding to pol II transcription complexes resolved on native gels (3). As shown in Fig. 2C, pol II elongation complexes (with label in the nascent RNA) were completely shifted upon addition of Gdown1. This shift in mobility was clearly distinct from that conferred by TFIIF and importantly, the Gdown1-based shift could not be reversed by the addition of TFIIF at Gdown1/TFIIF ratios similar to those used in the transcription assays. These results suggest that all of our pol II preparation is competent to interact with Gdown1.
Gdown1 did not completely block the ability of TFIIF to function in PIC formation (Fig. 2). This raised the question of whether Gdown1 can displace TFIIF from an already assembled PIC and, if so, whether this would compromise the ability of the PIC to initiate transcription. We therefore treated complete PICs assembled on the CMV promoter with a 20-fold excess (relative to total pol II) of Gdown1 and generated labeled RNAs with an initial addition of limiting NTPs. We did not observe any inhibition of RNA synthesis with Gdown1 challenge of the complete PIC (compare lanes 1 and 6 of Fig. 3). This might have been anticipated because TFIIF had already functioned to support PIC assembly in this case, and TFIIF is not needed for the initiation step itself (16). If Gdown1 had displaced TFIIF in the PICs in Fig. 3, lanes 6–10, we expected the resulting EECs to retain Gdown1. Gdown1 binding to free pol II (5) or to pol II in elongation complexes (3) is resistant to high salt concentrations. We therefore anticipated that EECs derived from PICs incubated with an excess of Gdown1 would retain any associated Gdown1 after 1.6 m KCl rinse and thus be resistant to TFIIF stimulation in a subsequent chase. However, we found that after high salt rinse, EECs that had been incubated with Gdown1 at the PIC stage were as responsive to TFIIF as complexes that had never seen Gdown1 (compare Fig. 3, lanes 4 and 5 with lanes 9 and 10). This result suggests that Gdown1 cannot associate with pol II in the PIC.
FIGURE 3.
Complete PICs challenged with Gdown1 show no defect at initiation and give rise to elongation complexes that are fully responsive to TFIIF after high salt rinse. PICs were prepared on the CMV template as described under “Experimental Procedures” and were incubated with 240 fmol of Gdown1 (20-fold excess relative to pol II and 3.3-fold excess relative to TFIIF used to assemble PICs) or buffer before initiation. After a 1-min pulse with limiting, labeled NTPs, EECs were high salt-rinsed and chased in the presence or absence of 166 fmol of TFIIF for 30 or 60 s as described under “Experimental Procedures.” The space between lanes 5 and 6 separates non-adjacent lanes within the same gel. Lengths are shown to the left for the size markers in the left-most two lanes. A schematic of the assay is shown to the right of the gel.
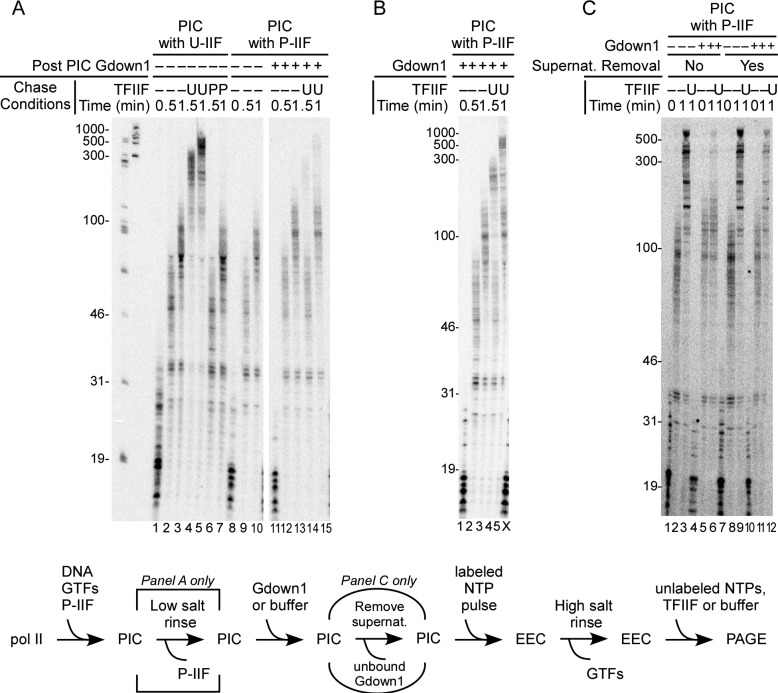
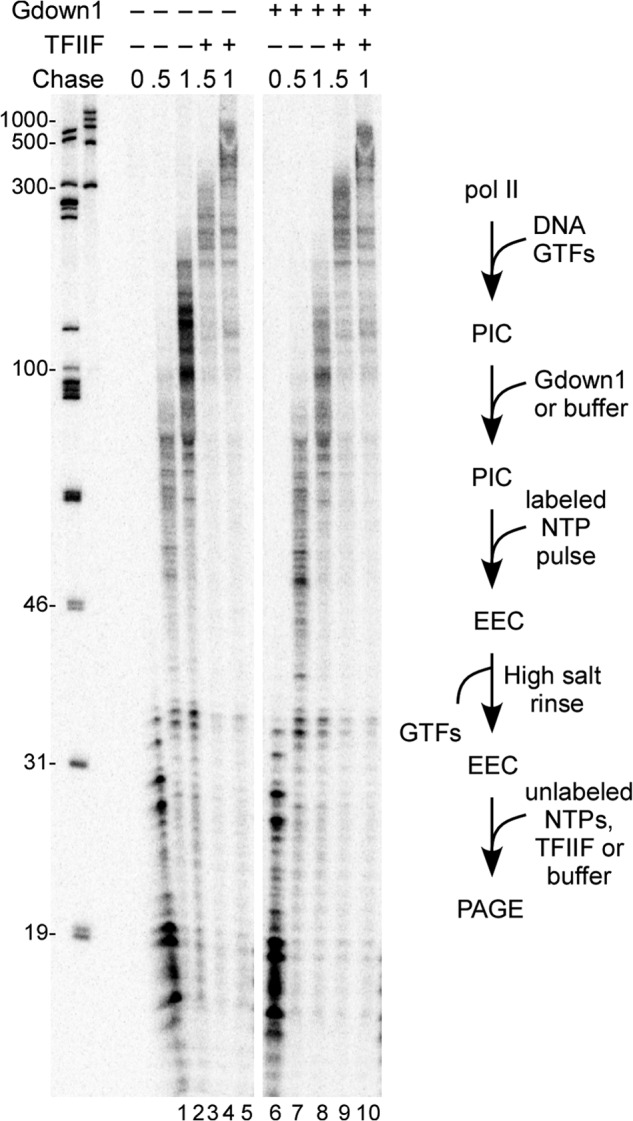
It could be argued that the failure of Gdown1 to associate with a preformed PIC might reflect some overall aspect of PIC structure, as opposed to a simple failure of Gdown1 to compete with TFIIF for pol II interaction. As noted earlier, TFIIF can be completely removed from the PIC without impairing initiation if the TFIIF was phosphorylated by CK2 (15, 16). Thus, by using P-IIF, we could test whether transcription-competent PICs that lack TFIIF are affected by incubation with Gdown1. Results of this assay are shown in Fig. 4. As expected (15), P-IIF supported transcript initiation (Fig. 4A, lane 8) but did not stimulate transcript elongation (Fig. 4A, compare lanes 2 and 3 and 6 and 7). In contrast to the results in Fig. 3, TFIIF-less PICs, which were incubated with Gdown1, gave rise to EECs that were non-responsive to TFIIF after high salt rinse (Fig. 4A, lanes 12–15). Thus, in the absence of TFIIF, Gdown1 can load into the transcription complex either in the PIC or at some point during early elongation prior to the rinse step.
FIGURE 4.
Loss of P-IIF from the PIC followed by challenge with Gdown1 results in inclusion of Gdown1 in the transcript elongation complex. PICs were prepared on the CMV template with 72 fmol of unphosphorylated (U) or CK2 phosphorylated (P) TFIIF per reaction as described under “Experimental Procedures.” A, PICs were low salt-rinsed (which removes only the P-IIF), incubated with a 20-fold excess of Gdown1 to pol II or with buffer, pulse-labeled, rinsed with high salt, and then chased with no addition, 166 fmol TFIIF, or P-IIF as described under “Experimental Procedures.” The schematic of the assay is shown below the panels. Note that the step of removing unbound Gdown1 from the PIC applies only to C. The white bar separates non-adjacent lanes within the same gel. The lengths of size markers are shown to the left of the panel. B, as in A, except that the PICs were not rinsed but used directly (in the assay schematic, the bracketed rinse step was removed). Lane X is the lower section of an adjacent lane, resulting from distortion in the gel. C, as described in A, except that the unbound Gdown1 in the supernatant (supernat.) was removed before the pulse. The schematic of the assay is shown below the panel. In this case, there was no low salt rinse step for the PIC.
In the earlier studies (16) and in the experiments for Fig. 4A that used P-IIF, a low salt rinse of the PIC was always included to guarantee complete removal of the P-IIF from the PIC prior to the start of transcription. We reasoned that because the interaction of P-IIF with pol II within the PIC is sufficiently weak to be disrupted by a single gentle wash, Gdown1 might be able to compete with P-IIF without a rinse step. The results of testing this idea are shown in Fig. 4B. In this case (lanes 4 and 5), the high salt-rinsed complexes that had been incubated with Gdown1 as PICs revealed a roughly equal partitioning between rapidly elongating complexes (which lacked Gdown1) and more slowly migrating complexes (which contained Gdown1). This result suggests that TFIIF can compete with Gdown1 for pol II association even when binding of TFIIF to pol II is weakened by phosphorylation of TFIIF.
In the reactions in Figs. 3 and 4, A and B, Gdown1 was added to a PIC and remained in the reaction during the pulse labeling step. Because Gdown1 binds tightly (and in preference to TFIIF) to elongation complexes, it was possible that in the complexes in Fig. 4A, lanes 12–15, Gdown1 did not bind to the PIC at all but instead entered the complex during the early steps in transcript elongation. To test this, we repeated the assay in which Gdown1 was incubated with PICs that had lost P-IIF, but in addition, we removed the Gdown1 before adding the NTP substrates for the pulse labeling. As shown in Fig. 4C, when these pulse-labeled complexes were rinsed with high salt and chased, they were responsive to TFIIF (compare lanes 8 and 9 and 11 and 12 of Fig. 4C). Thus, Gdown1 must have entered the transcription complexes during the pulse-labeling reactions in Fig. 4A, lanes 12–15, subsequent to the PIC.
We showed in Fig. 3 that when PICs containing TFIIF were challenged with Gdown1, Gdown1 did not apparently bind to those complexes, as judged by the effect of TFIIF addition after pulse-labeling and high salt rinse. However, the results in Fig. 4A show that Gdown1 does bind to TFIIF-less complexes during the pulse step with sufficient stability to survive high salt rinse because TFIIF was blocked from functioning during the chase of those complexes. This suggests that Gdown1 could be associating less stably with early elongation complexes when TFIIF is also present. In that case, it might also fail to bind sufficiently tightly to pol II to survive high salt rinse, which would lead to the result in Fig. 3, lanes 6–10. To test this idea, we performed the experiments shown in Fig. 5A, lanes 1–6. We again incubated Gdown1 with PICs followed by pulse labeling of the initial transcripts, but we proceeded directly to chase without any rinse step. (Note that no additional TFIIF was provided in these reactions since the TFIIF from the PIC assembly carried over into the chase.) In this case, a substantial fraction of the complexes that received Gdown1 elongated much more slowly than the controls (compare Fig. 5A, lanes 2 and 3 with lanes 5 and 6) consistent with the idea that Gdown1 can displace TFIIF beginning at the earliest stages of transcript elongation. To extend this idea, we repeated this experiment using a modified version of TFIIF (1–227 TFIIF) in which the largest subunit is truncated, retaining only 227 residues from its N terminus (Fig. 5B). In the absence of Gdown1 challenge, elongation was less stimulated than for complete TFIIF (Fig. 5B, lanes 8 and 9), as expected from earlier results (15). When Gdown1 was added to the PICs assembled with 1–227 TFIIF (Fig. 5B, lanes 10–12), the rate of elongation in the subsequent chase reaction was essentially the same as for complexes without TFIIF (compare with Fig. 1, lanes 1–3). In particular, note that the fraction of more rapidly elongating complexes in the 30-s control reaction with complete TFIIF was absent in the 30-s reactions with 1–227 TFIIF (compare lanes 5 and 11 of Fig. 5B). This result indicates that when TFIIF lacks the C-terminal segment of the largest subunit, it cannot compete significantly with Gdown1 for association with pol II once transcript elongation is under way. The potential significance of this last point is explored under “Discussion” below.
In lanes 7–12 of Fig. 5A, we repeated the test for Gdown1 association with the PIC, in this case using the no-rinse protocol as in lanes 1–6 of Fig. 5A. As we observed in Fig. 4C, incubation of Gdown1 with the PIC followed by removal of Gdown1 before the pulse labeling step did not result in detectable loss of responsiveness to TFIIF during the subsequent chase. Thus, we have no evidence that Gdown1 can stably associate with a pre-assembled PIC, regardless of the presence of TFIIF or the use of high salt rinsing in the analysis.
The results in Fig. 2 show that when Gdown1 is added in large excess over pol II prior to PIC formation, a small but reproducible fraction of transcriptionally active PICs is obtained. This raises the question of whether Gdown1 could remain associated with the PIC if it entered PIC assembly already in complex with pol II. Our approach to address this possibility was the same as in Fig. 3B, except that Gdown1 was incubated at 20-fold excess over total pol II prior to the addition of template and the other GTFs. As shown in Fig. 6, the transcriptionally active complexes from the pol IIG reactions were fully TFIIF-responsive after high salt rinse. This result is also consistent with the inability of Gdown1 to stably associate with the PIC.
FIGURE 6.
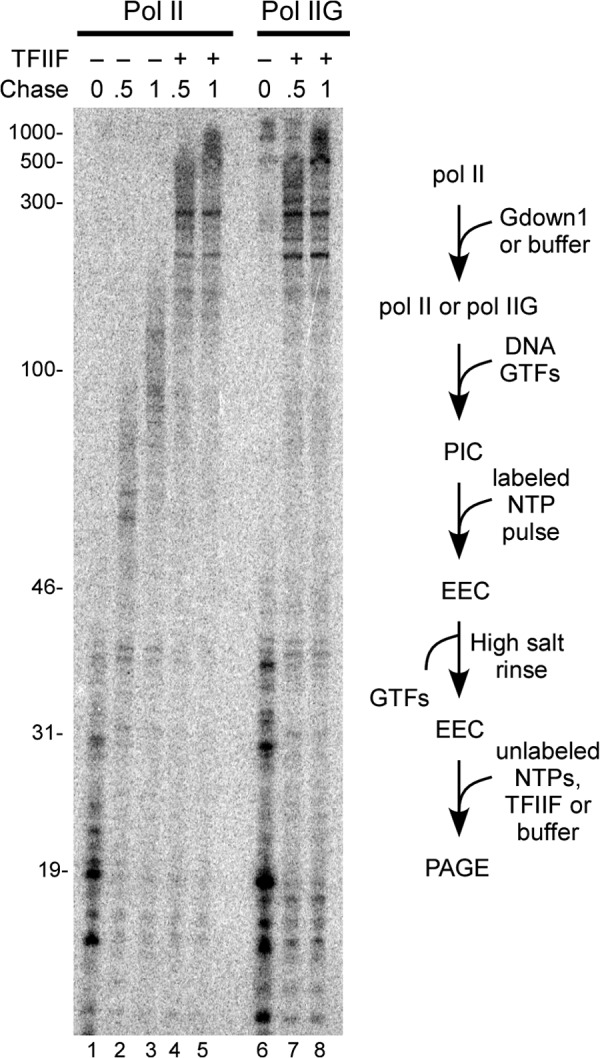
Transcriptionally active PICs formed with pol II(G) are fully stimulated by TFIIF during subsequent transcript elongation. Pol II was incubated with a 20-fold excess of Gdown1 or with buffer, followed by PIC assembly (containing 72 fmol of TFIIF), pulse-labeling, high salt rinse, and chase in the absence or presence of 166 fmol of TFIIF as described under “Experimental Procedures.” Lanes 6–8 contained 8× the standard reaction amount to increase signal for analysis. A schematic of the assay is shown to the right of the gel.
An alternative approach to exploring the interaction of Gdown1 with the PIC involves the use of premelted (“bubble”) templates. When the template is mispaired over the region that will be unwound upon open complex formation, PIC formation no longer requires TFIIE and TFIIH (20, 23, 24). For the experiment shown in Fig. 7, we assembled PICs on an Ad ML-based bubble template mismatched from −9 to +3 relative to transcription start, using pol II with either TBP, TFIIB, and TFIIF (A) or the complete set of GTFs (B and C). Gdown1 was either preincubated with pol II or incubated with the preformed PIC, in both cases at a 20-fold excess over pol II. The results are very similar to those seen with double stranded templates and the full set of GTFs. Pol IIG did support transcription at 18 ± 4% (n = 5) of the activity seen with the control when using the limited GTF set, whereas addition of Gdown1 to the preformed PIC did not inhibit transcription (A). Similar results were seen with the complete GTF set (B). These results do not support a significant role for TFIIH in the reduced activity of pol IIG in PIC formation. We also asked whether bubble template PIC formation with pol IIG was stimulated by providing a considerable excess of TFIIF (C). In this case, we used the full GTF set to support PIC assembly and an 8-fold excess of TFIIF over the normal amount. We found that the additional TFIIF increased the level of transcript with pol IIG to 44% of that seen with pol II alone (n = 3). These results reinforce the point made in the double-stranded template assays, namely that TFIIF can compete with Gdown1 in the context of PIC assembly.
DISCUSSION
Gdown1 quantitatively displaces TFIIF from both free pol II and pol II engaged in transcript elongation (3–5). This prevents TFIIF stimulation of elongation by pol IIG (3); most importantly, it could prevent initiation by pol IIG (4). This last point is difficult to reconcile with ChIP results which place Gdown1 at most pol II promoters, as well as biochemical studies which implicate Gdown1 in mediating the near-ubiquitous pause experienced by pol II in mammalian cells at ∼50 bases downstream of transcription start (3). We now show that although TFIIF is generally outcompeted by Gdown1 for interaction with pol II, this is not the case within the PIC. Once the PIC is assembled, TFIIF is not displaced by Gdown1. When PICs were incubated with Gdown1 but the Gdown1-containing supernatant was removed before NTP addition, complexes remained fully responsive to TFIIF during subsequent transcript elongation, even when the PICs lacked TFIIF. Thus, by this functional test, we cannot demonstrate any interaction of Gdown1 with the PIC. In addition, pol IIG does support some transcript initiation, apparently because TFIIF can function (albeit inefficiently) in the presence of Gdown1 during PIC formation. If TFIIF is removed from a PIC through the use of P-IIF, the transcription complex can quantitatively bind Gdown1 shortly after initiation (Fig. 4).
The relative ability of Gdown1 and TFIIF to interact with pol II within the PIC is clearly very different from the interaction of these factors with either free or transcriptionally engaged polymerase. A cryo-EM study (5) showed that although Gdown1 binds to free pol II over a broad area of the polymerase surface, the strongest interactions occur on the jaw domain of Rpb1 and the Rpb5 subunit on either side of the central catalytic cleft (see also (4)). The regions of Gdown1-pol II interaction generally overlap with areas of TFIIF-pol II interaction (5), but the tightest TFIIF-pol II interaction appears to occur on the lobe domain of Rpb2 (25). Thus, although many pol II binding surfaces are in common between Gdown1 and TFIIF, the most important sites are not identical. The PIC is a unique environment in terms of factor access to pol II because DNA is present as a fully double-stranded molecule located outside of the catalytic cleft. DNA in that position could block the ability of Gdown1 to associate simultaneously with the jaw domain and Rpb5. In this context, it is also worth noting that the central charged domain of the largest subunit of yeast TFIIF has been reported to interact with the jaw domain of yeast pol II (26). The analogous charged domain in human TFIIF is absent in 1–227 TFIIF (15). This may account for the apparent failure of this truncated version of TFIIF to compete significantly with Gdown1 for association with pol II in the early stages of transcript elongation, in contrast to the effect of complete TFIIF (Fig. 5B).
In considering how pol IIG might give rise to some functional PICs, the results of a recent structural study on the assembly of the mammalian PIC are informative (18). Those authors describe an early step in PIC assembly consisting of a complex of TBP, TFIIB, and pol II (along with TFIIA). In that complex, DNA has not yet assumed a fixed location relative to pol II and the GTFs, and pol II associates with the nascent PIC primarily through one domain of TFIIB. Pol IIG may also be capable of forming an initial pol IIG·TBP·TFIIB·template complex because Gdown1 is not expected to inhibit pol II/TFIIB interactions (5), and the template may not be stably positioned to interfere with Gdown1 binding on either side of the central cleft (18). Jishage et al. (4) demonstrated the existence of a similar complex containing pol IIG, TBP, and TFIIB in mobility shift experiments. He et al. (18) indicate that once TFIIF joins the pol II·TBP·TFIIB complex, DNA adopts a single position. It seems plausible that TFIIF could also interact with some of the pol IIG·TBP·TFIIB complexes to drive completion of PIC formation, although this reaction is not favored. The reconfiguring of the template that accompanies TFIIF binding could destabilize Gdown1-pol II interaction in those complexes that do complete PIC formation, consistent with our results in Figs. 4–6. This view of PIC assembly is also consistent with the fact that addition of a large excess of TFIIF leads to an increase in active PIC formation in the presence of pol IIG (Fig. 7) (4).
Although Gdown1 does not functionally associate with the PIC, even in the absence of TFIIF, Gdown1 binds tightly to elongation complexes in the presence of high salt and displaces TFIIF from those complexes. At what point after initiation does the switch to the preference for Gdown1 binding to pol II occur? The results in Figs. 4–6 show that the answer depends to some extent on the assay method. In our initial studies, we relied on the salt-resistant association of Gdown1 with free pol II and the salt-sensitive binding of TFIIF to pol II. Gdown1 cannot associate with early elongation complexes bearing TFIIF in a salt-resistant form because high salt rinse of those complexes leaves them fully responsive to TFIIF during subsequent chase (Fig. 3). When the same experiment was done with PICs that lack TFIIF, Gdown1 did load during the pulse since none of the salt-rinsed EECs responded to TFIIF during the chase (Fig. 4A). Gdown1 loaded into some but not all complexes assembled with P-IIF as judged by the high salt rinse assay (Fig. 4B), consistent with the weak association of P-IIF with the PIC. All of these results indicate that although Gdown1 is fully competent to load into the transcription complex shortly after transcription start, this association is competitive with TFIIF. In the assay in Fig. 5A, EECs containing TFIIF were directly challenged with Gdown1, without any rinsing step. In this case, Gdown1 clearly prevented TFIIF from acting in some of the complexes during chase. The pol II elongation complex undergoes several transitions during the early stages of transcript elongation (16, 20, 27), and it will be important to determine whether any of these coincide with the shift in pol II binding preference from TFIIF to Gdown1.
Gdown1 is important in driving pausing (3), but its antagonism with TFIIF (3–5) raised the question of how Gdown1 can be incorporated into the transcription complex. Our results provide at least a partial resolution of this paradox. The ability of pol IIG to support transcription in vitro is significant, even though this process is inefficient because it indicates that pol IIG should be competent to begin transcription in the cell. Additional mechanisms will presumably be important in allowing efficient participation of Gdown1 in transcription. CK2 phosphorylation of TFIIF facilitates loading of Gdown1 early in transcript elongation (Fig. 4). TFIIF can be extensively phosphorylated in vivo (10, 28–32). Some of the phosphorylation on TFIIF purified from nuclear extracts appears to result from CK2 (32), consistent with the fact that among the GTFs, TFIIF is a preferred CK2 substrate in vitro (33). The presence of CK2 at selected promoters has been documented previously (34).
Gdown1 was originally described as a factor that renders transcription dependent on the Mediator complex (1). This suggests that Mediator is likely to be involved in regulating transcription by pol IIG in the cell. The mechanisms through which Mediator might act to facilitate pol IIG function are not known, but it is relevant that TFIIF serves to specifically align pol II with Mediator (35, 36). This effect of TFIIF depends on the interaction of an activator with Mediator (35), consistent with the earlier observation that activators can drive structural changes in Mediator (37). Thus, Mediator may facilitate the otherwise weak ability of TFIIF to support PIC assembly despite the presence of Gdown1.
As a final point, it is interesting to consider the broader role of promoter-proximal pausing by pol II that is driven in part by Gdown1. Although poised polymerases provide an attractive mechanism for rapid induction of productive transcription in response to regulatory signals, such polymerases could also play an important role in maintaining promoter regions in a nucleosome-free state (38). A genome-wide survey of Gdown1 and pol II locations showed that genes expressed at low levels tend to have higher ratios of Gdown1 to polymerase near the promoter as compared with more highly expressed genes (3). This consistent with the idea that extended pausing by pol II just downstream of the initiation site, mediated by Gdown1, is particularly important in maintaining infrequently used promoters in an accessible, potentially active state. Because pol II is often found at enhancer elements as well as promoters (see for example, Ref. 39), one might anticipate that Gdown1 could also play an important role in keeping such elements accessible.
Acknowledgment
We thank Jeff Cooper for technical assistance in the purification of Gdown1.
This work was supported, in whole or in part, by National Institutes of Health Grant GM35500 (to D. H. P.). This work was also supported by National Science Foundation Grant 1121210 (to D. S. L.) and American Heart Association Postdoctoral Fellowship 12POST12040106 (to J. G.).
- pol II
- RNA polymerase II
- pol IIG
- RNA polymerase II containing the Gdown1 subunit
- EEC
- early elongation complex
- PIC
- preinitiation complex
- GTF
- general transcription factor
- P-IIF
- TFIIF factor phosphorylated with casein kinase 2
- CK2
- casein kinase 2
- Ad ML
- adenovirus major late
- nt
- nucleotide(s)
- TBP
- TATA box binding protein.
REFERENCES
- 1. Hu X., Malik S., Negroiu C. C., Hubbard K., Velalar C. N., Hampton B., Grosu D., Catalano J., Roeder R. G., Gnatt A. (2006) A mediator-responsive form of metazoan RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 103, 9506–9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roginski R. S., Mohan Raj B. K., Birditt B., Rowen L. (2004) The human GRINL1A gene defines a complex transcription unit, an unusual form of gene organization in eukaryotes. Genomics 84, 265–276 [DOI] [PubMed] [Google Scholar]
- 3. Cheng B., Li T., Rahl P. B., Adamson T. E., Loudas N. B., Guo J., Varzavand K., Cooper J. J., Hu X., Gnatt A., Young R. A., Price D. H. (2012) Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell 45, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jishage M., Malik S., Wagner U., Uberheide B., Ishihama Y., Hu X., Chait B. T., Gnatt A., Ren B., Roeder R. G. (2012) Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol. Cell 45, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Y. M., Chang J. W., Wang C. H., Lin Y. C., Wu P. L., Huang S. H., Chang C. C., Hu X., Gnatt A., Chang W. H. (2012) Regulation of mammalian transcription by Gdown1 through a novel steric crosstalk revealed by cryo-EM. EMBO J. 31, 3575–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nechaev S., Adelman K. (2008) Promoter-proximal pol II: When stalling speeds things up. Cell Cycle 7, 1539–1544 [DOI] [PubMed] [Google Scholar]
- 7. Gilmour D. S. (2009) Promoter proximal pausing on genes in metazoans. Chromosoma 118, 1–10 [DOI] [PubMed] [Google Scholar]
- 8. Zhou Q., Li T., Price D. H. (2012) RNA Polymerase II Elongation Control. Annu. Rev. Biochem. 81, 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hahn S. (2004) Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11, 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sopta M., Carthew R. W., Greenblatt J. (1985) Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 260, 10353–10360 [PubMed] [Google Scholar]
- 11. Price D. H., Sluder A. E., Greenleaf A. L. (1989) Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell Biol. 9, 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Izban M. G., Luse D. S. (1992) Factor-stimulated RNA polymerase-II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 267, 13647–13655 [PubMed] [Google Scholar]
- 13. Kephart D. D., Wang B. Q., Burton Z. F., Price D. H. (1994) Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J. Biol. Chem. 269, 13536–13543 [PubMed] [Google Scholar]
- 14. Lei L., Ren D., Finkelstein A., Burton Z. F. (1998) Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol. Cell Biol. 18, 2130–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Újvári A., Pal M., Luse D. S. (2011) The functions of TFIIF during initiation and transcript elongation are differentially affected by phosphorylation by casein kinase 2. J. Biol. Chem. 286, 23160–23167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Čabart P., Újvári A., Pal M., Luse D. S. (2011) TFIIF is not required for initiation by RNA polymerase II but it is essential to stabilize TFIIB in early transcription complexes. Proc. Natl. Acad. Sci. U.S.A. 108, 15786–15791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grünberg S., Warfield L., Hahn S. (2012) Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat. Struct. Mol. Biol. 19, 788–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He Y., Fang J., Taatjes D. J., Nogales E. (2013) Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murakami K., Elmlund H., Kalisman N., Bushnell D. A., Adams C. M., Azubel M., Elmlund D., Levi-Kalisman Y., Liu X., Gibbons B. J., Levitt M., Kornberg R. D. (2013) Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342, 1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pal M., Ponticelli A. S., Luse D. S. (2005) The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell 19, 101–110 [DOI] [PubMed] [Google Scholar]
- 21. Ujvári A., Luse D. S. (2006) RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 13, 49–54 [DOI] [PubMed] [Google Scholar]
- 22. Maldonado E., Drapkin R., Reinberg D. (1996) Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 274, 72–100 [DOI] [PubMed] [Google Scholar]
- 23. Parvin J. D., Sharp P. A. (1993) DNA topology and a minimal set of basal factors for transcription by RNA polymerase-II. Cell 73, 533–540 [DOI] [PubMed] [Google Scholar]
- 24. Pan G., Greenblatt J. (1994) Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J. Biol. Chem. 269, 30101–30104 [PubMed] [Google Scholar]
- 25. Eichner J., Chen H. T., Warfield L., Hahn S. (2010) Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29, 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sainsbury S., Niesser J., Cramer P. (2013) Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493, 437–440 [DOI] [PubMed] [Google Scholar]
- 28. Burton Z. F., Killeen M., Sopta M., Ortolan L. G., Greenblatt J. (1988) RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol. Cell Biol. 8, 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flores O., Maldonado E., Reinberg D. (1989) Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J. Biol. Chem. 264, 8913–8921 [PubMed] [Google Scholar]
- 30. Yonaha M., Aso T., Kobayashi Y., Vasavada H., Yasukochi Y., Weissman S. M., Kitajima S. (1993) Domain structure of a human general transcription initiation factor, TFIIF. Nucleic Acids Res. 21, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kitajima S., Chibazakura T., Yonaha M., Yasukochi Y. (1994) Regulation of the human general transcription initiation factor TFIIF by phosphorylation. J. Biol. Chem. 269, 29970–29977 [PubMed] [Google Scholar]
- 32. Rossignol M., Keriel A., Staub A., Egly J. M. (1999) Kinase activity and phosphorylation of the largest subunit of TFIIF transcription factor. J. Biol. Chem. 274, 22387–22392 [DOI] [PubMed] [Google Scholar]
- 33. Cabrejos M. E., Allende C. C., Maldonado E. (2004) Effects of phosphorylation by protein kinase CK2 on the human basal components of the RNA polymerase II transcription machinery. J. Cell Biochem. 93, 2–10 [DOI] [PubMed] [Google Scholar]
- 34. Lewis B. A., Sims R. J., 3rd, Lane W. S., Reinberg D. (2005) Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell 18, 471–481 [DOI] [PubMed] [Google Scholar]
- 35. Bernecky C., Grob P., Ebmeier C. C., Nogales E., Taatjes D. J. (2011) Molecular Architecture of the Human Mediator-RNA Polymerase II-TFIIF Assembly. PLoS Biol. 9, e1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernecky C., Taatjes D. J. (2012) Activator-mediator binding stabilizes RNA polymerase II orientation within the human mediator-RNA polymerase II-TFIIF assembly. J. Mol. Biol. 417, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer K. D., Lin S. C., Bernecky C., Gao Y., Taatjes D. J. (2010) p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 17, 753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilchrist D. A., Dos Santos G., Fargo D. C., Xie B., Gao Y., Li L., Adelman K. (2010) Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koch F., Fenouil R., Gut M., Cauchy P., Albert T. K., Zacarias-Cabeza J., Spicuglia S., de la Chapelle A. L., Heidemann M., Hintermair C., Eick D., Gut I., Ferrier P., Andrau J. C. (2011) Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18, 956–963 [DOI] [PubMed] [Google Scholar]