Background: Epithelial-mesenchymal transition (EMT) is a key process in embryonic development and cancer metastasis.
Results: Sp1 activates miR-200 transcription in epithelial cells and prevents EMT.
Conclusion: miR-200 family members require Sp1 to drive basal expression and maintain an epithelial state.
Significance: Defining the mechanisms controlling the epithelial state has implications for understanding early differentiation and for designing interventions to prevent cancer metastasis.
Keywords: Epithelial mesenchymal transition, Metastasis, MicroRNA, Sp1, Transcription, ZEB transcription factor, miR-200
Abstract
Epithelial-mesenchymal transition (EMT) is required for the specification of tissues during embryonic development and is recapitulated during the metastatic progression of tumors. The miR-200 family plays a critical role in enforcing the epithelial state with their expression lost in cells undergoing EMT. EMT can be mediated by activation of the ZEB1 and ZEB2 (ZEB) transcription factors, which repress miR-200 expression via a self-reinforcing double negative feedback loop to promote the mesenchymal state. However, it remains unclear what factors drive and maintain epithelial-specific expression of miR-200 in the absence of EMT-inducing factors. Here, we show that the transcription factor Specificity Protein 1 (Sp1) binds to the miR-200b∼200a∼429 proximal promoter and activates miR-200 expression in epithelial cells. In mesenchymal cells, Sp1 expression is maintained, but its ability to activate the miR-200 promoter is perturbed by ZEB-mediated repression. Reduction of Sp1 expression caused changes in EMT-associated markers in epithelial cells. Furthermore, we observed co-expression of Sp1 and miR-200 during mouse embryonic development wherein miR-200 expression was only lost in regions with high ZEB expression. Together, these findings indicate that miR-200 family members require Sp1 to drive basal expression and to maintain an epithelial state.
Introduction
Epithelial cells represent a mature stage of cellular differentiation; in the adult, the vast majority of epithelial cells remain epithelial until they die. Yet epithelial cells can retain the option of a change in differentiation state, which can be activated in the adult during wound healing or during the malignant progression of epithelial cell-derived cancers (carcinomas) (1). This change of differentiation state, known as epithelial-mesenchymal transition (EMT),4 has an important role during embryological development, where it enables cell migrations and tissue remodeling, and establishes various cellular lineages in the developing tissues and organs (2). The EMT process is reversible both in embryos and in cancer, and cells cultured in vitro can be switched between epithelial and mesenchymal states by manipulations of EMT-regulating microRNAs and/or transcription factors (3). Reversible EMT-like changes also occur during ES cell differentiation (4) and iPS cell establishment (5–7), and may also be important in allowing the establishment of cancer metastases (8). Thus the molecular controls that determine whether cells reside in the epithelial or mesenchymal state are of great interest.
Several transcription factors are able to prompt epithelial cells to undergo EMT, including ZEB1, ZEB2, Snail, Slug, and Twist (9–14). Interestingly, most of these factors are repressors and are reported to bind the same DNA motif, CACCTG, sometimes referred to as the E-box or Z-box (although the term E-box is also used to refer to the less specific motif CANNTG, which also binds a variety of other factors) (15). Key genes affected by the EMT-promoting factors include E-cadherin (9–11, 16, 17) and the two loci encoding the miR-200 family members (18–21).
The members of the miR-200 family of microRNAs are expressed in epithelial cells and prevent EMT by targeting ZEB1 and ZEB2 (18–21). ZEB1 and ZEB2 reciprocally repress the miR-200 genes (21, 22), creating a double negative feedback loop that provides to some extent a gene logic circuit that is capable of switching between either of two stable but interchangeable states (epithelial and mesenchymal) (3, 23). While it is clear that factors such as ZEB1 can repress miR-200 gene expression, allowing the mesenchymal gene expression program to be established, we were interested to know what drives miR-200 expression in epithelial cells. Because miR-200 expression is highly specific to epithelial cells, it is possible that one or more epithelial-specific activators may be important in driving this expression. However, our investigations reported here indicate that expression of the miR-200b∼200a∼429 gene is driven primarily by a ubiquitous activating transcription factor, Sp1, that is expressed in nearly all cells and tissues (24). This finding is consistent with the proposition that the epithelial state is a default state (25, 26), while the mesenchymal state requires active intervention by inducible regulators. Such a situation has ramifications for understanding the mechanisms controlling early differentiation and for designing interventions to prevent cancer metastasis.
EXPERIMENTAL PROCEDURES
Cell Culture and Generation of Stable Cell Lines
All cell lines were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS). The MDCK-Pez and MDCK-vector stable cell lines were generated as previously described (27).
Extraction of RNA and PCR Analysis
Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer's instructions. Messenger RNA and microRNA analysis was carried out as previously described with normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 snRNA levels, respectively (18). The primers used for mRNA qPCR are listed in supplemental Table S1.
Generation of miR-200b∼200a∼429 Promoter Constructs
The miR-200b∼200a∼429 −321/+19 promoter construct was generated as previously described (22). Mutations to the AP-2 and Sp1 binding sites were made in this construct using the QuikChange multi-site-directed mutagenesis kit (Stratagene). A −399/+31 E-cadherin promoter fragment (28) was PCR amplified from MCF-7 genomic DNA and cloned into the NheI and BglII sites of pGL3 basic. The primers for mutagenesis and cloning are shown in supplemental Table S2.
Transfection and Reporter Assays
For reporter assays, 6 × 104 cells were plated in 24-well plates and co-transfected with 200 ng of miR-200b∼200a∼429 promoter firefly luciferase reporter plasmids, 100 ng of pRL-TK Renilla plasmid (Promega) and in some cases 100 ng of expression vectors (pcDNA3.1, Sp1, or AP-2α) using Lipofectamine 2000 (Invitrogen). After 48 h of incubation, cells were lysed in Passive Lysis Buffer (Promega), and luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) using the GloMax Multi Detection System Luminometer (Promega). All reporter assays are shown as relative luciferase activities (averaged ratios of firefly luciferase/Renilla ± S.E.) and combined from at least three separate experiments. The pcDNA3.1-Sp1 expression vector was kindly supplied by Maciej Pietrzak of the Polish Academy of Sciences. The pCMX-AP-2α was donated by Nicholas Saunders of Diamantina Institute, University of Queensland, Australia. For siRNA experiments, cells were transfected in suspension with 20 nm control, Sp1 (Stealth siRNAs, Invitrogen), ZEB1, and ZEB2 (22) siRNAs using Lipofectamine RNAiMAX (Invitrogen). Cells were harvested 72 h later for downstream processing or retransfected when prolonged knockdown of these factors was required.
Chromatin Immunoprecipitation
ChIP assays were conducted with 1 × 106 MCF-7 cells per reaction cross-linked with 1% formaldehyde as previously described (29). Cells were lysed for 10 min, diluted, and sonicated on ice water (30 s on and off intervals for 18 min) on the Diagenode Bioruptor sonicator. Immunoprecipitations were performed overnight with 1 μg of Sp1 (Santa Cruz sc-H225) or ZEB1 (Santa Crux E20X), and captured for 2 h with protein A and protein G beads (Invitrogen, Dynal magnetic beads). Bead and immune complexes were washed of nonspecific binding three times with RIPA buffer and then twice in TE buffer before elution for 2 h at 68 °C with vortexing (Eppendorf Thermomixer). Supernatants were collected, and DNA collected using phenol-chloroform extraction. Real-time PCR of miR-200b∼200a∼429 and E-cadherin promoter regions were performed using primers listed in supplemental Table S3.
Immunofluorescent Staining for E-cadherin and Sp1
MDCK and MCF10A cells were transfected with siRNA as described above, plated onto fibronectin-coated chamber slides (BD Biosciences), and stained for E-cadherin and Sp1 as previously described (18). Nuclei were observed by co-staining with 4′6,6-diamidino-2-phenylindole (DAPI). Cells were visualized on an Olympus IX81 microscope, and pictures were taken using a Hamamatsu Orca camera. Images were analyzed with Xcellence software (Olympus).
In Situ hybridization
E13.5 embryos were fixed in 4% formaldehyde for 24 h and then were embedded in paraffin. Sagittal sections (4 μm) were placed on polysine glass slides for analysis. For in situ hybridization, sections were de-waxed by washing in xylene for 15 min, rehydrated through graded series of ethanol solution for 5 min, each and rinsed in phosphate-buffered saline with 0.1% Tween-20 (PBS-T) twice for 5 min. Tissues were permeabilized by treatment with proteinase K at 6.7 μg/ml in buffer (50 mm Tris, pH 7.5, 5 mm EDTA) for 5 min. The reaction was stopped by incubating sections in 4% formaldehyde for 15 min and rinsed in PBS 3× for 5 min. Sections were pre-washed in 1-methylimidazole buffer and fixed with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide as described previously (30). Sections were acetylated to reduce background staining by incubating for 10 min in 0.25% acetic anhydride in 0.1 m triethanolamine and rinsed 3× for 5 min in PBS. Sections were air-dried after dehydration by washing in increasing ethanol gradient for 5 min each. Probes (Exiqon, double-DIG-labeled LNA probes for miR-200b, scrambled control, and snRNA-U6 at 2 μl/slide) were denatured at 90 °C before dilution in 23 μl of hybridization buffer (Roche) and applied to the slide. Sections were covered with glass coverslips, sealed with rubber cement, and incubated overnight at 53 °C to allow hybridization of probe to internal microRNA. Coverslips were removed, and slides were washed in 5× saline-sodium citrate buffer (SSC, pH 7.0) for 5 min, twice 5 min in 1× SSC, and twice 5 min in 0.2× SSC all at 53 °C. Sections were rinsed in PBS, washed with levamisole buffer (0.24 mg/ml levamisole, 0.1 m Tris, pH 7.5, 0.15 m NaCl) and blocked (1/5th fetal calf serum, 1/5th 10% Roche blocking powder in maleate buffer, 3/5th maleate buffer) for 1 h at room temperature. Alkaline phosphatase-conjugated anti-DIG was applied to sections at 1:1000 dilution for 4 h at room temperature. Sections were then rinsed in PBST and color developed in buffer (0.1 m Tris, pH 9.5, 0.1 m NaCl, 0.05 m, MgCl2, 0.24 mg/ml levamisole, 1% Tween-20, 20 μl NBT/BCIP (Roche)) until complete. Reactions were stopped by washing in TE pH 8.0 and sections dried, dipped in xylene, and mounted in xylene-based mounting medium. Images were captured on the Hamamatsu Nanozoomer.
Immunohistochemistry
Embryos were fixed, and sections cut as described above. Sections were de-waxed by washing in xylene 3× for 5 min and rehydrated by washing in 100% ethanol twice for 10 min, 95% ethanol twice for 10 min, MQ-H2O twice for 5 min, and PBS for 5 min. For antigen unmasking, slides were boiled in 10 mm sodium citrate buffer (pH 6.0) for 10 min and cooled for 20 min. Once cooled, sections were washed 3× in water for 5 min, bleached in 1% hydrogen peroxide for 10 min, and rinsed 3× in water for 5 min. Sections were blocked in 10% horse serum at room temperature before antibodies were applied (1:400 ZEB1 (31), Sp1 1:200 (sc-H225X; Santa Cruz)). Slides were rinsed in PBS before secondary antibody was applied (1:500 biotin α-rabbit). The ABC and DAB kits (Vector Laboratories) were used to visualize ZEB1 and Sp1 expression. Sections were counterstained for 3 min in 1% methyl green, rinsed twice in water, dehydrated, and mounted in xylene-based mounting medium. Images were captured on the Hamamatsu Nanozoomer.
RESULTS
Sp1, but Not AP-2α, Activates the miR-200b∼200a∼429 Proximal Promoter
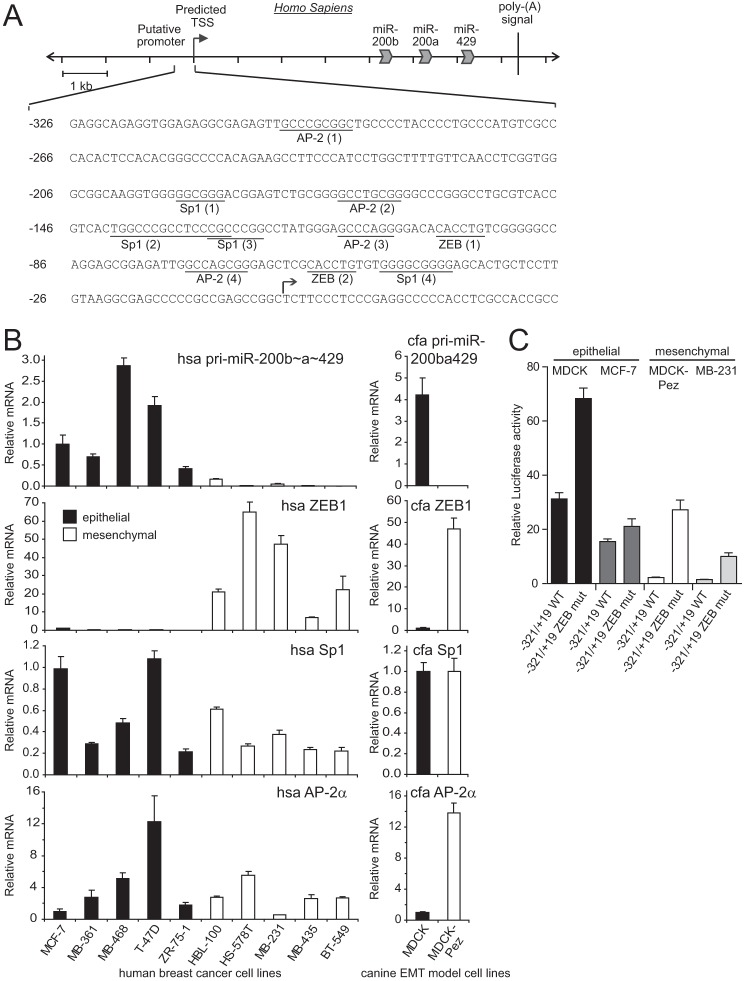
The expression of the miR-200 family is epithelial specific with the miR-200b∼200a∼429 cluster regulated by a promoter region located ∼4 kb upstream of the sequence encoding miR-200b (22). Using reporter gene assays, we have previously identified a −321/+19 bp segment of the miR-200b promoter (hereafter called miR-200b promoter) that is sufficient to drive strong epithelial-specific expression (Fig. 1, A and C and Ref. 22). This specificity of expression appears to be largely governed by the presence of repressive binding sites for the ZEB transcription factors, which when mutated allow expression of the miR-200b promoter in mesenchymal cells to near epithelial levels (Fig. 1C and Ref. 22). These data suggest that the miR-200b promoter may be positively regulated by factors present in both epithelial and mesenchymal cells, but whose activity is perturbed by ZEB binding in mesenchymal cells.
FIGURE 1.
The miR-200b∼200a∼429 minimal promoter and its activity in epithelial and mesenchymal cell lines. A, nucleotide sequence of the −321/+19 minimal proximal miR-200b∼200a∼429 promoter with predicted Sp1 and AP-2 transcription factor binding sites relative to the transcription start site (TSS) are indicated. The putative binding sites for Sp1, AP-2, and ZEB sites are underlined and are numbered in order of occurrence from the 5′-end. B, expression of the miR-200b∼200a∼429 primary transcript (Pri-200ba429), ZEB1, Sp1, and AP-2α in various epithelial and mesenchymal cell lines. C, activity of the −321/+19 miR-200b wild type and ZEB binding site mutant promoters in epithelial and mesenchymal cells. Data are expressed as relative luciferase activity ± S.E. from three independent experiments relative to the activity of the promoter-less reporter set to a value of 1.
To identify factors likely to be involved in transcriptional activation of the miR-200b promoter, we searched for potential transcription factor binding sites using the TRANSFAC database (32). This analysis indicated the presence of several Activator Protein 2 alpha (AP-2α) and Specificity Protein 1 (Sp1) binding sites within the proximal promoter (Fig. 1A). Examination of Sp1 and AP-2α levels in a panel of breast cancer cell lines and in an MDCK EMT model system (27), showed they were expressed at broadly similar levels in epithelial and mesenchymal cells, in contrast to the reciprocal cell type-specific expression of the miR-200b primary transcript and ZEB1 in these cell types (Fig. 1B).
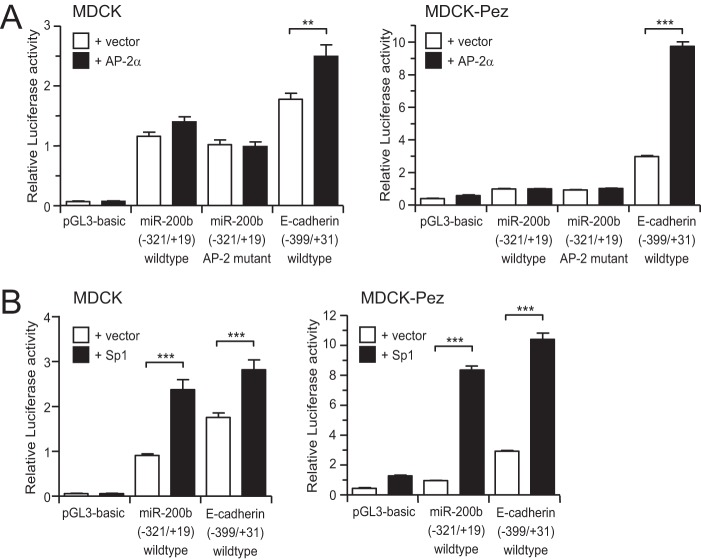
We initially selected AP-2α as a candidate factor for further investigation, as AP-2α has been reported in several contexts to activate expression of another epithelial-specific gene, E-cadherin (33–36). To determine whether AP-2α could activate the miR-200b promoter, we co-transfected the miR-200b promoter luciferase reporter along with an AP-2α expression vector into either epithelial canine MDCK cells or its mesenchymal derivative, MDCK-Pez cells (described in Ref. 27). As shown in Fig. 2A, although ectopic expression of AP-2α activated the E-cadherin promoter, it was unable to activate the miR-200b promoter in either cell type. To rule out the possibility that AP-2α activity may already be maximal in these cells and not further enhanced by exogenous AP-2α, we mutated each of the three predicted AP-2α binding sites in the miR-200b promoter. Mutation of these sites had no effect on promoter activity indicating that AP-2α was not responsible for activity of the miR-200b gene (Fig. 2A).
FIGURE 2.
Sp1, but not AP-2α, activates the miR-200 promoter. Wild type and AP-2 site mutant −321/+19 miR-200b promoter or 399/+31 E-cadherin promoter reporters were co-transfected with control or AP-2α expression vectors (A) or Sp1 expression vector (B) into epithelial MDCK and mesenchymal MDCK-Pez cell lines. Data are expressed as relative luciferase activity + S.E. from three independent experiments relative to the wild type −321/+19 miR-200b reporter. Statistical significance (**, p < 0.01; ***, p < 0.001) was determined relative to indicated controls transfection using unpaired two-tailed Student's t test.
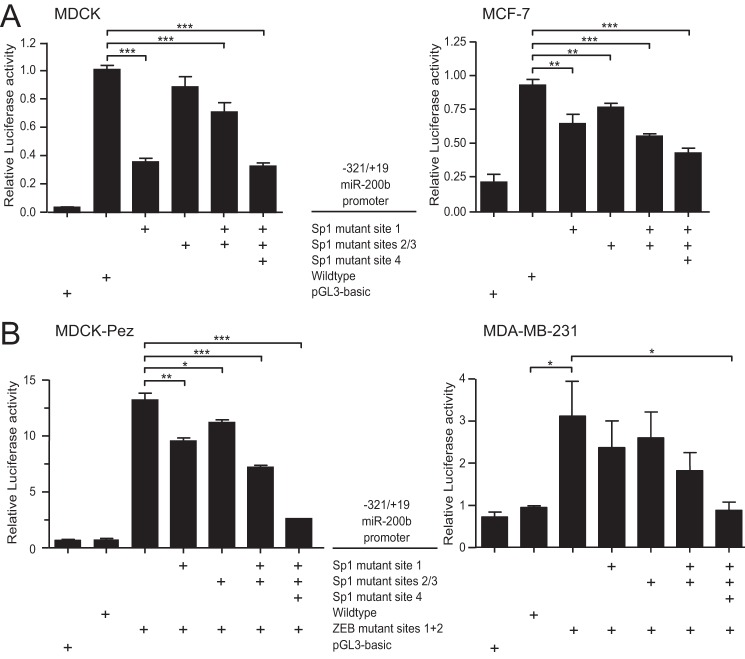
In contrast to AP-2α, ectopic expression of Sp1 significantly increased miR-200b promoter and E-cadherin activity in epithelial and mesenchymal cells (Fig. 2B). To verify this was a direct effect on the miR-200b promoter we mutated the predicted Sp1 binding sites and examined the effect on activity of the promoter in epithelial MDCK and MCF-7 cells. Mutation of each site reduced promoter activity (Fig. 3A), while mutation of all four sites produced the strongest reduction in activity, suggesting the sites act additively in activating expression. These results are consistent with the progressive loss of activity previously observed with successively truncated forms of the promoter (22).
FIGURE 3.
Intact Sp1 binding sites are required for miR-200b promoter activity in epithelial and mesenchymal cells. The −321/+19 wild type, ZEB, and Sp1 mutant miR-200b promoter reporters were transiently transfected into epithelial MDCK or MCF-7 (A) or mesenchymal MDCK-Pez or MDA-MB-231 (B) cells. Data are expressed as relative luciferase activity + S.E. from three independent experiments relative to the wild type −321/+19 miR-200b reporter. Statistical significance (*, p < 0.05; **, p < 0.01; ***, p < 0.001) was determined relative to indicated controls using unpaired two-tailed Student's t test.
We next assessed whether Sp1 in mesenchymal cells can activate the miR-200b promoter if binding of repressive factors, such as ZEB (ZEB1 or ZEB2), are prevented by mutation of the ZEB binding sites. We found that activity of the promoter lacking ZEB binding sites in mesenchymal MDCK-Pez and MDA-MB-231 cells was dependent on the Sp1 binding sites in the same way as it was in epithelial cells (Fig. 3B). These data demonstrate that Sp1 is able to drive expression of the miR-200b promoter in both epithelial and mesenchymal cells, and suggest that Sp1 activity on the miR-200b promoter in mesenchymal cells is disrupted by ZEB-mediated repression through the ZEB binding sites.
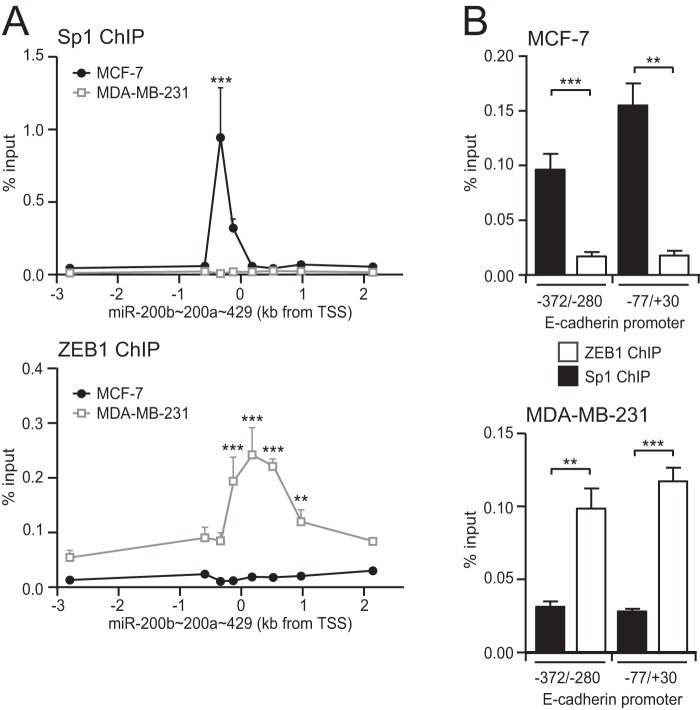
Sp1 and ZEB1 Binding to the miR-200b Promoter May Be Mutually Exclusive
To examine the binding of endogenous Sp1 to the miR-200b promoter, we performed ChIP PCR analysis on multiple regions spanning ∼4 kb of the miR-200b∼200a∼429 locus. In epithelial MCF-7 cells, a peak of Sp1 binding was observed coincident with the location of the −321/+19 miR-200b proximal promoter, confirming endogenous Sp1 specifically binds to this region (Fig. 4A). In contrast, there was no enrichment of Sp1 binding within this region in mesenchymal MDA-MB-231 cells (Fig. 4A) despite the expression of Sp1 in these cells (Fig. 1B). These results suggest that binding of Sp1 with the miR-200b promoter in mesenchymal cells may be prevented by ZEB1 binding to the nearby E-box sites. To examine this, we performed ChIP-PCR for ZEB1 over the same regions and found that ZEB1 specifically interacted within the −321/+19 miR-200b proximal promoter in MDA-MB-231 cells, but not in MCF-7 cells (Fig. 4A). Together, these data suggest that the binding of ZEB1 to the miR-200b promoter prevents binding of Sp1. Interestingly, this pattern of Sp1 and ZEB1 binding was also observed at the E-cadherin proximal promoter (Fig. 4B), suggesting an interplay between these factors in controlling epithelial gene expression may be more widespread.
FIGURE 4.
Reciprocal binding of Sp1 and ZEB1 to the miR-200b promoter. A, ChIP assays were performed in epithelial MCF-7 and mesenchymal MDA-MB-231 cells for binding of Sp1 and ZEB1 to the miR-200b∼200a∼429 promoter. PCR primers shown in supplemental Table S3 were used to amplify various regions in the vicinity of the TSS. Data are shown as mean % input DNA control ± S.E. of combined results from at least three experiments. Significant differences between antibody binding in MCF-7 and MDA-MB-231 cells are analyzed by 2-way ANOVA with Bonferroni post-tests, **, p < 0.01; ***, <0.001. B, enrichment of Sp1 and ZEB1 binding at two regions within the E-cadherin promoter in MCF-7 and MDA-MB-231 cells. Primers for the −372/−280 E-cadherin promoter region are from Ref. 70. Data are shown as mean % input DNA control ± S.E. of combined results from three experiments. Significant differences between indicated pairs are analyzed by unpaired two-tailed Student's t test, **, p < 0.01; ***, p < 0.001.
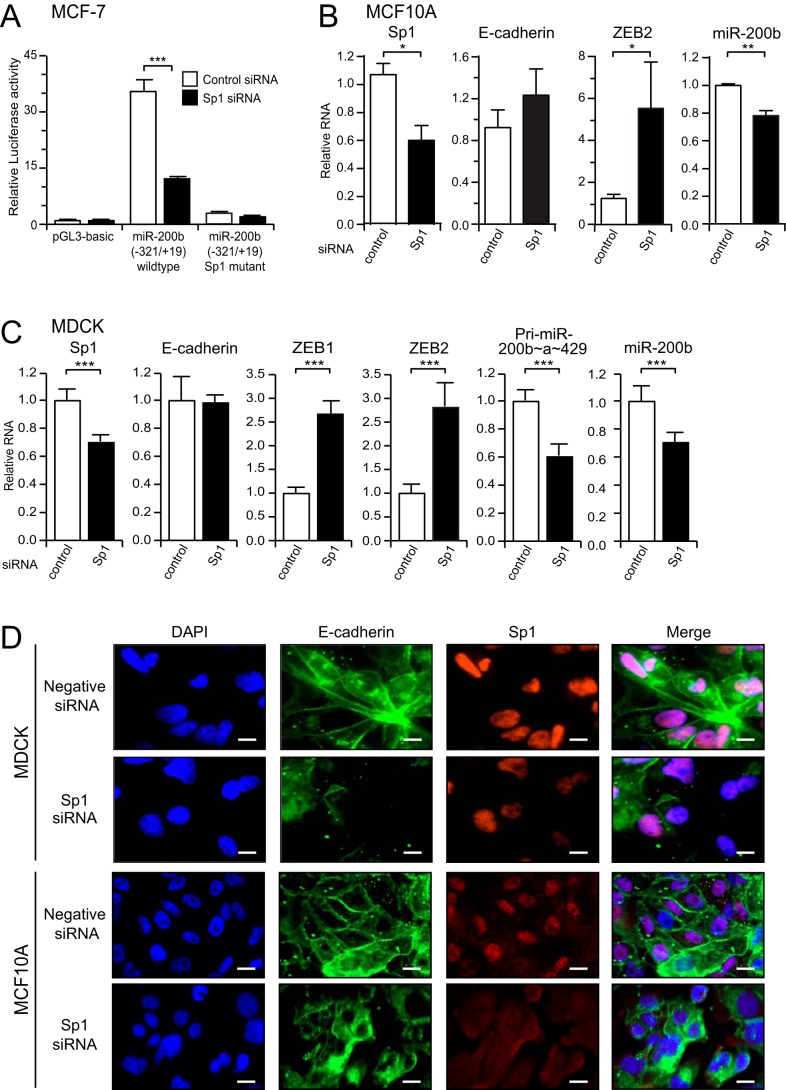
Reduction in Sp1 Levels Decreases miR-200 Levels and Causes Changes in EMT Marker Expression
Having shown that Sp1 binds to the miR-200b promoter, we next wanted to confirm whether endogenous Sp1 activates miR-200b promoter activity and expression in epithelial cells. To do this, we first co-transfected an Sp1 siRNA along with the wild type and Sp1 mutant miR-200b promoter reporter constructs into MCF-7 cells. Knockdown of Sp1 significantly reduced promoter activity of the wild type but not the Sp1 mutant construct (Fig. 5A), indicating Sp1 may be required for the basal expression of the miR-200b∼200a∼429 gene. As miR-200 is a critical enforcer of the epithelial phenotype, we reasoned that loss of Sp1 may trigger a reduction in miR-200 levels and induction of EMT. To test this hypothesis, we utilized two epithelial cell lines (MCF10A and MDCK), which are sensitive to undergoing EMT. Knockdown of Sp1 in either cell line by ∼40% resulted in a reduction of miR-200b expression and up-regulation of the mesenchymal markers ZEB1 and/or ZEB2 (Fig. 5, B and C). Although we did not observe a change in E-cadherin mRNA in the total population of cells, we found by immunofluorescence microscopy that cells with reduced Sp1 levels also exhibited loss or internalization of junctional E-cadherin (Fig. 5D). These data indicate that reduction in Sp1 levels leads to down-regulation of miR-200 and changes in EMT marker expression.
FIGURE 5.
Sp1 maintains miR-200 levels and the epithelial state of MDCK and MCF10A cells. A, activity of the −321/+19 miR-200b promoter reporters in MCF-7 cells after co-transfection with control or Sp1 siRNA. Data are expressed as relative luciferase activity ± S.E. from three independent experiments relative to the activity of the promoter-less reporter. Statistical significance (***, p < 0.001) was determined relative to indicated controls transfection using unpaired two-tailed Student's t test. Quantitative-PCR analysis of miR-200 and EMT markers in MCF10A (B) or MDCK (C) cells following transfection with control or Sp1 siRNAs for 6 or 3 days. Data are expressed as the mean relative to control siRNA + S.E. from three independent experiments. Statistical significance (*, p < 0.05; **, p < 0.01; ***, p < 0.001) was determined relative to indicated controls transfection using unpaired two-tailed Student's t test. D, immunofluorescence staining of MDCK or MCF10A cells for E-cadherin (green) or Sp1 (red) after treatment with control or Sp1 siRNAs for 3 days. DAPI staining (blue) was used to visualize nuclei. A merged image of the three is shown on the right hand panel. Scale bars indicate 10 μm.
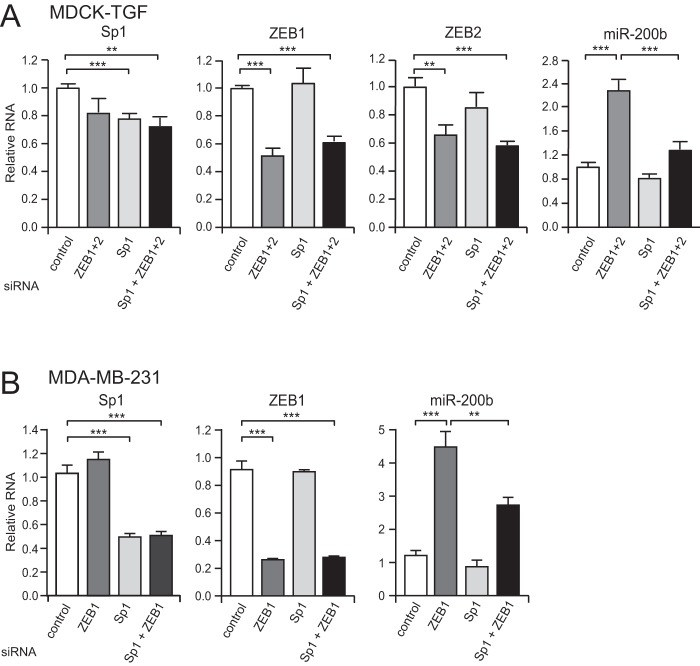
Sp1 Activates miR-200b Expression in the Absence of ZEB Binding in Mesenchymal Cells
The above reporter experiments suggest Sp1 can activate the miR-200b promoter in mesenchymal cells when ZEB binding is abrogated by ZEB site mutation. To confirm that endogenous Sp1 can drive expression of miR-200b in mesenchymal cells, we knocked down Sp1 concurrently with ZEB and examined their effect on miR-200 expression. Treatment of either MDA-MB-231 cells or TGF-β-induced MDCK cells (MDCK-TGF) (3) with ZEB siRNAs caused a significant increase in miR-200b levels as expected (Fig. 6, A and B). This de-repression and induction of miR-200b was markedly reduced after co-transfection with an Sp1 siRNA. These data indicate that Sp1 is a basal activator of miR-200 expression in both epithelial and mesenchymal cells, but whose activity is perturbed by the presence of the ZEB transcriptional repressors in mesenchymal cells.
FIGURE 6.
Sp1 activates miR-200b expression in the absence of ZEB binding in mesenchymal cells. Quantitative-PCR analysis of markers following transfection of MDCK-TGF (A) and MDA-MB-231 (B) cells with control, ZEB, or Sp1 siRNAs for 9 days. Data are expressed as the mean relative to control siRNA ± S.E. from three (MDCK-TGF) or six (MDA-MB-231) independent experiments. Statistical significance (**, p < 0.01; ***, p < 0.001) was determined relative to indicated controls transfection using unpaired two-tailed Student's t test.
miR-200b and Sp1 Are Co-expressed and Inversely Correlated with ZEB1 during Mouse Embryonic Development
miR-200 and its homolog, miR-8, have been shown to facilitate many developmental processes including eye formation and adipogenesis (37), body size in Drosophila (38), olfactory biogenesis in rats and zebrafish (39), osmoregulation in zebrafish (40), and eye and cranial cartilage development in Xenopus (41). To determine the relationship between miR-200, Sp1, and Zeb1 during mouse embryonic development we surveyed their expression in various embryonic tissues at E13.5 using sagittal formalin-fixed paraffin embedded (FFPE) sections by in situ hybridization for miR-200b and immunohistochemistry for Sp1 and Zeb1. As expected from reports indicating epithelial-specific expression of miR-200, miR-200b was expressed in epithelial tissues of the stomach, duodenum, kidney, olfactory epithelium, cochlea, lungs, and skin (Fig. 7). In these organs, we observed a clear co-expression of miR-200b with Sp1. Although Sp1 is considered a ubiquitous protein, we found that it was not expressed in all tissues, and an example being the forebrain, which shows negative staining for Sp1, as well as miR-200b. Consistent with previous studies (18–21), Zeb1 and miR-200b expression was found to be mutually exclusive in the embryonic tissues surveyed (Fig. 7). Together, these data are supportive of our in vitro findings demonstrating that miR-200 requires Sp1 to drive basal gene expression.
FIGURE 7.
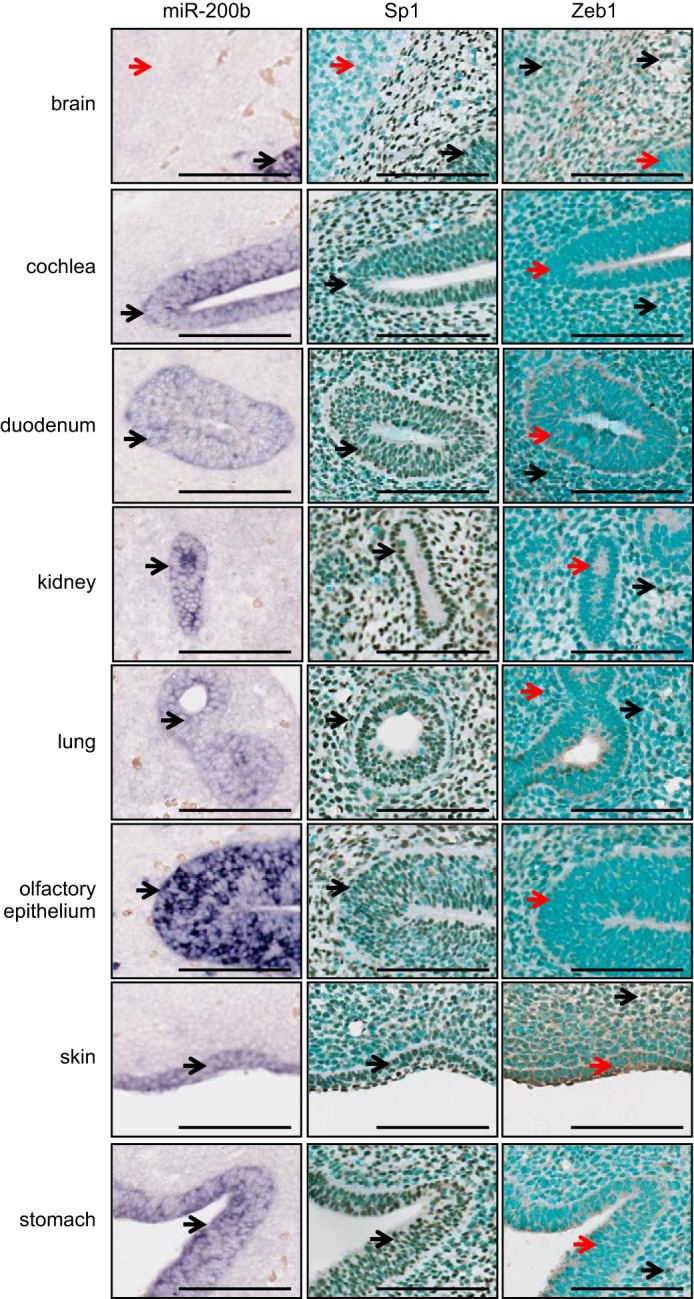
MiR-200b and Sp1 are co-expressed and inversely correlated with Zeb1 during mouse embryonic development. Paraffin sections of E13.5 mouse embryos were assayed for miR-200 expression by in situ hybridization using LNA probes, or Sp1 or Zeb1 expression using immunohistochemistry. Scale bars indicate 100 μm. Black arrows indicate positive staining, and red arrows indicate negative staining regions.
DISCUSSION
The ability of cells to transition between epithelial and mesenchymal states underlies developmental processes, pluripotency, and tumor progression and is tightly regulated by numerous signaling pathways and downstream effectors. In recent years, the miR-200 family have been proven to be powerful enforcers of the epithelial state. While various signaling pathways and mesenchymal transcription factors are known to repress miR-200 expression during EMT, it remains unclear what factors drive its expression in epithelial cells and whether they contribute to reversal of EMT (or MET). By analysis of the epithelial-specific miR-200b∼200a∼429 promoter (22), we identified Sp1 as an essential activator of miR-200 expression. We find Sp1 induces miR-200 expression through multiple sites while reduction of its expression can result in a reduction in epithelial and gain in mesenchymal properties.
These findings present intriguing implications for our understanding of how epithelial cell plasticity is controlled. It is well known that autocrine signaling pathways (such as TGF-β and PDGF) are able to drive EMT and act to maintain cells in a mesenchymal state (42–44). Mechanistically, one pathway that contributes to this process is the TGF-β/ZEB/miR-200 signaling network, whereby TGF-β stimulates ZEB production to effect miR-200 loss (3, 21, 23). Upon sustained TGF-β exposure, a self-reinforcing tripartite loop can be initiated between these factors, which enhance autocrine TGF-β signaling and maintains cells within a mesenchymal state (3). Significant perturbations of any one of these factors can cause a break in “mesenchymal” signaling with cells reverting to an epithelial phenotype (3). These previous studies suggested that perhaps cells normally reside in an “epithelial default” state (25, 26), which is stable but able to be altered by factors that initiate mesenchymal signaling. However, the mechanistic evidence for these observations has not been defined. Our finding here that the basal transcription factor, Sp1, is a primary driver of miR-200 expression fits well with the “epithelial default” model of epithelial plasticity. We show that Sp1 maintains basal miR-200 expression in both epithelial and mesenchymal cells, but in mesenchymal cells this capacity is likely overruled by competition with transcriptional repressors such as ZEB. In addition to miR-200, Sp1 has previously been shown to be an important transcriptional activator of E-cadherin (45), with re-expression of Sp1 increasing E-cadherin levels and suppressing metastasis in a lung adenocarcinoma model (46). Thus, Sp1 may promote an epithelial state by activating the expression of several key epithelial enforcing genes.
Sp1 has also been shown in some cases to enhance the expression of mesenchymal factors. For example, Sp1 can support TGF-β-induced EMT through activation of Snail expression (47), and can also acts as a cofactor with the TGF-β effector Smad complexes (48). Sp1 can also contribute to the up-regulation of MMP9 and Id-1 genes in mesenchymal cells (49, 50). A recent analysis of global cis regulatory elements (CREs) during EMT found Sp1 sites are among a select group of CREs frequently observed in genes that are differentially regulated during EMT (51). These findings reflect a complex role for Sp1 in EMT and suggest that its function may be context dependent and regulated by its interaction with other cofactors.
Sp1 is widely expressed in almost every cell type in the developing mouse; however, its expression level varies considerably across tissues (52). Mice lacking Sp1 expression die at day 11 of gestation through cell autonomous defects indicating its essential role in early development (53). We investigated miR-200 expression in relation to Sp1 and Zeb1 expression is several tissues from E13.5 mouse embryos and found Sp1 and miR-200 expression were coincident in all tissues tested, whereas miR-200 was lost in Zeb1-expressing cells. The inverse correlation of miR-200 and Zeb1 are consistent with the observations made in olfactory (39), palate (54), and inner ear (55) tissues as well as at the invasive front of colorectal cancer (56). Thus, we propose that Sp1 maintains basal expression of miR-200 in most cell types, and the absence of miR-200 is primarily triggered by the expression of ZEB and/or other repressing factors.
In addition to Sp1, the miR-200 family have been reported to be positively regulated by the p53/p63/p73 family (57–59). p53 family members have been shown to bind to regions outside of the minimal epithelial miR-200b∼200a-429 promoter and contribute to its activation (58, 59). Similarly, the miR-200c∼141 promoter is also activated by p53, with p53 loss causing reduction of miR-200c expression and EMT (57). We have found that Sp1 can also activate the miR-200c∼141 promoter; however this effect may be indirect as binding of endogenous Sp1 has not been detected in its promoter region. Interestingly, in breast cancer, p53 reduction or mutation can augment loss of miR-200 levels, but penetrance is often incomplete and is dependent on activation of other signaling pathways (such as HGF signaling via MET) for a full EMT (60, 61). This would be in keeping with the notion that activation of mesenchymal signaling pathways are needed to sustain repression of miR-200. Sp1 and p53 have also been reported to interact (62, 63) and may cooperatively enforce miR-200 expression in epithelial cells. Sp1 also plays roles in the maintenance of methylation-free CpG islands (53, 64) and may prevent DNA hyper-methylation of the miR-200 promoters, which contributes to their repression during EMT (3, 65, 66). Furthermore, the expression of the miR-200b∼200a∼429 transcript can also be stimulated through an alternative downstream promoter and an upstream enhancer element (67, 68), which may cooperatively bind Sp1 to enhance the activity of the epithelial-specific miR-200b promoter. Very recently, an Sp1-related family member KLF5 has also been shown to activate miR-200 expression and maintain epithelial characteristics of keratinocyte and breast cell lines (69). Together, these studies suggest that the Sp1/KLF family play important roles in enforcing miR-200 expression and the epithelial phenotype. Furthermore, these data lend support to the hypothesis that the epithelial phenotype is maintained as the default cellular state, and that generation of a mesenchymal state requires active intervention by inducible regulators. These findings have implications for our understanding of developmental and pathological scenarios that involve cell plasticity.
Supplementary Material
Acknowledgments
We thank Maciej Pietrzak and Nicholas Saunders for the Sp1 and AP-2α expression vectors, respectively, and members of the Goodall laboratory for helpful discussions.
This work was supported by grants from the National Health and Medical Research Council of Australia (APP1008440) and by a Cancer Council of South Australia Fellowship (to P. A. G.).

This article contains supplemental Tables S1–S3.
- EMT
- epithelial-mesenchymal transition
- MET
- mesenchymal-epithelial transition
- ES
- embryonic stem
- iPS
- induced pluripotent stem
- ZEB
- zinc finger E-box-binding protein
- Sp1
- specificity protein 1
- TSS
- transcription start site.
REFERENCES
- 1. Brabletz T. (2012) To differentiate or not–routes towards metastasis. Nat. Rev. Cancer 12, 425–436 [DOI] [PubMed] [Google Scholar]
- 2. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 3. Gregory P. A., Bracken C. P., Smith E., Bert A. G., Wright J. A., Roslan S., Morris M., Wyatt L., Farshid G., Lim Y. Y., Lindeman G. J., Shannon M. F., Drew P. A., Khew-Goodall Y., Goodall G. J. (2011) An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell 22, 1686–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eastham A. M., Spencer H., Soncin F., Ritson S., Merry C. L., Stern P. L., Ward C. M. (2007) Epithelial-mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res. 67, 11254–11262 [DOI] [PubMed] [Google Scholar]
- 5. Samavarchi-Tehrani P., Golipour A., David L., Sung H. K., Beyer T. A., Datti A., Woltjen K., Nagy A., Wrana J. L. (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 [DOI] [PubMed] [Google Scholar]
- 6. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., Qin D., Feng S., Song H., Yang D., Zhang B., Zeng L., Lai L., Esteban M. A., Pei D. (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 7. Liu X., Sun H., Qi J., Wang L., He S., Liu J., Feng C., Chen C., Li W., Guo Y., Qin D., Pan G., Chen J., Pei D., Zheng H. (2013) Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat. Cell Biol. 15, 829–838 [DOI] [PubMed] [Google Scholar]
- 8. Tsai J. H., Donaher J. L., Murphy D. A., Chau S., Yang J. (2012) Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. (2001) The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7, 1267–1278 [DOI] [PubMed] [Google Scholar]
- 10. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 11. Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 12. Savagner P., Yamada K. M., Thiery J. P. (1997) The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Biol. 137, 1403–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R. A. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 [DOI] [PubMed] [Google Scholar]
- 14. Eger A., Aigner K., Sonderegger S., Dampier B., Oehler S., Schreiber M., Berx G., Cano A., Beug H., Foisner R. (2005) DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 24, 2375–2385 [DOI] [PubMed] [Google Scholar]
- 15. Peinado H., Olmeda D., Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 16. Grooteclaes M. L., Frisch S. M. (2000) Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19, 3823–3828 [DOI] [PubMed] [Google Scholar]
- 17. Hajra K. M., Chen D. Y., Fearon E. R. (2002) The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
- 18. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 19. Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Korpal M., Lee E. S., Hu G., Kang Y. (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., Goodall G. J. (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68, 7846–7854 [DOI] [PubMed] [Google Scholar]
- 23. Gregory P. A., Bracken C. P., Bert A. G., Goodall G. J. (2008) MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle 7, 3112–3118 [DOI] [PubMed] [Google Scholar]
- 24. Wierstra I. (2008) Sp1: emerging roles–beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 372, 1–13 [DOI] [PubMed] [Google Scholar]
- 25. Frisch S. M. (1997) The epithelial cell default-phenotype hypothesis and its implications for cancer. Bioessays 19, 705–709 [DOI] [PubMed] [Google Scholar]
- 26. Polyak K., Weinberg R. A. (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9, 265–273 [DOI] [PubMed] [Google Scholar]
- 27. Wyatt L., Wadham C., Crocker L. A., Lardelli M., Khew-Goodall Y. (2007) The protein tyrosine phosphatase Pez regulates TGFβ, epithelial-mesenchymal transition, and organ development. J. Cell Biol. 178, 1223–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bussemakers M. J., Giroldi L. A., van Bokhoven A., Schalken J. A. (1994) Transcriptional regulation of the human E-cadherin gene in human prostate cancer cell lines: characterization of the human E-cadherin gene promoter. Biochem. Biophys. Res. Commun. 203, 1284–1290 [DOI] [PubMed] [Google Scholar]
- 29. Lim Y. Y., Wright J. A., Attema J. L., Gregory P. A., Bert A. G., Smith E., Thomas D., Lopez A. F., Drew P. A., Khew-Goodall Y., Goodall G. J. (2013) Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J. Cell Sci. 126, 2256–2266 [DOI] [PubMed] [Google Scholar]
- 30. Pena J. T., Sohn-Lee C., Rouhanifard S. H., Ludwig J., Hafner M., Mihailovic A., Lim C., Holoch D., Berninger P., Zavolan M., Tuschl T. (2009) miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods 6, 139–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costantino M. E., Stearman R. P., Smith G. E., Darling D. S. (2002) Cell-specific phosphorylation of Zfhep transcription factor. Biochem. Biophys. Res. Commun. 296, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Batsché E., Muchardt C., Behrens J., Hurst H. C., Crémisi C. (1998) RB and c-Myc activate expression of the E-cadherin gene in epithelial cells through interaction with transcription factor AP-2. Mol. Cell Biol. 18, 3647–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. West-Mays J. A., Sivak J. M., Papagiotas S. S., Kim J., Nottoli T., Williams T., Fini M. E. (2003) Positive influence of AP-2α transcription factor on cadherin gene expression and differentiation of the ocular surface. Differentiation 71, 206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sumigama S., Ito T., Kajiyama H., Shibata K., Tamakoshi K., Kikkawa F., Williams T., Tainsky M. A., Nomura S., Mizutani S. (2004) Suppression of invasion and peritoneal carcinomatosis of ovarian cancer cells by overexpression of AP-2α. Oncogene 23, 5496–5504 [DOI] [PubMed] [Google Scholar]
- 36. Schwartz B., Melnikova V. O., Tellez C., Mourad-Zeidan A., Blehm K., Zhao Y. J., McCarty M., Adam L., Bar-Eli M. (2007) Loss of AP-2α results in deregulation of E-cadherin and MMP-9 and an increase in tumorigenicity of colon cancer cells in vivo. Oncogene 26, 4049–4058 [DOI] [PubMed] [Google Scholar]
- 37. Kennell J. A., Gerin I., MacDougald O. A., Cadigan K. M. (2008) The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 15417–15422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hyun S., Lee J. H., Jin H., Nam J., Namkoong B., Lee G., Chung J., Kim V. N. (2009) Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139, 1096–1108 [DOI] [PubMed] [Google Scholar]
- 39. Choi P. S., Zakhary L., Choi W. Y., Caron S., Alvarez-Saavedra E., Miska E. A., McManus M., Harfe B., Giraldez A. J., Horvitz H. R., Schier A. F., Dulac C. (2008) Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron 57, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flynt A. S., Thatcher E. J., Burkewitz K., Li N., Liu Y., Patton J. G. (2009) miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J. Cell Biol. 185, 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gessert S., Bugner V., Tecza A., Pinker M., Kühl M. (2010) FMR1/FXR1 and the miRNA pathway are required for eye and neural crest development. Dev. Biol. 341, 222–235 [DOI] [PubMed] [Google Scholar]
- 42. Oft M., Peli J., Rudaz C., Schwarz H., Beug H., Reichmann E. (1996) TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 10, 2462–2477 [DOI] [PubMed] [Google Scholar]
- 43. Jechlinger M., Sommer A., Moriggl R., Seither P., Kraut N., Capodiecci P., Donovan M., Cordon-Cardo C., Beug H., Grünert S. (2006) Autocrine PDGFR signaling promotes mammary cancer metastasis. J. Clin. Invest. 116, 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scheel C., Eaton E. N., Li S. H., Chaffer C. L., Reinhardt F., Kah K. J., Bell G., Guo W., Rubin J., Richardson A. L., Weinberg R. A. (2011) Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145, 926–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y. N., Lee W. W., Wang C. Y., Chao T. H., Chen Y., Chen J. H. (2005) Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 24, 8277–8290 [DOI] [PubMed] [Google Scholar]
- 46. Hsu T. I., Wang M. C., Chen S. Y., Yeh Y. M., Su W. C., Chang W. C., Hung J. J. (2012) Sp1 expression regulates lung tumor progression. Oncogene 31, 3973–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choi J., Park S. Y., Joo C. K. (2007) Transforming growth factor-β1 represses E-cadherin production via slug expression in lens epithelial cells. Invest. Ophthalmol. Vis. Sci. 48, 2708–2718 [DOI] [PubMed] [Google Scholar]
- 48. Jungert K., Buck A., von Wichert G., Adler G., König A., Buchholz M., Gress T. M., Ellenrieder V. (2007) Sp1 is required for transforming growth factor-β-induced mesenchymal transition and migration in pancreatic cancer cells. Cancer Res. 67, 1563–1570 [DOI] [PubMed] [Google Scholar]
- 49. Jordà M., Olmeda D., Vinyals A., Valero E., Cubillo E., Llorens A., Cano A., Fabra A. (2005) Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J. Cell Sci. 118, 3371–3385 [DOI] [PubMed] [Google Scholar]
- 50. Jordà M., Vinyals A., Marazuela A., Cubillo E., Olmeda D., Valero E., Cano A., Fabra A. (2007) Id-1 is induced in MDCK epithelial cells by activated Erk/MAPK pathway in response to expression of the Snail and E47 transcription factors. Exp. Cell Res. 313, 2389–2403 [DOI] [PubMed] [Google Scholar]
- 51. Venkov C., Plieth D., Ni T., Karmaker A., Bian A., George A. L., Jr., Neilson E. G. (2011) Transcriptional networks in epithelial-mesenchymal transition. PloS ONE 6, e25354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saffer J. D., Jackson S. P., Annarella M. B. (1991) Developmental expression of Sp1 in the mouse. Mol. Cell Biol. 11, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marin M., Karis A., Visser P., Grosveld F., Philipsen S. (1997) Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89, 619–628 [DOI] [PubMed] [Google Scholar]
- 54. Shin J. O., Nakagawa E., Kim E. J., Cho K. W., Lee J. M., Cho S. W., Jung H. S. (2012) miR-200b regulates cell migration via Zeb family during mouse palate development. Histochem. Cell Biol. 137, 459–470 [DOI] [PubMed] [Google Scholar]
- 55. Hertzano R., Elkon R., Kurima K., Morrisson A., Chan S. L., Sallin M., Biedlingmaier A., Darling D. S., Griffith A. J., Eisenman D. J., Strome S. E. (2011) Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis. PLoS Genet. 7, e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paterson E. L., Kazenwadel J., Bert A. G., Khew-Goodall Y., Ruszkiewicz A., Goodall G. J. (2013) Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia 15, 180–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chang C. J., Chao C. H., Xia W., Yang J. Y., Xiong Y., Li C. W., Yu W. H., Rehman S. K., Hsu J. L., Lee H. H., Liu M., Chen C. T., Yu D., Hung M. C. (2011) p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 13, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim T., Veronese A., Pichiorri F., Lee T. J., Jeon Y. J., Volinia S., Pineau P., Marchio A., Palatini J., Suh S. S., Alder H., Liu C. G., Dejean A., Croce C. M. (2011) p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 208, 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Knouf E. C., Garg K., Arroyo J. D., Correa Y., Sarkar D., Parkin R. K., Wurz K., O'Briant K. C., Godwin A. K., Urban N. D., Ruzzo W. L., Gentleman R., Drescher C. W., Swisher E. M., Tewari M. (2012) An integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. Nucleic Acids Res. 40, 499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Herschkowitz J. I., Zhao W., Zhang M., Usary J., Murrow G., Edwards D., Knezevic J., Greene S. B., Darr D., Troester M. A., Hilsenbeck S. G., Medina D., Perou C. M., Rosen J. M. (2012) Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc. Natl. Acad. Sci. U.S.A. 109, 2778–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Knight J. F., Lesurf R., Zhao H., Pinnaduwage D., Davis R. R., Saleh S. M., Zuo D., Naujokas M. A., Chughtai N., Herschkowitz J. I., Prat A., Mulligan A. M., Muller W. J., Cardiff R. D., Gregg J. P., Andrulis I. L., Hallett M. T., Park M. (2013) Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc. Natl. Acad. Sci. U.S.A. 110, E1301–E1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Borellini F., Glazer R. I. (1993) Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J. Biol. Chem. 268, 7923–7928 [PubMed] [Google Scholar]
- 63. Lagger G., Doetzlhofer A., Schuettengruber B., Haidweger E., Simboeck E., Tischler J., Chiocca S., Suske G., Rotheneder H., Wintersberger E., Seiser C. (2003) The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol. Cell Biol. 23, 2669–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brandeis M., Frank D., Keshet I., Siegfried Z., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H. (1994) Sp1 elements protect a CpG island from de novo methylation. Nature 371, 435–438 [DOI] [PubMed] [Google Scholar]
- 65. Vrba L., Jensen T. J., Garbe J. C., Heimark R. L., Cress A. E., Dickinson S., Stampfer M. R., Futscher B. W. (2010) Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE 5, e8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Davalos V., Moutinho C., Villanueva A., Boque R., Silva P., Carneiro F., Esteller M. (2012) Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31, 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Attema J. L., Bert A. G., Lim Y. Y., Kolesnikoff N., Lawrence D. M., Pillman K. A., Smith E., Drew P. A., Khew-Goodall Y., Shannon F., Goodall G. J. (2013) Identification of an Enhancer That Increases miR-200b∼200a∼429 Gene Expression in Breast Cancer Cells. PloS ONE 8, e75517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wee E. J., Peters K., Nair S. S., Hulf T., Stein S., Wagner S., Bailey P., Lee S. Y., Qu W. J., Brewster B., French J. D., Dobrovic A., Francis G. D., Clark S. J., Brown M. A. (2012) Mapping the regulatory sequences controlling 93 breast cancer-associated miRNA genes leads to the identification of two functional promoters of the Hsa-mir-200b cluster, methylation of which is associated with metastasis or hormone receptor status in advanced breast cancer. Oncogene 31, 4182–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang B., Zhang Z., Xia S., Xing C., Ci X., Li X., Zhao R., Tian S., Ma G., Zhu Z., Fu L., Dong J. T. (2013) KLF5 activates microRNA 200 transcription to maintain epithelial characteristics and prevent induced epithelial-mesenchymal transition in epithelial cells. Mol. Cell Biol. 33, 4919–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wiklund E. D., Bramsen J. B., Hulf T., Dyrskjøt L., Ramanathan R., Hansen T. B., Villadsen S. B., Gao S., Ostenfeld M. S., Borre M., Peter M. E., Ørntoft T. F., Kjems J., Clark S. J. (2011) Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer 128, 1327–1334 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.