Background: Rhomboid proteases are ubiquitous, and their role in Archaea has not been explored.
Results: We generated a rhomboid deletion mutant that displayed a glycosylation defect.
Conclusion: Deletion of a rhomboid protease gene altered S-layer glycoprotein N-glycosylation.
Significance: This work provides structural characterization of a novel oligosaccharide bound to H. volcanii S-layer glycoprotein and relates a rhomboid protease with the protein glycosylation process.
Keywords: Archaea, Glycoprotein, Glycosylation, Mass Spectrometry (MS), Rhomboid Protease, Halophiles
Abstract
Rhomboid proteases occur in all domains of life; however, their physiological role is not completely understood, and nothing is known of the biology of these enzymes in Archaea. One of the two rhomboid homologs of Haloferax volcanii (RhoII) is fused to a zinc finger domain. Chromosomal deletion of rhoII was successful, indicating that this gene is not essential for this organism; however, the mutant strain (MIG1) showed reduced motility and increased sensitivity to novobiocin. Membrane preparations of MIG1 were enriched in two glycoproteins, identified as the S-layer glycoprotein and an ABC transporter component. The H. volcanii S-layer glycoprotein has been extensively used as a model to study haloarchaeal protein N-glycosylation. HPLC analysis of oligosaccharides released from the S-layer glycoprotein after PNGase treatment revealed that MIG1 was enriched in species with lower retention times than those derived from the parent strain. Mass spectrometry analysis showed that the wild type glycoprotein released a novel oligosaccharide species corresponding to GlcNAc-GlcNAc(Hex)2-(SQ-Hex)6 in contrast to the mutant protein, which contained the shorter form GlcNAc2(Hex)2-SQ-Hex-SQ. A glycoproteomics approach of the wild type glycopeptide fraction revealed Asn-732 peptide fragments linked to the sulfoquinovose-containing oligosaccharide. This work describes a novel N-linked oligosaccharide containing a repeating SQ-Hex unit bound to Asn-732 of the H. volcanii S-layer glycoprotein, a position that had not been reported as glycosylated. Furthermore, this study provides the first insight on the biological role of rhomboid proteases in Archaea, suggesting a link between protein glycosylation and this protease family.
Introduction
Intramembrane-cleaving proteases (I-CLiPs) hydrolyze proteins localized in lipid bilayers (1). Their activity exposes and/or releases functional domains of substrate proteins serving as a regulatory mechanism for diverse signaling pathways (2). Based on their catalytic mechanism, I-CliPs are grouped into three classes: the S2P metalloproteases; the GxGD-type aspartyl proteases, which include presenilin/γ-secretase and signal peptide peptidases; and the rhomboid family of serine proteases (2). Rhomboid proteases (Rho)4 are conserved in the three domains of life and have been implicated in a variety of processes, including epidermal growth factor signaling in Drosophila melanogaster (3), mitochondrial dynamics in yeast (4, 5), and apicomplexan parasite invasion (6, 7). The relevance of Rho in the physiology of prokaryotes has been poorly investigated. The AarA rhomboid protease from the pathogenic bacterium Providencia stuartii cleaves the N-terminal extension of TatA, a membrane-bound component of the twin arginine protein translocation pathway. Processing of TatA activates the translocation process, allowing the export of an unknown quorum-sensing signal (8). In Mycobacteria, Rho null mutants display impaired biofilm formation and increased antibiotic sensitivity (9). Rho homologs are widely represented in archaeal genomes, and the predicted proteins contain the typical multispanning transmembrane domains (TMDs) and the amino acid residues involved in catalysis (Ser-His dyad), suggesting that they are probably functional enzymes (10). However, archaeal Rho remain uncharacterized, and their function has not yet been addressed.
Archaea predominate in environments lethal to most cells, including extreme temperatures, pH values, and salinity. Haloarchaea flourish in habitats containing high salt concentrations (2–5 m NaCl). The only barrier between the cytoplasmic membrane and the harsh environment is the S-layer, a highly organized proteinaceous, two-dimensional crystalline array of one or more proteins that self-assemble around the entire cell surface. S-layer proteins are often glycosylated, and they are thought to play a critical role in the interaction of these microorganisms with the environment (11, 12). In the haloarchaeon Haloferax volcanii, the S-layer glycoprotein is an 81.7-kDa protein containing both N- and O-glycosidic bonds. O-Glycosylation of this protein occurs through binding of glucosyl (1→2) galactose disaccharides to a C-terminal threonine cluster (13, 14). Of the seven putative N-glycosidic sequons found within the S-layer glycoprotein, it was determined that Asn-13 and Asn-83 are modified by a pentasaccharide comprising two hexoses, two hexuronic acids, and a methyl ester of hexuronic acid (15, 16). It was also determined that the sequon at Asn-370 is not modified (15). The aforementioned pentasaccharide became a model to study the protein N-glycosylation process in Archaea (17), and it has also been found decorating H. volcanii flagellins (18). Interestingly, growth at different salt concentrations leads to alterations in H. volcanii S-layer glycoprotein N-glycosylation. In high salt, S-layer glycoprotein Asn-13 and Asn-83 are modified by the pentasaccharide. However, when cells were grown in low salt, substantially less pentasaccharide was detected, whereas a distinct tetrasaccharide composed by three hexoses (one of them sulfated) and a rhamnose, which is absent in cells grown at high salinity, was found at Asn-498. Thus, in response to changes in environmental salinity, H. volcanii modulates not only the N-linked glycans decorating the S-layer glycoprotein but also the sites of such posttranslational modification (19). Recently, it has been reported that two different glycosylation pathways participate in the synthesis of the tetra- and pentasaccharide (20).
To explore the biological role of Rho in Archaea, we constructed a null mutant of a zinc finger-Rho (RhoII) in the model haloarchaeon H. volcanii. The mutant evidenced reduced motility and increased sensitivity to novobiocin as well as a different electrophoretic pattern of glycoproteins compared with the parental strain. We report for the first time the presence of N-acetylglucosamine N-linked oligosaccharides in the S-layer glycoprotein of H. volcanii and show that these sugar chains are shorter in the strain deficient in the RhoII protease. Furthermore, we provide information on the structure and composition of this novel oligosaccharide and show that it is linked to Asn-732, a putative glycosylation site where no modification had been reported so far.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Strains, plasmids, and primers used in this study are listed in Table 1. H. volcanii strains were grown in 18% (w/v) MGM or CA medium5 at 42 °C and 150 rpm. For motility assays, H. volcanii strains were stab-inoculated in 0.25% agar CA plates and grown at 42 °C for 2–3 days. Motility was determined by measuring the diameter of the swimming ring, using the ImageJ program.
TABLE 1.
Strains, plasmids, and primers used in this study
Restriction sites are denoted in lowercase type.
| Name | Description | Reference/Source |
|---|---|---|
| Plasmids | ||
| pKS505 | Gurken TMD in pKS29; ampr | Ref. 8 |
| pKS506 | LacY-TM2 in pKS29; ampr | Ref. 8 |
| pKS507 | Dm Spitz TMD in pKS29; ampr | Ref. 8 |
| pKS508 | PstI TatA aa1–50 into pKS29; ampr | Ref. 8 |
| pTA131 | Ampr; pBluescript II containing Pfdx-pyrE2 | Ref. 26 |
| pMIG1 | pTA131 containing rhoII-flanking regions | This study |
| pTA963 | AmpR, overexpression vector with His6 tag, pyrE2 and hdrB markers, and pHV2 origin. | Ref. 45 |
| pMCF1 | pTA963 with region 207 bp upstream of Hvo_0727 start codon and 103 bp downstream of Hvo_0726 stop codon, cloned between the ApaI and BamHI sites. | This study |
| Strains | ||
| E. coli DH5α | E. coli F− φ80dlacZΔM15 (lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 thi-1 gyrA96 relA1 | Invitrogen |
| E. coli GM33 | LAM-, IN(rrnD-rrnE)1, F− dam-3 sup-85 (Am) | Ref. 46 |
| E. coli Rossetta (DE3) | F_ ompT [Ion] hsdSB (rB mB) (an E. coli B strain) with DE3, a prophage carrying the T7 RNA polymerase gene | Novagen |
| E. coli MG1655 | ΔglpEGR::kan | |
| H. volcanii H26 | H. volcanii DS70 ΔpyrE2 | Ref. 28 |
| H. volcanii MIG1 | H26 ΔrhoII | This study |
| Primers | ||
| HindFupRho2 | 5′-AGaagcttTTCATCCCGCAGACGT-3′ | This study |
| RevUpsRho2ecoRI | 5′-TgaattcATTACCAACGCCTTA-3′ | This study |
| BamFdownRho2 | 5′-AggatccGAGATGCGGCCGGCCCGTC-3′ | This study |
| RevdownRHO2Xba | 5′-AtctagaTCTCAGTCTTCGTACT-3′ | This study |
| Hvo0727KOverify5′ | 5′-TACGAGTACGACAAGGACAT-3′ | This study |
| Hvo0727KOverify3′ | 5′-TAAGTGTACGCGGTTCGCCA-3′ | This study |
| 7 S F | 5′-CCAACGTGGAAGCCTCGTC-3′ | Ref. 21 |
| 7 S R | 5′-GGTGGTCCGCTGCTCACTTC-3′ | Ref. 21 |
| FwOperonRhoII | 5′-GTGATGAGGACCGTCTTTTG-3′ | This study |
| RvOperonRhoII | 5′-CGCGTAATCTCGCATTTCTC-3′ | This study |
Escherichia coli was grown in Luria-Bertani medium (LB), with ampicillin (100 μg ml−1) when needed. E. coli was transformed by the CaCl2 method (22). To induce the synthesis of chimeric substrates, the cultures (A600 = 0.4–0.7) were incubated with 0.1 m isopropyl 1-thio-β-d-galactopyranoside for 3 h.
Cell Fractionation
H. volcanii cultures were grown to an A600 = 1–1.5, and cells were harvested by centrifugation (10,000 × g 10 min, 4 °C). Cell pellets were suspended in 50 mm HCl-Tris, 2 m NaCl (pH 7.5) and disrupted with an ultrasonic processor (3 × 30 s, 80 W). Lysates were clarified by centrifugation (17,000 × g for 20 min at 4 °C), and membranes were pelleted by centrifugation (70,000 × g for 1 h at 4 °C), washed with the same buffer, and recentrifuged for 30 min. Membrane fractions were suspended in 1× SDS-PAGE loading buffer containing 0.1% (w/v) SDS and 0.05 m DTT, incubated for 10 min at 70 °C. Samples used for oligopeptide analysis were further treated with 10 mm iodoacetamide and incubated at room temperature for 30 min in darkness.
Cell extracts of E. coli cells harboring recombinant plasmids that encoded chimeric Rho substrates (23) were obtained as follows. Cells were harvested by centrifugation (10,000 × g for 10 min at 4 °C), and pellets were suspended in 20 mm HCl-Tris (pH 7.5), 200 mm NaCl, 1 mm EDTA, 5% (v/v) glycerol, 1.5 μm pepstatin, and 1 mg ml−1 lysozyme. Cells were disrupted, and lysates were clarified as described above. The supernatants were used as a source of Rho substrates.
Protease Assay
E. coli MG1655 harboring the plasmids with the heterologous substrates were used to prepare cell extracts. These preparations were incubated with H. volcanii membrane fractions in 0.2% (w/v) dodecyl maltoside, 50 mm HCl-Tris (pH 7.5), 1.2 m NaCl, and 1 mm EDTA (final volume 75 μl) at 37 °C for 16 h. After incubation, trichloroacetic acid (TCA) was added to a final concentration of 10% (v/v), and samples were incubated on ice for 30 min, centrifuged (17,000 × g, 15 min), and washed twice with cold 80% (v/v) acetone. Pellets were air-dried and suspended in Laemmli (24) sample buffer.
SDS-PAGE and Western Blotting
Samples were applied onto polyacrylamide gels containing 0.1% SDS. After electrophoresis, proteins were visualized with colloidal Coomassie Brilliant Blue G-250 or periodic acid-Schiff staining (25).
For Western blotting, gels were transferred to PVDF membranes and blocked with TBST buffer (25 mm Tris-HCl, pH 7.4, 0.01% (v/v) Tween 20) with 5% (w/v) skimmed milk. Membranes were incubated with 1:3000 anti-His antibody in blocking buffer, washed with TBST (3 × 10 min), and incubated with 1:10,000 alkaline phosphatase-conjugated anti-mouse IgG for 2 h. After washing (TBST, 3 × 10 min), blots were developed with 0.33 mg ml−1 nitro blue tetrazolium and 0.01 mg ml−1 5-bromo-4-chloro-3-indolyl-phospate.
Construction of an H. volcanii rhoII In-frame Knock-out Mutant
The knock-out constructs were generated as described elsewhere (26). In brief,800 bp flanking each end of the rhoII gene were PCR-amplified and sequentially cloned into the EcoRI/HindIII (upstream region) and the BamHI/XbaI (downstream region) sites of the haloarchaeal suicide vector pTA131. The construct described above (pMIG1) was first amplified in E. coli DH5α and then passed through E. coli GM33 (dam−) to obtain non-methylated plasmid, which was then transformed into H. volcanii H26 using the polyethylene glycol (PEG) method.5 A single homologous recombination event between one of the flanking regions on the knock-out construct and the chromosome (pop-in) was selected for by growth on CA medium, which lacks uracil. Recombinants were next grown in liquid 18% MGM with two passages to fresh medium to allow for a second recombination event that would result in excision of the plasmid from the chromosome (pop-out). Liquid cultures were then transferred to CA plates with uracil (10 μg ml−1) and 5-FOA (50 μg ml−1) and incubated at 42 °C for 5–10 days. Colonies were screened by PCR, using the Hvo0727KOverify5′ and Hvo0727KOverify3′ primers, followed by sequencing of the amplicon to confirm that the chromosomal replacement event had occurred.
Generation of a Construct Containing the Complete rhoII/endV Operon and Its Regulatory Sequences
The chromosomal region comprising 207 bp upstream of the rhoII start codon and 113 bp downstream of the endV stop codon was amplified by PCR, using genomic H. volcanii H26 DNA as a template. The amplicon (2037 bp) was cloned in the TOPO-Blunt vector (Invitrogen), digested with ApaI and BamHI, and subcloned into pTA963 previously digested with the same enzymes. The ligation product was transformed into E. coli DH5α and then passed through E. coli GM33 and transformed into H. volcanii H26 or MIG1 as described above.
Real-time Quantitative PCR
RNA was extracted as described previously (27), concentration was determined spectrophotometrically, and integrity was assessed by gel electrophoresis in glioxal gels. Contaminant proteins and genomic DNA were eliminated by phenol/chloroform/isoamylic alcohol (25:24:1, v/v/v) extraction and digestion with RQ1 DNase, respectively. The absence of contaminating DNA was assessed by PCR amplification using 7SF and 7SR primers. First strand cDNA was synthesized by incubating 2–5 μg of genomic DNA-free RNA and 0.5 μg of the indicated primer at 70 °C (5 min) and on ice (5 min). Then 0.5 mm dNTPs, 2.5 μl of 10× RQ1 RT buffer, and 200 units of Moloney murine leukemia virus reverse transcriptase were added (final volume of 25 μl) and incubated at 37 °C for 1 h, and the reaction was stopped at 70 °C (5 min).
Real-time qPCR was performed using a StepOneTM real-time PCR system (Applied Biosystems). Each 20-μl real-time qPCR reaction mixture contained 2 μl of a 1:20 dilution of the corresponding cDNA, a 0.5 μm concentration of each primer, and 10 μl of 2× qPCR reaction mix with SYBR Green as the detection and ROX as a the normalizing dye. The PCR conditions consisted of a denaturation cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 55 °C for 1 min, and a dissociation cycle at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. The melting curve generated at the end of real-time PCR cycles was analyzed to confirm the absence of nonspecific double-stranded DNA-SYBR Green hybrids. Real-time qPCR data analysis was performed using the gene expression function of the StepOneTM software version 2.1 (Applied Biosystems) by means of the comparative CT method (28). The expression levels of target genes were normalized against that of the 7 S RNA gene (endogenous control), and cDNA from H. volcanii H26 was used as a reference sample. Statistical analyses of data were performed using an unpaired two-tailed Student's t test (95% confidence).
Deglycosylation of S-layer Glycoprotein
The protein band corresponding to the S-layer glycoprotein was cut out from the gel, frozen for 3 h, and washed (mixing for 30 min) with (a) acetonitrile, (b) 20 mm NaHCO3, pH 7, and (c) acetonitrile. The gel pieces were dried, and the N-glycans were released by incubation with PNGase F (20 milliunits) (New England Biolabs Inc., Beverly, MA) overnight at 37 °C in 20 mm NaHCO3, pH 7 (30 μl). The gel pieces were thoroughly washed, and supernatants were removed and dried. Glycans were filtered through an Ultrafree McFilter (Mr 5000), dried, resuspended in 0.1% (v/v) formic acid (20 μl), and left at room temperature for 40 min. Finally, the sample was dried and suspended in water.
Acid Hydrolysis of the Released Oligosaccharides
A sample of the released oligosaccharides obtained after PNGase F treatment was hydrolyzed with 2 m trifluoroacetic acid for 2 h at 100 °C. The acid was eliminated by evaporation; the hydrolysate was dried and resuspended in water for HPAEC-PAD analysis.
Analysis of Oligosaccharide and Monosaccharide Composition of S-layer Glycoprotein by HPAEC-PAD
For HPAEC analysis, a DX-500 Dionex BioLC system (Dionex Corp.) with a pulse amperometric detector was used. The following columns and conditions were employed: (a) for oligosaccharides, Carbopack P-200 column equipped with a P-200 precolumn; gradient elution with 100 mm NaOH, 0–70 mm sodium acetate for 35 min; the flow rate was 0.45 ml min−1; (b) for monosaccharide analysis, Carbopack P-20 column equipped with a P-20 precolumn; (i) for neutral and amino sugars, a 16 mm NaOH isocratic program was used, and (ii) for acidic sugars, a 25 mm NaOH, 80 mm NaAcO isocratic program was used; flow rate, 0.5 ml min−1.
Glycoprotein Digestion
The protein band corresponding to the S-layer glycoprotein was cut out from the gel and washed with acetonitrile. The gel pieces were reduced with 10 mm DTT in 50 mm NH4HCO3 at 55 °C for 30 min. They were further washed with acetonitrile and alkylated with 55 mm IAA in 50 mm NH4HCO3 for 20 min at room temperature in darkness. After washing with 50 mm NH4HCO3 for 10 min and with acetonitrile for 5 min, they were dried in a SpeedVac. The gel slices were rehydrated with 20 ng μl−1 trypsin (Sigma) in 40 mm NH4HCO3, 9% acetonitrile and incubated at 37 °C overnight. The peptides were extracted by sonication with 50% acetonitrile in 1% TFA, and supernatant was taken to dryness. In another case, further digestion was carried upon the addition of 20 ng μl−1 Glu-C (V8) protease (Promega) in 50 mm NH4HCO3, pH 8, at 37 °C overnight. After incubation, samples were dried in a SpeedVac.
Mass Spectrometry Analysis
Matrices and calibrating chemicals were purchased from Sigma-Aldrich. Measurements were performed using an Ultraflex II TOF/TOF mass spectrometer equipped with a high performance solid-state laser (λ = 355 nm) and a reflector. The system is operated by the Flexcontrol version 2.4 software package (Bruker Daltonics GmbH, Bremen, Germany). Samples were irradiated with a laser power of 25–50% and measured in the linear and the reflectron modes, in positive and negative ion modes.
Laser-induced Dissociation Tandem Mass Spectrometry (LID-MS/MS) Analysis in the MALDI-TOF/TOF-MS/MS Instrument
The Ultraflex II MALDI-TOF/TOF MS spectrometer was used. For all experiments using the tandem time-of-flight LIFT mode, the ion source voltage was set at 8.0 kV with a precursor ion mass window of 3 Da. Precursor ions generated by LID were accelerated at 19.0 kV in the LIFT cell. The reflector voltage was set at 29.5 kV.
Sample Preparation
Prior to the analysis of the oligosaccharides by UV-MALDI-TOF MS, remnants of impurities were removed using a biphasic microcolumn consisting of C-18 phase and Dowex 50X-H+ resin. The sample was incubated on the column for 15 min and eluted with 500 μl of water and dried.
For glycomic analysis, 2,5-dihydroxybenzoic acid was used as matrix. The samples were loaded onto a ground steel plate (Bruker Daltonics GmbH) using the sandwich method. Mass spectra were the sum of 100–300 single laser shots, depending on the sample conditions.
For glycopeptide analysis, the glycopeptide mixtures obtained after protease digestion were purified by cotton HILIC SPE microtips as described elsewhere (29). The enriched glycopeptide mixtures were resuspended in 50% (v/v) acetonitrile in 1% (v/v) formic acid. The samples were loaded onto an AnchorChip target (Bruker Daltonics GmbH) as 50% mixtures with (3 μg μl−1) α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 1% TFA.
Spectrum Calibration
External calibration reagents were used (commercial proteins bradykinin 1–7, Mr 757.399; angiotensin I, Mr 1296.685; renin substrate, Mr 1758.933; and insulin β-chain, Mr 3494.6506) with α-cyano-4-hydroxycinnamic acid as matrix in positive and negative ion mode and with β-cyclodextrin (cycloheptaamylose, Mr 1135.0) and γ cyclodextrin (cyclooctaamylose, Mr 1297.1) with norharmane as matrix, in positive and negative ion mode.
RESULTS
Haloarchaeal Rho Display Distinctive Features and Can Process Heterologous Substrates
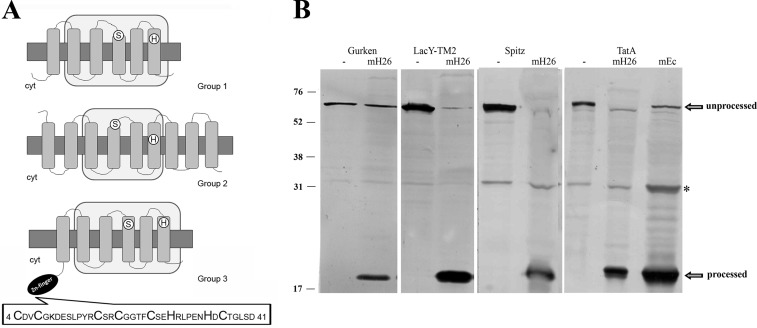
To examine the occurrence of Rho in haloarchaea, a compilation of the proteases predicted in sequenced genomes was performed and manually verified using the BLAST program, available on the NCBI website. The in silico analysis showed that haloarchaea contain two or three sequences related to Rho except for the haloalkaliphilic archaeon Natrialba magadii, which encodes five Rho homologs. Analysis of the translated polypeptides evidenced the serine (GXSG) and histidine residues typical of the catalytic dyad of this protease family (Fig. 1A) (30) as well as the conserved asparagine and histidine residues, which have been proposed to contribute to oxyanion hole formation (31, 32). Recently, Baker and Urban (33) reported the occurrence of four primary sequence keystone regions in which both packing and hydrogen-bonding interactions are used to stabilize the structure of the E. coli Rho, GlpG. These regions are conserved in haloarchaeal Rho, suggesting that these enzymes are probably stabilized by the same molecular interactions as in their bacterial counterparts (not shown). The core region of bacterial Rho consists of six TMDs, whereas the eukaryotic members of this family have an additional TMD helix, either N- or C-terminal to the core six (34). Based on the SMART program (35, 36), different topologies were predicted for the haloarchaeal Rho; thus, in this work, they were classified into three groups (Fig. 1A). Group 1 is represented by the typical six-TMD proteins (HQ2329A, rrnAC2647, Nmag_2518, Hlac_0158, and NP3724A) homologous to E. coli GlpG. This type of Rho was not identified in H. volcanii and Halobacterium sp. Group 2 includes proteins with 8–12 TMDs (Hlac_1491, rrnAC0415, Nmag_3579, HQ1869A, Hvo_1474, NP2230A, and VNG0858C), which contain a Rho domain and extra TMDs of unknown function. Group 3 includes six TMD homologs in which the Rho domain is preceded by an N-terminal AN-1 zinc finger motif and occurs in all of the haloarchaea examined (Hvo_0727, Hlac_0599, VNG_0361, HQ1233A, rrnAC0675, Nmag_1128, and NP1156A). The H. volcanii Hvo_0727 zinc finger shows the typical pattern CX2CX9,12CX1,2CX4CX2HX5HXC described in the conserved domain database (Fig. 1A, inset). The occurrence of the AN1 zinc finger-Rho arrangement has been reported in haloarchaea and some methanogens (37). In this work, we expanded the search and found that this combination only occurs in members of Euryarchaeota, including all of the haloarchaea and Archaeoglobus fulgidus, Archaeoglobus veneficus, Ferroglobus placidus, Methanococcoides burtonii, Methanohalophilus mahii, Methanocelea paludicola, Methanohalobium evestigatum, and the uncultured methanogenic archaeon RC-1. We performed a combined domain search with the SMART program (35, 36), which showed that the association of N-terminal zinc finger (B-box) domains with Rho is conserved in Actynobacteria and that C-terminal RBZ and DHHC zinc fingers exist in plant and fungus Rho.
FIGURE 1.
Haloarchaeal Rho display conserved active site residues and membrane associated Rho activity. A, the translated amino acid sequences of Rho homologs were introduced in the TMHMM and SMART programs, and the retrieved topologies and domain architectures are schematically represented. Top, E. coli GlpG protein, representing Group 1. Middle, Hvo_1474, (Group 2). Bottom, Hvo_0727 (Group 3). Gray rectangles, TMDs; light gray rectangles, Rho domains; white circles, positions of active site serine and histidine; black ellipse, cytoplasmic zinc finger motif. The sequence of the zinc finger motif is shown at the bottom. Cyt, cytoplasm. B, cleavage of specific Rho substrates by H. volcanii solubilized membrane fractions. Cell extracts of E. coli MG1655 transformed with plasmid pKS505 (Gurken), pKS506 (LacY-TMD2), pKS507 (Spitz), or pKS508 (TatA) were incubated with (mH26) or without (−) solubilized membranes of H. volcanii H26. As a cleavage site control, cell extract of pKS508 incubated with E. coli Rossetta solubilized membranes (mEc) is shown on the right. After incubation, samples were TCA-precipitated and used in Western blotting experiments with anti-His antibody. The bands corresponding to unprocessed and processed substrates are pointed out by arrows. *, a nonspecific protein from E. coli extracts that cross-reacts with the anti-His antibody. Migration of molecular mass markers (kDa) is indicated at the left.
To verify whether the predicted H. volcanii Rho were active enzymes, we measured the hydrolytic activity of solubilized membrane protein samples on chimeric protein substrates that contained the TMD of known Rho targets (23). All substrates tested, bearing the TMD of D. melanogaster Gurken and Spitz proteins, E. coli LacYTMD2, and P. stuartii TatA protein, were processed in the presence of H. volcanii membranes (Fig. 1B). Furthermore, the products were of the same size as those generated in the presence of E. coli solubilized membranes, suggesting that substrate cleavage probably occurred in the same region in both organisms. Considering that the hydrolytic activity of this protease family is highly specific, this result strongly evidenced the presence of active rhomboid-like proteases in H. volcanii.
Construction and Phenotypic Characterization of a rhoII Deletion Mutant
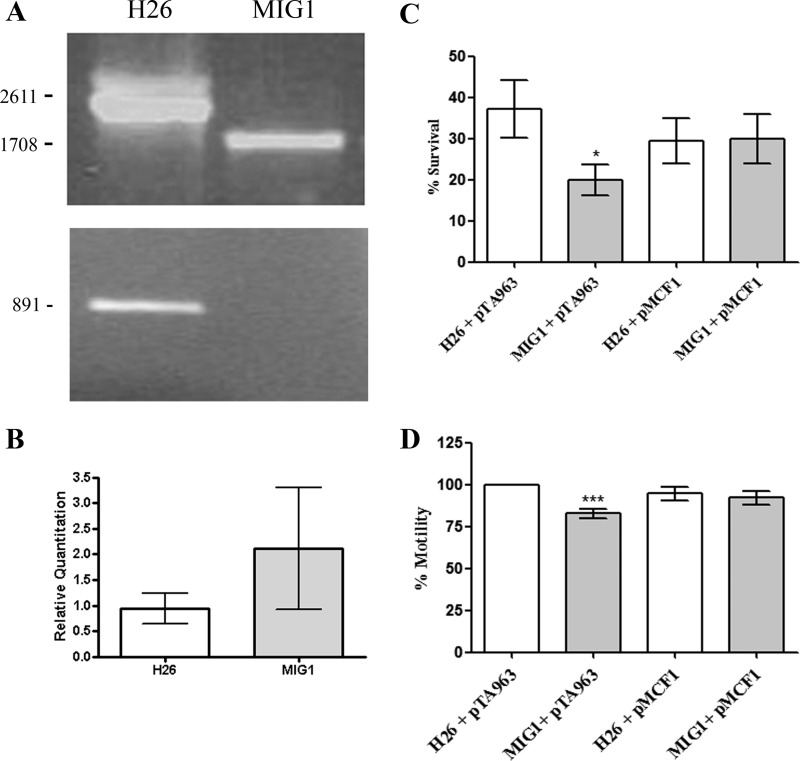
To gain insight into the function of RhoII (Hvo_0727), an in-frame rhoII deletion mutant strain (denoted as MIG1) was constructed in H. volcanii by means of the “pop in-pop out” method (26). Gene elimination from the chromosome was confirmed by PCR using primers external to the deletion construct (Hvo0727Koverify5′ and Hvo0727Koverify3′), which produced amplicons of 2611 and 1708 bp in the parent (H26) and MIG1 strains, respectively (Fig. 2A, top). Additionally, absence of rhoII in MIG1 was evidenced by PCR, using primers that amplified the full-length rhoII gene (891 bp) (Fig. 2A, bottom). The rhoII sequence is 1 bp from Hvo_0726, which encodes a putative endonuclease (EndV), suggesting that these genes probably form an operon. To rule out polar effects of the rhoII deletion on the downstream gene, endV mRNA was measured by real-time qPCR. According to our results, the expression of endV was not affected by the rhoII deletion (Fig. 2B).
FIGURE 2.
Deletion of rhoII, phenotypic characterization, and complementation of the mutant. A, gene deletion was confirmed by colony-PCR using primers positioned upstream (Hvo0727KOverify5′) and downstream (Hvo0727KOverify3) the rhoII knock-out assembly (top) or with primers specific for rhoII coding sequence (NdeIfwRho2/Rho2RevNOSTOPHind; bottom). Amplicon size (bp) is indicated on the left. B, expression of the endV gene was quantified in H26 and MIG1 strains by real-time qPCR, as described under “Experimental Procedures.” C, fresh cultures of each strain were inoculated (A600 = 0.01) in CA medium containing 0.125 μg μl−1 novobiocin. After 24 h, A600 was measured, and the results were expressed as a percentage of the A600 of control tubes inoculated in CA without antibiotic. Data on this graph correspond to means of three independent experiments, performed in duplicate. *, value significantly different (p < 0.05) from that observed for H26 pTA963. D, single colonies were stab-inoculated in 0.25% agar CA medium, and after a 48-h incubation at 42 °C, motility halos were measured. The data were expressed as a percentage of the diameter of the H26 pTA963 strain. The plotted data correspond to three independent experiments in which 8–11 individual colonies of each strain were analyzed. ***, value significantly different from the H26 pTA963 strain (p < 0.01). Error bars, S.D.
The rhoII gene was deleted successfully, indicating that the encoded protein is not essential for viability of H. volcanii under standard laboratory conditions. MIG1 was identical to the parent H26 strain with regard to colony morphology, growth rate under various conditions (MGM and minimal medium; 30, 37 and 42 °C; 1.5 and 4.8 m NaCl), and hydrolytic activity against heterologous Rho substrates. Because H. volcanii encodes two Rho homologs (Hvo_0727 and Hvo_1474; Fig. 1A), the possibility that the protein product of Hvo_1474 could complement, at least partially, the rhoII deletion cannot be ruled out. However, MIG1 showed increased sensitivity to novobiocin (Fig. 2C) and a slight but reproducible reduction in motility on 0.25% CA-agar plates (Fig. 2D). Attempts to complement MIG1 with a copy of rhoII in trans were unsuccessful because no recombinant protein or mRNA could be detected when the rhoII gene was cloned in several haloarchaeal expression vectors and introduced into the mutant strain. Preliminary experiments indicated that rhoII and endV are transcribed as a bicistronic mRNA; thus, failure to detect rhoII mRNA and protein from the plasmids could be due to instability of the mRNA containing only the rhoII sequence. Therefore, we generated a PCR product that comprised both genes as well as the predicted promoter and terminator sequences and cloned it in the pTA963 vector (26) previously digested with ApaI and BamHI. This combination of restriction enzymes releases the ptnA promoter and renders a promoterless vector; therefore, the resulting plasmid (pMCF1) would express the rhoII/endV operon from its own promoter. When introduced in the MIG1 strain, this construct could successfully complement the observed phenotypes (Fig. 2, C and D). No significant differences regarding novobiocin resistance or motility were observed between the parent strain harboring the empty vector and that harboring pMCF1. These results indicate that the observed differences are due to the absence of rhoII.
Deficiency in RhoII Affects Glycosylation of the S-layer Glycoprotein in H. volcanii
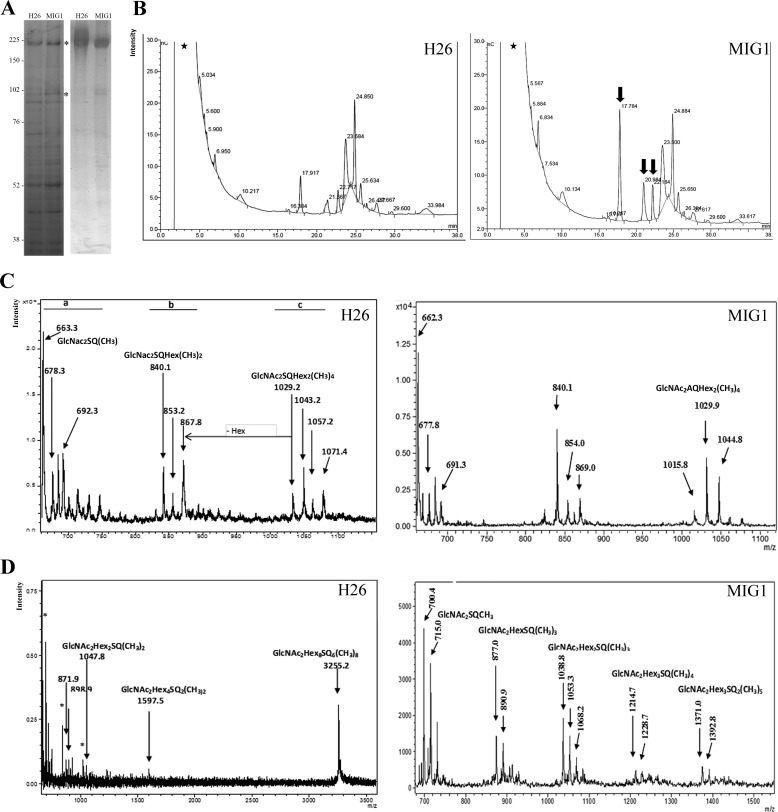
To identify putative substrates of RhoII, we compared SDS-PAGE profiles of membrane proteins in H. volcanii H26 and MIG1. Coomassie Brilliant Blue G-250 staining evidenced two polypeptides of 190 and 98 kDa that were enriched in the MIG1 strain (Fig. 3A, left). These polypeptides were identified as the S-layer glycoprotein and a putative periplasmic substrate-binding protein of an ABC transporter (Hvo_0062), respectively, based on MALDI-TOF mass spectrometry analysis. At first glance, these two proteins did not seem to have a functional link. The putative periplasmic substrate-binding protein was predicted as a soluble polypeptide because it was unlikely that it could be a Rho substrate. Examination of the amino acid sequence of Hvo_0062 evidenced two putative N-glycosylation sites, Asn-198 and -524 (predicted with the NetNGlyc 1.0 server available on the CBS website. Consistent with this observation, periodic acid-Schiff staining confirmed that both polypeptides were glycosylated in the WT and MIG1 strains (Fig. 3A, right). Based on the similarity in electrophoretic mobility, the 98-kDa polypeptide may correspond to an unidentified glycoprotein of H. volcanii that had been previously reported by other groups (38, 39). In the parent strain, the S-layer glycoprotein showed a smeared band with apparent higher molecular masses than the protein synthesized by MIG1. The smeared band probably reflected polypeptides with a different degree of posttranslational modification(s) (i.e. glycosylation). Taking into account these observations, we speculated that the deletion of rhoII may cause a defect on protein glycosylation. To test this hypothesis, we analyzed the N-linked oligosaccharide chains bound to the S-layer protein derived from WT and MIG1 strains. Membrane proteins extracted from both strains were subjected to SDS-PAGE, and the S-layer polypeptides were excised from the gel and digested with PNGase F, and the products were analyzed by HPAEC-PAD. Fig. 3B shows that hydrolysis with this glycosidase released various products in both strains, evidencing the presence of N-acetylglucosamine N-linked oligosaccharides in the H. volcanii S-layer glycoprotein. Interestingly, the S-layer glycoprotein synthesized by MIG1 was enriched in oligosaccharides that migrated with lower retention times (17.784, 20.984, and 22.184 min) compared with the parent strain, supporting our hypothesis that N-glycosylation is affected in MIG1.
FIGURE 3.
Effect of rhoII deletion on S-layer glycoprotein glycosylation. A, membrane fractions H. volcanii H26 and MIG were fractionated on a polyacrylamide gel (8%) and stained with Coomassie Brilliant Blue G-250 (left) or periodic acid-Schiff (right). The migration of molecular mass markers is indicated on the left. The bands corresponding to proteins identified by MALDI-TOF MS are indicated with asterisks. B, HPAEC-PAD chromatograms of the PNGase F-digested S-layer glycoprotein extracted from H26 and MIG1 strains. Arrows, enriched peaks in the MIG1 strain. C, MALDI-TOF MS analysis of the oligosaccharides released by PNGase F from the S-layer glycoprotein of H. volcanii H26 (left) and MIG1 (right), performed in the linear negative ion mode. D, MALDI-TOF MS analysis of the oligosaccharides released by PNGase F from the S-layer glycoprotein of H. volcanii WT (left) or MIG1 (right) spectrum performed in the linear positive ion mode. Asterisks, signals corresponding to matrix; stars, void volume.
Structure Determination of the Novel Oligosaccharide Associated with the S-layer Glycoprotein
Considering the lack of reports on glycans bound by N-acetylglucosamine to the S-layer glycoprotein of H. volcanii and in an effort to evaluate the effect of rhoII deletion on the S-layer glycoprotein glycosylation, a detailed structural characterization of the novel oligosaccharide was performed. MALDI-TOF MS analysis of the total enzymatically released oligosaccharide fraction was performed for the WT and mutant strains in the positive and negative polarity.
When the spectrum of the total enzymatically released oligosaccharide fraction from the WT was recorded using 2,5-dihydroxybenzoic acid as matrix in the negative linear mode (Fig. 3C, left), three clusters of ions were clearly observed: cluster a from m/z 650 to m/z 760, cluster b from m/z 820 to m/z 890, and cluster c, from m/z 1020 to m/z 1080. Cluster a presented a major peak at m/z 663.3 (calc. m/z 664.2002, C23H40N2O18S−) attributed to the [M − H]− ion corresponding to a GlcNAc2SQ trisaccharide bearing a methyl group. Furthermore, ions at m/z 678.3 (calc. m/z 678.2002) and m/z 692.3 (calc. m/z 692.2002) were attributed to the same oligosaccharide differing in the number of methyl groups (Δ14).
Regarding cluster b, it showed a major peak at m/z 840.1 (calc. m/z 839.2609, C30H51N2O23S−) ascribed to a GlcNAc2SQHex tetrasaccharide structure carrying two methyl groups. Accordingly, ions at m/z 853.2 and m/z 867.8 differed in the number of methyl substitutions. In addition, cluster c presented a peak at m/z 1029.2 (calc. m/z 1029.345, C38H65N2O28S−) corresponding to a GlcNAc2SQHex2 pentasaccharide structure bearing four methyl groups. Also ions at m/z 1043.2, m/z 1057.2, and m/z 1071.4 differed in the number of methyl substituents.
The deprotonated species [M − H]− corresponding to ion m/z 1029.2 was selected as precursor ion and subjected to laser-induced LID-MS/MS analysis in the MALDI-TOF/TOF-MS/MS instrument (data not shown). The main signal at m/z 875.6 (calc. m/z 875.4) was attributed to a 0,3X1α cleavage of the sulfoquinovose (6-deoxy-6-sulfoglucose; SQ) unit bearing a methyl group, and signal at m/z 257.7 (calc. m/z 257.0) was diagnostic for the presence of the methyl sulfoquinovosyl unit. In addition, ion at m/z 696.9 (calc. m/z 696.4) corresponded to the loss of the non-substituted hexose unit (Δ179) from ion m/z 875.6. In accordance, the presence of the terminal non-substituted hexose was confirmed by ions at m/z 180.2 (calc. m/z 179.0) and ion at m/z 104.4 (calc. m/z 104.1; 2,5X2β). On the other hand, ion at m/z 897.7 (calc. m/z 897.3, 2,5X2γ) confirmed the presence of three methyl groups in another hexose unit, one of them in position 2. A branched GlcNAc residue linked to a non-substituted hexose was confirmed by ion at m/z 363.5 (calc. m/z 364.1). When the same analysis was performed with the oligosaccharide fraction from M1G1 mutant, a similar spectrum was obtained.
In the positive ion mode (Fig. 3D), the WT strain showed a main ion at m/z 3255.2 (calc. m/z 3255.7591, C108H182N2Na3O93S6) consistent with a GlcNAc2Hex2(SQHex)6Na2 oligosaccharide structure bearing eight methyl groups as [M + Na]+. In addition, signal at m/z 1597.5 (calc. m/z 1597.4131, C54H91N2Na2O45S2) corresponded to a GlcNAc2Hex2(SQHex)2 species bearing two methyl groups, and signal at m/z 1047.8 (calc. m/z 1047.2927, C36H61N2Na2O28S) corresponded to a GlcNAc2Hex2SQ structure bearing two methyl groups. This signal correlates with ion m/z 1029.2 detected in the negative ion mode (Fig. 3C, left) differing in the number of methyl substituents. In the low molecular weight range, signal at m/z 898.9 (calc. m/z 899.2477; C31H53N2Na2O23S) was attributed to a GlcNAc2HexSQ structure bearing two methyl groups, and signal at m/z 871.9 (calc. m/z 871.2242; C29H49N2Na2O23S) was attributed to a GlcNAc2HexSQ structure carrying one methyl substituent. When MALDI-TOF/TOF-MS/MS of ion at m/z 3255.2 was performed, very few signals could be detected (data not shown), among them m/z 2926.0 (calc. m/z 2926.7; C98H165N2Na3O83S5) due to the 0,3X11α cleavage of the last SQ unit bearing a methyl group as described in the negative ion mode and signal at m/z 2755.8 (calc. m/z 2755.6391; C90H154N2Na3O79S5) corresponding to a GlcNAc2Hex2(SQHex)5 structure carrying seven methyl groups after the loss of the two acetyl groups (2 × 42 mu). In addition, whereas ion at m/z 1415.4 (calc. m/z 1414.3238; C46H77N2Na3O39S2) was attributed to GlcNAc2Hex(SQHex)2 bearing two methyl substituents after the loss of an acetyl group, signal at m/z 927.7 (calc. m/z 927.274; C31H56N2NaO26S) correlated with GlcNAc2Hex(SQHex) bearing one methyl group after the loss of the two acetyl groups (data not shown). At this point, the presence of an N-linked oligosaccharide bearing at least six SQ-Hex units linked to a di-N-acetylglucosamine-dihexose backbone was confirmed.
Interestingly, the MIG1 mutant did not show the peak corresponding to GlcNAc2Hex2(SQHex)6 that was observed in the WT in the positive ion mode, but the highest mass signal evidenced at m/z 1392.8 could be attributed to GlcNAc2Hex3SQ2 bearing five methyl groups after the loss of two acetyl groups (2 × 42) (Fig. 3D, right). These results indicate that the MIG1 strain lacks the repeating units of SQ-Hex observed for the oligosaccharide described in this work, confirming that the MIG1 has a defect in protein glycosylation.
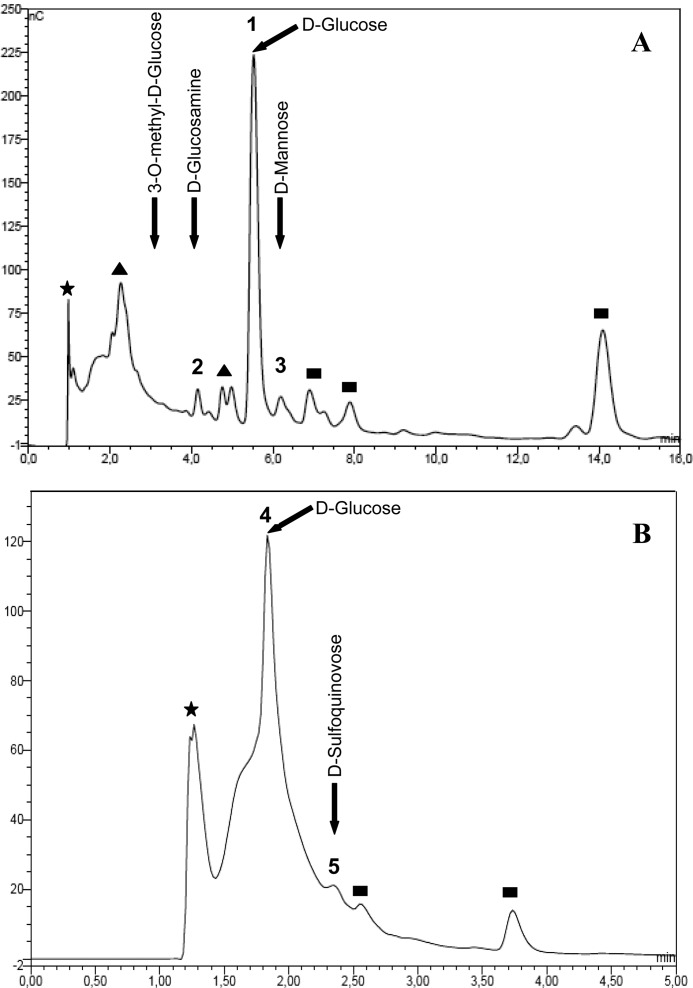
To determine the sugar components, the PNGase-released oligosaccharides from the parent strain were hydrolyzed with 2 n TFA and subjected to HPAEC-PAD under standard conditions for neutral and amino sugars (Fig. 4A). The analysis showed glucose (peak 1) as a main component and two minor signals coincident with glucosamine (peak 2) and mannose (peak 3). Notably, other two peaks, one of them eluting very near the void volume (Rt 2.27 min), suggesting the presence of substituted monosaccharides, and the other one eluting with Rt 14.1 min, corresponding to a product of partial hydrolysis (not shown), were also detected. Accordingly, when the same sample was analyzed under conditions for acidic carbohydrates (Fig. 4B), a peak corresponding to the partial hydrolysis product (Rt 1.3 min) and a main unresolved peak corresponding to neutral monosaccharides (peak 4) were detected. In addition, a small signal coincident with an authentic sample of SQ was evidenced (peak 5).
FIGURE 4.
Monosaccharide composition of the novel oligosaccharide. A, HPAEC-PAD profile of neutral sugars obtained after acid hydrolysis of the oligosaccharides released by PNGase F treatment of H. volcanii H26 S-layer glycoprotein (Fig. 3B). B, acidic sugars obtained as in A. Standards are indicated by arrows. Filled stars, void volume; filled triangles, undetermined substituted monosaccharides; filled squares, products of partial hydrolysis.
The SQ-containing Oligosaccharide Is Bound to Position Asn-732 of the S-layer Glycoprotein
The S-layer glycoprotein is glycosylated at positions Asn-13, Asn-83, and Asn-498, whereas the putative glycosylation site Asn-370 is not modified. However, the possible modification of Asn-274, Asn-279, and Asn-732 has not been reported so far (19). Therefore, it was important to determine the binding site(s) of this novel oligosaccharide. For that purpose, the S-layer glycoprotein was digested with Glu-C alone or with trypsin plus Glu-C and the peptide products were examined by MALDI-TOF MS.
MALDI-TOF MS analysis in the positive ion mode of the micro-SPE-enriched fraction obtained from the WT S-layer glycoprotein after Glu-C digestion showed two major signals. Ion at m/z 3153.01 (calc. m/z 3152.1713), based on manual interpretation, was ascribed to a GlcNAc2Hex2NaSQ oligosaccharide bearing three methyl groups linked to the 725DTTTSSDNATDTTTTTDGPTE745 peptide (calc. m/z 2131.8633) containing the Asn-732 glycosylation site. In accordance, signal at m/z 2154.08 (calc. m/z 2153.8633) matched with the sodium adduct of the corresponding peptide moiety (Table 2).
TABLE 2.
Glycopeptides observed in MALDI-TOF MS analysis of proteolytic digests of the S-layer from H. volcanii
| Peptide residues | Glycosylation site | Theoretical Peptidea [M+H]+ | Observed glycopeptide (m/z) | Theoretical glycopeptide (m/z) | Glycan modification (m/z) |
|---|---|---|---|---|---|
| 3–14 | Asn-13 | 1296.5703 | 2184.32 | 2184.79 | Hex2(HexA)3Na(CH3) |
| 65–90 | Asn-83 | 2644.3213 | 3349.13 | 3348.48 | Hex(HexA)3(CH3) |
| 65–95 | Asn-83 | 3189.5546 | 3550.50 | 3549.62 | HexHexA Na |
| 492–515 | Asn-498 | 2435.2413 | 3146.99 | 3146.41 | HexSO4Hex2Rha |
| 725–745 | Asn-732 | 2131.8633 | 3153.01 | 3152.17 | GlcNAc2Hex2SQ Na (CH3)3 |
| 725–745 | Asn-732 | 2131.8633 | 3617.00 | 3616.23b | GlcNAc2Hex3SQ2Na2 (CH3)4 + H2O |
| 725–745 | Asn-732 | 2131.8633 | 4135.97 | 4136.39 | GlcNAc2Hex5SQ3Na (CH3)5 + H2O |
| 725–745 | Asn-732 | 2260.9059 | 4266.15 | 4265.43 | GlcNAc2Hex5SQ3Na (CH3)5 + H2O |
a Theoretical peptides are based on in silico digest of the known protein sequence.
b As [M + Na]+.
When MALDI-TOF/TOF-MS/MS of the parent ion at m/z 3153 was performed (data not shown), a main signal at m/z 1308.61 (calc. m/z 1309.4044) corresponded to the DTTTSSDNAT glycopeptide containing the Asn-732 (a10 cleavage-H2O-COOH) carrying a truncated diglucosamine moiety. Other peptide fragment ions, such as m/z 1883.58 (calc. m/z 1883.7625; b19 cleavage), m/z 718.23 (calc. m/z 719.28625; x7-H2O-COOH), and m/z 387.97 (calc. m/z 387.1637; z4 cleavage), confirmed the glycopeptide structure.
Double digestion with trypsin plus Glu-C, followed by micro-SPE enrichment and MALDI-TOF MS analysis in the positive ion mode, provided signals corresponding to peptides containing Asn-13, Asn-83, and Asn-498 bearing the previously described penta- and tetrasaccharides (Fig. 5A and Table 2). Interestingly, another important signal at m/z 3617.00 (calc. m/z 3616.2262) was detected. This signal matched with the sodiated adduct of the Asn-732-containing peptide (725DTTTSSDNATDTTTTTDGPTE745) linked to the GlcNAc2Hex3SQ2Na2 oligosaccharide bearing four methyl groups in accordance with the novel glycopeptide detected after Glu-C digestion and with the oligosaccharide determined after PNGase F hydrolysis.
FIGURE 5.
Determination of the glycosylation site of the SQ-containing oligosaccharide. A, MALDI-TOF MS analysis in the m/z 1000–4000 range of the glycopeptides obtained by trypsin plus Glu-C digestion of the S-layer glycoprotein. B, MALDI-LID-MS/MS of ion m/z 3617.43. C, MALDI-TOF MS in the m/z 2000–5500 range of the glycopeptides analyzed in A. Filled circles with diamonds, methyl sulfoquinovose; open circles, dimethyl hexose; open squares, N-acetylglucosamine; filled circles, hexose.
When MALDI-TOF/TOF-MS/MS of the parent ion at m/z 3617.00 was performed (Fig. 5B), signal at m/z 3124.17 (calc. m/z 3124.15) corresponded to the peptide moiety linked to GlcNAc2Hex2SQNa bearing one methyl group, and signal at m/z 2848.88 (calc. m/z 2848.0549) matched with the a19 peptide fragment linked to the same oligosaccharide. In addition, signal at m/z 2664.62 (calc. m/z 2664.0643) was attributed to the peptide moiety linked to a dehydrated fragment composed of GlcNAc2Hex, and ion at m/z 2335.27 (calc. m/z 2335.8633) corresponded to the peptide moiety linked to one GlcNAc unit. Other fragment ions, such as m/z 1978.94 (calc. m/z 1978.5653), corresponding to the sodiated adduct of a peptide fragment (DNATD) linked to the proposed oligosaccharide, and m/z 376.49 (calc. m/z 376.1821), accounting for the GlcNAc-Asn unit, confirmed the novel structure.
Finally, in an attempt to detect glycopeptides of a larger molecular weight, the spectrum was acquired in the range m/z 2000–5500 (Fig. 5C). It must be noted that, although the main ions detected were the ones described above, other diagnostic signals appeared. Notably, in the high mass range region, the ion at m/z 4135.97 (calc. m/z 4136.3905) corresponded to the 725DTTTSSDNATDTTTTTDGPTE745 peptide (calc. m/z 2131.8633) linked to a GlcNAc2Hex2Na(SQHex)3 oligosaccharide bearing five methyl groups and a water unit. In accordance, signal at m/z 4266.15 (calc. m/z 4265.4331) matched with the 724EDTTTSSDNATDTTTTTDGPTE745 peptide containing the Asn-732 site (calc. m/z 2260.9059) linked to the GlcNAc2Hex2Na(SQHex)3 oligosaccharide bearing five methyl groups and a water unit (Table 2). On the basis of the data obtained, we were able to identify the Asn-732 as the N-glycosylation site of the novel oligosaccharide in the S-layer glycoprotein of H. volcanii.
DISCUSSION
Although the structure, mechanism, and biochemical function of rhomboid proteases as intramembrane proteases have been extensively studied, the cellular roles of these enzymes as well as their natural substrates remain largely undetermined. Particularly, there are no reports on the characterization of Rho from Archaea. In this work, in silico examination of complete genomes showed that haloarchaea encode Rho-like proteases with the canonical six-TMD architecture as well as with unique features that may include the presence of extra C-terminal segments or N-terminal AN-1 zinc fingers fused to the Rho domain (Fig. 1A). The presence of zinc finger domains fused to Rho is conserved in many euryarchaea (including all of the haloarchaea with a sequenced genome), actinomycetes, and some plant and fungus Rho. The significance of this domain combination remains elusive and is worthy of investigation. Zinc fingers usually bind one or more Zn2+ atoms and display finger-like protrusions that can interact with target molecules. The binding specificity and the function of these domains are very diverse, and although they were first described as DNA-binding domains, they can also interact with lipids, proteins, and RNA (40). In Archaea, AN1-Zn fingers are also linked to other membrane proteases, such as transglutaminase-like thiol peptidases and zinc-dependent metallopeptidases, the chaperone DNAJ, and other TMD-containing proteins of unknown function. Based on this observation, a role in membrane-associated proteolysis and/or regulation of protein folding or stability was proposed for these zinc fingers (37).
H. volcanii encodes two Rho homologs designated by us rhoI and rhoII. Consistently, membrane fractions of this model haloarchaeon displayed hydrolytic activity against known Rho substrates (Fig. 1B). The deletion of rhoII from the chromosome was successful, indicating that this protease is not essential for viability of H. volcanii under standard laboratory conditions. The MIG1 strain did not evidence alterations in growth compared with the WT and showed no differences with respect to hydrolysis of Rho substrates (not shown). Because H. volcanii encodes for two Rho homologs (Fig. 1A), we cannot rule out the possibility that RhoI could complement RhoII activity. However, the sensitivity to the antibiotic novobiocin was higher in MIG1 than in the WT strain (Fig. 2C). Similarly, an increased sensitivity to novobiocin has also been reported for mycobacterial Rho null mutants (9). In addition, cell motility was slightly reduced in the mutant strain (Fig. 2D). Although the observed phenotypes were mild, they were consistent and highly reproducible. Because the rhoII gene could not be expressed from plasmid vectors, to validate the observed phenotypes, a construct bearing the complete operon and its regulatory sequences was generated. Preliminary proteomics data confirmed that RhoII is expressed in the complemented strain (data not shown). Transformation of MIG1 with this construct successfully complemented the observed phenotypes. Taking into consideration that there were no alterations in endV expression in MIG1 (Fig. 1B) and that the WT strain bearing either the empty plasmid or the plasmid containing the complete operon showed no differences, it can be concluded that the differences in novobiocin resistance and motility were due to the absence of rhoII.
In an attempt to detect RhoII substrates, we compared the SDS-PAGE profiles of the parental and mutant strains. Protein staining evidenced two differential bands (Fig. 3A) that were identified as the S-layer glycoprotein and a periplasmic subunit of an ABC transporter. Detection of periplasmic components of ABC transporters in archaeal membrane preparations has been reported previously (41), and their presence is probably due to interaction with integral membrane subunits. Sequence analysis showed that the ABC component contained two glycosylation sites, and its glycosylated nature was confirmed by glycan-specific staining (Fig. 3A). The majority of Rho substrates that have been identified are one-TMD membrane proteins; however, it has been reported that mammalian Rho RHBDD1 is able to cleave the multitransmembrane protein TSAP6 (42). Considering that the proteins that were detected as differential were glycoproteins and that the ABC transporter is a soluble protein (probably not a Rho substrate), we hypothesized that MIG1 may be affected in protein glycosylation. Moreover, proteins with reduced glycosylation tend to be stained more intensely with Coomassie Brilliant Blue G-250 because the dye can reach the protein more easily (43) than in their glycosylated counterparts. This could account for the differences in intensity, observed in the protein gels.
In order to test this hypothesis, we compared the glycans associated with the S-layer glycoprotein, a very abundant membrane protein that has been extensively used as a model to study protein glycosylation (20) in H. volcanii. We show, for the first time, the presence of N-linked oligosaccharides hydrolyzed by PNGase F in the S-layer glycoprotein of H. volcanii, because after the enzymatic treatment, the HPAEC-PAD profile clearly showed the presence of released oligosaccharides. Interestingly, oligosaccharides with shorter retention times accumulated in MIG1, indicating that in this strain, N-glycosylation was affected (Fig. 3B). The main ions obtained by MALDI-TOF analysis of the released material from both strains could be attributed to a novel GlcNAc2Hex2(SQHex)6Na2 (m/z 3255.2) for the WT and GlcNAc2Hex3SQ2Na2 for MIG1, confirming that the glycan bound to the S-layer glycoprotein was truncated in the mutant strain. Although in this work we generated a construct that complemented the other observed phenotypes, complementation of the rhoII deletion effect on glycosylation was not possible. The pMCF1 plasmid has the pyrE gene as an auxotrophic selection marker; therefore, the complemented strain had to be grown in medium lacking uracil. When H. volcanii was grown in CA medium without uracil, the material released by PNGase digestion was very scarce, preventing us from further characterizing the bound oligosaccharides.
The S-layer glycoprotein from H. volcanii has been reported to bear a pentasaccharide of known composition at positions Asn-13 and Asn-83 as well as a sulfated tetrasaccharide at position Asn-379, which is present in low salt conditions (19). However, there have been no reports on N-acetylglucosamine N-linked long chain oligosaccharides for this protein. This prompted us to go deeper into determining the structure of the novel glycan detected in this work. By MALDI-TOF MS analysis of the released oligosaccharides in the negative ion mode, three clusters of ions attributed to the presence of a GlcNAc2SQ structure substituted with one or two hexosyl units were detected. Differences in the degree of methyl substitution of these structures account for the detected microheterogeneity. Furthermore, LID-MS/MS spectra showed a diagnostic fragment ion corresponding to the methylsulfoquinovose moiety assuring the proposed structure. In addition, MALDI-TOF MS analysis in the positive ion mode showed a GlcNAc2Hex2(SQHex)6 oligosaccharide. The fact that glucose was observed as the major neutral component in the HPAEC-PAD analysis after acid hydrolysis suggests a repeating SQ-Glc unit. This new structural feature, a repeating methyl-SQ-Hex disaccharide, has not been reported so far.
The N-glycosylation site of the novel oligosaccharide was also determined. MALDI-MS analysis of digested glycopeptides obtained from the S-layer glycoprotein revealed that the novel oligosaccharide is linked to the Asn-732 glycosylation site.
Fig. 6 shows a schematic representation of the H. volcanii S-layer glycosylation pattern, including the novel oligosaccharide described in this work. It has been speculated that a high degree of glycosylation may be protective against the harsh environmental conditions of S. acidocaldarius in its natural habitats (pH 2–3 and 75–80 °C) (32). In addition, an excess of negative surface charge is thought to stabilize proteins in high salt conditions, possibly via the formation of an energetically favorable protein-water-salt hydration network, as proposed to explain the highly acidic nature of the N-glycans decorating the Halobacterium salinarum S-layer glycoprotein (14, 44). Accordingly, in the S-layer glycoprotein of H. volcanii, the presence of a long N-linked oligosaccharide chain with at least six sulfoquinovose-containing units would also provide a negative surface and may improve resistance against hydrolytic cleavages.
FIGURE 6.
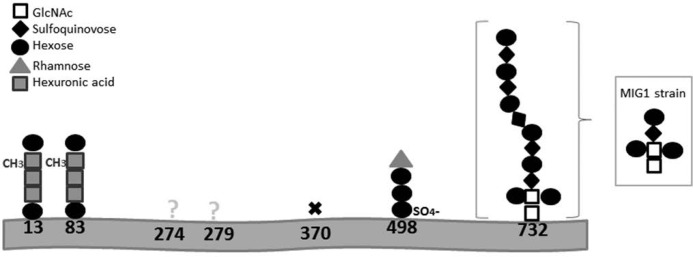
Schematic depiction of the N-glycosylation of the H. volcanii S-layer glycoprotein. The identities of the different sugar residues are given in the top left. X, no glycosylation. Inset, N-linked oligosaccharide found in MIG1 strain.
The data presented in this work represent the first study on archaeal Rho and suggest a link between this proteases and protein N-glycosylation. Determining this link will be the focus of future investigations. In addition, this work contributes to the fundamental knowledge of microbial glycobiology, describing the structure of a novel SQ-containing oligosaccharide. Moreover, our data indicate that this glycan is linked through GlcNAc to the Asn-732 site of the S-layer glycoprotein of H. volcanii, a glycosylation site that had not been studied previously. The identification of specific RhoII substrates and studies on the biosynthesis and assembly of the oligosaccharide described in this work will provide valuable insights into the N-glycosylation process and its regulation in Archaea.
Acknowledgments
We thank Dr. M. Pohlschröder (University of Pennsylvania) for plasmid pTA131, Dr. A. Large (University of Birmingham, UK) for H. volcanii H26, Dr. K. Strisovsky (Academy of Sciences, Czech Republic) for the chimeric substrates constructs and E. coli MG1655 strain, and Dr. D. Schleheck (University of Konstanz, Germany) for an authentic sample of sulfoquinovose.
This work was supported by Consejo Nacional de Investigaciones Científicas y Técnicas Grants PIP-1783 and 1548, Universidad Nacional de Mar del Plata Grant Exa 454/09 (to R. E. D. C.), Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) Grant PICT-01824 (to M. I. G.), and Universidad de Buenos Aires Grant 20020100100517 (to A. C.). The Ultraflex II (Bruker) TOF/TOF mass spectrometer was supported by ANPCyT Grant PME 125.
M. Dyall-Smith, Protocols for Haloarchael Genetics in The Halo Handbook.
- Rho
- rhomboid protease(s)
- PNGase F
- peptide-N-glycosidase F
- TMD
- transmembrane domain
- qPCR
- quantitative PCR
- HPAEC
- high performance anion exchange chromatography
- PAD
- pulsed amperometric detection
- LID
- laser-induced dissociation
- SQ
- sulfoquinovose (6-deoxy-6-sulfoglucose).
REFERENCES
- 1. Wolfe M. S., Kopan R. (2004) Intramembrane proteolysis: theme and variations. Science 305, 1119–1123 [DOI] [PubMed] [Google Scholar]
- 2. Urban S. (2009) Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 7, 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee J. R., Urban S., Garvey C. F., Freeman M. (2001) Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107, 161–171 [DOI] [PubMed] [Google Scholar]
- 4. Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S. (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 [DOI] [PubMed] [Google Scholar]
- 5. McQuibban G. A., Saurya S., Freeman M. (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423, 537–541 [DOI] [PubMed] [Google Scholar]
- 6. Baker R. P., Wijetilaka R., Urban S. (2006) Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. 2, e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baxt L. A., Baker R. P., Singh U., Urban S. (2008) An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev. 22, 1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stevenson L. G., Strisovsky K., Clemmer K. M., Bhatt S., Freeman M., Rather P. N. (2007) Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc. Natl. Acad. Sci. U.S.A. 104, 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kateete D. P., Katabazi F. A., Okeng A., Okee M., Musinguzi C., Asiimwe B. B., Kyobe S., Asiimwe J., Boom W. H., Joloba M. L. (2012) Rhomboids of Mycobacteria: characterization using an aarA mutant of Providencia stuartii and gene deletion in Mycobacterium smegmatis. PloS One 7, e45741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maupin-Furlow J. A., Gil M. A., Humbard M. A., Kirkland P. A., Li W., Reuter C. J., Wright A. J. (2005) Archaeal proteasomes and other regulatory proteases. Curr. Opin. Microbiol. 8, 720–728 [DOI] [PubMed] [Google Scholar]
- 11. Eichler J., Adams M. W. (2005) Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69, 393–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jarrell K. F., Jones G. M., Nair D. B. (2010) Biosynthesis and role of N-linked glycosylation in cell surface structures of archaea with a focus on flagella and s layers. Int. J. Microbiol. 2010, 470138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sumper M., Berg E., Mengele R., Strobel I. (1990) Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172, 7111–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mengele R., Sumper M. (1992) Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 267, 8182–8185 [PubMed] [Google Scholar]
- 15. Abu-Qarn M., Yurist-Doutsch S., Giordano A., Trauner A., Morris H. R., Hitchen P., Medalia O., Dell A., Eichler J. (2007) Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 374, 1224–1236 [DOI] [PubMed] [Google Scholar]
- 16. Magidovich H., Yurist-Doutsch S., Konrad Z., Ventura V. V., Dell A., Hitchen P. G., Eichler J. (2010) AglP is a S-adenosyl-l-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii. Mol. Microbiol. 76, 190–199 [DOI] [PubMed] [Google Scholar]
- 17. Calo D., Kaminski L., Eichler J. (2010) Protein glycosylation in Archaea: sweet and extreme. Glycobiology 20, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 18. Tripepi M., You J., Temel S., Önder Ö., Brisson D., Pohlschröder M. (2012) N-Glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J. Bacteriol. 194, 4876–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guan Z., Naparstek S., Calo D., Eichler J. (2012) Protein glycosylation as an adaptive response in Archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ. Microbiol. 14, 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaminski L., Guan Z., Yurist-Doutsch S., Eichler J. (2013) Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. MBio 4, e00716–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paggi R. A., Madrid E. A., D'Alessandro C. P., Cerletti M., De Castro R. E. (2010) Growth phase-dependent biosynthesis of Nep, a halolysin-like protease secreted by the alkaliphilic haloarchaeon Natrialba magadii. Lett. Appl. Microbiol. 51, 36–41 [DOI] [PubMed] [Google Scholar]
- 22. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23. Strisovsky K., Sharpe H. J., Freeman M. (2009) Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell 36, 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Dubray G., Bezard G. (1982) A highly sensitive periodic acid-silver stain for 1,2-diol groups of glycoproteins and polysaccharides in polyacrylamide gels. Anal. Biochem. 119, 325–329 [DOI] [PubMed] [Google Scholar]
- 26. Allers T., Ngo H. P., Mevarech M., Lloyd R. G. (2004) Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nieuwlandt D. T., Palmer J. R., Armbruster D. T., Kuo Y. P., Oda W., Daniels C. J. (1995) in Archaea: A Laboratory Manual (Robb F. T., Place A. R., Sowers K. R., Schreier H. J., DasSarma S., Fleischmann E. M., eds) pp. 161–162, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Wong M. L., Medrano J. F. (2005) Real-time PCR for mRNA quantitation. BioTechniques 39, 75–85 [DOI] [PubMed] [Google Scholar]
- 29. Selman M. H., Hemayatkar M., Deelder A. M., Wuhrer M. (2011) Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal. Chem. 83, 2492–2499 [DOI] [PubMed] [Google Scholar]
- 30. Lemberg M. K., Menendez J., Misik A., Garcia M., Koth C. M., Freeman M. (2005) Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 24, 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lemieux M. J., Fischer S. J., Cherney M. M., Bateman K. S., James M. N. (2007) The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc. Natl. Acad. Sci. U.S.A. 104, 750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shlay J. C., Bartsch G., Peng G., Wang J., Grunfeld C., Gibert C. L., Visnegarwala F., Raghavan S. S., Xiang Y., Farrough M., Perry H. E., Kotler D., El-Sadr W. M. (2007) Long-term body composition and metabolic changes in antiretroviral naive persons randomized to protease inhibitor-, nonnucleoside reverse transcriptase inhibitor-, or protease inhibitor plus nonnucleoside reverse transcriptase inhibitor-based strategy. J. Acquir. Immune Defic. Syndr. 44, 506–517 [DOI] [PubMed] [Google Scholar]
- 33. Baker R. P., Urban S. (2012) Architectural and thermodynamic principles underlying intramembrane protease function. Nat. Chem. Biol. 8, 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Urban S. (2006) Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 20, 3054–3068 [DOI] [PubMed] [Google Scholar]
- 35. Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Letunic I., Doerks T., Bork P. (2009) SMART 6: recent updates and new developments. Nucleic Acids Res. 37, D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burroughs A. M., Iyer L. M., Aravind L. (2011) Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol. Biosyst. 7, 2261–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhu B. C., Drake R. R., Schweingruber H., Laine R. A. (1995) Inhibition of glycosylation by amphomycin and sugar nucleotide analogs PP36 and PP55 indicates that Haloferax volcanii β-glucosylates both glycoproteins and glycolipids through lipid-linked sugar intermediates: evidence for three novel glycoproteins and a novel sulfated dihexosyl-archaeol glycolipid. Arch. Biochem. Biophys. 319, 355–364 [DOI] [PubMed] [Google Scholar]
- 39. Konrad Z., Eichler J. (2002) Protein glycosylation in Haloferax volcanii: partial characterization of a 98-kDa glycoprotein. FEMS Microbiol. Lett. 209, 197–202 [DOI] [PubMed] [Google Scholar]
- 40. Laity J. H., Lee B. M., Wright P. E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 [DOI] [PubMed] [Google Scholar]
- 41. Palmieri G., Balestrieri M., Peter-Katalinić J., Pohlentz G., Rossi M., Fiume I., Pocsfalvi G. (2013) Surface-exposed glycoproteins of hyperthermophilic Sulfolobus solfataricus P2 show a common N-glycosylation profile. J. Proteome Res. 12, 2779–2790 [DOI] [PubMed] [Google Scholar]
- 42. Wan C., Fu J., Wang Y., Miao S., Song W., Wang L. (2012) Exosome-related multi-pass transmembrane protein TSAP6 is a target of rhomboid protease RHBDD1-induced proteolysis. PloS One 7, e37452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meyer B. H., Zolghadr B., Peyfoon E., Pabst M., Panico M., Morris H. R., Haslam S. M., Messner P., Schäffer C., Dell A., Albers S. V. (2011) Sulfoquinovose synthase: an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol. Microbiol. 82, 1150–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kandiba L., Aitio O., Helin J., Guan Z., Permi P., Bamford D. H., Eichler J., Roine E. (2012) Diversity in prokaryotic glycosylation: an archaeal-derived N-linked glycan contains legionaminic acid. Mol. Microbiol. 84, 578–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allers T., Barak S., Liddell S., Wardell K., Mevarech M. (2010) Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marinus M. G. (1973) Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol. Gen. Genet. 127, 47–55 [DOI] [PubMed] [Google Scholar]