Background: ClpC/Hsp93 is a chloroplast molecular chaperone whose function is essential for plant viability.
Results: Chloroplast ClpC localized in both the stromal and envelope-membrane fractions associates with the Clp proteolytic core.
Conclusion: ClpC functions primarily as the chaperone component of the chloroplast Clp protease.
Significance: Learning how ClpC contributes to key functions of the Clp protease is crucial to understanding the importance of this essential enzyme for chloroplast biology.
Keywords: Chloroplast, Molecular Chaperone, Protease, Protein Degradation, Protein Translocation
Abstract
The molecular chaperone ClpC/Hsp93 is essential for chloroplast function in vascular plants. ClpC has long been held to act both independently and as the regulatory partner for the ATP-dependent Clp protease, and yet this and many other important characteristics remain unclear. In this study, we reveal that of the two near-identical ClpC paralogs (ClpC1 and ClpC2) in Arabidopsis chloroplasts, along with the closely related ClpD, it is ClpC1 that is the most abundant throughout leaf maturation. An unexpectedly large proportion of both chloroplast ClpC proteins (30% of total ClpC content) associates to envelope membranes in addition to their stromal localization. The Clp proteolytic core is also bound to envelope membranes, the amount of which is sufficient to bind to all the similarly localized ClpC. The role of such an envelope membrane Clp protease remains unclear although it appears uninvolved in preprotein processing or Tic subunit protein turnover. Within the stroma, the amount of oligomeric ClpC protein is less than that of the Clp proteolytic core, suggesting most if not all stromal ClpC functions as part of the Clp protease; a proposal supported by the near abolition of Clp degradation activity in the clpC1 knock-out mutant. Overall, ClpC appears to function primarily within the Clp protease, as the principle stromal protease responsible for maintaining homeostasis, and also on the envelope membrane where it possibly confers a novel protein quality control mechanism for chloroplast preprotein import.
Introduction
Molecular chaperones and proteases are integral components of the quality control processes active within the crowded and dynamic protein environment of all living cells. Chaperones are a large diverse group of proteins involved in many structural functions and active cellular processes including protein folding/unfolding and complex assembly/disassembly (1). Certain chaperones also work in concert with proteases, recognizing polypeptide substrates and unfolding them prior to degradation (2). Proteases themselves are equally crucial to various cellular activities both in maintaining proteostasis and during major cellular events like cell division and differentiation. Chaperones and proteases also perform vital protective and restorative roles during and after periods of stress. Polypeptides that increasingly lose their native structure or become otherwise damaged must be targeted for degradation before they reach cytotoxic levels (3).
Clp/Hsp100 proteins are a class of molecular chaperones within the broader family of AAA+ (ATPases associated with diverse cellular activities) proteins. They are found in a wide range of organisms from bacteria to mammals and facilitate a diverse array of cellular functions. Most Clp/Hsp100 proteins also function as the regulatory component of Clp proteases. The model Clp protease in Escherichia coli is a two-component enzyme comprised of an oligomeric Hsp100 (either ClpA or ClpX) complex bound to a proteolytic core. The proteolytic core is composed of two opposing heptameric rings of ClpP that together create a central cavity housing the proteolytic active sites (4). The core is flanked on one or both sides by a single hexameric ring of the Hsp100 partner (5). The Hsp100 complex selects substrates and unfolds them, translocating the proteins through the narrow entrance of the Clp proteolytic core into the central chamber (6, 7). Once bound to the active sites, the protein substrate is rapidly degraded to small peptide fragments that eventually diffuse out of the core complex. The substrate specificity of the Hsp100 partner can also be modified by different adaptor proteins such as ClpS that recognize motifs at either the N or C termini of the targeted substrates (8).
The Clp protein family in vascular plants is far more diverse than in other organisms, with the majority of these Clp proteins localized in the chloroplast (9). All chloroplast Clp proteins are constitutively expressed and most are abundant in leaves compared with other tissues (9, 10). Plant chloroplasts contain four distinct Hsp100 proteins (ClpC1, ClpC2, ClpD, and ClpB3) and yet little is known about their specific chaperone activities or substrate specificity. The closely related ClpC ortholog in the cyanobacterium Synechococcus elongatus has various chaperone characteristics in vitro including the prevention of protein aggregation, and resolubilization/refolding of aggregated polypeptides (11). It also associates to the ClpP3/R proteolytic core to form the principle Clp protease in cyanobacteria and whose function is essential for cell viability (12, 13). A homologous Clp proteolytic core also exists in plant chloroplasts but one that consists of 11 different subunits (14). This core complex comprises two distinct heptameric rings, one with the ClpP3–6 subunits (P-ring) and the other with ClpP1 and ClpR1–4 (R-ring) (15). Also peripherally attached to the P-ring are two accessory proteins, ClpT1 and -T2, that are involved in core assembly (16) and possibly substrate recognition (17). All chloroplast Hsp100 proteins with the exception of ClpB3 contain the conserved motifs in the C terminus necessary for association to the Clp proteolytic core (18). How much the different Hsp100 proteins contribute to the Clp proteolytic activity in chloroplasts, however, remains unknown although a structural association between ClpC and the Clp proteolytic core has been demonstrated (19–21).
Genetic studies have clearly shown the crucial role of chloroplast ClpC and the Clp protease for plant viability (15, 22–27). Putative substrates for the chloroplast Clp protease have been identified and range from various metabolic enzymes to regulatory proteins involved in homeostatic functions such as chloroplast gene expression, RNA maturation, protein synthesis and recycling, and tetrapyrrole synthesis (15, 28, 29). Apart from its presumed involvement in Clp proteolysis, ClpC has also been implicated in the chloroplast import of cytosolic preproteins by its close proximity to Tic110, one of the principle subunits of the Tic complex (Translocon at the inner envelope membrane) (30, 31). This together with the finding that ClpC stably binds transit peptides in vitro (32) has suggested that it might function as the driving force for preprotein import through the Tic complex (33). Despite this, evidence for the direct involvement of ClpC in preprotein import and processing in chloroplasts remains elusive.
Although ClpC was first shown to be essential for normal chloroplast function many years ago (34), little is still known about its specific characteristics, the interaction between the two paralogs, ClpC1 and ClpC2, or their contribution to Clp proteolytic activity. The amino acid sequences of the two mature ClpC proteins are more than 90% identical (35), and overexpression of ClpC2 can fully complement the loss of ClpC1 in Arabidopsis (25), suggesting both perform similar, if not identical functions in the chloroplast. Mutagenesis studies have shown that loss of ClpC1 in Arabidopsis causes significant phenotypic changes, the most prominent being slower growth rates, leaf chlorosis, and impaired photosynthetic activity (36–38), whereas loss of ClpC2 produces little or no effect (37, 39). In this study, we have determined many of the important features of the chloroplast ClpC proteins in Arabidopsis thaliana including their relative abundance, localization, and impact upon Clp proteolytic activity. We show that ClpC localized in both the stroma and envelope membrane appears to function in association with the Clp proteolytic core, revealing new dimensions to the functional importance of this essential molecular chaperone in chloroplasts.
EXPERIMENTAL PROCEDURES
Plant Growth Conditions
Seeds of Arabidopsis thaliana wild type (ecotype Columbia-0), and clpC1 knock-out (36) and clpC2 knockdown (39) T-DNA insertion lines were sown in a 1:5 perlite/soil mixture after vernalization at 4 °C for at least 48 h. All plants were grown either in individual pots or as lawns under the following standard conditions: 8 h photoperiod with white light (about 150 μmol photons m−2 s−1), 23/18 °C day/night temperatures, and 65% relative air humidity.
Immunoblotting from Leaf Protein Extracts
Total cell proteins were isolated from leaves from wild type, clpC1 and clpC2 mutant plants as previously described (15). The protein concentration of each final sample extract was determined using the Bradford protein assay (Bio-Rad). Samples were then loaded, based on equal protein content, on pre-cast 3–8% Tris acetate NuPAGE gels (Invitrogen). Following electrophoretic separation, proteins were transferred to supported nitrocellulose (Bio-Rad) using an Xcell® blotting apparatus (Invitrogen). For detecting the different Clp proteins, specific polyclonal antibodies were used as previously detailed (16, 36, 39). Primary antibodies were detected with the horseradish peroxidase-linked, anti-rabbit IgG secondary antibody from donkey and visualized by enhanced chemiluminescence (ECL Advance, GE Healthcare). Chemiluminescent signals were detected and quantified using the ChemiGenius 2 imaging system (Syngene) and associated software. To ensure correct loading, samples were separated on additional gels and stained with Coomassie Brilliant Blue G-250 to detect the relative amount of Rubisco2 large subunit.
Fractionation of Stromal, Thylakoid Membrane, and Envelope Membrane Proteins
Intact chloroplasts were isolated from wild type Arabidopsis, and clpC1 and clpC2 mutant plants according to Sjögren et al. (15). Fractionation of stromal, thylakoid membrane, and envelope membrane proteins was performed as previously described (40). The protein concentration of the stroma and envelope membrane fractions was determined using the Bradford protein assay (Bio-Rad), whereas the Chl concentration of the leaf and thylakoid membrane fraction was measured according to Porra et al. (41). Samples from each fraction along with one from whole leaves were separated on pre-cast 3–8% Tris acetate gels or linear 12% BisTris NuPAGE gels (Invitrogen) depending on the size range of the proteins being examined. Following separation, proteins were transferred to a supported nitrocellulose membrane (Bio-Rad) using an Xcell blotting apparatus (Invitrogen). Amounts of ClpC1, ClpC2, total ClpC, ClpD, and ClpP6 were then determined by immunoblotting with specific antibodies as described above, as were the control proteins for the stromal (small subunit of Rubisco, SSU), envelope membrane (Tic110), and thylakoid membrane (Lhcb2) fractions. Detection and quantification of antibody signals were done as described above.
Protein Complex Separation by colorless native-PAGE
Protein complexes from stromal fractions were separated by colorless native-PAGE as previously detailed (16). After separation, all proteins were transferred to supported nitrocellulose using a semi-dry electrophoretic cell (Trans-Blot, Bio-Rad). The Clp proteolytic core (325 kDa) was detected using antibodies specific for three different subunits of the core complex: ClpP6, ClpR2, and ClpT1 (15). Detection and quantification of antibody signals were done as described above.
Protein Complex Separation by Blue Native-PAGE
Protein complexes from the envelope membrane fraction were separated by blue native-PAGE as previously described (42) with the following modifications. The envelope membrane sample was pelleted by centrifugation at 130,000 × g for 30 min and then solubilized in 25 mm BisTris/HCl, pH 7.5, 1.5% (w/v) n-dodecyl β-d-maltoside, 20% (w/v) glycerol, and 0.5 m aminocaproic acid. The sample was incubated on ice for 10 min and then centrifuged at 20,000 × g for 20 min to pellet any residual insoluble material. Prior to loading, 0.1 volumes of sample buffer (100 mm BisTris/HCl, pH 7.5, 0.5 m aminocaproic acid, 30% (w/v) sucrose, and 50 mg/ml of Brilliant Serva Blue G-250) was added to both stroma and envelope membrane samples. Samples were separated on 4–13% polyacrylamide gradient gels as previously described (16, 42) and then transferred to supported nitrocellulose using a semi-dry electrophoretic cell (Trans-Blot, Bio-Rad). The Clp proteolytic core and subcomplexes (P- and R-rings) were detected using antibodies specific for different subunits: ClpP4, ClpP6, ClpR3, and ClpR4 (15). Detection and quantification of antibody signals were done as described above.
Purification of Recombinant Clp Proteins
The Arabidopsis CLPP4 and CLPP5 gene sequences (excluding the region coding for the chloroplast transit peptide) were commercially synthesized (Invitrogen), with the codon sequences optimized for E. coli protein overexpression. Restriction sites were also included at the 5′ and 3′ ends of both genes (NcoI/BamHI for CLPP4, NdeI/KpnI for CLPP5) to facilitate cloning into the pACYC Duet expression vector (Novagen), along with the sequence for a His6 tag at the 3′ end to aid purification. The ClpP4 and ClpP5 proteins were overexpressed in E. coli BL21-STAR cells (Invitrogen) and purified by sequential affinity and gel filtration chromatography as previously described (11). The purified ClpP4 and ClpP5 were stored in 20 mm Tris-HCl, pH 7.5, 75 mm NaCl, 1 mm DTT, and 20% glycerol (w/v).
The DNA sequence coding for the mature Arabidopsis ClpD protein was PCR amplified from a full-length cDNA clone and ligated into the pCDF expression vector (Novagen). Restriction sites NcoI and NotI were included at the 5′ and 3′ ends, respectively, to facilitate cloning along with a His6 tag at the 3′ end to enable later protein purification. The ClpD protein was overexpressed in E. coli BL21-CodonPlus (Stratagene) as previously described (11), but formed inclusion bodies that were resolubilized in 6 m urea, 20 mm Tris-Cl, pH 8, 300 mm NaCl, 40 mm imidazole, and 0.5 mm DTT. The soluble ClpD was then purified by sequential Ni2+ affinity and gel filtration chromatography according to Andersson et al. (11). Recombinant Synechococcus ClpC was purified as previously described (11).
Chloroplast Protein Import Experiments
Chloroplasts from 14-day-old plants were isolated according to Aronsson and Jarvis (43, 44) and used for the import assays. Template DNA from Arabidopsis cDNA clones for the precursors of the Rubisco small subunit, the subunit II of CF0 of the photosynthetic ATPase, and plastocyanin were amplified using M13 primers and used for in vitro transcription/translation using a coupled TnT system (Promega) based on rabbit reticulocyte lysate containing [35S]methionine and T7 RNA polymerase (43, 44). Chloroplast protein import reactions were performed as described by Aronsson and Jarvis (43, 44). Briefly, each 200 μl of import assay contained 107 chloroplasts, 5 mm MgATP, translation mixture not exceeding 10% of the total volume, and was carried out in white light (100 μmol photons m−2 s−1) at 25 °C for various time periods. Samples were resolved on 12% SDS-PAGE gels. The gels were fixed, exposed to x-ray film, and then quantified using ImageQuant software (Molecular Dynamics).
Protein Degradation Assay
Intact chloroplasts from wild type, clpC1 and clpC2 mutant plants were isolated as previously described (15). The number of intact chloroplasts in each preparation was determined by phase-contrast microscopy (Olympus BX50) using a hemocytometer. Each intact chloroplast sample was diluted to a final concentration of 1.5 × 106 chloroplasts μl−1 with 5 mm Mg-ATP, 2.5 mm phosphocreatine, 50 mg ml−1 of creatine phosphokinase, 0.33 m sorbitol, 5 mm MgCl2, 10 mm NaHCO3, and 20 mm HEPES/NaOH, pH 8. Chloroplasts were incubated for 3 h under white light (about 60 μmol of photons m−2 s−1) at 25 °C. Reactions were stopped by adding 5 volumes of rupture buffer (10 mm MgCl2, 20 mm HEPES/NaOH, pH 7.6), and then centrifuged at 20,000 × g for 10 min to separate the thylakoid membranes from the stroma/envelope membrane fraction. Protein concentration of the stromal/envelope membrane fraction was determined using the BCA protein assay (Pierce Chemicals). Protein samples were separated by denaturing-PAGE on either 3–8% Tris acetate (for EF-Ts, HSP90, RH3, and Tic110) or 12% BisTris (for Tic55, Tic40, and Tic20) gels. The amount of each protein substrate was detected by either staining with Coomassie Brilliant Blue G-250 (EF-Ts, HSP90, and RH3) or by immunoblotting using a specific antibody (Tic110, Tic40, Tic20, and Tic55). Quantifications were performed using the ChemiGenius 2 imaging system (Syngene) and associated software.
RESULTS
Relative Abundance of ClpC1, ClpC2, and ClpD during Leaf Development
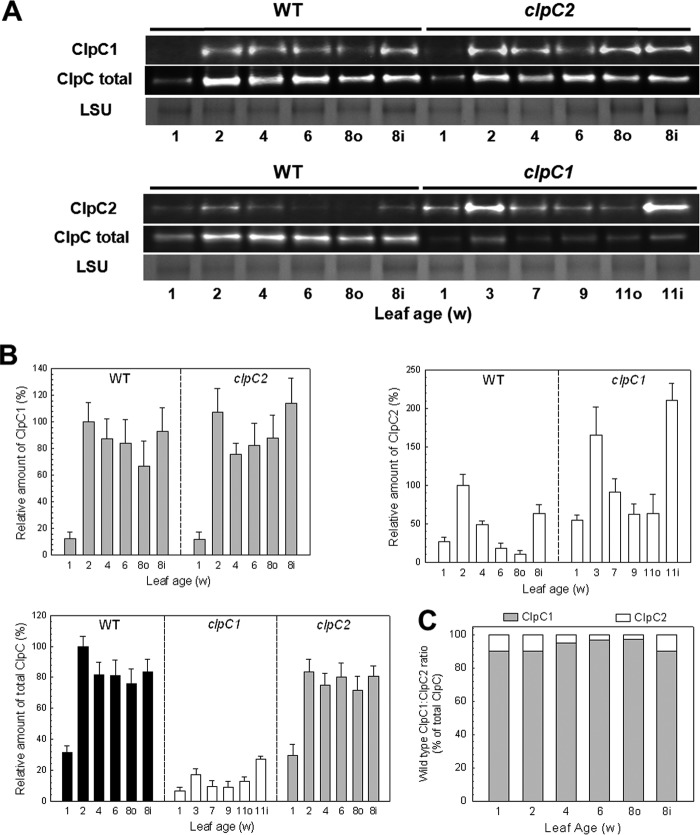
Despite the existence of two ClpC paralogs in chloroplasts, essentially nothing is known about their relative characteristics. To address this issue, we first examined the relative amounts of ClpC1 and -C2 in Arabidopsis leaves during vegetative growth, from cotyledons (1 week old) to mature leaves (8 weeks old). Included in the analysis were leaves from wild type plants and those from clpC1 knock-out and clpC2 knockdown (<10% of wild type levels of ClpC2 remaining; 39) mutant lines. Due to its slow growth phenotype (36), clpC1 mutant leaves were taken at the same developmental age as the wild type and clpC2 mutant. Antibodies were used to detect either the amount of total ClpC protein or more specifically that of ClpC1 or -C2. As shown in Fig. 1, A and B, the amount of both ClpC1 and ClpC2 in wild type Arabidopsis was relatively low in cotyledons but increased significantly in the first leaves (2 weeks). As the leaves matured, ClpC1 levels declined to about 60% of the 2-week level, whereas that for ClpC2 dropped more dramatically to only 10% of the 2-week level. The comparatively high level of both ClpC paralogs in the youngest leaves was also observed in the first leaves from the second rosette (inner leaves 8 weeks, 8i). In the clpC2 mutant, the near complete loss of ClpC2 had a minor effect on ClpC1 levels, with the amount of ClpC1 protein in the different leaves being very similar to that in the wild type except for the levels in the 4–8-week-old leaves being somewhat higher. In terms of total ClpC, the loss of ClpC2 had little overall effect consistent with the lack of any significant phenotypic changes in the clpC2 mutant line (39). The level of ClpC2 in the clpC1 mutant also followed the pattern observed in wild type leaves except that the overall amount of ClpC2 was about 2-fold higher in each leaf. In terms of the total amount of ClpC (Fig. 1B), the amount in the clpC2 mutant was only slightly lower than that in the wild type, suggesting that ClpC1 constitutes the majority of ClpC protein. In contrast, the total ClpC content in clpC1 leaves was relatively low despite induction of the ClpC2 protein, consistent with the more severe phenotype previously observed for this mutant (36–38).
FIGURE 1.
ClpC protein abundance during vegetative growth. A, relative amounts of ClpC1, ClpC2, and total ClpC protein in cotyledons and developing leaves from Arabidopsis wild type (WT), and clpC1 and clpC2 mutants. Leaves were compared at the same developmental ages, which was 1 week old for all cotyledons and 2–8 weeks old (for WT and clpC2 mutant) or 3–11 weeks (clpC1 mutant) for developing leaves from the first rosette. Leaves from the second rosette (inner, i) were also taken at the same time as the mature leaves from the first rosette (outer, o). Total cell extracts were isolated from each sample and separated by denaturing-PAGE loaded on the basis of equal protein content. Proteins were visualized by immunoblotting using antibodies specific for ClpC1, ClpC2, or total ClpC. B, quantification of the relative amounts of ClpC1 (gray bars), ClpC2 (white bars), and total ClpC protein (black bars) in wild type Arabidopsis, and clpC1 and clpC2 mutants. Quantifications were normalized to the value from 2-week-old wild type leaves, which were set to 100%. Values shown are averages ± S.D. (n = 3). C, estimation of the relative proportions of ClpC1 and ClpC2 in wild type Arabidopsis during vegetative growth. LSU, large subunit.
We next calculated the actual ratio of ClpC1 to ClpC2 protein in wild type leaves knowing that the amounts of ClpC1 in the clpC2 mutant and ClpC2 in the clpC1 mutant as determined using the paralog-specific antibodies were equivalent to the same levels as measured with the total ClpC antibody (Fig. 1B). Then for each leaf, the proportion of ClpC1 between the wild type and clpC2 mutant, and ClpC2 between the wild type and clpC1 mutant as determined using the paralog-specific antibodies could be used to ascertain how much each paralog contributed to the total amount of ClpC protein. It should be noted that the levels of ClpC1 protein were adjusted to account for the residual amount of ClpC2 in the clpC2 knockdown line. As shown in Fig. 1C, ClpC1 constitutes by far the bulk of ClpC protein in Arabidopsis chloroplasts, ranging from 90% in cotyledons and up to more than 97% in mature leaves.
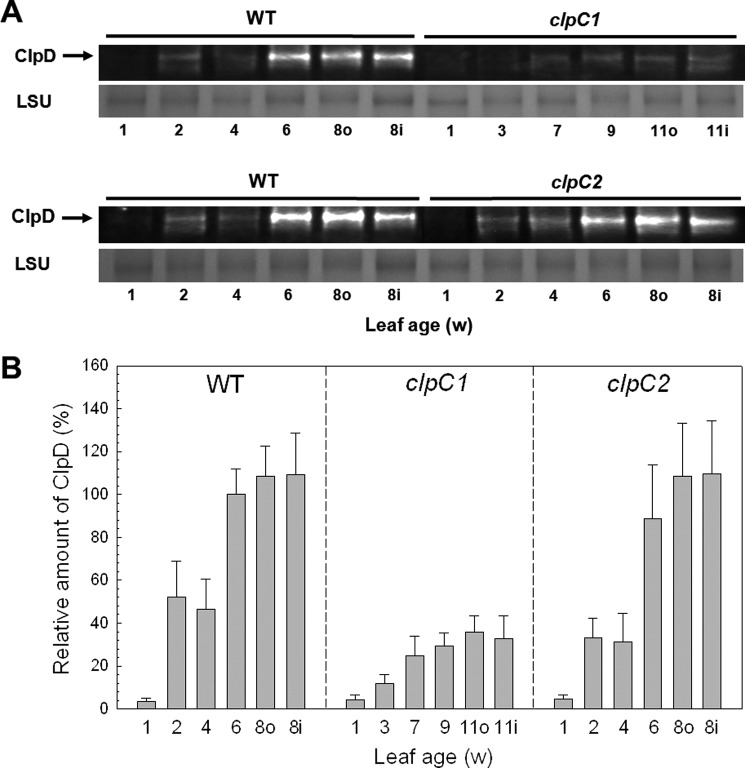
In addition to ClpC, we also examined the level of the closely related paralog ClpD during vegetative growth (Fig. 2, A and B). As for ClpC, ClpD content in cotyledons was relatively low but increased severalfold in the first leaves (2 weeks old). The amount of ClpD then doubled in 6-week-old leaves compared with those from 4 weeks and remained at this level to maturity; it also stayed at this high level in the first leaves from the second rosette. The same profile for ClpD content was also observed in leaves from the clpC2 mutant, suggesting loss of ClpC2 has little or no effect on ClpD levels. In contrast, loss of ClpC1 had a severe effect on ClpD accumulation as leaves matured, with less than half the wild type levels in the various aged leaves (Fig. 2).
FIGURE 2.
Abundance of ClpD during vegetative growth. A, relative amounts of ClpD in cotyledons and developing leaves from Arabidopsis wild type (WT), clpC1 and clpC2 mutants. Leaves were compared at the same developmental ages, which was 1 week old for all cotyledons and 2–8 weeks old (for WT and clpC2 mutant) or 3–11 weeks (clpC1 mutant) for developing leaves from the first rosette. Leaves from the second rosette (inner, i) were also taken at the same time as the mature leaves from the first rosette (outer, o). Total cell extracts were isolated from each sample and separated by denaturing-PAGE loaded on the basis of equal protein content. Proteins were visualized by immunoblotting using antibodies specific for ClpD. B, quantification of the relative amounts of ClpD in wild type Arabidopsis (black bars), clpC1 (white bars), and clpC2 (gray bars) mutants. Quantifications were normalized to the value from 6-week-old wild type leaves, which were set to 100%. Values shown are averages ± S.D. (n = 3). LSU, large subunit.
Both ClpC1 and ClpC2 Associate to the Envelope Membrane
Besides being a stromal protein, ClpC also associates with the envelope membrane (30, 31) although its relative distribution between these two intrachloroplastic locations remains unclear. To address this point, we isolated intact chloroplasts from wild type Arabidopsis and then isolated the stromal, envelope membrane, and thylakoid membrane fractions. The purity of each fraction was determined by immunoblotting using antibodies specific for proteins exclusively located in either the stroma (SSU), the envelope membranes (Tic110), or thylakoid membranes (Lhcb2), neither of which showed any visible contamination in the opposing fractions (Fig. 3A). Using first the antibody for total ClpC, it was clear that the ClpC protein exists both in the stroma and attached to the envelope membrane, consistent with previous studies (30, 31, 45, 46). Using the ClpC1- and -C2-specific antibodies also revealed that the ClpC paralogs were present in both fractions. In the case of ClpC2, an additional, slightly larger protein was also detected in the stromal fraction, which given the high specificity of the antibodies probably derives from an unknown modified form of the ClpC2 protein. The localization of the closely related ClpD was also examined and shown to be exclusively in the stroma. No Clp proteins were found associated to thylakoid membranes as previously shown (9).
FIGURE 3.
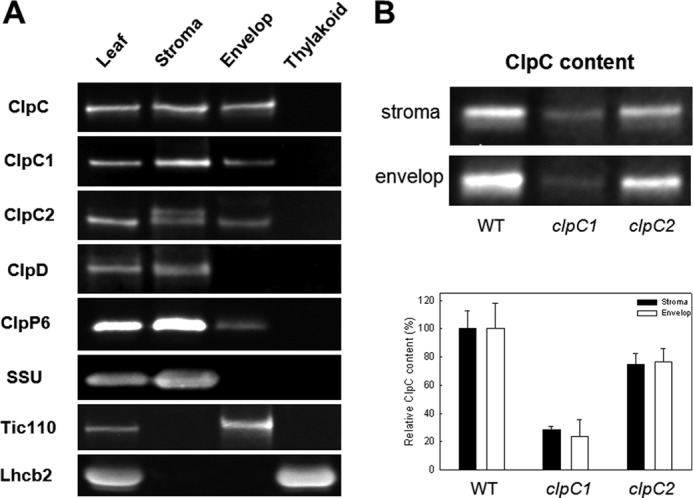
Intrachloroplastic location of Arabidopsis ClpC, ClpD, and ClpP6 proteins. A, intact chloroplasts were purified from Arabidopsis leaves and separated into stroma, envelope membranes, and thylakoid membranes fractions. Each fraction along with a whole leaf extract was separated by denaturing-PAGE, with the ClpC, ClpD, and ClpP6 proteins detected by immunoblotting. The purity of each fraction was also determined by immunoblotting using antibodies specific for marker proteins exclusively located in the stroma (small subunit of Rubisco, SSU), envelope membranes (Tic110), or thylakoid membranes (Lhcb2). Shown is a representative of three independent replicates. B, relative amount of ClpC in stromal and envelope membrane fractions from wild type Arabidopsis, and clpC1 and clpC2 mutants. Each fraction was separated by denaturing-PAGE and the total amount of ClpC in each was determined by immunoblotting. Quantifications were done on three replicates, with the values shown as averages ± S.E.
To accurately quantify distribution of the ClpC paralogs between the stroma and envelope membranes, the amounts of ClpC1 and -C2 in each fraction were normalized to the amounts of the marker proteins, SSU and Tic110. More specifically, the level of SSU in the stroma was normalized to that detected in the whole leaf extract, as was the amount of Tic110 in the envelope membrane fraction. By adjusting the amounts of ClpC proteins by the same ratio, an average of 30% (±3.5% S.D., n = 3) of the total ClpC is associated to the envelope membrane, with no significant difference in the distribution of ClpC1 and ClpC2.
We next tested if the distribution of ClpC proteins to the stroma and envelope membranes varied in either of the clpC1 or clpC2 mutant lines. As shown in Fig. 3B, the wild type proportion of ClpC1 in stroma and envelope membranes remained unchanged in the clpC2 mutant, as did the ratio of ClpC2 in the clpC1 mutant. This suggests that both ClpC proteins have similar affinities to the envelope membranes and that this does not change in the absence of the other paralog. This also shows that there is no greater affinity of either ClpC paralog to the different intrachloroplastic locations.
Clp Proteolytic Core Attached to the Envelope Membrane
Due to the high proportion of ClpC attached to the envelope membrane and its known association with the Clp proteolytic core (19–21), we also examined if any Clp core complex was also bound to the envelope membrane. Using the ClpP6 subunit as a marker protein, an average of 8% (±2.2% S.D., n = 3) of the ClpP6 protein was attached to the envelope membranes, small but significantly more than the stromal protein contamination in this fraction (<2%) as determined from the stromal marker protein SSU.
We next examined which oligomeric state(s) the ClpP6 protein attached to the envelope membrane exists given that it is known that at least half of the Clp proteolytic core subunits in the stroma exist in their respective heptameric P- and R-rings (15, 16, 28). To examine this in more detail, we also examined another subunit within the P-ring (ClpP4) and two from the R-ring (ClpR3 and ClpR4). As can be seen in Fig. 4, only the proteolytic core complex associates to the envelope membranes, with none of the heptameric rings typically observed in the stroma. It should also be noted that no monomers of the four subunits were observed in either the stroma and envelope membrane fractions (data not shown), which for the stroma is consistent with earlier studies (15, 16, 28).
FIGURE 4.
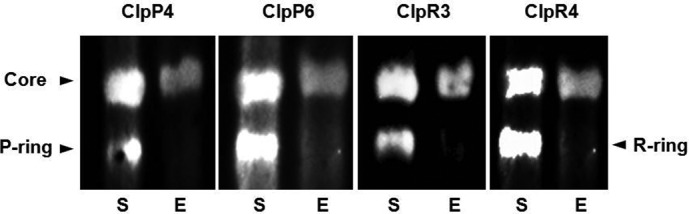
Association of the Clp proteolytic core to the envelope membranes in wild type Arabidopsis. Stromal (30 μg of protein, S) and envelope membrane (15 μg of protein, E) fractions from wild type Arabidopsis were separated by blue native-PAGE. The Clp proteolytic core and subcomplexes (P- and R-rings) were detected using antibodies specific for different subunits: ClpP4, ClpP6, ClpR3, and ClpR4.
Varied Amounts of the Clp Proteolytic Core in the clpC1 and clpC2 Mutants
As an Hsp100 protein, chloroplast ClpC has long been presumed to function as a chaperone both alone and as the regulatory partner within the Clp proteolytic complex despite the lack of supportive experimental evidence. In regards to its role within the chloroplast Clp protease, we examined how the loss of ClpC affected the Clp proteolytic core complex. For this, we isolated stromal proteins from wild type Arabidopsis and the two clpC mutants and then resolved the native protein complexes by colorless native-PAGE. The amount of the Clp proteolytic core was determined using antibodies specific for marker subunits of each subcomplex (i.e. ClpP6 for the P-ring, ClpR2 for R-ring, and ClpT1 for the two extrinsic ClpT proteins). As shown in Fig. 5A, the amount of stromal Clp proteolytic core was 2.5-fold higher in the clpC1 mutant compared with the wild type, most likely as a compensatory response to the significant loss of its chaperone partner. In contrast, the level of Clp proteolytic core decreased in the clpC2 mutant by 10–20%, which correlated to the decrease in total ClpC protein from wild type levels in this knockdown line.
FIGURE 5.
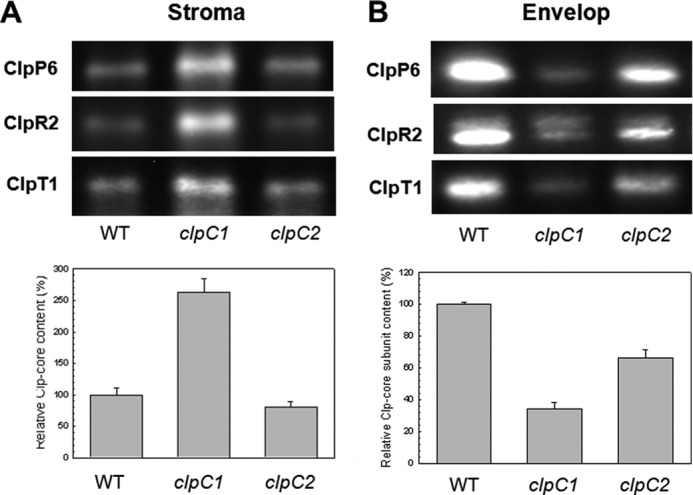
Amount of the Clp proteolytic core in wild type and clpC1 and clpC2 mutants. Stromal (A) and envelope membrane (B) fractions from wild type Arabidopsis (WT), and clpC1 and clpC2 mutants was separated by native- and denaturing-PAGE, respectively, on the basis of equal protein content. The relative amount of Clp proteolytic core in each fraction was visualized and quantified using antibodies specific for each core component: ClpP6 for the P-ring, ClpR2 for the R-ring, and ClpT1 for the extrinsically bound proteins. Values shown are averages from triple replicates with the three Clp-core specific antibodies ClpP6, ClpR2, and ClpT1 ± S.E. (n = 9), with values for the wild type core complex set to 100%.
We next examined the amount of Clp proteolytic core subunits associated to the envelope membranes in the two mutant lines (Fig. 5B). In the clpC1 mutant, the amount of all three subunits decreased to 30% of the wild type levels, suggesting a loss of Clp proteolytic core attached to the envelope membranes despite the large induction of the different subunits within the mutant leaves. This also confirms that the amount of Clp core subunits detected in the envelope membrane fraction was not due to stromal protein contamination. The fact that the drop in envelope membrane-bound Clp core in clpC1 matched that of total ClpC indicates that the core complex associates to the envelope membrane only via ClpC binding; an arrangement fully consistent with the chaperone partner being responsible for substrate recognition and binding. This conclusion is further supported in the clpC2 mutant, in which the Clp proteolytic core subunits attached to the envelope membrane decreased by the same extent as total ClpC (about 70% of wild type levels).
Quantification of the Chaperone and Proteolytic Components of the Clp Protease
To further investigate the contribution of ClpC to Clp degradation activity in chloroplasts, we next determined the relative amounts of both chaperone and proteolytic components. The level of each native Clp protein in the stroma was determined using known amounts of recombinant protein and plotted as a percentage of total stromal protein (Fig. 6, A and B). Of the Hsp100 proteins, the combined amount of ClpC1/C2 constitutes 0.24% of the total stromal protein, whereas ClpD is an order of magnitude lower at 0.02%. In terms of the number of molecules, these percentages for ClpC and ClpD equate to 26 and 2 nmol/g of total stroma protein, respectively. However, because Hsp100 chaperones function as hexameric complexes, the number of functional units would be 4.3 nmol of ClpC hexamer and 0.4 nmol of ClpD hexamer per g of total stroma protein.
FIGURE 6.
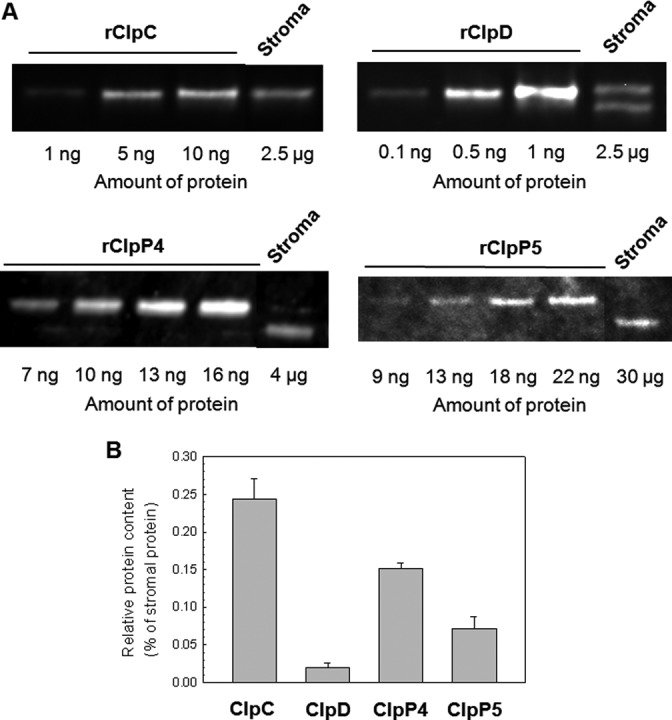
Relative amount of Clp proteins in stroma of wild type Arabidopsis. A, different amounts of recombinant ClpC (rClpC), ClpD (rClpD), ClpP4 (rClpP4), and ClpP5 (rClpP5) were separated by denaturing-PAGE along with a stromal protein extract from wild type Arabidopsis. Each Clp protein was detected by immunoblotting using a specific polyclonal antibody. Shown is a representative replicate of three that were quantified (B) and the average relative amount of each Clp protein in wild type stroma calculated.
We then determined the relative amount of Clp proteolytic core using two of the subunits as marker proteins, ClpP4 and ClpP5. It should be noted that we also attempted to include ClpP3 and ClpP6 in this analysis but their recombinant forms were poorly expressed in E. coli. As shown in Fig. 6, ClpP4 (0.15%) was clearly more abundant than ClpP5 (0.08%), consistent with an earlier proposed ratio of three ClpP4 and two ClpP5 subunits within the P-ring of the core complex (14, 15). A more recent study has proposed the opposite ratio (i.e. 2 ClpP4: 3 ClpP5; see Ref. 17) but the experimental error of this estimate does not exclude the ratio supported by the above quantification. Using the above percentages we then calculated the number of ClpP4 and ClpP5 molecules to 60 ± 3 and 31 ± 7 nmol/g of total stromal protein, respectively. Based on each of these amounts, we then estimated the relative content of the Clp proteolytic core assuming a ClpP4/ClpP5 stoichiometry of 3:2 per core, which came to about 18 nmol/g of total stromal protein. However, it has been shown that a large proportion of the Clp core subunits in the stroma exist in smaller oligomers, which in the case of ClpP4/5 is the P- and P/T1-rings (15–17). Although the appearance of these subcomplexes could be due to dissociation of the core complex during sample isolation and/or buffer conditions during electrophoresis, they consistently occur in similar proportions when using variations in such conditions and in independent studies using different gel conditions (16, 17, 28). We have previously shown that in wild type Arabidopsis the ratio of core complex to the combined amounts of P- and P/T1-rings is about 1:1 (16), which would then halve the estimate of the Clp proteolytic core content in the stroma to 9 nmol/g of stromal protein. Comparing this value to that of ClpC and ClpD suggests that in the stroma there is an excess of Clp proteolytic core complexes relative to the Hsp100 hexameric partners, which would be consistent with both ClpC and ClpD functioning exclusively as part of a Clp protease complex.
From the above results, it is also possible to estimate the amount of ClpC and Clp proteolytic core attached to the envelope membranes. As shown in Fig. 3A, an average of 30% of the total ClpC in chloroplasts is associated to the envelope membranes. Given that the stromal content of ClpC is 4.3 nmol/g of total stromal protein, then about 1.3 nmol of ClpC/g of stromal protein would be expected on the envelope membrane. As for the Clp proteolytic core, 8% of the total amount in the stroma associates to the envelope membranes, which would be 1.4 nmol/g of total stromal protein. Given that only the Clp proteolytic core is attached to the envelope membrane (Fig. 4), these estimates would suggest therefore that there is a sufficient number of Clp proteolytic cores to bind to all the ClpC hexamers associated to the envelope membranes.
Possible Involvement of the Clp Protease in Tic Subunit Turnover
Despite the relatively large proportion of ClpC bound to the envelope membranes, little is still known about its exact function. Its association to Tic110 has long suggested a role in chloroplast protein import, possibly as the motor protein driving precursor translocation through the Tic import complex (33, 47). The occurrence of the Clp proteolytic core on the envelope membrane in association with ClpC forces us to consider the possible role of a Clp protease during protein import. Besides the fact that a specific protease (i.e. signal processing peptidase) has already been identified for cleaving the transit peptide of imported precursor proteins (48), the mechanics of how the Clp protease operates makes it unsuitable for such a processing function. A more plausible role for an envelope membrane-bound Clp protease would be the turnover of one or more subunits of the Tic complex. To test this possibility, we examined the degradation rate of several Tic subunits in close proximity to ClpC, namely Tic110, Tic40, Tic20, and Tic55. By comparing the degradation rates of these proteins in wild type chloroplasts relative to that in chloroplasts from transgenic lines with only 5–10% Clp proteolytic activity remaining (i.e. clpP6 antisense lines or clpR1 knock-out mutant), the possible involvement of the Clp protease could be ascertained. As shown in Fig. 7, significant degradation of all subunits except Tic20 was observed in the wild type during the 3-h time course. However, this degradation rate was unaffected in the clpP6 antisense or clpR1 mutant lines suggesting that an envelope-bound Clp protease is not responsible for the turnover of these proteins.
FIGURE 7.
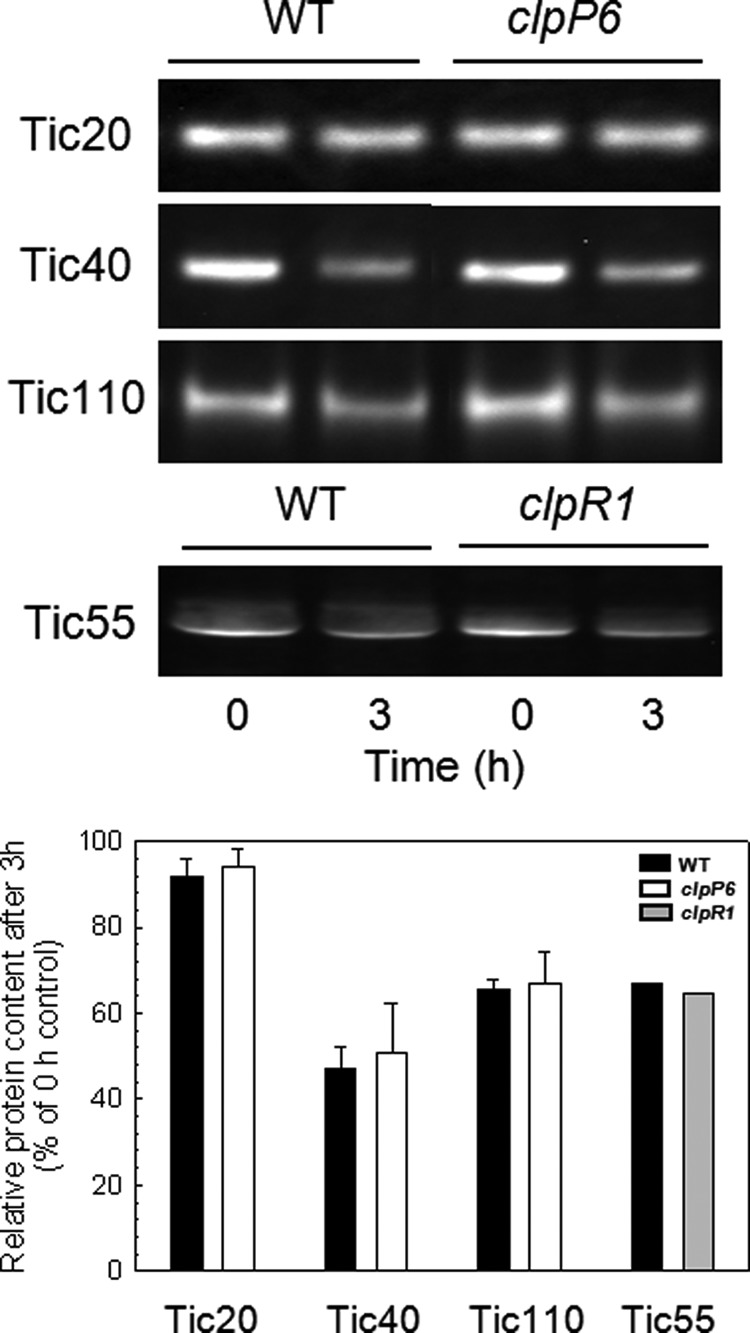
Turnover of Tic protein subunits in the inner envelope membranes. Degradation of the inner envelope membrane Tic subunits Tic110, Tic55, Tic 40, and Tic20 in chloroplasts of wild type Arabidopsis and clpP6 antisense lines or clpR1 knock-out mutant. Equal amounts of intact chloroplasts were incubated in the presence of light and ATP, with envelope membrane fractions isolated at 0 and 3 h. The two protein fractions were separated by denaturing-PAGE and each of the four Tic subunits were detected by immunoblotting with specific antibodies. The relative amount of the different Tic proteins was quantified at each time point and normalized to the 0 h control, which was set to 100%. Values shown are averages ± S.D. (n = 3).
Effect of the Clp Protease on Chloroplast Protein Import
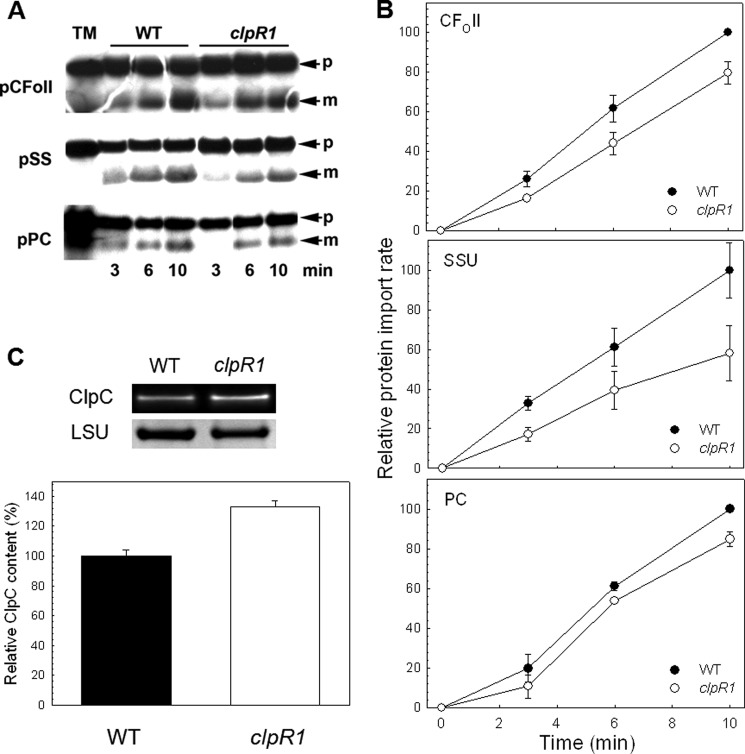
Given that the Clp protease is not involved in Tic subunit turnover or precursor protein processing, the question remains as to its function on envelope membranes. We next examined if loss of the Clp proteolytic core directly affected chloroplast protein import. For this, we determined the import efficiency of several in vitro translated preproteins into chloroplasts isolated from wild type Arabidopsis and the clpR1 knock-out mutant. As shown in Fig. 8, A and B, a small but significant decrease in the rate of import was observed in the clpR1 mutant for all three preproteins tested compared with the wild type. This decrease in import efficiency was not due to a lower level of ClpC in the mutant chloroplasts (Fig. 8C); ClpC content was in fact higher in the clpR1 line suggesting a possible compensatory response to the loss of Clp proteolytic activity.
FIGURE 8.
Chloroplast protein import assays. A, import of different precursor proteins into wild type Arabidopsis and clpR1 mutant chloroplasts: the Rubisco small subunit (pSS), the subunit II of CFO of the photosynthetic ATPase (pCFoII), and plastocyanin (pPC). Import proceeded for 3, 6, and 10 min, as indicated, and then samples were analyzed by denaturing-PAGE and fluorography. TM, translation mixture; p, precursor protein; m, mature protein. B, quantification of the import data shown in A. Mature protein bands observed in A were quantified using ImageQuant software, and then the data expressed as percentages of the value for the final wild type time point. Values shown are mean ± S.E. derived from at least three assays. C, amount of total ClpC in leaves of wild type Arabidopsis and clpR1 mutant. Whole cell extracts were isolated from 2-week-old leaves and separated by denaturing-PAGE on the basis of equal protein content. Proteins were visualized by immunoblotting and quantifications were normalized to the wild type value, which were set to 100%. Values shown are averages ± S.D. (n = 3).
Contribution of ClpC to Chloroplast Clp Proteolytic Activity
Because ClpC is thought to be the principle chaperone component of the chloroplast Clp protease, we next examined whether reduction of ClpC content also affected the degradation rate of stromal protein substrates. Performing the proteolytic assay with intact chloroplasts from wild type Arabidopsis and the clpC1 and clpC2 mutants, we first analyzed the degradation rate of the elongation factor-Ts (EF-Ts). Consistent with our earlier observations (15), more than 50% of EF-Ts was degraded in wild type chloroplasts over the 3-h time course of the assay; a degradation that was dependent on light and ATP (Fig. 9, A and B). In the clpC1 mutant, which has a greatest reduction in total ClpC content, the degradation rate of EF-Ts was almost completely inhibited, with only a 10% loss of EF-Ts by the end of the assay. In comparison, the degradation rate of EF-Ts was essentially unaffected in the clpC2 mutant (Fig. 9B), which in relationship to the clpC1 mutant has only a minor reduction in total ClpC content. A similar trend was also observed for two other substrates of the Clp protease, heat shock protein 90 (HSP90) and RNA helicase 3 (RH3). As shown in Fig. 8C, degradation of both HSP90 and RH3 were greatly reduced in the clpC1 mutant compared with the wild type, but they were essentially unaffected in the clpC2 mutant.
FIGURE 9.
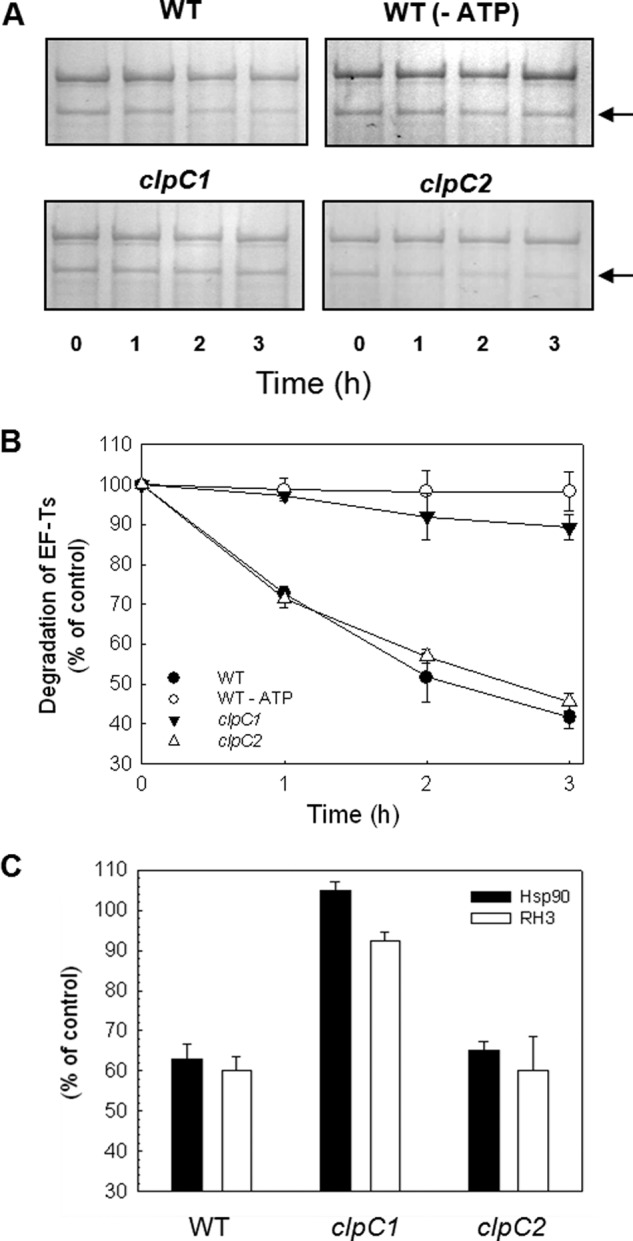
Degradation of specific stromal proteins in clpC1 and clpC2 mutants. A, equal amounts of intact chloroplasts from wild type (WT), and clpC1 and clpC2 mutants were incubated 0–3 h in the presence of light and ATP, with the exception of the wild type control samples incubated in darkness without light (WT −ATP). Isolated stroma from different time points where separated by SDS-PAGE and proteins were visualized with Coomassie Blue staining. Three identified Clp substrate bands were quantified during the time course, with the results shown in the figure being representative of the average for one substrate (EF-Ts, indicated by the black arrow). B, degradation rate for EF-Ts in wild type, wild type control (−ATP), and clpC1 and -C2 mutants. Values shown are average ± S.E. (n = 3), where protein content from the 0-h sample was set to 100%. C, degradation of heat shock protein 90 (Hsp90) and RNA helicase 3 (RH3) in chloroplasts of wild type, and clpC1 and -C2 mutants after a 3-h incubation. Values shown are average ± S.E. (n = 3), where protein content from the 0-h sample was set to 100%.
DISCUSSION
ClpC is a member of the Hsp100 family of molecular chaperones and is the ortholog in photosynthetic organisms to the E. coli ClpA. In contrast to most cyanobacteria and algae, vascular plants generally contain two closely related clpC genes encoding near identical proteins, ClpC1 and ClpC2 (35, 49). Their high sequence similarity suggests that ClpC1 and ClpC2 are functionally indistinct, which is supported by the ability of overexpressed ClpC2 to complement the chlorotic phenotype of the Arabidopsis clpC1 mutant (25). The main difference between the two ClpC paralogs, therefore, appears to be their differential expression, with ClpC1 being by far the most abundant throughout vegetative growth. That ClpC2 accumulates mostly in younger leaves might simply be due to a need for higher total ClpC content in the more metabolically active chloroplasts. Despite their similarity, loss of the more abundant ClpC1 in the clpC1 mutant only increases ClpC2 content 2–3-fold. The fact that the clpC1 mutant exhibits a constant chlorotic phenotype suggests that the capacity of the clpC2 gene to be up-regulated is limited to the amount of ClpC2 observed in the clpC1 mutant.
Like the ClpC proteins, ClpD content in cotyledons is relatively low but increases severalfold in the first true leaves. As for both ClpC paralogs, ClpD has the conserved motifs in the C terminus for ClpP association, namely the P-loop and the recently identified R-motif specific for ClpR-containing proteolytic cores (18). However, given the amino acid differences in the N-terminal domain, it is likely that ClpD targets a different set of protein substrates to that by ClpC. Whether ClpD functions as a chaperone independent of the Clp protease remains uncertain, although there is more than sufficient stromal Clp proteolytic core for it to function exclusively within the protease. The fact that loss of ClpC1 caused a significant reduction in ClpD content also suggests a possible structural interaction between the two Hsp100 proteins, a phenomenon we are now investigating. It should be noted that a recent study showed Arabidopsis ClpD, like ClpC, could interact with the transit peptide of a preprotein in vitro (50), although a role for ClpD in the chloroplast protein import process in vivo would seem questionable given its lack of envelope membrane association.
ClpC is located both in the stroma and attached to the inner envelope membrane. Although this membrane ClpC has long been believed to constituent only a small fraction of the total chloroplast ClpC content (31), a recent proteomic study has estimated it to be over 50% (46). We have shown that a significant proportion of ClpC does indeed associate to the envelope membrane, 30% of the total ClpC in chloroplasts. The discrepancy between our estimate and that from the proteomic study could be due to the latter having a greater stromal protein contamination in the envelope membrane fractions (10%; 46). There is also no significant difference in the distribution of ClpC1 and ClpC2 between the stroma and envelope membranes, again supporting that both ClpC paralogs are functionally indistinguishable.
The large proportion of ClpC attached to envelope membranes suggests it contributes more than previously assumed to the overall importance of ClpC in Arabidopsis chloroplasts. On the envelope membrane, ClpC binds to Tic110 (30, 31), an integral subunit of the Tic translocon complex, which in cooperation with the Toc complex (situated in the outer envelope membrane) imports the bulk of nuclear-encoded polypeptides destined for the chloroplast. Tic110 function is essential for chloroplast protein import (38) and is thought to form a protein translocation pore (51), mediate binding of preproteins exiting the Tic complex and recruit various molecular chaperones including ClpC, Hsp70, and Hsp60 (52–54). Although the exact function of ClpC bound to Tic110 is unknown, it is generally thought to act as a “motor” protein driving transport of precursor proteins via ATP hydrolysis through the Tic complex (30, 31, 55). The current model for protein import suggests the transit peptide of preproteins emerging from the membrane channel binds to Tic110, which then recruits Tic40 (53, 56). The preprotein is transferred to ClpC, which then completes the import of the preprotein into the stroma via Tic40-stimulated ATP hydrolysis (56). The functioning of ClpC, however, in such a central role as a motor protein in the protein import process remains inconclusive. Studies on import rates in the clpC1 mutant have shown highly variable effects with different preproteins, ranging from 0 to 50% decreases relative to those in the wild type (36–38, 54, 57), but none proportional to the loss of total ClpC content (i.e. 70%). This inconsistency might be explained by the recent discovery of a second possible import system involving a stromal Hsp70 system at the inner envelope membrane (54, 58). It is also known that the Hsp100 protein ClpB can cooperate with the Hsp70/Hsp40 chaperones in bacteria and yeast to optimize resolublization/refolding of denatured and aggregated polypeptides (59–61), and it is plausible that a similar bi-chaperone system of ClpC/Hsp70/Tic40 might be driving preprotein import via the Tic complex inside chloroplasts.
Given the existence of the Clp proteolytic core attached to the envelope membrane, the function of membrane-bound ClpC must now be re-evaluated. Quantifications suggest that all the ClpC associated to the envelope membrane presumably in the functional hexameric state could have the Clp proteolytic core attached. Assuming all envelope ClpC is bound to the Tic complex, then the role of a Clp protease in the chloroplast protein import pathway needs to be considered. Even ignoring the defined role of signal processing peptidase in cleaving the transit peptide from imported preproteins (48), the proteolytic action of the Clp protease makes it unsuitable for such a specific processing function. Those Tic subunits in close proximity to ClpC, namely Tic110, Tic40, and Tic55, are also not degraded by the Clp protease; a function that might instead be performed by the recently discovered FtsH11 and FtsH12 proteases also bound to the envelope membranes (46). It is equally interesting that no degradation of Tic20 was observed in contrast to the other Tic subunits. It has recently been proposed that Tic20 forms a channel within the inner envelope membrane independent of the main Tic110 translocation pore and thus imports a different subset of preproteins (62). If so, then the differences in degradation rates observed in this study could reflect the existence of two such distinct translocons and that they might be differentially regulated.
Given the above, the question remains as to the exact function of an envelope Clp protease associated to the Tic complex. One intriguing possibility is that the ClpC via its association to the Clp proteolytic core confers a protein quality control mechanism for screening preproteins exiting the Tic complex, in particular during the processing of their transit peptides and subsequent refolding. Although thousands of chloroplast proteins derive from nuclear genes, little is yet known about the efficiency of their import into chloroplasts, especially in the most metabolically active chloroplasts within young leaves when import rates would be most rapid, or their refolding to the mature conformation after removal of the transit peptide. In general, all nascent polypeptides must quickly and efficiently attain their native structure following synthesis. Failure to do so leads to extensive protein misfolding that can have devastating effects on cellular processes due to either loss of normal protein function or a cytotoxic gain of function (63). Misfolding can occur in newly synthesized polypeptides for various reasons, such as from genetic mutation and mistranslation. Chaperones perform a crucial surveillance role in screening for such biosynthetic errors, conferring an essential quality control that maintains the integrity of the proteome. In the cytosol, Hsp70 chaperones interact with the linear polypeptide chain during translation to prevent premature misfolding. Once synthesis is complete, Hsp70 initiates correct protein folding with the aid of additional chaperones including those of the Hsp60 and Hsp90 classes. If misfolding occurs, however, Hsp70 redirects the nascent polypeptide to the ubiquitin-mediated protein degradation pathway, removing these damaged proteins before they adversely affect cellular homeostasis (63).
Although the exact details of how cytosolic Hsp70 detects such aberrant polypeptides remains unclear, it typically occurs post-translationally once folding is initiated. But what about those proteins destined for the chloroplast? Because these preproteins are kept in an unfolded state by Hsp70 to maintain import competency, any biosynthetic mistakes within these polypeptides are likely to go undetected by the cytosolic protein quality control systems. Indeed, the folding of most chloroplast-destined preproteins usually occurs once they exit the Tic complex and it is only at this stage that the potential for misfolding would first be detected. As such, the ability to effectively remove the aberrant preprotein by degradation would have the advantage of preventing prolonged impairment of the import capacity of the Tic complex if the preprotein remains bound, and avoid any potential interference of chloroplast functions if the damaged polypeptide is inadvertently released. Having the Clp proteolytic core attached to the ClpC bound to the Tic complex could well confer such a protein quality control system to newly imported polypeptides. Because the occurrence of such damaged preproteins would presumably be low, the small but significant decrease in import efficiency in the clpR1 mutant would be consistent with such a function for an envelope membrane Clp protease. It would also explain why unprocessed preproteins accumulate in Arabidopsis mutants with significantly reduced Clp proteolytic activity (24).
Despite little experimental evidence, ClpC and ClpD have long been presumed to function as chaperones both independently and within the Clp protease. ClpC association to the chloroplast Clp proteolytic core was implied by co-immunoprecipitation of certain ClpP paralogs using ClpC-specific antibodies (19–21) and the ability of purified chloroplast ClpC to promote proteolysis by E. coli ClpP in vitro (34). Determining the relative amounts of each component is an important step in discerning between these two distinct chaperone activities. The constitutive level of ClpC in the stroma is an order of magnitude higher than that of ClpD. That ClpD content during leaf maturation is inversely proportional to that of ClpC2 suggests a switch in protein substrate specificity, one which appears to continue during leaf senescence and certain stress regimes (9, 64). When calculating the amount of intact Clp proteolytic core, it appears double that of hexameric ClpC and ClpD combined, suggesting that most if not all of these stromal Hsp100 chaperones are functioning within the Clp protease. Indeed, the assembly of the Clp proteolytic core from the pool of P- and R-rings, which appears to be regulated by the accessory ClpT proteins (16) might well be determined by the availability of stromal ClpC and ClpD. The importance of ClpC in the stromal Clp protease was further demonstrated by the dramatic loss of Clp proteolytic activity in the clpC1 mutant. What little degradation remained in the clpC1 mutant was almost certainly due to the small amount of ClpC2 participating in a fraction of active Clp protease. Moreover, the severalfold increase of the Clp proteolytic core in the clpC1 mutant was almost certainly a compensatory response to the near-abolition of Clp degradation activity in the stroma. Involvement of ClpC2 in Clp proteolysis was also supported by the slight reduction in degradation of the three marker substrates in the clpC2 mutant, which was proportional to the loss of total ClpC protein. Altogether, the two chloroplast ClpC paralogs in Arabidopsis appear to be functionally indistinct and to act solely as the chaperone partner for the stromal Clp protease. They also both associate to the envelope membrane and likely participate in the chaperone-driven preprotein translocation through the Tic complex, as well as potentially conferring a novel quality control capacity on preprotein processing/folding.
This work was supported by grants from The Swedish Research Council (VR) and Stiftelsen Olle Engkvist Byggmästare (to A. K. C.).
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SSU
- small subunit of Rubisco
- EF
- elongation factor.
REFERENCES
- 1. Ellis R. J. (2005) in Molecular Chaperones and Cell Signalling (Henderson B., Pockley A. G., eds) pp. 3–21, University Press, Cambridge [Google Scholar]
- 2. Wickner S., Maurizi M. R., Gottesman S. (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science 286, 1888–1893 [DOI] [PubMed] [Google Scholar]
- 3. Vierstra R. D. (1996) Proteolysis in plants: mechanisms and functions. Plant Mol. Biol. 32, 275–302 [DOI] [PubMed] [Google Scholar]
- 4. Wang J., Hartling J. A., Flanagan J. M. (1997) The structure of ClpP at 2.3-Å resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456 [DOI] [PubMed] [Google Scholar]
- 5. Grimaud R., Kessel M., Beuron F., Steven A. C., Maurizi M. R. (1998) Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273, 12476–12481 [DOI] [PubMed] [Google Scholar]
- 6. Kim Y. I., Levchenko I., Fraczkowska K., Woodruff R. V., Sauer R. T., Baker T. A. (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 8, 230–233 [DOI] [PubMed] [Google Scholar]
- 7. Ortega J., Lee H. S., Maurizi M. R., Steven A. C. (2002) Alternating translocation of protein substrates from both ends of ClpXP protease. EMBO J. 21, 4938–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirstein J., Molière N., Dougan D. A., Turgay K. (2009) Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. 7, 589–599 [DOI] [PubMed] [Google Scholar]
- 9. Zheng B., Halperin T., Hruskova-Heidingsfeldova O., Adam Z., Clarke A. K. (2002) Characterization of chloroplast Clp proteins in Arabidopsis: localization, tissue specificity and stress responses. Physiol. Plant. 114, 92–101 [DOI] [PubMed] [Google Scholar]
- 10. Peltier J.-B., Ripoll D. R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K. J. (2004) Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279, 4768–4781 [DOI] [PubMed] [Google Scholar]
- 11. Andersson F. I., Blakytny R., Kirstein J., Turgay K., Bukau B., Mogk A., Clarke A. K. (2006) Cyanobacterial ClpC/HSP100 protein displays intrinsic chaperone activity. J. Biol. Chem. 281, 5468–5475 [DOI] [PubMed] [Google Scholar]
- 12. Stanne T. M., Pojidaeva E., Andersson F. I., Clarke A. K. (2007) Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J. Biol. Chem. 282, 14394–14402 [DOI] [PubMed] [Google Scholar]
- 13. Andersson F. I., Tryggvesson A., Sharon M., Diemand A. V., Classen M., Best C., Schmidt R., Schelin J., Stanne T. M., Bukau B., Robinson C. V., Witt S., Mogk A., Clarke A. K. (2009) Structure and function of a novel type of ATP-dependent Clp protease. J. Biol. Chem. 284, 13519–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peltier J. B., Emanuelsson O., Kalume D. E., Ytterberg J., Friso G., Rudella A., Liberles D. A., Söderberg L., Roepstorff P., von Heijne G., van Wijk K. J. (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sjögren L. L., Stanne T. M., Zheng B., Sutinen S., Clarke A. K. (2006) Structural and functional insights into the chloroplast ATP-dependent Clp protease in. Arabidopsis. Plant Cell 18, 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sjögren L. L., Clarke A. K. (2011) Assembly of the chloroplast ATP-dependent Clp protease in Arabidopsis is regulated by the ClpT accessory proteins. Plant Cell 23, 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olinares P. D., Kim J., Davis J. I., van Wijk K. J. (2011) Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23, 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tryggvesson A., Ståhlberg F. M., Mogk A., Zeth K., Clarke A. K. (2012) Interaction specificity between the chaperone and proteolytic components of the cyanobacterial Clp protease. Biochem. J. 446, 311–320 [DOI] [PubMed] [Google Scholar]
- 19. Desimone M., Weiß-Wichert C., Wagner E., Altenfeld U., Johanningmeier U. (1997) Immunochemical studies on the Clp-protease in chloroplasts: evidence for the formation of a ClpC/P complex. Bot. Acta 110, 234–239 [Google Scholar]
- 20. Sokolenko A., Lerbs-Mache S., Altschmied L., Herrmann R. G. (1998) Clp protease complexes and their diversity in chloroplasts. Planta 207, 286–295 [DOI] [PubMed] [Google Scholar]
- 21. Halperin T., Ostersetzer O., Adam Z. (2001) ATP-dependent association between subunits of Clp protease in pea chloroplasts. Planta 213, 614–619 [DOI] [PubMed] [Google Scholar]
- 22. Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. (2001) The chloroplast clpP gene encoding a proteolytic subunit of ATP-dependent protease and is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42, 264–273 [DOI] [PubMed] [Google Scholar]
- 23. Kuroda H., Maliga P. (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425, 86–89 [DOI] [PubMed] [Google Scholar]
- 24. Rudella A., Friso G., Alonso J. M., Ecker J. R., van Wijk K. J. (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18, 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng B., MacDonald T. M., Sutinen S., Hurry V., Clarke A. K. (2006) A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224, 1103–1115 [DOI] [PubMed] [Google Scholar]
- 26. Kovacheva S., Bédard J., Wardle A., Patel R., Jarvis P. (2007) Further in vivo studies on the role of the molecular chaperone Hsp93, in plastid protein import. Plant J. 50, 364–379 [DOI] [PubMed] [Google Scholar]
- 27. Koussevitzky S., Stanne T. M., Peto C. A., Giap T., Sjögren L. L., Zhao Y., Clarke A. K., Chory J. (2007) An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63, 85–96 [DOI] [PubMed] [Google Scholar]
- 28. Stanne T. M., Sjögren L. L., Koussevitzky S., Clarke A. K. (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem. J. 417, 257–268 [DOI] [PubMed] [Google Scholar]
- 29. Nishimura K., Asakura Y., Friso G., Kim J., Oh S. H., Rutschow H., Ponnala L., van Wijk K. J. (2013) ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25, 2276–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akita M., Nielsen E., Keegstra K. (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelopee membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16, 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosano G. L., Bruch E. M., Ceccarelli E. A. (2011) Insights into the CLP/HSP100 chaperone system from chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 286, 29671–29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flores-Pérez Ú., Jarvis P. (2013) Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 1833, 332–340 [DOI] [PubMed] [Google Scholar]
- 34. Shanklin J., DeWitt N. D., Flanagan J. M. (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7, 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adam Z., Adamska I., Nakabayashi K., Ostersetzer O., Haussuhl K., Manuell A., Zheng B., Vallon O., Rodermel S. R., Shinozaki K., Clarke A. K. (2001) Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 125, 1912–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sjögren L. L., MacDonald T. M., Sutinen S., Clarke A. K. (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136, 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Constan D., Froehlich J. E., Rangarajan S., Keegstra K. (2004) A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136, 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kovacheva S., Bédard J., Patel R., Dudley P., Twell D., Ríos G., Koncz C., Jarvis P. (2005) In vivo studies on the roles of Tic110, Tic40, and Hsp93 during chloroplast protein import. Plant J. 41, 412–428 [DOI] [PubMed] [Google Scholar]
- 39. Park S., Rodermel S. R. (2004) Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Block M. A., Tewari A. K., Albrieux C., Maréchal E., Joyard J. (2002) The plant S-adenosyl-l-methionine:Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur. J. Biochem. 269, 240–248 [DOI] [PubMed] [Google Scholar]
- 41. Porra R. J., Thompson W. A., Kriedemann P. E. (1998) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 42. Randelj O., Rassow J., Motz C. (2007) Separation of proteins by blue native electrophoresis. Methods Mol. Biol. 390, 417–427 [DOI] [PubMed] [Google Scholar]
- 43. Aronsson H., Jarvis P. (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529, 215–220 [DOI] [PubMed] [Google Scholar]
- 44. Aronsson H., Jarvis R. P. (2011) Rapid isolation of Arabidopsis chloroplasts and their use for in vitro protein import assays. Methods. Mol. Biol. 774, 281–305 [DOI] [PubMed] [Google Scholar]
- 45. Froehlich J. E., Wilkerson C. G., Ray W. K., McAndrew R. S., Osteryoung K. W., Gage D. A., Phinney B. S. (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Proteome Res. 2, 413–425 [DOI] [PubMed] [Google Scholar]
- 46. Ferro M., Brugière S., Salvi D., Seigneurin-Berny D., Court M., Moyet L., Ramus C., Miras S., Mellal M., Le Gall S., Kieffer-Jaquinod S., Bruley C., Garin J., Joyard J., Masselon C., Rolland N. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell Proteomics 9, 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwenkert S., Soll J., Bölter B. (2011) Protein import into chloroplasts: how chaperones feature into the game. Biochim. Biophys. Acta 1808, 901–911 [DOI] [PubMed] [Google Scholar]
- 48. Richter S., Lamppa G. K. (1999) Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J. Cell Biol. 147, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gottesman S., Squires C., Pichersky E., Carrington M., Hobbs M., Mattick J. S., Dalrymple B., Kuramitsu H., Shiroza T., Foster T., Clark W. P., Ross B., Squires C. L., Maurizi M. R. (1990) Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 87, 3513–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bruch E. M., Rosano G. L., Ceccarelli E. A. (2012) Chloroplastic Hsp100 chaperones ClpC2 and ClpD interact in vitro with a transit peptide only when it is located at the N-terminus of a protein. BMC Plant Biol. 12, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Balsera M., Goetze T. A., Kovács-Bogdán E., Schürmann P., Wagner R., Buchanan B. B., Soll J., Bölter B. (2009) Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulphide bridge. J. Biol. Chem. 284, 2603–2616 [DOI] [PubMed] [Google Scholar]
- 52. Kessler F., Blobel G. (1996) Interaction of the protein import and folding machineries in the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 93, 7684–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Inaba T., Li M., Alvarez-Huerta M., Kessler F., Schnell D. J. (2003) atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278, 38617–38627 [DOI] [PubMed] [Google Scholar]
- 54. Su P.-H., Li H. M. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jackson-Constan D., Akita M., Keegstra K. (2001) Molecular chaperones involved in chloroplast protein import. Biochim. Biophys. Acta 1541, 102–113 [DOI] [PubMed] [Google Scholar]
- 56. Chou M. L., Chu C. C., Chen L. J., Akita M., Li H. (2006) Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175, 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chu C-C., Li H. M. (2012) The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo. Plant Physiol. 158, 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi L.-X., Theg S. M. (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22, 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Glover J. R., Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 [DOI] [PubMed] [Google Scholar]
- 60. Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., Bukau B. (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U.S.A. 96, 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mogk A., Tomoyasu T., Goloubinoff P., Rüdiger S., Röder D., Langen H., Bukau B. (1999) Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18, 6934–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kovács-Bogdán E., Benz J. P., Soll J., Bölter B. (2011) Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 11, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McClellan A. (2012) in eLS, John Willey & Sons, Ltd., Chichester, 10.1002/9780470015902.a0020886.pub2 [DOI] [Google Scholar]
- 64. Nakabayashi K., Ito M., Kiyosue T., Shinozaki K., Watanabe A. (1999) Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol. 40, 504–514 [DOI] [PubMed] [Google Scholar]