Background: Adenoviruses (AdV) are broadly employed as gene delivery vectors.
Results: Copy numbers of all AdV proteins were measured, and the release of proteins upon heat stress investigated.
Conclusion: The viral protease plays a distinct role in the segmented release of AdV proteins.
Significance: Our characterization by mass spectrometry provides new insight in HAdV disassembly during entry into host cells.
Keywords: Adenovirus, Mass Spectrometry (MS), Proteomics, Virus Entry, Virus Structure
Abstract
Using high-resolution MS-based proteomics in combination with multiple protease digestion, we profiled, with on average 90% sequence coverage, all 13 viral proteins present in an human adenovirus (HAdV) vector. This in-depth profile provided multiple peptide-based evidence on intrinsic protease activity affecting several HAdV proteins. Next, the generated peptide library was used to develop a targeted proteomics method using selected reaction monitoring (SRM) aimed at quantitative profiling of the stoichiometry of all 13 proteins present in the HAdV. We also used this method to probe the release of specific virus proteins initiated by thermal stimulation, mimicking the early stage of HAdV disassembly during entry into host cells. We confirmed the copy numbers of the most well characterized viral capsid components and established the copy numbers for proteins whose stoichiometry has so far not been accurately defined. We also found that heating HAdV induces the complete release of the penton base and fiber proteins as well as a substantial release of protein VIII and VI. For these latter proteins, maturational proteolysis by the adenoviral protease leads to the differential release of fragments with certain peptides being fully released and others largely retained in the AdV particles. This information is likely to be beneficial for the ongoing interpretation of high resolution cryoEM and x-ray electron density maps.
Introduction
Adenoviruses (AdVs)7 are large (∼150 MDa protein content), double-stranded DNA viruses found in all vertebrates (1). Human AdVs (HAdVs) are normally associated with mild infections but can cause potentially severe disease in immunocompromised individuals. First isolated in the early 1950s, HAdV has served as a model virus to elucidate the molecular basis of viral structure, replication, and pathogenesis (2–4), with notable contributions to the field of eukaryotic molecular biology (5, 6). Most importantly, HAdVs are broadly employed in molecular biotechnology as gene delivery vectors and DNA-based vaccine vehicles (7, 8). Nevertheless, these applications are partly hampered by the immune response elicited by the capsid proteins, as well as the difficulty in targeting specific cell types. Structure-based redesign for vector cell targeting and alteration of antigenic properties is therefore needed, but this requires a detailed knowledge of the HAdV structure. The recently reported crystal structure (2) and high-resolution cryoelectron microscopy reconstruction (3) of intact HAdV have provided a more detailed molecular model of the structure and assembly of the HAdV particle (Fig. 1); however, a complete and accurate assignment of all capsid proteins is still lacking. The structure of the outer capsid is mostly well defined, but the stoichiometry and structure of the inner capsid and core proteins, many of them interacting with the DNA, remains elusive. HAdV assembles from 13 distinct proteins to form an icosahedral capsid (9). The main components are the 240 trimers of protein II (hexon) that comprise the faces and edges of the capsid structure. Polypeptide III (penton) is present in 60 copies and forms 12-penton base complexes centered on the 12 vertices of the icosahedron. Extending outward from each penton base are 12 polypeptide IV (fiber) trimers (10). The locations, interactions, and stoichiometries of these three proteins are revealed in the crystal structure and EM models. However, HAdV particles contain 10 additional proteins, for which precise structural information remains unclear. Four minor cement proteins are primarily associated with the capsid shell proteins IIIa, VI, VIII, and IX. The genomic core of the virion is associated with six additional proteins, i.e. V, VII, Mu, IVa2, terminal protein, and the 23K maturational protease. An amazing attribute of one of these cement proteins, VI, is that it serves several other crucial functions in the virus life cycle (11–13). Table 1 summarizes key characteristics of the proteins known to be present in HAdV particles. Developing a method to determine the exact stoichiometry for all of the HAdV proteins is likely to be beneficial to improving the x-ray and EM models of HAdV. MS-based proteomics techniques have emerged as the method of choice to characterize the protein content of a given biological sample (14–16). Recent developments in new analyzers (17–19) and fragmentation methods (20, 21) dramatically increased MS sensitivity, accuracy, and sequencing speed. Consequently, the characterization of several thousands of proteins expressed in complex biological samples (e.g. mammalian cells or tissues) is achievable employing two-dimensional chromatography strategies or even in a single shotgun LC-MS analysis (22, 23). A variety of methods have focused on the relative quantification of protein expression, defining protein level changes in two (or more) samples or conditions. Although typical MS-based methods can accurately detect expression changes of thousands of proteins, the estimation of their absolute abundance (e.g. essential to define the stoichiometry of a protein in a protein complex) remains a much more challenging task. The most commonly used approach relies on reference peptides that contain heavy isotope labels and are further chemically identical to the original peptides (so-called AQUA peptides (24)). Relying on a subset of specific peptides, absolute protein quantification can be best approached by targeted proteomics strategies. Selected reaction monitoring (SRM) has emerged as a method to quantitatively assess the abundance of not only small molecules but also a targeted set of proteins. SRM assays are typically performed by LC-MS/MS employing triple quadrupole mass spectrometers. In this approach, the selectivity of two mass filters is applied to target a specific peptide via the combination of its precursor ion m/z and multiple diagnostic collision-induced dissociation fragment ions (named peptide transitions), along with the specific peptide retention time. This setup allows targeting of specific peptides even in complex samples and removes most of the background chemical noise, increasing both sensitivity and dynamic range (25). To design an SRM assay, the transitions specific for each quantified peptide needs to be extracted from shotgun MS/MS spectra or, if available, from public repositories (26–28). Here, we employed a combination of shotgun and targeted proteomics to characterize the HAdV (Ad5F35) proteome (Fig. 2). We first used high resolution LC-MS/MS analysis, employing three different proteases (trypsin, chymotrypsin, and Lys-N (29)) to improve coverage of all proteins present in the HAdV particles, with the aim to evaluate intrinsic adenovirus proteinase activity and to select the best peptide candidates for downstream SRM assays. Next, we developed a SRM assay to define the copy number of each of the 13 detected HAdV proteins and further applied this method to monitor the changes occurring in HAdV upon heat-induced virion disassembly that mimics a key step in host infection (11, 30–32).
FIGURE 1.
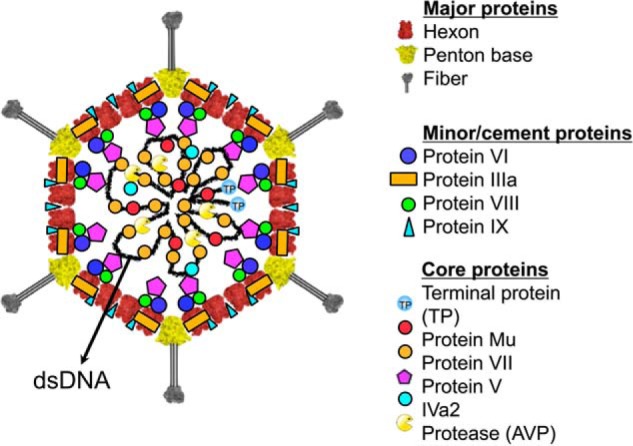
Structural model of adenovirus (51). Schematic diagram of HAdV showing the structural proteins for which the localization in the virion is known. The major capsid proteins are the hexon, penton base, and fiber. The capsid is stabilized on the outside by the minor protein IX and IIIa (A. Reddy and P. Nemerow, personal communication). The remaining minor capsid proteins, VI and VIII, have been localized to the inner surface of the capsid. The core proteins, TP, Mu, V, and VII, are associated with the double-stranded DNA genome in the interior of the virion. The presence and locations of AVP and the protein IVa2 are currently ambiguous.
TABLE 1.
Detailed features on each of the 13 HAdV (Ad5F35) proteins
These features include gene name, Uniprot accession ID, number of residues, and molecular mass. Next, the previously reported localization in the virion and protein copy number were given as reported in Refs. 2, 3, 45. The last column provides the copy numbers measured in this study, whereby the average is reported over two replicates.
| Polypeptide | Uniprot ID | Residues | MW (kDa) | Location | Protein copy number (2, 3, 42) | Measured protein copy number |
|---|---|---|---|---|---|---|
| Hexon | P04133 | 952 | 107.9 | Coat | 720 | 821 ± 40 |
| Penton | P12538 | 571 | 63.3 | Vertex | 60 | 60 |
| Fiber | Q67733 | 323 | 35.3 | Vertex | 36 | 36 ± 1 |
| IIIa | P12537 | 585 | 65.2 | Cement | 68 ± 2 | 57 ± 6 |
| VIII | P24936 | 227 | 24.7 | Cement | 127 ± 3 | 102 ± 11 |
| VI | P24937 | 250 | 27 | Cement | 342 ± 4 | 359 ± 24 |
| IX | P03281 | 140 | 14.4 | Cement | 247 ± 2 | 676 ± 122 |
| VII | P68951 | 198 | 22 | Core | 833 ± 19 | 527 ± 44 |
| V | P24938 | 366 | 41.3 | Core | 157 ± 1 | 148 ± 15 |
| TP | P04499 | 653 | 74.6 | Core | 2 | 5 ± 1 |
| μ | P14269 | 80 | 8.8 | Core | ∼104 | 290 ± 18 |
| AVP | P03253 | 204 | 23.1 | Core | 15 ± 5 | 7 ± 1 |
| IVa2 | P03271 | 449 | 50.9 | 7 ± 1 | 5 ± 1 |
FIGURE 2.
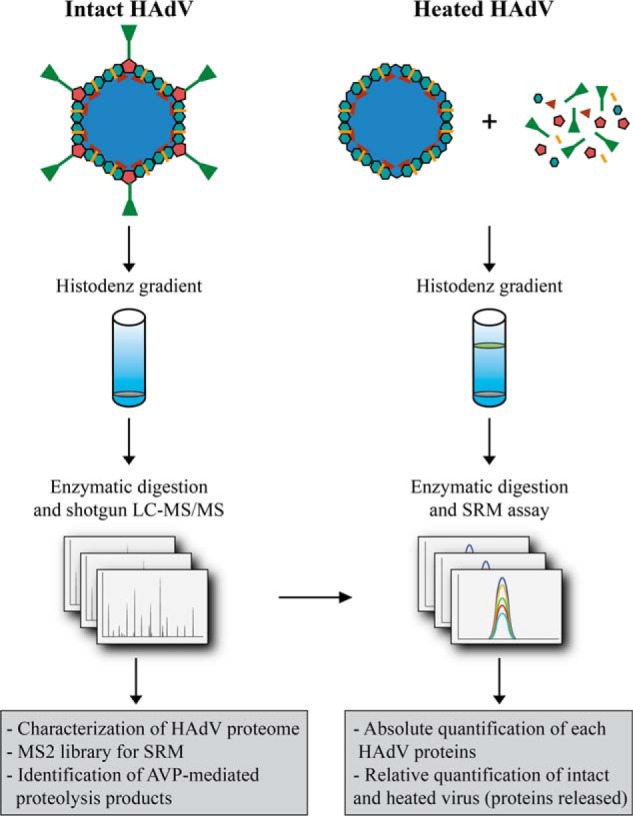
Schematic of the MS-based workflow combining shotgun and targeted approaches to characterize HAdV (Ad5F35). HAdV from intact and heat-stressed particles were purified by HistoDenz density gradients. A first analysis was performed by combining shotgun LC-MS/MS analysis with multiple proteases for protein digestion. This analysis was used to identify all HAdV proteins with high sequence coverage, identify AVP cleavage sites, and build a spectral library from which we selected the peptides employed in the SRM assay. Using these assays, we quantitatively profiled the stoichiometry of all 13 proteins present, as well as their (partial) release upon heat stress.
EXPERIMENTAL PROCEDURES
Adenovirus Preparation
Replication-defective HAdV type 5 pseudotyped with the HAdV type 35 fiber (Ad5F35) was purified as described previously (33) using cesium chloride density gradients. The virus was dialyzed into DX10/10 buffer (40 mm Tris, pH 8.1, 500 mm NaCl, 10% glycerol, 10% ethylene glycol, 2% sucrose, and 1% mannitol). Partial capsid disassembly was achieved using thermal perturbation (34, 35). Briefly, Ad5F35 (5 mg) was diluted in 7.5 mm HEPES, 50 mm NaCl, pH 7.4, divided into 6 × 500 μl aliquots, and incubated at 55 °C for 15 min. The released proteins were isolated from the intact particles and viral cores by density sedimentation on discontinuous histodenz gradients (30–80%).
Capsid Disassembly Measured by DNA Exposure Following Thermal Treatment
For DNA exposure experiments, 5 μg of Ad5F5 (HAdV5) or Ad5F35, in 10 μl A195 buffer, was mixed with 5 μl of Pico Green (Invitrogen), diluted 1:20 in A195 buffer and transferred to a 96-well RT-PCR plate (Bio-Rad). Triplicate samples were heated for 1 min in a Bio-Rad CFX96 real-time PCR system from 37–55 °C in 0.5 °C increments. Data collection was done with Channel 1/FAM settings (450–490 nm excitation and 510–530 nm emission). Measurements of capsid disassembly via immunoblot with antibodies to protein VI or fiber was done as described previously (34, 35).
Protein Extraction and Digestion
For LC-MS analysis, purified HAdV particles were resuspended in 50 mm ammonium bicarbonate, 5% (w/v) sodium deoxycholate (Sigma Aldrich) and heated at 90 °C for 5 min to disrupt the virus particles. Proteins (∼200 μg) for each enzymatic digestion were first reduced using dithiothreitol (DTT, Sigma Aldrich) for 30 min at 56 °C and then alkylated by iodoacetamide (Sigma Aldrich) for 30 min in the dark. Samples were diluted such that the final concentration of sodium deoxycholate was 0.5%, followed by enzymatic digestion overnight at 37 °C employing three different proteases in parallel. Trypsin (Promega) and chymotrypsin (Roche Applied Science) were added in a substrate/enzyme ratio of 50:1 (w/w), whereas LysN (U-Protein Express, The Netherlands) was added in a substrate/enzyme ratio of 75:1. Digestion was quenched by acidification with trifluoroacetic acid (TFA, Sigma Aldrich) to a final concentration of 0.5%. Sodium deoxycholate removal was performed by liquid-liquid extraction. Briefly, samples were diluted two times with ethyl acetate, vortexed for 30 s, and then centrifuged at 20,000 × g for 1 min. The peptide-containing aqueous phase at the bottom of the Eppendorf tubes was recovered and desalted by solid phase extraction (Sep-pack Vac C18 cartridges, Waters). Desalted peptides were eluted in 80% acetonitrile, dried in a SpeedVac, and then resuspended in 10% formic acid solution. Sample digests from all three proteases were used for a shotgun experiment, whereas solely tryptic samples were analyzed by SRM and shotgun label-free quantification analyses.
Shotgun LC-MS Analysis
Sample digests from the three proteases were analyzed separately by a single LC-MS/MS run. We performed nanoflow LC-MS/MS using either an LTQ-Orbitrap XL coupled to an Agilent 1200 HPLC system (Agilent Technologies), an LTQ-Orbitrap Elite, and Orbitrap Q-exactive mass spectrometers (Thermo Electron, Bremen, Germany) coupled to a EASY nLC 1000 system (Thermo Fisher Scientific). Approximately 40 ng of digested HAdV was delivered to a trap column (ReproSil C18, (Dr. Maisch, GmbH, Ammerbuch, Germany); 20 mm × 100 μm inner diameter, packed in-house) at 5 μl/min in 100% solvent A (0.1 m acetic acid in water). Next, peptides were eluted from the trap column onto an analytical column (ReproSil-Pur C18-AQ (Dr. Maisch, GmbH, Ammerbuch, Germany); 40 cm in length, 50-μm inner diameter, packed in-house) at ∼100 nl/min in a 60 min or 3 h gradient from 0 to 40% solvent B (0.1 m acetic acid in 8:2 (v/v) acetonitrile/water). The eluent was sprayed via distal coated emitter tips. The mass spectrometers were operated in data-dependent mode, automatically switching between MS and MS/MS. In the LTQ-Orbitrap XL and LQT-Orbitrap Elite, the full-scan MS spectra (from m/z 350 to 1500) were acquired in the Orbitrap analyzer with a full width at half maximum resolution of 30,000 and 60,000 at 400 m/z. After the survey scans, the 10 most intense precursor ions at a threshold above 5000 were selected for MS/MS. Peptide fragmentation was carried out using a decision tree performed by collision-induced dissociation or electron transfer dissociation, depending on their charge state and mass. In both of the fragmentation methods employed, the fragment ions readout is in the ion trap analyzer. In the Q-exactive orbitrap full-scan MS spectra (from m/z 350 to 1500) were acquired with a full width at half maximum resolution of 35,000 at 400 m/z. After the survey scans, the 10 most intense precursor ions at an intensity threshold >4200 were selected for MS/MS. Peptide fragmentation was carried out by higher collisional dissociation with a normalized collision energy of 25.
Bioinformatic Analysis
MS raw data from the shotgun LC-MS/MS analyses was processed by Proteome Discoverer (version 1.3, Thermo Electron). Peptide identification was performed with Mascot 2.3 (Matrix Science) against a concatenated forward-decoy UniPROT database, including the HAdV protein sequences and supplemented with all of the frequently observed contaminants in MS. The reversed sequences (decoy) were created for each protein entry present in the forward database and were employed to calculate the false discovery rate (36) and therefore to adjust the filtering criteria. The following parameters were used: 50 ppm precursor mass tolerance, 0.6 Da (for collision-induced dissociation and electron transfer dissociation spectra) and 0.05 Da (for higher collisional dissociation spectra) fragment ion tolerance. To evaluate the adenovirus proteinase activity, we allowed the identification of peptides containing one nonspecific cleavage and a maximum of two missed cleavages. Carbamidomethyl cysteine was allowed as fixed modification, whereas oxidized methionine and protein N-terminal acetylation were set as variable modifications. Finally, results were filtered using the following criteria: (i) mass deviations of ±6 p.p.m., (ii) Mascot ion score of at least 20, (iii) a minimum of six amino acid residues per peptide, and (iv) position rank 1 in Mascot search. As a result, we obtained peptide false discovery rates below 1% for each of the three peptide mixtures analyzed.
Label-free Quantification
Label-free quantification of protein abundances was performed by calculating the area under the curve for each precursor ion identified as a peptide mapping to one of the pVI or pVIII proteolytic products. The area under the curves for these specific peptides were calculated by Proteome Discoverer (version 1.3, Thermo Electron) and used to compare the peptides abundance in the heated versus intact HAdV to calculate the ratios.
SRM Analysis
For each of the HAdV proteins, we aimed to select up to three peptides based on the following criteria: (i) absence of nonspecific or missed cleavages, (ii) absence of methionine residues, and (iii) fragmentation efficiency. SRM transitions were created using Skyline software (37) from the Orbitrap Q-exactive higher collisional dissociation spectral library. Synthetic stable isotope-labeled peptides with a C-terminal 15N-, and 13C-labeled arginine or lysine residue (>99 atom % isotopic enrichment) were purchased from Thermo Fisher Scientific (Ulm, Germany). For each synthetic peptide, 100 pmol were spiked into 50 μg of HAdV protein digests before LC-MS quantification. A total of 33 peptides and 210 transitions were employed for the whole assay. The LC-MS/MS was performed using a TSQ Vantage triple quadrupole (Thermo, San Jose, CA) coupled to an EASY nLC 1000 system (Thermo Fisher Scientific). Approximately 40 ng of digested HAdV are delivered to a trap column (ReproSil C18, (Dr. Maisch, GmbH, Ammerbuch, Germany); 20 mm × 100 μm inner diameter, packed in-house) at 5 μl/min in 100% solvent A (0.1 m acetic acid in water). Next, peptides eluted from the trap column onto an analytical column (ReproSil-Pur C18-AQ (Dr. Maisch, GmbH, Ammerbuch, Germany); 40-cm length, 50-μm inner diameter, packed in-house) at ∼100 nl/min in a 60-min gradient from 0 to 40% solvent B (0.1 m acetic acid in 8:2 (v/v) acetonitrile/water). The triple quadrupole mass analyzer setup was configured to perform MS/MS scans triggered on each target peptide according to their scheduled retention time window (3 min). Q1 and Q3 peak width set at 0.7 Da, and the cycle time corresponds to 1.8 s. The triple quadrupole collision energy was calculated according to the following equations: CE = 0.03× (m/z) + 2.905 and CE = 0.038× (m/z) + 2.281 (CE indicates collision energy and m/z indicates mass to charge ratio) for doubly and triply charged precursor ions, respectively. The area for each peptide peak was calculated by using Skyline software (37). The quantification was then performed by comparing the area for each of the endogenous peptides, in comparison with the area of the corresponding heavy standard peptide added to the HAdV digest in known amount.
RESULTS
In-depth HAdV (Ad5F35) Proteome Profiling by Shotgun LC-MS/MS
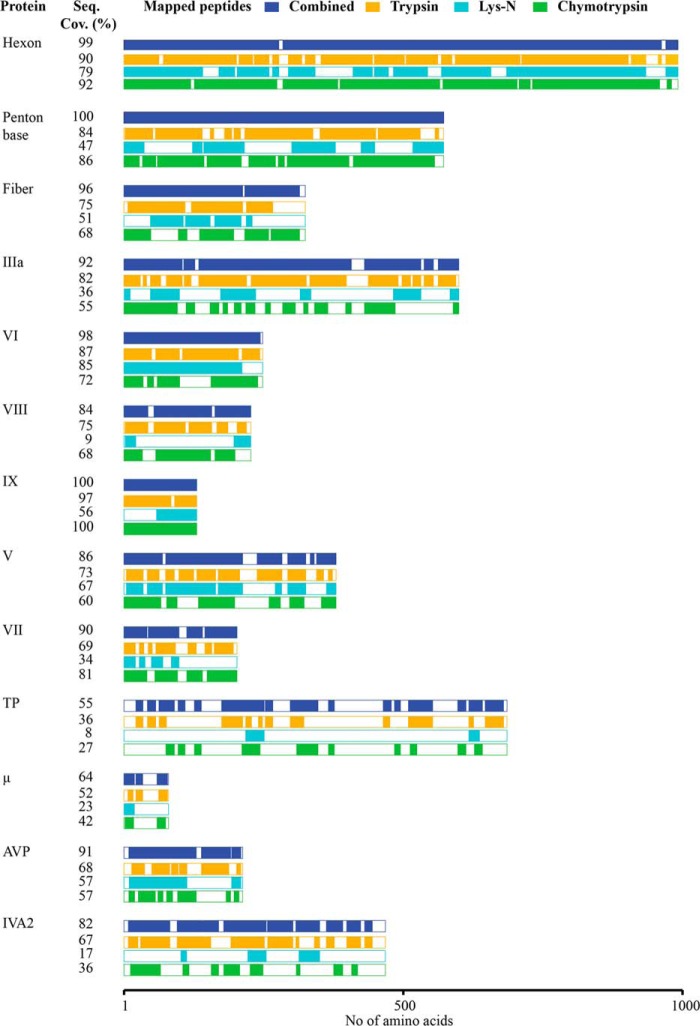
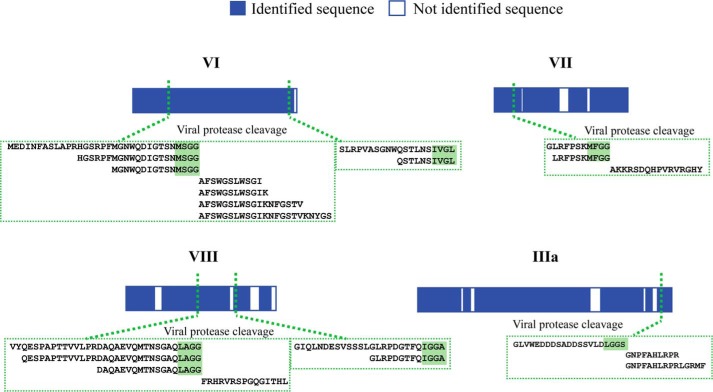
The first aim in this study was to extensively identify and sequence the proteins present in HAdV particles using shotgun high resolution LC-MS/MS analysis. Despite the moderate complexity (supposedly 12–13 proteins) and middling dynamic range in protein abundance (ranging from 2 to ∼850 copies) in the HAdV proteome, we observed that full protein coverage could not be reached for each protein of HAdV when employing exclusively the commonly used protease trypsin. This issue was related to the primary structure of certain proteins or protein domains. In some HAdV proteins, the low frequency of arginine (Arg) and lysine (Lys) results in long and hydrophobic tryptic peptides, while in other proteins, the high frequency of R residues results in small, hydrophilic tryptic peptides that are all easily missed in LC-MS/MS analysis. To improve sequence coverage of all HAdV viral proteins, we employed a multiprotease digestion strategy (38, 39) from which the results are summarized in Fig. 3 and supplemental Fig. 1, and supplemental Table 1. We were able to confidently identify 13 HAdV proteins with an average sequence coverage of 90%, achieving near full sequencing (99%) for four of these (hexon, penton base, VI, and IX). Of the three enzymes employed, trypsin provided the highest coverage, with average sequence coverage of 73%, compared with 65 and 44%, respectively for chymotrypsin and LysN. The strategy of using multiple proteases clearly improved the comprehensiveness of our analysis, leading to an average increase of 15% sequence coverage with the highest increases for the terminal protein IVa2 and the AVP (Fig. 3). Importantly, the use of multiple proteases was highly beneficial in the characterization of AVP proteolytic cleavage sites in the different capsid proteins. AVP plays a pivotal role in AdV maturation, processing multiple procapsid preproteins to generate infectious virus particles. HAdV precursor proteins targeted by AVP include pIIIa, pVI, pVII, pVIII, pX, and pTP (40–43). Accordingly, we identified AVP-mediated cleavage events in the proteins pVI, pVIII, pVII, and pIIIa, identifying multiple peptides from each of them using either tryptic, chymotryptic, and LysN digestion (Fig. 4). Detecting these cleavage sites in separate experiments using complementary proteases indicates the likelihood that they are present in HAdV.
FIGURE 3.
High-resolution MS-based proteomics profiling in combination with multiple protease digestion allowed sequence coverage of on average 90% for the HAdV proteome. Representation of the 13 HAdV proteins identified in the shotgun LC-MS/MS analyses. The sequence coverage (%) along with the specific area of the protein sequenced is reported for each of the employed enzymes trypsin (orange), Lys-N (light blue), and chymotrypsin (green), as well as the cumulative coverage (dark blue).
FIGURE 4.
Identified AVP cleavage sites in four HAdV (Ad5F35) proteins. Representation of the four HAdV proteins (bars) for which direct evidence of AVP cleavage could be found. For each of the six cleavage sites, the set of confidently identified peptides deriving from multiple protease digestions are highlighted in the green boxes.
Determination of the Stoichiometry of Proteins in HAdV (Ad5F35)
To define the stoichiometry for each of the viral proteins we developed a targeted SRM assay, relying on reference peptides that incorporate heavy isotopes (AQUA peptides (24)). The selection of the peptides employed in this assay was based on their chemo-physical properties and fragmentation efficiency retrieved from our shotgun experiments (see “Experimental Procedures” and supplemental Fig. 1). We could select a total of 33 peptides to quantify the 13 identified HAdV proteins, which corresponded to a total of 210 transitions monitored. Protein quantification was performed in two biological replicates, with a peptide coefficient of variability within 5%, underlining the high intrinsic accuracy of the method (supplemental Table 2). To calculate the protein copy number of the different proteins in the virion, we normalized the SRM quantification using the known copy number of the penton base protein (60 copies) as a reference, for which the exact stoichiometry has been accurately established with radiolabeling experiments (44) and high-resolution crystal structure and EM models (2, 3). The copy numbers that we experimentally determined for each of the HAdV proteins, averaged over two independent biological replicates, as well as the literature reported copy numbers are provided in Fig. 5 and Table 1. Notably, the copy numbers determined by SRM were in good agreement with the copy numbers reported for the seven HAdV proteins whose copy numbers have been most accurately established (hexon, penton base, fiber, IIIa, VI, VIII, and V). The copy number of most of the other viral proteins is either not available or has been only roughly estimated. For this set of proteins, our measurements suggested that the copy numbers of protein VII and AVP were ∼40% lower compared with the previously estimated values. We obtain the first firm experimental evidence for the presence of five to six copies of the protein IVa2, a molecule crucial in packaging of the viral genome (45). Proteins μ, TP, and IX were, according to our data, unexpectedly more abundant than estimated. Unfortunately, for TP and IX, we observed significant variability between the values for the three individual SRM peptides (supplemental Table 2), hampering the ability to make an accurate estimate. In these cases, the accuracy of SRM quantification might be affected by incomplete digestion of the target protein, partial artifactual modification, and/or partial loss of the synthetic standard during the analysis. Ideally, including more peptides for the quantification of each protein would make the SRM quantification more comprehensive and improve the identification of these outlier peptides. However, considering the strict parameters to take into account when selecting SRM peptide candidates (e.g. absence of missed cleavages and/or amino acids that are prone to oxidation, etc.), this might be practically unfeasible for some proteins. An extreme example was provided by μ, a small protein (4 kDa) for which we could, based on the criteria mentioned above, select and quantify only one peptide.
FIGURE 5.
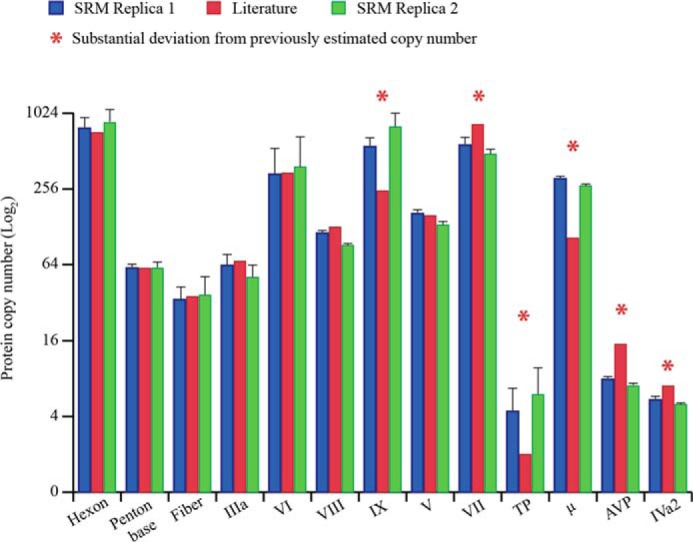
Stoichiometry of the proteins in HAdV (Ad5F35) particles. The bar chart reports the copy number (log2) for each of the HAdV proteins obtained by the SRM assay in two independent biological replicates (blue and red bars) and the estimated copy number known from literature (green bars). The error bars indicate the relative S.D. calculated from the quantitative values, retrieved by each of the SRM peptides employed for each protein.
Heat-induced Release of Proteins in HAdV (Ad5F35)
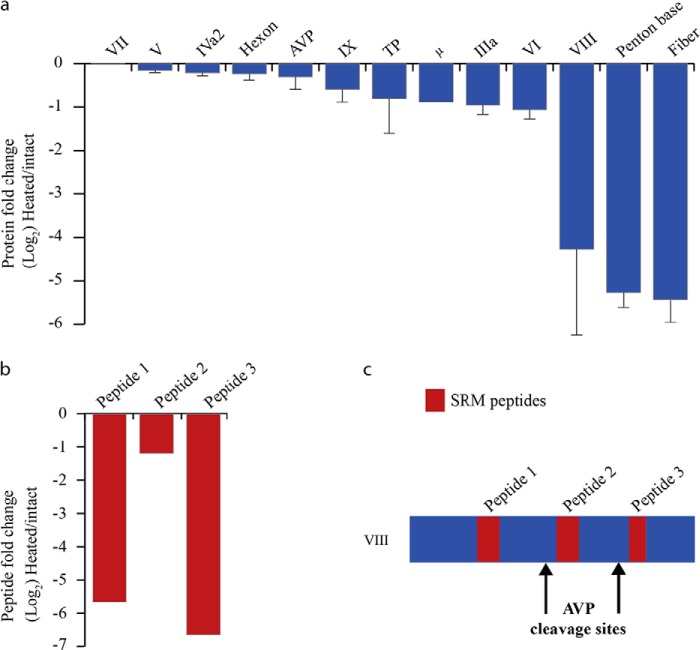
HAdV host infection is a process that involves a complex series of molecular events that can be divided in three main steps: (i) the binding of fiber and penton base proteins to host cell receptors, (ii) virus internalization through receptor-mediated endocytosis, triggering partial capsid disassembly to facilitate protein VI-mediated escape from the low pH endosomal compartments, and (iii) the transfer of viral genome into the host nucleus. Monitoring in vivo of the structural changes taking place upon virus internalization has been challenging. However, several methods have been developed to mimic these structural changes in vitro. One of the most established method employs the thermal treatment of HAdV particles at 55 °C, which induces the release of proteins at the capsid vertices (i.e. the penton base and fiber protein), generating a structure that has been proposed to persist in the acidic environment of the endosome and is sufficiently stable enough to protect the virus genome during its journey to the nucleus (11, 30–32). The best conditions to perform the thermostability assay were determined by monitoring the DNA accessibility on adenovirus type 5 capsids equipped with either the type 5 or type 35 fiber protein at different temperatures (Fig. 6). The results indicate both Ad5F5 (HAdV5) and Ad5F35 exhibit maximal accessibility to a DNA-binding fluorescent dye (Pico Green) at 55 °C and that this correlates very well with the release/degradation of protein VI and fiber during the earliest stage of cell infection (46). To accurately define which proteins are released upon heat treatment, and to what extent, we applied our SRM method to measure the difference in protein abundance between the intact and the thermally treated HAdV particles (Fig. 2). Our data revealed that the penton base and fiber proteins had been completely released in the heated particles (Fig. 7a and Table 2). We also found that proteins VI, VIII, μ, and IIIa are substantially released (by ∼50%), whereas the hexon was released to a lower extent (Fig. 7a and Table 2). All three peptides monitored for the cement protein VI showed a 40–60% decrease in the SRM quantification, indicating that ∼50% of protein VI is released upon heating. Similarly, all three monitored peptides for protein IIIa decreased by ∼50%. Interestingly, two of three peptides monitored for the cement protein VIII reduced as much as the penton base and fiber (i.e. more than 30-fold), in sharp contrast the third peptide monitored was only reduced by a factor of 2 (Fig. 7b). As we described above, pVIII contains two AVP cleavage sites (Figs. 4 and 7c). Therefore, the N and C termini, as well as the core domain, could better be considered as distinct entities, which may play different roles during host infection or virus particle assembly. Each of the three VIII peptides monitored by SRM belong to three distinct VIII fragments resulting from the AVP activity (Fig. 7c). To validate further this apparent differential release of VIII fragments, the heat-induced differential release of the three VIII fragments was additionally analyzed by label free quantification using LC-MS/MS. Comparing the precursor ion intensity of each identified peptide relative to each fragment of VIII clearly confirmed the SRM results (Table 3 and supplemental Table 3). We found a similar behavior for VI, in which the amount of N-terminal fragment released was six times higher compared with the VI core domain. The VI N-terminal fragment was also shown to be released in complex with peripentonal hexons (52). The partially released proteins VI, VIII, and IIIa have been proposed to be located near the capsid vertices and to interact with the penton base proteins (2, 3), which may explain their partial release upon heating. Release of some of these proteins, particularly the putative membrane lytic VI, is critical for the endosomal lysis and release of partially disassembled virions into the cytoplasm and their eventual transport into nucleus (11, 47).
FIGURE 6.
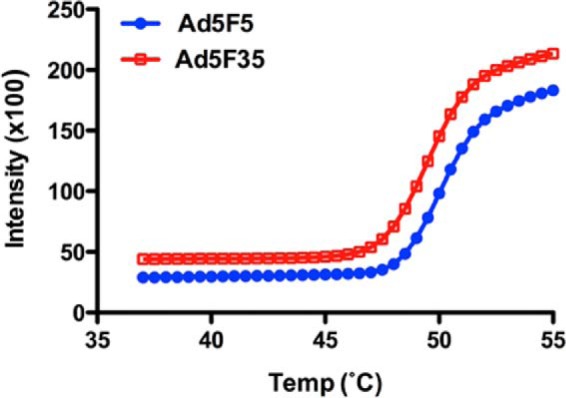
Thermally induced disassembly of AdV capsids. Ad5F5 (HAdV5) and Ad5F35 were heated to the indicated temperatures in the presence of Pico green. Capsid disassembly, as measured by dye accessibility to the viral genome, was measured in relative fluorescence units. Data are the mean ± S.D. from triplicate samples. From the data, it appears that Ad5F5 and Ad5F35 disassemble with similar kinetics, and both reach a plateau at ∼55 °C.
FIGURE 7.
Quantification of the released HAdV proteins upon heat stress. a, the fold change in the abundance of HAdV proteins upon heat stress are shown. The y axis represents the protein ratios of heat-stressed/intact virus in log2 scale, and the error bars represent the relative S.D. b, fold change in the abundance of various segments of protein VIII, generated by the HAdV protease: peptides 1 and 3 are fully released, whereas peptide 2 is only marginally released. c, the protein domain in which these peptides reside suggest that only the N- and C-terminal segment of protein VIII are completely released.
TABLE 2.
The percentage of protein released from intact HAdV upon mild thermal stress
The amount of released protein was calculated averaging over the three peptides included per protein in the SRM assay. For proteins V, TP, AVP, and IVa2, only two peptides could be used. For the 4-kDa μ protein, only a single peptide could be selected. See supplemental Table 2 for detailed results and statistical information on each individual peptide used in the SRM assay.
| Polypeptide | Released amount (%) |
|---|---|
| Hexon | 15.5 |
| Penton | 97.3 |
| Fiber | 98.3 |
| IIIa | 41.6 |
| VIII | 85 |
| VI | 51 |
| IX | 32.8 |
| VII | 0 |
| V | 10.3 |
| TP | 32.2 |
| m | 41.3 |
| AVP | 19.5 |
| IVa2 | 13.9 |
TABLE 3.
HAdV proteins VI and VIII exhibit a segmented release upon heat stress
For proteins VI and VIII, evidence was found both in the SRM assays as well as in the label-free quantification that selected regions were differentially released, whereby the terminal ends were more released then the core part of the proteins. These terminal stretches correspond to regions generated by the ADV protease. The table shows the amount of protein segment released by comparing the precursor areas for each of the identified peptides mapping to a specific segment of protein VI and VIII. For details, see also supplemental Table 3.
| Protein domain | Released amount (%) |
|---|---|
| VI N-terminal | 94.8 |
| VI core | 66 |
| VIII N-terminal | 98.6 |
| VIII core | 82.7 |
| VIII C-terminal | 99.7 |
DISCUSSION
Here, we used different MS-based approaches to profile in-depth the HAdV proteome, to define the stoichiometry of its proteins and to monitor changes occurring during heat-initiated disassembly mimicking in vitro infection models. Nearly full sequencing of the HAdV proteome was achieved, which required three complementary proteases for enzymatic digestions. The multiprotease approach allowed us to obtain an average increase of 70% sequence coverage compared with a previously reported HAdV proteome analysis by two-dimensional LC-MS (48) and to identify unambiguously the presence of three additional proteins (IVa2, X, and TP) as components of the HAdV, which are present at relatively low copy numbers. We also confidently identified a total of six AVP cleavage sites in four HAdV proteins using peptide-based evidence from different protease digests. We next used this in-depth proteome map of HAdV to develop a SRM based-method to define the stoichiometry of each protein in HAdV. We were able to select the optimal peptide candidates to be employed in the SRM assay based on their chemo-physical properties and fragmentation efficiency. The SRM based quantification of the copy numbers of each individual HAdV protein correlated well with the copy number extracted from crystal and EM-derived models and structures (2, 3). Our data complemented these models, providing better estimates on copy numbers of inner (cement and core) HAdV proteins. Another key aspect of our approach is that it can be applied to accurately define the protein abundance under two different conditions. For instance, one of the most intriguing questions concerns the elucidation of the structural rearrangements taking place in the virus upon its internalization by the host cells. Because performing this analysis in vivo represents a challenge, in vitro models are typically used to mimic these structural changes. One of the applied models is based on the release of the capsid vertices upon thermal stress (11, 30–32). We employed our SRM method to compare protein levels in intact and heated viruses. The data clearly demonstrated the release of the capsid vertices observed earlier (30, 49) and revealed the extent to which additional capsid proteins are released. Moreover, we showed evidence for a strong release of the terminal domains of pVI and pVIII generated by AVP mediated cleavage, suggesting a possible role of these protein fragments in escape from the endosome to the host cytosol. We suggest that our approach could be further employed to monitor structural changes in different in vitro models and possibly also in in vivo experiments, ultimately leading to a better understanding of the mechanisms involved in the infection process. We believe that this work opens up a better avenue for characterizing virus by MS, thereby allowing a useful comparison to structural models as well as correlation with events occurring in virus infection.
Supplementary Material
This work was supported by the Netherlands Proteomics Centre and by the European Community's Seventh Framework Programme FP7/2007-2013 through PRIME-XS Project Grant Agreement 262067 (to M. B., S. D. P., J. S., and A. J. R. H.).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (50) with the data set identifier PXD000591.

This article contains supplemental Tables 1–3 and Fig. 1.
- AdV
- adenovirus
- HAdV
- human AdV
- TP
- terminal protein
- AVP
- adenovirus proteinase
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Davison A. J., Benko M., Harrach B. (2003) Genetic content and evolution of adenoviruses. J. Gen. Virol. 84, 2895–2908 [DOI] [PubMed] [Google Scholar]
- 2. Reddy V. S., Natchiar S. K., Stewart P. L., Nemerow G. R. (2010) Crystal structure of human adenovirus at 3.5 Å resolution. Science 329, 1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu H., Jin L., Koh S. B., Atanasov I., Schein S., Wu L., Zhou Z. H. (2010) Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329, 1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nemerow G. R. (2000) Cell receptors involved in adenovirus entry. Virology 274, 1–4 [DOI] [PubMed] [Google Scholar]
- 5. Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. (1977) An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 [DOI] [PubMed] [Google Scholar]
- 6. Berget S. M., Moore C., Sharp P. A. (1977) Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U.S.A. 74, 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xin K. Q., Sekimoto Y., Takahashi T., Mizuguchi H., Ichino M., Yoshida A., Okuda K. (2007) Chimeric adenovirus 5/35 vector containing the clade C HIV gag gene induces a cross-reactive immune response against HIV. Vaccine 25, 3809–3815 [DOI] [PubMed] [Google Scholar]
- 8. Sharma A., Tandon M., Bangari D. S., Mittal S. K. (2009) Adenoviral vector-based strategies for cancer therapy. Curr. Drug Ther. 4, 117–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burnett R. M. (1985) The structure of the adenovirus capsid. II. The packing symmetry of hexon and its implications for viral architecture. J. Mol. Biol. 185, 125–143 [DOI] [PubMed] [Google Scholar]
- 10. Liu H., Wu L., Zhou Z. H. (2011) Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy. J. Mol. Biol. 406, 764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiethoff C. M., Wodrich H., Gerace L., Nemerow G. R. (2005) Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79, 1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGrath W. J., Ding J., Didwania A., Sweet R. M., Mangel W. F. (2003) Crystallographic structure at 1.6-A resolution of the human adenovirus proteinase in a covalent complex with its 11-amino-acid peptide cofactor: insights on a new fold. Biochim. Biophys. Acta 1648, 1–11 [DOI] [PubMed] [Google Scholar]
- 13. Wodrich H., Guan T., Cingolani G., Von Seggern D., Nemerow G., Gerace L. (2003) Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 22, 6245–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Altelaar A. F., Munoz J., Heck A. J. (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 [DOI] [PubMed] [Google Scholar]
- 15. Mann M., Kulak N. A., Nagaraj N., Cox J. (2013) The coming age of complete, accurate, and ubiquitous proteomes. Mol. Cell 49, 583–590 [DOI] [PubMed] [Google Scholar]
- 16. Bensimon A., Heck A. J., Aebersold R. (2012) Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 81, 379–405 [DOI] [PubMed] [Google Scholar]
- 17. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Second T. P., Blethrow J. D., Schwartz J. C., Merrihew G. E., MacCoss M. J., Swaney D. L., Russell J. D., Coon J. J., Zabrouskov V. (2009) Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 81, 7757–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andrews G. L., Simons B. L., Young J. B., Hawkridge A. M., Muddiman D. C. (2011) Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600). Anal. Chem. 83, 5442–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 22. Cristobal A., Hennrich M. L., Giansanti P., Goerdayal S. S., Heck A. J., Mohammed S. (2012) In-house construction of a UHPLC system enabling the identification of over 4000 protein groups in a single analysis. Analyst 137, 3541–3548 [DOI] [PubMed] [Google Scholar]
- 23. Nagaraj N., Kulak N. A., Cox J., Neuhauser N., Mayr K., Hoerning O., Vorm O., Mann M. (2012) System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell Proteomics 10.1074/mcp.M111.013722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Picotti P., Aebersold R. (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 [DOI] [PubMed] [Google Scholar]
- 26. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 27. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prince J. T., Carlson M. W., Wang R., Lu P., Marcotte E. M. (2004) The need for a public proteomics repository. Nat. Biotechnol. 22, 471–472 [DOI] [PubMed] [Google Scholar]
- 29. Taouatas N., Heck A. J., Mohammed S. (2010) Evaluation of metalloendopeptidase Lys-N protease performance under different sample handling conditions. J. Proteome Res. 9, 4282–4288 [DOI] [PubMed] [Google Scholar]
- 30. Russell W. C., Valentine R. C., Pereira H. G. (1967) The effect of heat on the anatomy of the adenovirus. J. Gen. Virol. 1, 509–522 [DOI] [PubMed] [Google Scholar]
- 31. Pérez-Berná A. J., Ortega-Esteban A., Menéndez-Conejero R., Winkler D. C., Menéndez M., Steven A. C., Flint S. J., de Pablo P. J., San Martín C. (2012) The role of capsid maturation on adenovirus priming for sequential uncoating. J. Biol. Chem. 287, 31582–31595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Russell W. C. (2009) Adenoviruses: update on structure and function. J. Gen. Virol. 90, 1–20 [DOI] [PubMed] [Google Scholar]
- 33. Reddy V. S., Natchiar S. K., Gritton L., Mullen T. M., Stewart P. L., Nemerow G. R. (2010) Crystallization and preliminary X-ray diffraction analysis of human adenovirus. Virology 402, 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith J. G., Nemerow G. R. (2008) Mechanism of adenovirus neutralization by human α-defensins. Cell Host Microbe 3, 11–19 [DOI] [PubMed] [Google Scholar]
- 35. Moyer C. L., Nemerow G. R. (2012) Disulfide-bond formation by a single cysteine mutation in adenovirus protein VI impairs capsid release and membrane lysis. Virology 428, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 37. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swaney D. L., Wenger C. D., Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng M., Taouatas N., Cappadona S., van Breukelen B., Mohammed S., Scholten A., Heck A. J. (2012) Protease bias in absolute protein quantitation. Nat. Methods 9, 524–525 [DOI] [PubMed] [Google Scholar]
- 40. Anderson C. W. (1990) The proteinase polypeptide of adenovirus serotype 2 virions. Virology 177, 259–272 [DOI] [PubMed] [Google Scholar]
- 41. Mangel W. F., Baniecki M. L., McGrath W. J. (2003) Specific interactions of the adenovirus proteinase with the viral DNA, an 11-amino-acid viral peptide, and the cellular protein actin. Cell Mol. Life Sci. 60, 2347–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weber J. (1976) Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J. Virol. 17, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Webster A., Russell S., Talbot P., Russell W. C., Kemp G. D. (1989) Characterization of the adenovirus proteinase: substrate specificity. J. Gen. Virol. 70 (Pt 12), 3225–3234 [DOI] [PubMed] [Google Scholar]
- 44. van Oostrum J., Burnett R. M. (1985) Molecular composition of the adenovirus type 2 virion. J. Virol. 56, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Christensen J. B., Byrd S. A., Walker A. K., Strahler J. R., Andrews P. C., Imperiale M. J. (2008) Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J. Virol. 82, 9086–9093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greber U. F., Willetts M., Webster P., Helenius A. (1993) Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75, 477–486 [DOI] [PubMed] [Google Scholar]
- 47. Moyer C. L., Wiethoff C. M., Maier O., Smith J. G., Nemerow G. R. (2011) Functional genetic and biophysical analyses of membrane disruption by human adenovirus. J. Virol. 85, 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chelius D., Hühmer A. F., Shieh C. H., Lehmberg E., Traina J. A., Slattery T. K., Pungor E., Jr (2002) Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J. Proteome Res. 1, 501–513 [DOI] [PubMed] [Google Scholar]
- 49. Rexroad J., Wiethoff C. M., Green A. P., Kierstead T. D., Scott M. O., Middaugh C. R. (2003) Structural stability of adenovirus type 5. J. Pharm. Sci. 92, 665–678 [DOI] [PubMed] [Google Scholar]
- 50. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nemerow G. R., Pache L., Reddy V., Stewart P. L. (2009) Insights into adenovirus host cell interactions from structural studies. Virology 384, 380–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Snijder J., Benevento M., Moyer C. L., Reddy V., Nemerow G. R., Heck A. J. R. (2014) The cleaved N-terminus of pVI binds peripentonal hexons in mature adenovirus. J. Mol. Biol. 10.1016/j.jmb.2014.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.